Summary
The CAP superfamily member, CRISPLD2, has previously been shown to be associated with nonsyndromic cleft lip and palate (NSCLP) in human populations and to be essential for normal craniofacial development in the zebrafish. Additionally, in rodent models, CRISPLD2 has been shown to play a role in normal lung and kidney development. However, the specific role of CRISPLD2 during these developmental processes has yet to be determined. In this study, it was demonstrated that Crispld2 protein localizes to the orofacial region of the zebrafish embryo and knockdown of crispld2 resulted in abnormal migration of neural crest cells (NCCs) during both early and late time points. An increase in cell death after crispld2 knockdown as well as an increase in apoptotic marker genes was also shown. This data suggests that Crispld2 modulates the migration, differentiation, and/or survival of NCCs during early craniofacial development. These results indicate an important role for Crispld2 in NCC migration during craniofacial development and suggests involvement of Crispld2 in cell viability during formation of the orofacies. genesis 53:660–667, 2015.
Keywords: neural crest, birth defects, early development, migration
INTRODUCTION
CRISPLD2, a member of the CAP superfamily of proteins (Cysteine-rich secretory proteins, Antigen 5 and Pathogenesis-related 1 proteins), was first isolated as a gene induced by glucocorticoids during lung development in the rat (initially named Lgl1; Kaplan et al., 1999). Further characterization of this gene suggested a role in kidney development as well as links to retinoic acid signaling during both lung and kidney development (Nadeau et al., 2010; Quinlan et al., 2007). CRISPLD2 is predicted to be a secreted glycoprotein containing an N-terminal SCP domain consisting of hydrophobic amino acids and two C-terminal LCCL cysteine-rich domains (Oyewumi et al., 2003). The functions of these domains are unknown. Homozygous Crispld2 knockout mice are embryonic lethal, precluding the study of many early developmental programs. While heterozygous mice appear normal, they have abnormal lung development with delayed alveolar maturation and disorganization of lung elastin fibers trapped in the interstitium (Lan et al., 2009). This data led Oyewumi et al. to conclude that CRISPLD2 functions to regulate epithelial–mesenchymal interactions in the lung (Oyewumi et al., 2003).
We have previously shown and others have confirmed an association between CRISPLD2 and nonsyndromic cleft lip/palate (NSCL/P) (Chiquet et al., 2007; Letra et al., 2010; Mijiti et al., 2015; Shen et al., 2011; Shi et al., 2010). We have also shown that Crispld2 is expressed in the craniofacial region of both mouse and zebrafish embryos and that knockdown of crispld2 in zebrafish results in craniofacial abnormalities (Chiquet et al., 2007; Yuan et al., 2012). Craniofacial development depends on the correct formation, migration and differentiation of cranial neural crest cells (NCCs) during early embryogenesis. Cranial NCCs first form as a single layer of neuroepithelial cells which undergo epithelial to mesenchymal transition (EMT) prior to migration into the craniofacies. This transition in the NCCs is associated with the loss of cell–cell attachment, breakdown of basal lamina, and an increase in mobility, which allows for the correct migration of these cells into the presumptive cranial region of the embryo. Epithelial–mesenchymal interactions/transitions also play important roles in gastrulation, lung and cardiac development as well as cancer progression, and metastasis (Feng et al., 2010; Kang and Svoboda, 2005; Micalizzi et al., 2010; Sanders and Prasad, 1989). Recent studies have shown a role for CRISPLD2 in cell migration, apoptosis, and wound healing (Zhang et al., 2015) and have suggested a role for CRISPLD2 in endometriosis (Yoo et al., 2014), cytokine modulation in airway smooth muscle cells (Himes et al., 2014), binding of lipid A/lipopolysaccharide to regulate endotoxin function through interaction with the LCCL-domains (Vasarhelyi et al., 2014; Wang et al., 2009), and in septic shock (Wang et al., 2013). In this work, we show that knockdown of crispld2 affects migration of cranial NCCs and cell viability during zebrafish craniofacial development.
