Abstract
Mouse mammary tumor virus (MMTV) induces breast cancer in mice in the absence of known virally-encoded oncogenes. Tumorigenesis by MMTV is thought to occur primarily through insertional mutagenesis, leading to the activation of cellular proto-oncogenes and outgrowth of selected cells. Here we investigated whether MMTV encodes microRNAs (miRNAs) and/or modulates host miRNAs that could contribute to tumorigenesis. High throughput small RNA sequencing analysis of MMTV-infected cells and MMTV-induced mammary tumors demonstrates that MMTV does not encode miRNAs. However, infected tissues have altered levels of several host miRNAs, including increased expression of members of the oncogenic miRNA cluster, miR-17-92. Notably, similar changes in miRNA levels have been previously reported in human breast cancers. Combined, our results demonstrate that virally encoded miRNAs do not contribute to MMTV-mediated tumorigenesis, but that changes in specific host miRNAs in infected cells may contribute to virus replication and tumor biology.
Keywords: Retroviruses, Mouse mammary tumor virus (MMTV), Breast cancer, Virus-host interactions, miRNAs, miR-17-92 cluster.
Introduction
Mouse mammary tumor virus (MMTV) is an oncogenic, complex retrovirus that causes breast cancer and T-cell lymphomas in mice (reviewed in (Dudley et al., 2016; Ross, 2010)). MMTV is transmitted both as endogenous proviruses (Mtvs) and exogenous viruses (Callahan et al., 1982; Imai et al., 1994; Stewart et al., 2000), yet tumors are induced primarily by exogenous viruses transmitted from mothers to progeny through breast milk (Bittner, 1936). Large amounts of virus in the milk allow infection of gut-associated dendritic cells followed by transmission to T and B cells in the Peyer’s patches (Beutner et al., 1994; Courreges et al., 2007; Golovkina et al., 1992; Held et al., 1993). Expression of the sag gene product on antigen-presenting cells leads to stimulation and amplification of MMTV-expressing lymphocytes, which are required for efficient infection of the mammary gland, the most permissive tissue for virus production (Finke and Acha-Orbea, 2001; Golovkina et al., 1998; Zhu et al., 2004). MMTV is a known insertional mutagen. High levels of virus replication in permissive mammary epithelial cells results in relatively random insertions near the vicinity of a specific subset of growth promoting protooncogenes, e.g., Wnt1, Fgf3, and Notch4, activating their expression (reviewed in (Dudley et al., 2016)). This activation results in clonal expansion of cells carrying proviruses, and ultimately in tumor formation (Callahan and Smith, 2000; Dudley et al., 2016; Ross, 2010). Since MMTV does not encode a known oncogene, other MMTV-encoded factors may be involved in breast cancer induction. MicroRNAs (miRNAs) are small regulatory (~21-24 nucleotide (nt)) non-coding RNAs that are involved in diverse biological processes, including cancer (reviewed in (Di Leva et al., 2014)). Although the non-acute retrovirus bovine leukemia virus (BLV) encodes miRNAs that may contribute to their oncogenic capacity (Gillet et al., 2016; Kincaid et al., 2012), whether MMTV encodes miRNAs is unknown.
MicroRNAs are generated from structured longer primary transcripts (pri-miRNA) that are cleaved via a series of endonucleases (reviewed in (Kim et al., 2009)). The RNA-induced silencing complex (RISC) binds the final mature miRNA product and scans mRNAs (Bartel, 2009). Docking to mRNAs via partial sequence complementarity results in decreased translation and steady state levels of targeted transcripts (Bartel, 2004 and 2009). Although most miRNAs derive from a monocistronic locus, greater than one-third of human miRNAs exist in clusters and are transcribed as polycistrons (Altuvia et al., 2005; Bartel, 2004). A single type of miRNA can have numerous mRNA targets (Bartel, 2009). Some miRNAs are oncogenic (oncomiRs), act as tumor suppressors, or both, depending on context (Esquela-Kerscher and Slack, 2006; Hede, 2010; Pekarsky and Croce, 2010; Zeitels et al., 2014). Of note, miRNAs also play a role in tumors associated with virus infection (Gillet et al., 2016; Gottwein et al., 2007; Linnstaedt et al., 2010; Skalsky et al., 2007; Zhao et al., 2009).
