For undiagnosed patients and those with rare diseases, the affected individuals themselves are an especially critical source of phenotyping information. These patients live with their condition and develop explicit and implicit knowledge about it, whether from multiple clinician evaluations or from other families and patients experiencing diagnosis for similar conditions. From these interactions, they develop a lexicon of relevant terms; these terms are frequently in plain language, but can also include clinical terms. In exceptional cases, patients’ self-phenotyping and investigations have led to clinical diagnosis; for example, Jill Viles, despite skepticism from doctors, managed to not only diagnose herself and her family members but also to discover fundamental biology and improved management of autosomal dominant Emery–Dreifuss muscular dystrophy1. Further, some phenotypes are not readily observed clinically but can be documented by the patient or family; for instance, the Might family (NGLY1) observed that their baby did not produce tears when crying, and they used this feature to help identify other families with the same disease.
Human genetics and precision medicine aim to understand the relationship between genetic variants and diseases. Whole-exome and whole-genome sequencing have transformed the ability to comprehensively characterize genetic variants. Although whole-exome and whole-genome sequencing have led to the discovery of many novel disease-associated genes, the diagnostic yield in patients without a clear clinical diagnosis has been 11–25%2. With approximately 30,000–100,000 called variants in a typical exome or ~4.5 million variants in a typical genome, multiple candidate genes remain after a bioinformatic analysis; additional methods and data are needed to refine the list of candidates.
The Human Phenotype Ontology (HPO) was created to enable ‘deep phenotyping’, that is, capture of symptoms and phenotypic findings using a logically constructed hierarchy of phenotypic terms3. The HPO has become the de facto standard for representing clinical phenotype data to inform diagnoses for rare genetic diseases by the 100,000 Genomes Project, the NIH Undiagnosed Diseases Program (UDP) and Undiagnosed Diseases Network (UDN), and thousands of other clinics, laboratories, tools and databases3–5. The computable phenotypic profiles (sets of terms) of individual patients allow imperfect or ‘fuzzy’ matching against the phenotypic profiles of known diseases and model organisms; this matching is based on proximity of terms in the hierarchy and on term specificity3,6,7. In comparison to using whole-exome or whole-genome sequencing and clinical data alone, we have shown that HPO-based phenotyping improves molecular diagnosis in the UDP by 10–20%8.
One of the bottlenecks is that deep phenotyping can be time-consuming and miss key phenotypic features not observed clinically. Patients could contribute computable phenotype data themselves; however, the ‘terminology gap’ between medical professionals and patients is a hindrance. The terminology gap has also limited patient participation both in research studies and in clinical phenotyping9. Current patient vocabularies (such as the Consumer Health Vocabulary Initiative; see URLs) provide broad consumer equivalents for clinical findings, medical procedures and equipment, but are not well integrated with research terminologies; thus, they provide neither the structure nor the coverage required for translational research and diagnostic tools. While consumer vocabularies and clinical survey instruments can be mapped to HPO terms, they tend to provide only high-level phenotype terms that are unsuitable for use in clinical genetics contexts. Finally, lay synonyms would be very useful to improve information retrieval for patients from the literature.
To address these issues, we sought to make the HPO capable of capturing patient-generated phenotypic profiles for use in diagnostic and patient community settings (registries, forums, clinics and patient websites). To achieve this, we systematically added patient-centered synonyms throughout the HPO by manually reviewing many sources and knowledgebases, including Wikipedia, MedlinePlus, the Mayo Clinic, Online Mendelian Inheritance in Man (OMIM), the Elements of Morphology and patient forums such as the WebMD message board, as well as other ontologies, terminologies and specialty texts. We also made synonym creation across terms consistent. For example, ‘increased bone density in 2nd toe bone’ and ‘increased bone density in 3rd toe bone’, etc., were added as synonyms for ‘sclerosis of the 2nd toe phalanx’ and ‘sclerosis of the 3rd toe phalanx’, respectively. The synonyms are classified as exact, broad, narrow or related according to Open Biological and Biomedical Ontology Foundry (see URLs) convention. Synonyms are classified as exact if they can be used interchangeably in computational algorithms without loss of precision. For 40% of the terms, there exists no reasonable non-clinical language, for example, ‘anomalous origin of left coronary artery from the pulmonary artery’ (HP:0011638). Other terms such as phenotypes involving the radius and ulna could not be succinctly distinguished in layperson terminology (for example, ‘forearm bone’). We also recognized relationships within the ontology when adding layperson synonyms: for example, ‘yellowing of the skin’ was added to the HPO class ‘jaundice’, but also to subclasses. ‘Intermittent jaundice’, for example, would have the lay synonym ‘intermittent yellowing of the skin’,