RESULTS AND DISCUSSION
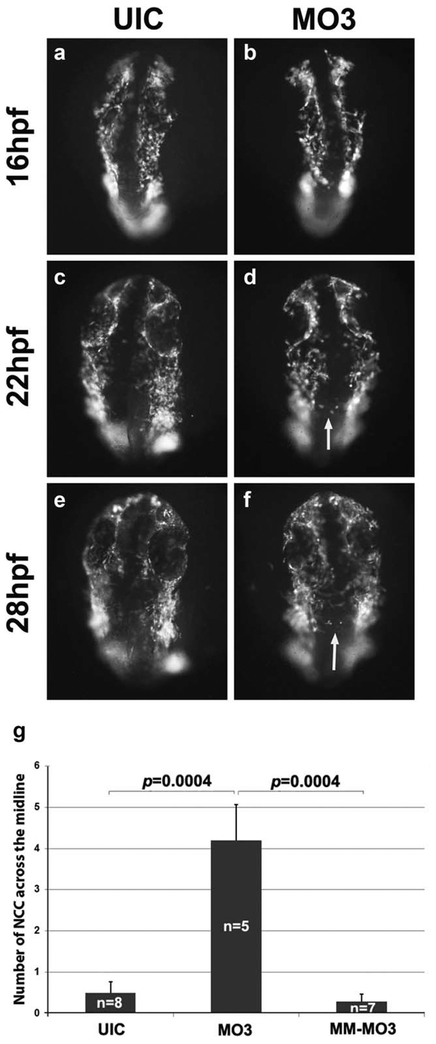
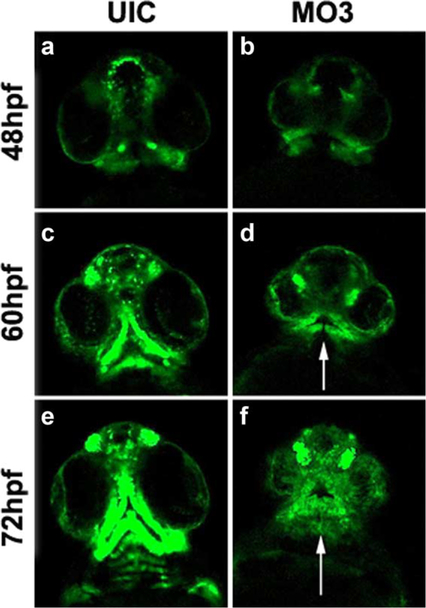
We have previously shown that knockdown of crispld2 in zebrafish embryos results in abnormal expression of NCC markers at 24 hpf causing severe orofacial deformities by 5 dpf (Yuan et al., 2012). To further understand the role of Crispld2 during craniofacial development, we evaluated pre-migratory and migrating NCCs in Crispdl2 knockdown embryos to determine the initial stage at which loss of Crispld2 results in abnormal phenotypes. To pinpoint the timing of Crispld2 function during orofacial development, a morpholino (MO3) directed against the start site of crispld2 (Yuan et al., 2012) was used to knockdown translation of Crispld2 protein in the Tg(−4.9sox10:EGFP) transgenic zebra-fish line. This transgenic line contains GFP downstream of a sox10 promoter/enhancer element that drives expression of GPF in pre-migratory and migrating NCCs. As shown in Figure 1, imaging data from MO3-injected embryos suggests that during early time points (16–28 hpf), the number of NCCs are decreased and move/migrate in a disorganized manner (Supporting Information Movie 1). In addition, NCCs were observed migrating across the midline (white arrows in Figure 1), which was not observed in control embryos. Quantification of the numbers of cells crossing the midline in MO3 injected, control mismatch MO3 (MM-MO3) shows a significant increase in the number of cells crossing the midline in MO3 injected embryos when compared with uninjected and control MM-MO3 injected embryos (Fig. 1g). These observations suggest that Crispld2 protein is needed at early developmental/differentiation stages for proper number, cell movement and/or migration of NCCs. At later time points (48–72 hpf), MO3-injected embryos have severely depleted numbers of NCCs that show abnormal migration and abnormal formation of the presumptive cartilage elements of the orofacial region (white arrows in Fig. 2, Supporting Information Movie 2). Taken together, this data suggests an essential role for Crispld2 in NCC survival and migration during both early and late craniofacial development. The loss of NCCs seen in our live imaging experiments could be the result of a decrease in cell proliferation, an increase in cell death or a combination of both.
FIG.1.
NCC migration is disorganized in MO3 morphants. Timelapse live cell imaging captures of sox10:GFP embryos showing migrating NCC cells at 16 (a, b), 22 (c, d), and 28 hpf (e, f). UIC (a, c, and e) and MO3-injected embryos (b, d, and f) showing a dorsal view of migrating NCCs. White arrows point to abnormally migrating NCCs. (g) Quantification of number of cells crossing the midline in UIC, MO3 and control MM-MO3 injected embryos.
FIG. 2.
Aberrant oral cartilage in MO3 morphants. Time-lapse live cell imaging captures of sox10:GFP embryos showing migrating NCC cells at 48 (a, b), 60 (c, d), and 72 hpf (e, f). UIC (a, c, and e) and MO3-injected embryos (b, d, and f) showing a ventral view of migrating NCCs. White arrows point to loss of normal migration and abnormal formation of presumptive cartilage elements.