Viral miRNAs are encoded by diverse DNA virus families, including herpesviruses, polyomaviruses, adenoviruses, anelloviruses, ascoviruses, and baculoviruses (Grundohoff and Sullivan, 2011; Kincaid and Sullivan,2012). Reported functions for these miRNAs include targeting host or viral transcripts to alter immune detection, apoptosis, latency, neurovirulence, and virus replication (reviewed in (Grundhoff and Sullivan, 2011; Kincaid and Sullivan, 2012; Skalsky and Cullen, 2010)). Although most viruses with RNA genomes lack encoded miRNAs, likely to protect their genomes from endonucleolytic degradation, some retroviruses, such as BLV and foamy viruses (FVs), encode miRNAs that utilize non-canonical biogenesis routes that avoid cleaving the genomic RNA (Grundhoff and Sullivan, 2011; Kincaid et al., 2012, 2014; Kincaid and Sullivan, 2012; Whisnant et al., 2014). MicroRNAs from both of these retroviral families encode miRNAs that mimic the seed sequence of host miRNAs with known oncogenic activities (Kincaid et al., 2012, 2014). The number of retroviral genera that encode viral miRNAs remains unknown.
In this study, we applied high-throughput small RNA sequencing to detect virus-encoded miRNAs in MMTV-infected mouse mammary cells as well as primary MMTV-infected mammary tumors. These results revealed no evidence for virally encoded miRNAs. In contrast, small RNA sequencing and Northern blot analyses demonstrated increased expression of a subset of host encoded miRNAs, including members of the oncogenic miR-17-92 cluster, in both MMTV-infected breast tumors and normal mammary glands. Together, these results suggest that MMTV infection alters host miRNA levels likely to play a role in virus replication and tumorigenesis.
Materials and Methods
Tumor Samples:
MMTV-induced mammary tumor RNAs from archived samples were tested in this study. The tumors were generated as described previously (Bhadra et al., 2005; Mustafa et al., 2000). Briefly, a cloned MMTV isolate, HYB-MTV (Shackleford and Varmus, 1988), was stably transfected into rat XC cells that are devoid of any endogenous Mtvs. Virus-producing cells (2 × 107) were injected intraperitoneally into inbred female BALB/c weanlings. Infected mice were subjected to continuous breeding to accelerate mammary tumorigenesis. Tumors were observed in most mice within 6 to 12 months following injection of virus-producing cells. Tumor-bearing mice were subsequently euthanized by CO2 asphyxiation, and the tumor tissue or mammary glands from uninfected or MMTV-infected mice were harvested for RNA isolation. All mouse handling and experimental procedures were approved by the University of Texas at Austin Institutional Animal Care and Use Committee.
Mm5MT Virus Stock Preparation:
Approximately 2 × 106 Mm5MT mammary tumor cells (ATCC; USA) were plated into two T175 flasks and, after ~24 hours, were induced overnight with 10−6 M dexamethasone (Sigma, USA) prepared in molecular biology grade 100% ethanol (Sigma). Supernatants from the hormone-induced cells were harvested for virus stock production at 24 hours post induction. Cellular debris was removed by centrifugation at 4,000 rpm for 10 min at 4°C followed by filtration through a 0.2 μm syringe filter. Aliquots of the filtered virus stock were stored at −80°C for subsequent use.
MMTV Infection of HC11 Cells:
Mouse mammary HC11 cells (passage 40) were plated at a density of 1×106 cells per T75 flask (Corning, USA). After ~ 24 hours, cells were infected with 2.5 ml of the Mm5MT virus stock using 10 mg/ml of DEAE dextran (Sigma). For mock infection, the same volume of media lacking virus was added to duplicate flasks. Infection was performed for 4 hours at 37°C in the presence of 5% CO2 with intermittent shaking every 30 minutes. Infection was stopped by the addition of fresh media, and cells were harvested for protein and RNA extraction at 48 hours post infection.