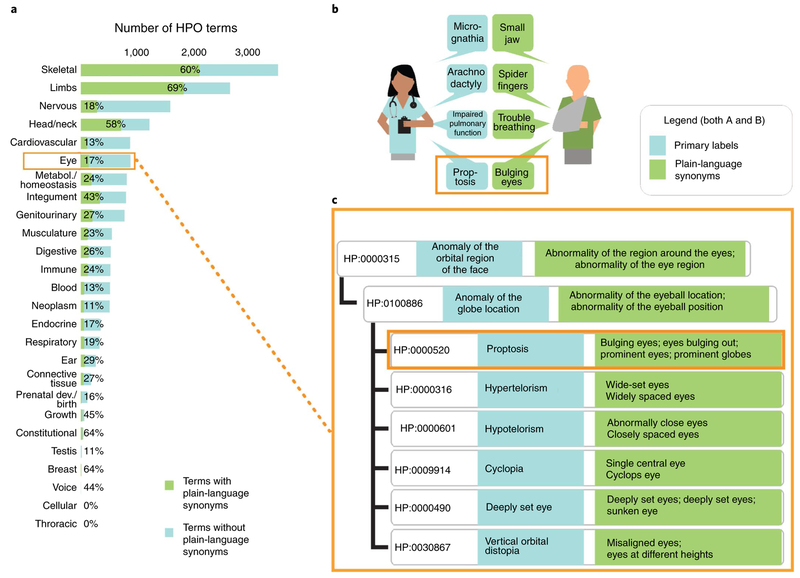
We reviewed the population and distribution of lay terms within the HPO and found that the major fraction of layperson synonyms were added to very specific terms (for example, ‘clinodactyly’ rather than ‘abnormality of the digits’), suggesting that they would be specific enough to be used diagnostically. In total, 36% of the HPO terms from the most recent release (see URLs) have at least one layperson synonym (4,547 of 12,623) (Fig. 1). 89% of the diseases (8,666 of 9,657) in the HPO database have at least one HPO annotation with a layperson synonym, and 60% of all disease annotations (73,932 of 122,120) are referring to HPO terms with lay translations. This further suggests that the laypersonHPO would be useful in a diagnostic setting despite incomplete coverage. Further, because the lay translation of the HPO uses the same logical infrastructure as the HPO, patient-generated phenotyping data can be readily combined with clinical phenotyping data in the context of variant prioritization to improve diagnostic rates as well as other analytics such as examining expressivity, penetrance and disease progression. Through a recently funded partnership with the Patient-Centered Outcomes Research Institute (PCORI), we will perform a formal evaluation of the layperson HPO. This will include an informatics comparison against the gold-standard HPO used in genomic diagnostics and usability and effective HPO profile generation in cohorts of patients with rare diseases.
Figure 1: HPO pipeline.
a, Coverage of HPO terms with plain-language synonyms (terms broken down by anatomical system), b, A physician and a patient describe a patient’s phenotype profile in different ways but with the same meaning. This constellation of diverse phenotypes is common in Marfan syndrome; each has a plain-language equivalent. c, A sub-branch of eye phenotypes within the HPO. Terms are structured rigorously, not only in terms of hierarchy (as shown) but also in terms of logical definitions (not shown).
We envision the layperson HPO as a resource that will allow patients and families to become more effective partners in translational research, empowering families to achieve an accurate diagnosis, as well as providing opportunities for people to improve the lives of others by increasing medical knowledge through their personal perspectives. The laypersonHPO will support efforts such as the patient-friendly self-phenotyping tool Phentypr (see URLs); MyGene2 (see URLs), which extracts lay synonyms from patient stories; the HPO browser (see URLs) for display; the National Organization for Rare Disorders (see URLs) in its registry tool; and navigation on the Genetic and Rare Diseases Information Center. Rare disease phenotyping data should be as computationally useful and as open aspossible (Findable, Accessible, Interoperable and Reusable, FAIR; see URLs). Therefore,we urge clinical geneticists, communities of patients with rare diseases, phenotyping tools,registries and consumer testing laboratories to help evaluate, adopt and contribute towardwhat we think is a critical resource for engaging patients in their own deep phenotyping. Finally, we envision that the layperson HPO will enable patients with rare diseases to share their phenotyping profiles openly on the web, even in forums such as Facebook. This will allow the use of informatics to support open querying for similar patients to improve diagnosis and for cohort and community identification globally.
Acknowledgements
Funding was provided by the US National Institutes of Health (NIH) Monarch Initiative (OD 5R24OD011883; N.A.V., L.C., T.G., C.M., J.A.M., P.N.R., K.S., S.K., M.A.H.) and Patient-Centered Outcomes Research Institute (PCORI) contract number ME-1511-33184 (N.A.V., I.H., C.B., K.S., J.A.M., M.A.H.).
Footnotes
Competing interests The authors declare no competing interests.
URLs
Consumer Health Vocabulary initiative, http://consumerhealthvocab.chpc.utah.edu/CHVwiki/; Open Biological and Biomedical Ontology Foundry, http://obofoundry.org/; Human Phenotype Ontology, http://purl.obolibrary.org/obo/hp.owl; Phentypr, https://phenotypr.com/; MyGene2, http://mygene2.org/; HPO browser, http://www.human-phenotype-ontology.org/; National Organization for Rare Disorders, https://rarediseases.org/; Genetic and Rare Diseases Information Center, https://rarediseases.info.nih.gov/; FAIR, https://www.force11.org/group/fairgroup/fairprinciples.
References
- 1.Epstein D The muscular dystrophy patient and Olympic medalist with the same genetic disorder. ProPublica (15 January 2016). [Google Scholar]
- 2.Yang Y et al. N. Engl. J. Med 369, 1502–1511 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Köhler S et al. Nucleic Acids Res. 45 (D1), D865–D876 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Posey JE et al. Genet. Med 18, 678–685 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bone WP et al. Genet. Med 18, 608–617 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Köhler S et al. Am. J. Hum. Genet 85, 457–464 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mungall CJ et al. Nucleic Acids Res. 45 (D1), D712–D722 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gall T et al. Front. Med 4, 62 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park MS, He Z, Chen Z, Oh S & Bian J JMIR Med. Inform 4, e41 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]