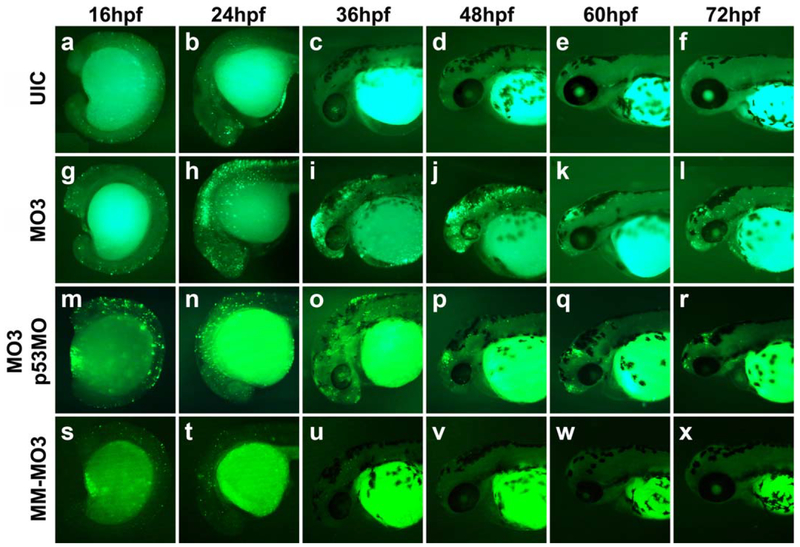
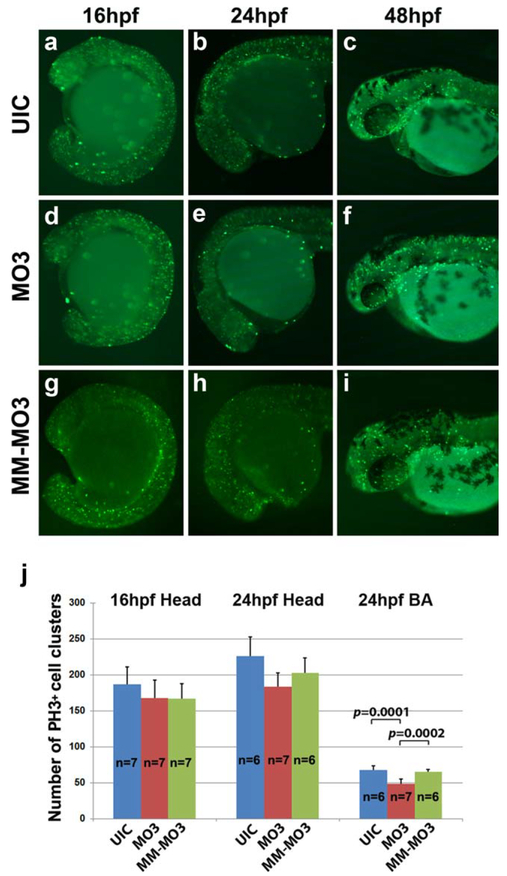
NCC markers are abnormally expressed in Crispld2 knockdown embryos and at later time points, these embryos exhibit a severe loss of palatal and lower jaw cartilage (Yuan et al., 2012). To better understand the effect of decreased Crispld2 protein on NCC marker genes and cartilage formation, the role of Crispld2 in cell viability was examined. TUNEL assays were carried out on Crispld2 knockdown embryos at multiple time points to determine if a reduction in Crispld2 protein resulted in increased cell death in these embryos as compared with uninjected embryos. Starting at 16 hpf, knockdown of Crispld2 resulted in a slight increase in TUNEL staining (Fig. 3g) compared with uninjected controls (UICs) (Fig. 3a) and control MM-MO3 injected embryos (Fig. 3s). From 24 to 48 hpf, Crispld2 knockdown embryos showed a marked increase in TUNEL positive cells (Fig. 3h–j) as compared with UICs (Fig. 3b–d) and control MM-MO3 injected embryos (Fig. 3t–v). By 60 hpf, the TUNEL positive cells decreased in the craniofacial region of MO-injected embryos compared with earlier time points; however, the TUNEL positive cells were continuously higher than in control embryos (Fig. 3e, k, w). Co-injection of crispld2 MO with p53 MO results in severe craniofacial defects (Yuan et al., 2012), suggesting that p53-mediated cell death is not responsible for this phenotype. To test this hypothesis, crispld2/p53 double knockdown embryos were assayed for TUNEL staining. While TUNEL staining in these embryos is reduced compared with crispld2 MO injected alone, there was a marked increase when compared with control embryos (Fig. 3m–r). To assess whether Crispld2 is involved in cell proliferation, immunohistochemistry was carried out using anti-phosphorylated histone H3 antibodies at multiple time points in uninjected, control MM-MO3 injected and MO3-injected embryos (Fig.4). At 16, 24, and 48 hpf, only a slight change in proliferating cells was observed. While the overall number of proliferating cells showed no significant increase or decrease in the head region of either 16 or 24 hpf embryos, there was a slight decrease in number of proliferating cells in the branchial arches (Fig. 4j and Supporting Information Fig. 1). Our cell death and proliferation assay data suggest that Crispld2 is not necessary for proliferation but likely plays a role in cell survival and/or in the ability to promote proper NCCs migration into the orofacial region. The increase in apoptosis seen in Crispld2 knockdown embryos (Fig. 3) suggests that cell survival is negatively affected when Crispld2 levels are decreased during embryogenesis. Whether Crispld2 plays a direct role in cell migration or this is an indirect effect of loss of cell viability has yet to be determined.