RNA Isolation:
RNA samples from normal mammary or tumor tissues and cultured cells were isolated using Trizol reagent (Invitrogen, USA) as recommended by the manufacturer. Extracted RNA samples were resuspended in nuclease-free buffer, quantitated by absorbance at 260 nm, and stored in 70% ethanol at −80°C until further use.
Small RNA Sequencing:
For mammary gland and mammary tumor samples, total RNA (with RNA Integrity Number (RIN values of ≥ 8) was processed for Illumina TruSeq small RNA library construction by Sengenics (Kuala Lumpur, Malaysia). RIN values (Agilent) are calculated based on the ratio of 28S and 18S ribosomal RNA to total RNA with values of 1 (low integrity) to 10 (high integrity) (Schroeder et al., 2006). For HC11 cell culture samples, total RNA (RIN values of 9.8-10.0 as measured by Agilent 2100 Bioanalyzer or 2200 TapeStation) harvested 48 hours post infection were processed for Illumina TruSeq small RNA library construction by Macrogen Inc. (Seoul, Korea). Briefly, a 3’ adaptor was ligated to the RNA molecules followed by hybridization of an RT primer to the adaptor. The 5’ adaptor was then ligated to total RNA, followed by reverse transcription of RNA in a first strand synthesis reaction. After first-strand cDNA synthesis, adaptor ligated molecules were amplified with adaptor-specific primers to enrich for the miRNA fraction of total RNA. Amplified material was then purified and size selected on a polyacrylamide gel. The mammary gland and mammary tumor libraries were sequenced using the Illumina HiSeq 2000 platform and approximately 20 million single-end reads were sequenced per sample with a 75-bp read length. The HC11 cell culture samples were sequenced using the Illumina HiSeq 2500 platform and approximately 100 million single-end reads were sequenced per sample with a 51-bp read length.
MicroRNA (miRNA) Sequencing Analysis:
Small RNA reads were pre-processed by trimming the adapter sequences and removing trimmed sequences shorter than 16 nucleotides with Cutadapt (version 1.4.2) (Martin, 2011). Reads were mapped with SHRiMP2 (version 2.2.3) (David et al., 2011) to the reference sequences consisting of miRBase release 21 annotated mouse miRNAs (Griffiths-Jones et al., 2006) and MMTV reference sequences ((HYB MTV (Shackleford and Varmus, 1988) clone sequenced in-house for in vivo samples or C3H provirus sequence AF228552.1 for cell culture experiments), MTV-6_X64554.1, MTV-8_M22028.1, MTV-8_X05400.1, MTV-8_X74073.1, MTV-8_M74713.1, MTV-9_M29600.1, MTV-9_M21800.1). Novel candidate microRNA identification was performed by converting the SHRiMP2 aligned sam files to miRDeep2 format, removing reads shorter than 20 nucleotides, and running the miRDeep2 pipeline with default parameters (Friedländer et al., 2011). Host microRNA quantification was performed using the mirUtils package (version 1.0.0-r27). Student’s t-test was performed to test for statistical differences between the tumor and mammary gland samples.
Data Availability:
RNA-seq data has been deposited through NCBI GEO (accession no GSE101333).
Northern Blot Analysis:
Northern blot analysis of miRNAs expressed in control (mammary gland RNA from pregnant and lactating female BALB/cJ mice), HYB-MTV-infected lactating mammary glands, and HYB MTV-induced mammary tumor RNA samples was performed as described previously (McClure et al., 2011). Sequences of probes for Northern blot hybridization are listed in Table S1.
Results
Virus-encoded miRNAs are undetectable in both an in vitro cell culture model and in vivo MMTV-induced mammary tumors.