FIG. 3.
Increased apoptosis is present in the head of MO3 injected embryos. Co-injection of MO3 with p53MO and whole mount TUNEL staining shows partial inhibition of MO3-induced apoptosis. Lateral view of UIC (a–f), MO3-injected embryos (g–l), MO3/p53MO co-injected embryos (m–r), and control MM-MO3-injected embryos (s–x), at 16 hpf (a, g, m, s), 24 hpf (b, h, n, t), 36 hpf (c, i, o, u), 48 hpf (d, j, p, v), 60 hpf (e, k, q, w), and 72 hpf (f, l, r, x).
FIG. 4.
Cell proliferation is unchanged in MO3 injected embryos. Whole mount immunohistochemistry with mitotic marker anti-phosphohistone H3 shows proliferating cells in developing embryos. UIC (a–c), MO3 (d–f), and control MM-MO3-injected embryos (g–i) embryos at 16 hpf (a, d, g), 24 hpf (b, e, h), and 48 hpf (c, f, i). (j) Quantification of number of proliferating cells in UIC, MO3, and control MM-MO3 injected embryos at 16 and 24 hpf.
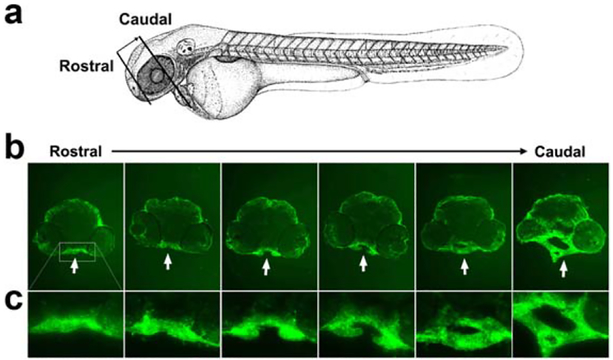
Knocking down crispld2 in the zebrafish results in a decrease of Crispld2 protein in whole embryo lysates as seen by western blot using an antibody directed against zebrafish Crispld2 and this knockdown subsequently led to the significant malformation of the zebrafish craniofacial region (Yuan et al., 2012). We utilized a zebra-fish anti-Crispld2 antibody to determine the localization of the protein in the craniofacial region of embryos. Figure 5 shows a series of consecutive rostral-to-caudal transverse sections through the zebrafish head at 48 hpf. Crispld2 protein is highly expressed in the oropharynx (see white arrows in B, shown higher magnification in C), particularly in the region exhibiting the most severe abnormalities after MO knockdown. Primary and secondary antibody controls confirm this specific staining (Supporting Information Fig. 2). At this same time point, severe loss of migrating NCCs is observed in this region (Fig. 2). Additional studies are needed to determine whether Crispld2 functions as a secreted molecule during craniofacial development as it is suggested to do in lung and kidney development (Nadeau et al., 2010; Quinlan et al., 2007).
FIG. 5.
Crispld2 is highly expressed in zebrafish oropharynx at 48 hpf. (a) Schematic presentation of the location and orientation of transverse sections from rostral to caudal. (b) Top panels show Crispld2 immunostaining through zebrafish oropharynx (bright green) from rostral to caudal at 20× magnification. (c) Enlarged images of oropharynx area (rectangle) of embryos shown in B.
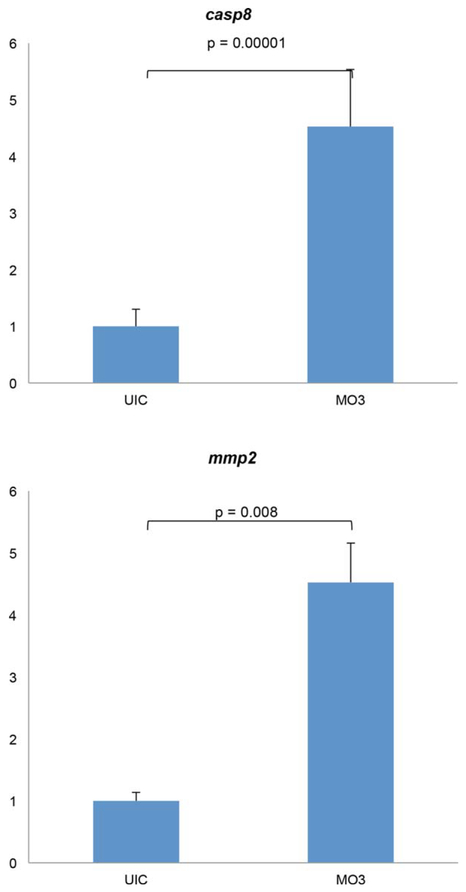
We have previously shown that p53-mediated cell death is not responsible for the severe craniofacial defects seen in Crispld2 knockdown embryos (Yuan et al., 2012). Here, we show an increase in apoptosis in the presence of reduced levels of Crispld2, even upon concurrent knockdown of p53 protein translation. Taken together, these results suggest that cell death resulting from a loss of Crispld2 is mediated by a p53-independent pathway. We next evaluated the expression levels of Caspase 8 (Casp8) and matrix metalloproteinase-2 (Mmp2), as additional cell death pathway genes, in crispld2 knockdown embryos using real-time quantitative PCR. Caspase 8 is a known mediato r of both apoptosis and necrosis and has been shown to act in both p53-dependent and -independent cell death pathways (Blander, 2014; Nikoletopoulou et al., 2013; Shalini et al., 2015). Mmp2 has also been shown to be involved in apoptosis (Mohammad and Kowluru, 2010; Roberts et al., 2002; Vorotnikova et al., 2004). Here, we show that expression levels of Casp8 and Mmp2 are significantly increased in embryos injected with crispld2 MO (Fig. 6), providing insights into potential pathways for the regulation of cell viability by Crispld2. Interestingly, both Casp8 and Mmp2 genes have been shown to play a role in craniofacial development (Ahi et al., 2014; Ekbote et al., 2014; Huang et al., 2011; Mosig et al., 2007; Smane et al., 2013). Moreover, MMP gene variants have been significantly associated with NSCL/P and shown as functionally relevant to this condition (Letra et al., 2007, 2012, 2014).
FIG. 6.
Cell death related gene expression is up-regulated in Crispld2 knockdown embryos. Both mmp2 and casp8 show marked increase in mRNA expression levels in knockdown embryos at 16 hpf. P-value less than 0.05 denotes statistical significance.
In summary, our results demonstrate an important role for Crispld2 in NCC migration, differentiation, and/or survival, and contribute to our better understanding of how this NSCLP-associated gene functions in normal and abnormal craniofacial development. As a secreted glycoprotein, Crispld2 is likely interacting with extracellular components essential for cell migration and loss of this important molecule results in cell death and/or abnormal migration of NCCs into the craniofacies. Additional studies are needed to determine specific downstream effectors of the Crispld2 pathway and fully elucidate the mechanism of function of Crispld2 during craniofacial development. Similarly to CASP8 and MMP2, any downstream pathway effectors should also be considered potential NSCLP candidate genes and should be tested for their roles in craniofacial development as well as analyzed in human NSCLP populations for association and linkage. Further characterization of Crispld2 and its role in craniofacial development will need to be carried out in stable zebrafish crispld2 mutant lines and/or conditional mouse mutants that eliminate Crispld2 specifically in the developing cranio-facial tissue.
METHODS
Zebrafish
Zebrafish (Danio rerio) were raised and housed following standard techniques (Westerfield, 1995). Fertilized eggs were obtained through in-tank breeding.
Morpholino and Human CRISPLD2 mRNA Injections
Zebrafish crispld2 antisense morpholino (MO3: TTGATGATTTCAGGCCCGGACTCTA) targeting the ATG site in exon1, control mismatch MO3 (MM-MO3: TTcATcATTTgAGcCCCcGACTCTA, mismatches in small caps) and p53 MO (GCGCCATTGCTTTGCAAGAATTG) were designed by GeneTools (Philomath, OR). MOs were diluted in nuclease-free water to a stock concentration of 65 mg/mL or 2 mM. Injections of MOs were diluted to 0.2 mg/mL for MO3 and MM-MO3 and 4.5 mg/mL for p53MO in Danieu buffer. A plasmid harboring the full-length human CRISPLD2 cDNA representing NCBI reference sequence NM_031476 was purchased from OriGene (#RC214912, OriGene Technologies.) The full length CRISPLD2 cDNA was subcloned into the pCS2 vector and CRISPLD2 mRNA was generated using the mMessage mMachine Sp6 kit (Ambion). mRNA was resuspended in nuclease-free water to 500 μg/mL stock concentration and diluted to 50 μg/mL in Danieu buffer for injection. One-cell embryos were injected with 1 nL of MOs and mRNA (Yuan et al., 2012).