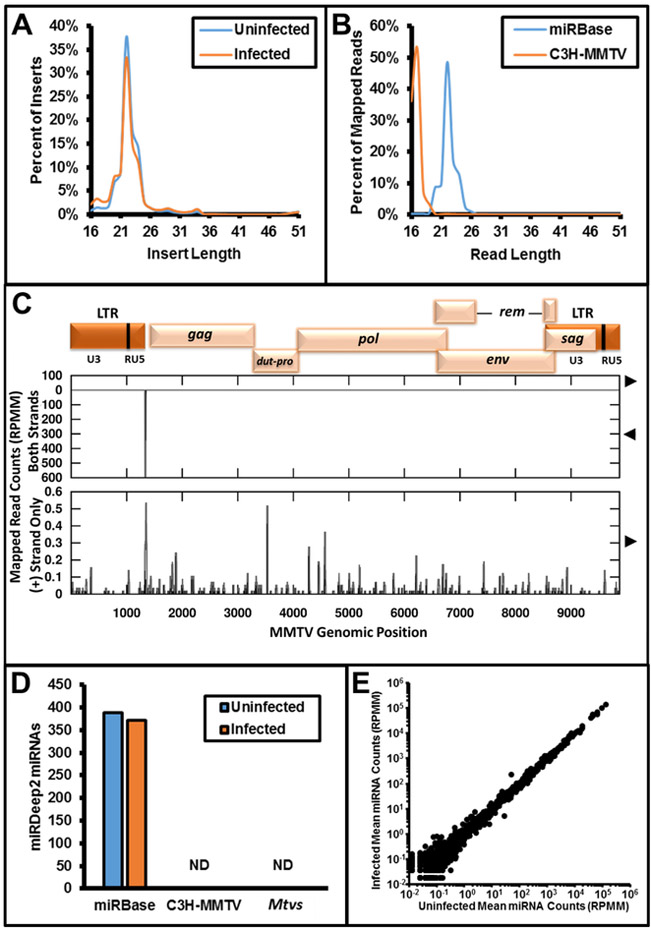
To determine whether MMTV expresses viral miRNAs during replication in an in vitro cell culture model, we infected mouse mammary cell line HC11 cells with C3H-MMTV and harvested total protein and RNA at 48 hours post infection. Western blot analysis confirmed expression of viral gene products, which verified active viral replication (Figure S1). Small RNA sequencing libraries were prepared using RNA harvested from infected and uninfected cells. Greater than 80 million single-end reads were produced with a modal insert length of 22 nucleotides after adaptor removal for both libraries, consistent with the expected size for libraries enriched for miRNAs (Figure 1A). We mapped the reads to miRBase release 21 as well as the exogenous C3H-MMTV provirus and the three endogenous proviruses of BALB/c mice (Mtv 6, 8, and 9). Although sporadic reads mapped to the C3H-MMTV provirus reference sequence, we did not observe any enrichment for reads in the size class of miRNAs as observed for the annotated miRBase host miRNAs (Figure 1B, 1C). We observed a distinct peak of reads mapping antisense to the viral genome at the position of the primer binding site with a modal length of 17 nucleotides. Because this region is complementary to host-specific transfer RNAs (tRNAs), we propose that these reads represent fragments of degraded host encoded tRNAs. Next, we used the miRDeep2 pipeline to identify signatures of miRNAs (including the presence of a predicted stem loop structure, clearly defined 5’ ends as processed by Dicer/Drosha, etc.). We recovered 371-388 miRBase annotated miRNAs per library, but no exogenous or endogenous MMTV-encoded miRNAs consistent with standard criteria were detected (Friedländer et al., 2011) (Figure 1D). No significant differences were observed between miRs detected in infected and uninfected normal mammary cells (Figure 1E). From these data, we conclude that C3H-MMTV does not express virally encoded miRNAs in an HC11 mammary culture model.
Figure 1.
Analysis of small RNA-seq libraries prepared from cultured HC11 cells infected with C3H-MMTV. A. Length of inserts after adaptor trimming for small RNA libraries prepared from uninfected and infected HC11 cells. B. Length of reads mapped to the indicated reference sequences (miRBase: miRBase release 21 annotated miRNA sequences, C3H MMTV: MMTV reference sequence GenBank accession AF228552.1) for small RNA library prepared from C3H-MMTV infected HC11 cells. C. Plots of read coverage and start sites for reads mapping to the C3H-MMTV reference sequence. At the top, a map of MMTV genomic organization is provided. On the Y-axis, coverage is plotted with gray lines, and read start counts are plotted with black impulses. Values above the X-axis represent the forward strand and below values represent reads mapping to the negative strand. Genomic position is indicated on the X-axis. Top graph presents summary of all reads mapping to the reference sequence. Bottom graph only depicts reads mapping to the sense strand of the viral reference sequence. D. Summary of miRDeep2 analysis of small RNA library prepared from C3H-MMTV-infected HC11 cells. Y-axis indicates the number of candidate miRNAs for the respective set of reference sequences [miRBase, C3H-MMTV, and Mtvs (see Methods for accession numbers)] indicated on the X-axis. ND indicates none detected. E. Quantification of miRBase annotated miRNAs in uninfected and infected HC11 samples as assayed by small RNA-seq. Counts for miRBase annotated microRNAs are given in reads per million mapped with one pseudocount (RPMM).