Time-Lapse Live Imaging of NCC Lineage
Zebrafish embryos were collected from transgenic reporter strain Tg(sox10:egfp). Total amount of 0.2 ng MO3 was injected into the developing embryos. UIC and MO Embryos were dechorionated manually at the 10-somite stage and then submerged in 0.8%–1.2% aga-rose in E3 media containing tricane (Sigma 886-86-2) and covered with cover slips. Time-lapse live embryo imaging was taken place using a compound microscope (Axiovert200M, AxioCam HrM [B&W], Axiovision Software) and a confocal microscope (Leica TCS SP5). Serial images were acquired from 16 to 72 hpf with 30 minutes interval with a 10× objective.
Fluorometric TUNEL Staining
The DeadEnd™ Fluorometric TUNEL System (Promega, Madison, WI) was used to detect apoptotic cells in zebrafish embryos injected with MO3, co-injected with MO3 and p53MO, MM-MO3 and UICs. Embryos were collected at 16, 24, 36, 48, 60, and 72 hpf and fixed in 4% PFA for 1 h at room temperature. Embryos were permeablized overnight at 4°C in 1% triton X 100 in 1× PBS. Following equilibration of embryos in equilibration buffer for 30 minutes at room temperature, embryos were incubated in Nucleotide/rTdT mix for 3 h at 37°C, with light protection. Embryos were washed three times for 10 minutes in 1× PBT. Imaging was performed using LAS Montage Module (Leica, Wetzlar, Germany).
Whole Mount Immunofluorescence Staining with Mitotic Marker Phosphohistone H3
Embryo collection, injection, fixation and permeabilization were performed same as described in TUNEL staining method section. After permeabilizing overnight in 1% TritonX 100/1× PBT, embryos were incubated for 2 h at room temperature in immunofluorescence (IF) blocking solution (10 mg/mL BSA, 10% Fetal Calf Serum, 0.1% Tween-20, 1% DMSO, in 1× PBS), followed by three times 15 minutes wash with IF blocking solution. Embryos were incubated in 1:200 dilution of anti-phosphohistone H3 antibody (Dr. Wagner) in IF blocking solution, washed 3 × 15 minutes with IF blocking solution. Embryos were incubated in 1:500 dilution of Alexa Fluor 488 donkey anti-rabbit IgG (#A21206, Life-Technologies, Eugene) in IF blocking solution. Embryos were washed three times in 1× PBT. Imaging was performed using LAS Montage Module (Leica, Wetzlar, Germany).
Immunohistochemical Staining
UIC embryos were collected at 48 hpf and fixed in 4% paraformaldehyde for 1h at room temperature. Embryos were incubated in 15% and 30% sucrose in PBS consecutively until sucrose has fully penetrated the embryos and all embryos have settled to the bottom. Embryos were embedded in O.C.T. compound overnight at −20°C, and 10 μm thick transverse sections were collected onto glass slides and dried for 3 h at room temperature. Samples were incubated for 1 h in blocking solution containing 0.1% BSA and 5% normal donkey sera in PBS. A custom polyclonal antibody against zebrafish Crispld2 was generated in rabbit (Yuan et al., 2012). Embryos were incubated for 3 h room temperature with rabbit anti-Crispld2 at 1:250 dilution in blocking solution, followed by three times 15 minutes washes with blocking solution. Alexa Fluor 488 donkey anti-rabbit IgG (#A21206, LifeTechnologies, Eugene) was used as secondary antibody at 1:500 dilution in blocking solution and 1 h incubation at room temperature. A compound microscope was used for imaging (Olympus BX51, SPOT RT camera).
mRNA Quantification
Total RNA was isolated from 16 hpf UIC and MO3 injected zebrafish embryos (embryos were de-yolked and the anterior one-third of the embryo was isolated) using TRIzol Reagent (Invitrogen) according to kit instructions. Total RNA was then reverse transcribed using SuperScript II reverse transcriptase (Invitrogen) using manufacturer’s instructions. On-demand TaqMan gene expression assays for zebrafish casp8 and mmp2 were obtained from Life Technologies (Foster City, CA) and quantitative real-time PCR was run on a ViiA7 sequence detection instrument (Life Technologies) using TaqMan chemistry. Relative changes in mRNA levels were assessed with the comparative cycle threshold (CT) and the 2−ddCt method (Pfaffl, 2001). Beta actin (actb1) was used as endogenous control. Reactions were run in triplicate. P-value less than 0.05 denotes statistical significance.
Supplementary Material
Acknowledgments
Contract grant sponsor: NIH R01DE11931 to JTH
Footnotes
Additional Supporting Information may be found in the online version of this article.
The authors have no conflict of interests.