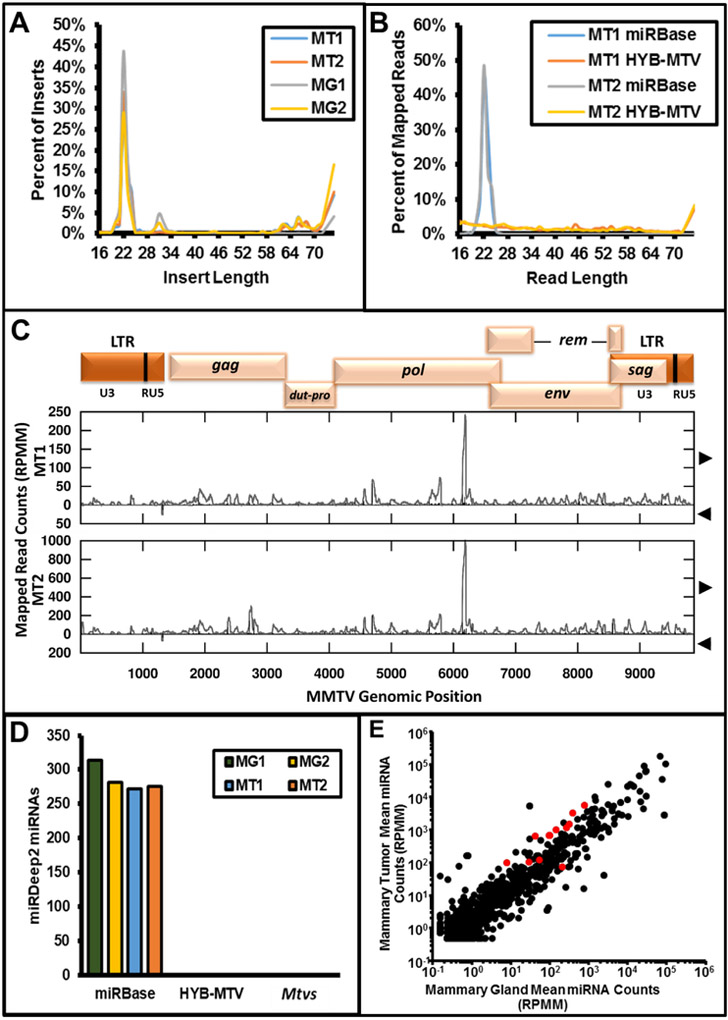
Next, we determined whether MMTV has the potential to express viral miRNAs in vivo. MMTV-induced mouse mammary tumors are permissive for MMTV replication (Dzuris et al., 1999; Tremblay et al., 1989), allowing high levels of infection (Callahan and Smith, 2000). In addition, because MMTV is not subject to superinfection resistance (Dzuris et al., 1999), cells are repeatedly re-infected both in vivo and in cell culture, but the higher levels of re-infection in vivo amplify virus production. Therefore, RNA preparations were obtained from two different mammary tumors derived from two BALB/c mice inoculated with the C3H-MMTV-related infectious clone (HYB-MTV; Shackleford and Varmus, 1988). For comparison, RNA from two BALB/c lactating mammary glands from two different mice was also used. Each sample yielded >20 million single-end reads with a modal insert length of 22 nucleotides, consistent with the expected size for libraries enriched for miRNAs (Figure 2A). We applied the same mapping and analysis workflow as outlined for the cell culture libraries above. Although sporadic reads covering most of the genome mapped to the HYB-MTV provirus reference sequence, we did not observe an enrichment for reads in the size class of miRNAs as observed for the annotated miRBase host miRNAs (Figure 2B, 2C). A discrete peak at position ~ nt 6100 within the pol region was detected on the positive strand for both mammary tumors (Figure 2C). The peak consisted of heterogeneous lengths and start sites, which may be attributable to a previously described internal MMTV promoter for RNA polymerase II (Miller et al., 1992). The miRDeep2 analysis recovered 272-275 miRBase annotated microRNAs per tumor sample, but no exogenous or endogenous MMTV-encoded miRNAs were identified (Figure 2D). Thus, in agreement with the earlier in silico analysis of the MMTV genome (Kincaid et al., 2012) and our analysis of in vitro infected cells, these results indicate that MMTV does not encode detectable miRNAs either in a HC11 mammary culture model or in vivo.