LITERATURE CITED
- Ahi EP, Kapralova KH, Palsson A, Maier VH, Gudbrandsson J, Snorrason SS, Jonsson ZO, Franzdottir SR. 2014. Transcriptional dynamics of a conserved gene expression network associated with craniofacial divergence in Arctic charr. Evodevo 5:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blander JM. 2014. A long-awaited merger of the pathways mediating host defence and programmed cell death. Nat Rev Immunol 14:601–618. [DOI] [PubMed] [Google Scholar]
- Chiquet BT, Lidral AC, Stal S, Mulliken JB, Moreno LM, Arcos-Burgos M, Valencia-Ramirez C, Blanton SH, Hecht JT. 2007. CRISPLD2: A novel NSCLP candidate gene. Hum Mol Genet 16:2241–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekbote AV, Danda S, Zankl A, Mandal K, Maguire T, Ungerer K. 2014. Patient with mutation in the matrix metalloproteinase 2 (MMP2) gene – a case report and review of the literature. J Clin Res Pediatr Endocrinol 6:40–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Di R, Tao F, Chang Z, Lu S, Fan W, Shan C, Li X, Yang Z. 2010. PDK1 regulates vascular remodeling and promotes epithelial-mesenchymal transition in cardiac development. Mol Cell Biol 30:3711–3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himes BE, Jiang X, Wagner P, Hu R, Wang Q, Klanderman B, Whitaker RM, Duan Q, Lasky-Su J, Nikolos C, Jester W, Johnson M, Panettieri RA Jr., Tantisira KG, Weiss ST, Lu Q. 2014. RNA-Seq transcriptome profiling identifies CRISPLD2 as a glucocorticoid responsive gene that modulates cytokine function in airway smooth muscle cells. PLoS One 9:e99625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Yokota T, Iwata J, Chai Y. 2011. Tgf-beta-mediated FasL-Fas-Caspase pathway is crucial during palatogenesis. J Dent Res 90:981–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang P, Svoboda KK. 2005. Epithelial-mesenchymal transformation during craniofacial development. J Dent Res 84:678–690. [DOI] [PubMed] [Google Scholar]
- Kaplan F, Ledoux P, Kassamali FQ, Gagnon S, Post M, Koehler D, Deimling J, Sweezey NB. 1999. A novel developmentally regulated gene in lung mesenchyme: Homology to a tumor-derived trypsin inhibitor. Am J Physiol 276:L1027–L1036. [DOI] [PubMed] [Google Scholar]
- Lan J, Ribeiro L, Mandeville I, Nadeau K, Bao T, Cornejo S, Sweezey NB, Kaplan F. 2009. Inflammatory cytokines, goblet cell hyperplasia and altered lung mechanics in Lgl1 +/− mice. Respir Res 10:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letra A, Silva RA, Menezes R, Astolfi CM, Shinohara A, de Souza AP, Granjeiro JM. 2007. MMP gene polymorphisms as contributors for cleft lip/palate: Association with MMP3 but not MMP1. Arch Oral Biol 52:954–960. [DOI] [PubMed] [Google Scholar]
- Letra A, Menezes R, Cooper M, Fonseca R, Tropp S, Govil M, Granjeiro J, Imoehl S, Mansilla M, Murray J, Castilla E, Orioli I, Czeizel AE, Ma L, Chiquet B, Hecht J, Vieira A, Marazita M. 2010. Crispld2 variants including a C471t silent mutation may contribute to nonsyndromic cleft lip with or without cleft palate. Cleft Palate Craniofac J 48(4):363–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letra A, Silva RM, Motta LG, Blanton SH, Hecht JT, Granjeirol JM, Vieira AR. 2012. Association of MMP3 and TIMP2 promoter polymorphisms with nonsyndromic oral clefts. Birth Defects Res A Clin Mol Teratol 94:540–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letra A, Zhao M, Silva RM, Vieira AR, Hecht JT. 2014. Functional significance of MMP3 and TIMP2 polymorphisms in cleft lip/palate. J Dent Res 93:651–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micalizzi DS, Farabaugh SM, Ford HL. 2010. Epithelialmesenchymal transition in cancer: Parallels between normal development and tumor progression. J Mammary Gland Biol Neoplasia 15:117–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijiti A, Ling W, Maimaiti A, Tuerdi M, Tuerxun J, Moming A. 2015. Preliminary evidence of an interaction between the CRISPLD2 gene and nonsyndromic cleft lip with or without cleft palate (nsCL/P) in Xinjiang Uyghur population, China. Int J Pediatr Otorhinolaryngol 79:94–100. [DOI] [PubMed] [Google Scholar]
- Mohammad G, Kowluru RA. 2010. Matrix metalloproteinase-2 in the development of diabetic retinopathy and mitochondrial dysfunction. Lab Invest 90:1365–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosig RA, Dowling O, DiFeo A, Ramirez MC, Parker IC, Abe E, Diouri J, Aqeel AA, Wylie JD, Oblander SA, Madri J, Bianco P, Apte SS, Zaidi M, Doty SB, Majeska RJ, Schaffler MB, Martignetti JA. 2007. Loss of MMP-2 disrupts skeletal and craniofacial development and results in decreased bone mineralization, joint erosion and defects in osteoblast and osteoclast growth. Hum Mol Genet 16:1113–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau K, Montermini L, Mandeville I, Xu M, Weiss ST, Sweezey NB, Kaplan F. 2010. Modulation of Lgl1 by steroid, retinoic acid, and vitamin D models complex transcriptional regulation during alveolarization. Pediatr Res 67:375–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoletopoulou V, Markaki M, Palikaras K, Tavernarakis N. 2013. Crosstalk between apoptosis, necrosis and autophagy. Biochim Biophys Acta 1833:3448–3459. [DOI] [PubMed] [Google Scholar]
- Oyewumi L, Kaplan F, Sweezey NB. 2003. Lgl1, a mesenchymal modulator of early lung branching morpho-genesis, is a secreted glycoprotein imported by late gestation lung epithelial cells. Biochem J 376:61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan J, Kaplan F, Sweezey N, Goodyer P. 2007. LGL1, a novel branching morphogen in developing kidney, is induced by retinoic acid. Am J Physiol Renal Physiol 293:F987–F993. [DOI] [PubMed] [Google Scholar]
- Roberts LM, Visser JA, Ingraham HA. 2002. Involvement of a matrix metalloproteinase in MIS-induced cell death during urogenital development. Development 129:1487–1496. [DOI] [PubMed] [Google Scholar]
- Sanders EJ, Prasad S. 1989. Invasion of a basement membrane matrix by chick embryo primitive streak cells in vitro. J Cell Sci 92(Pt 3):497–504. [DOI] [PubMed] [Google Scholar]
- Shalini S, Dorstyn L, Dawar S, Kumar S. 2015. Old, new and emerging functions of caspases. Cell Death Differ 22:526–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Liu RM, Yang L, Wu H, Li PQ, Liang YL, Xie XD, Yao T, Zhang TT, Yu M. 2011. The CRISPLD2 gene is involved in cleft lip and/or cleft palate in a Chinese population. Birth Defects Res A Clin Mol Teratol 91: 918–924. [DOI] [PubMed] [Google Scholar]
- Shi J, Jiao X, Song T, Zhang B, Qin C, Cao F. 2010. CRISPLD2 polymorphisms are associated with nonsyndromic cleft lip with or without cleft palate in a northern Chinese population. Eur J Oral Sci 118: 430–433. [DOI] [PubMed] [Google Scholar]
- Smane L, Pilmane M, Akota I. 2013. Apoptosis and MMP-2, TIMP-2 expression in cleft lip and palate. Stomatologija 15:129–134. [PubMed] [Google Scholar]
- Vasarhelyi V, Trexler M, Patthy L. 2014. Both LCCL-domains of human CRISPLD2 have high affinity for lipid A. Biochimie 97:66–71. [DOI] [PubMed] [Google Scholar]
- Vorotnikova E, Tries M, Braunhut S. 2004. Retinoids and TIMP1 prevent radiation-induced apoptosis of capillary endothelial cells. Radiat Res 161:174–184. [DOI] [PubMed] [Google Scholar]
- Wang ZQ, Xing WM, Fan HH, Wang KS, Zhang HK, Wang QW, Qi J, Yang HM, Yang J, Ren YN, Cui SJ, Zhang X, Liu F, Lin DH, Wang WH, Hoffmann MK, Han ZG. 2009. The novel lipopolysaccharide-binding protein CRISPLD2 is a critical serum protein to regulate endotoxin function. J Immunol 183: 6646–6656. [DOI] [PubMed] [Google Scholar]
- Wang T, Wang ZQ, Wang L, Yan L, Wan J, Zhang S, Jiang HQ, Li WF, Lin ZF. 2013. CRISPLD2 is expressed at low levels during septic shock and is associated with procalcitonin. PLoS One 8:e65743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M 1995. The zebrafish book: A guide for the laboratory use of zebrafish (Danio rerio), 3rd ed. Eugene, OR: M. Westerfield. [Google Scholar]
- Yoo JY, Shin H, Kim TH, Choi WS, Ferguson SD, Fazleabas AT, Young SL, Lessey BA, Ha UH, Jeong JW. 2014. CRISPLD2 is a target of progesterone receptor and its expression is decreased in women with endometriosis. PLoS One 9:e100481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Q, Chiquet BT, Devault L, Warman ML, Nakamura Y, Swindell EC, Hecht JT. 2012. Craniofacial abnormalities result from knock down of nonsyndromic clefting gene, crispld2, in zebrafish. Genesis 50: 871–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Sweezey NB, Kaplan F. 2015. LGL1 modulates proliferation, apoptosis, and migration of human fetal lung fibroblasts. Am J Physiol Lung Cell Mol Physiol 308:L391–L402. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.