Figure 2.
Analysis of small RNA-seq libraries prepared from uninfected mammary glands and HYB-MTV induced mammary tumors. A. Length of inserts after adaptor trimming for small RNA libraries prepared from two uninfected mammary glands and two HYB-MTV induced mammary tumors (MG = uninfected mammary gland, MT = infected mammary tumor). B. Length of reads mapped to the indicated reference sequences (miRBase: miRBase release 21 annotated miRNA sequences, HYB-MTV: MMTV reference sequence for small RNA library prepared from two HYB-MTV induced mammary tumors. C. Plots of read coverage and start sites for reads mapping to the HYB-MTV reference sequence for libraries prepared from two HYB-MTV induced mammary tumors. At the top, map of HYB-MTV genomic organization. On the Y-axis, coverage is plotted with gray lines and read start counts are plotted with black impulses. Values above the X-axis represent the forward strand and below values represent reads mapping to the negative strand. Genomic position is indicated on the X-axis. D. Summary of miRDeep2 analysis of small RNA libraries prepared from the two uninfected mammary glands and two HYB-MTV-induced mammary tumors. Y-axis indicates the number of candidate miRNAs for the respective set of reference sequences (miRBase, HYB-MTV, and Mtvs (see Methods for accession numbers)) indicated on the X-axis. ND indicates none detected. E. Quantification of miRBase annotated miRNAs in two uninfected mammary glands and two HYB-MTV induced mammary tumors samples as assayed by small RNA-seq. Mean counts for miRBase annotated microRNAs are given in reads per million mapped with one pseudocount (RPMM). Dots colored red indicate miRNAs that met the criteria for significant differential expression (RPMM >= 100 in at least one sample, and Student’s t-test p-value < 0.05).
MMTV–infected tissues show increased levels of specific members of the miR-17-92 cluster and decreased miR-10b.
We next determined whether differences in host miRNAs were observable between uninfected mammary gland and MMTV-infected tumor samples. We compared the normalized miRNA reads across libraries (lactating mammary gland samples (LMG1 and LMG2) with HYB-MTV-induced mammary tumors (HYB-MT1 and HYB-MT2). This analysis revealed that >200 known cellular miRNAs were detectable in mammary tissues (Supp. Data File 1). Only 12 (5%) of the readily detectable miRNAs (≥ 100 reads per million mapped (RPMM) in at least one library) were significantly differentially expressed (p value < 0.05, Student’s t-test) in the mammary tumors compared to the lactating mammary gland samples (Figure 2E, Table 1). Of these, 11 miRNAs were upregulated in the tumor samples ranging from 2.3x to 15x higher. Only a single miRNA, mmu-miR-381-3p, was significantly downregulated (2.9x less). We also observed some miRNAs with similar magnitude differences that did not meet the statistical cutoff. This is the result of greater variance in expression of those miRNAs between samples. Of note, many of the miRNAs with significantly different levels have been previously associated with altered expression in mammary tumors of mouse (Zhu et al., 2011) and human (Mogilyansky and Rigoutsos, 2013; van Haaften and Agami, 2010; Volinia et al., 2006) origin.
Table 1:
List of miRNAs differentially expressed in mammary tumors as determined by miRNASeq analysis. Expression values are rounded to the nearest RPMM.
| miRNA | LMG1 | LMG2 | HYB- MT-1 |
HYB- MT-2 |
p value (<0.05) |
Log2 Ratio of Means |
|---|---|---|---|---|---|---|
| mmu-mir-19a(mmu-miR-19a-3p) | 13 | 2 | 96 | 104 | 0.005 | 3.71 |
| mmu-mir-20a(mmu-miR-20a-5p) | 599 | 170 | 3474 | 3234 | 0.007 | 3.13 |
| mmu-mir-17(mmu-miR-17-5p) | 230 | 61 | 1020 | 996 | 0.010 | 2.79 |
| mmu-mir-183(mmu-miR-183-5p) | 1146 | 442 | 6021 | 5235 | 0.012 | 2.83 |
| mmu-mir-19b-2(mmu-miR-19b-3p) | 161 | 36 | 634 | 713 | 0.016 | 2.77 |
| mmu-mir-19b-1(mmu-miR-19b-3p) | 160 | 36 | 630 | 709 | 0.016 | 2.77 |
| mmu-mir-93(mmu-miR-93-5p) | 476 | 154 | 1368 | 1676 | 0.032 | 2.27 |
| mmu-mi r-429(mmu-mi R-429-3p) | 377 | 153 | 1034 | 1324 | 0.038 | 2.15 |
| mmu-mi r-542(mmu-mi R-542-3p) | 66 | 20 | 525 | 761 | 0.038 | 3.91 |
| mmu-mir-381(mmu-miR-381-3p) | 236 | 183 | 81 | 62 | 0.039 | −1.55 |
| mmu-mir-362(mmu-miR-362-5p) | 43 | 14 | 110 | 99 | 0.039 | 1.87 |
| mmu-mir-674(mmu-miR-674-3p) | 67 | 40 | 116 | 131 | 0.046 | 1.22 |
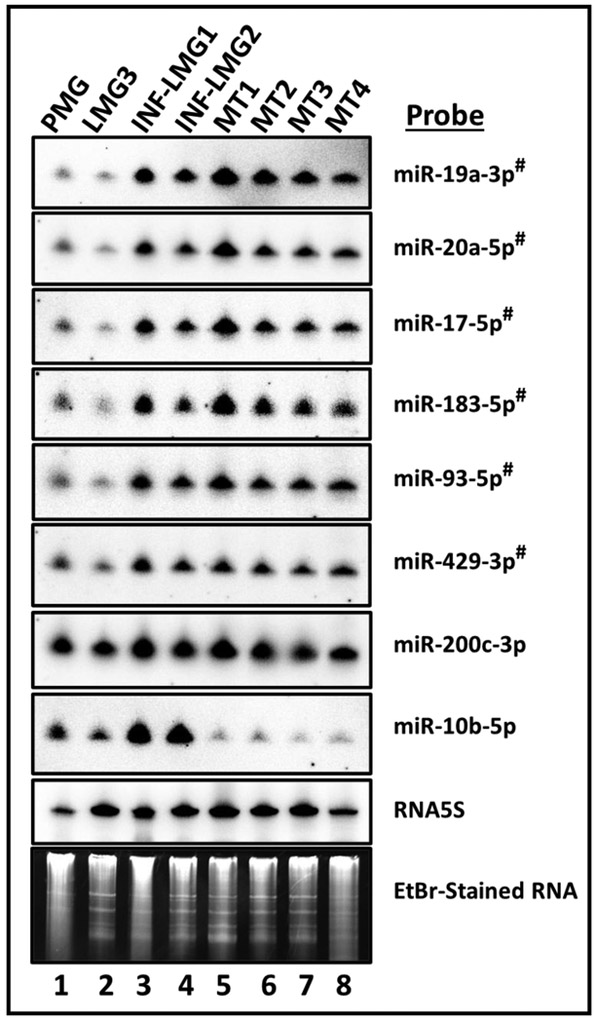
To independently confirm the differences in relative expression observed, Northern blot analysis was conducted for select miRNAs. RNA was harvested from four BALB/c MMTV-induced mammary tumor samples from four different mice (including MT1 and MT2 used for miRNA-seq analysis) together with two MMTV-infected lactating mammary gland samples from BALB/c mice (INF-LMG1 and INF-LMG2). RNA was also examined from two uninfected normal mammary tissues, pregnant (PMG) and lactating mammary gland (LMG3) tissues. An increase was observed in the expression of several members of the miR-17-92 cluster in both infected tumor and non-tumor samples (miRs 17, 19, and 20; Figure 3). Since the small number of samples used for the RNA-seq experiments provide lower statistical power, we also tested miR-200c-3p and miR-10b-5p, miRNAs that trended as differentially expressed in our small RNA-seq analysis, but did not attain statistical significance (p ~0.08) (Supp. Data File 1). Except for miR-200c-3p, these combined results confirm the trends observed in our high throughput RNA-seq analysis. Furthermore, for 6 of the 8 miRNAs analyzed (miR-19a-3p, miR-20a-5p, miR-17a-5p, miR-183a-5p, miR-93-5p, miR-429-3p), our findings demonstrate that similar changes occur in both the tumor and infected lactating tissues (Figure 3 and Table 2). These findings are consistent with a model whereby MMTV infection alters host miRNA levels during mammary gland development, and that some changes persist through tumor appearance.
Figure 3.
Differential expression of specific miRNAs in uninfected pregnant (PMG) and lactating (LMG3) mammary glands versus MMTV-infected (INF-LMG1/2) and tumor mammary gland (MT1, 2, 3 or 4) samples as assessed by Northern blot assays. The miRNAs that were observed to be differentially-expressed in a statistically significant (p<0.05) manner by RNASeq analysis are indicated with # superscript. PMG/LMG, pregnant and lactating mammary glands, respectively; INF-LMG1 and INF-LMG2, infected mammary gland samples 1 and 2; MT1-4, HYB-MTV-induced mammary tumor samples 1-4. MT1 and MT2 are the tumor samples assayed by miRNASeq analysis. EtBr; is ethidium bromide stain of low molecular weight RNA, which serves as a loading control.
Table 2:
Comparison of differentially expressed miRNAs in MMTV-induced mammary tumors as determined by miRNASeq and Northern blot analyses.
| RNASeq Analysis | Northern Blot Analysis | ||
|---|---|---|---|
|
Differentially Expressed
miRNAs in BALB/c Mammary Tumors (MaTu) |
MMTV- Infected LMG |
MMTV-induced
MaTu Tumor |
|
| 1 | mmu-mir-19a(mmu-miR-19a-3p) Upregulated |
Upregulated | Upregulated |
| 2 | mmu-mir-20a(mmu-miR-20a-5p) Upregulated |
Upregulated | Upregulated |
| 3 | mmu-mir-17(mmu-miR-17-5p) Upregulated |
Upregulated | Upregulated |
| 4 | mmu-mir-183(mmu-miR-183-5p) Upregulated |
Upregulated | Upregulated |
| 5 | mmu-mir-19b-2(mmu-miR-19b-3p) Upregulated |
Not tested | Not tested |
| 6 | mmu-mir-19b-1(mmu-miR-19b-3p) Upregulated |
Not tested | Not tested |
| 7 | mmu-mir-93(mmu-miR-93-5p) Upregulated |
Upregulated | Upregulated |
| 8 | mmu-mir-429(mmu-miR-429-3p) Upregulated |
Upregulated | Upregulated |
| 9 | mmu-mir-542(mmu-miR-542-3p) Upregulated |
Not tested | Not tested |
| 10 | mmu-mir-381(mmu-miR-381-3p) Down-regulated |
Not tested | Not tested |
| 11 | mmu-mir-362(mmu-miR-362-5p) Upregulated |
Not tested | Not tested |
| 12 | mmu-mir-674(mmu-miR-674-3p) Upregulated |
Not tested | Not tested |
| 13 | mmu-mir-200c(mmu-miR-200c-3p) Upregulated (p < 0.080)1 |
No change | No change |
| 14 | mmu-mir-10b(mmu-miR-10b-5p) Down-regulated (p < 0.084)1 |
Upregulated | Down-regulated |
Values were not significant.
LMG = lactating mammary gland.
Discussion
The number of retroviruses that encode miRNAs remains unknown. Previous results have shown that the retroviruses bovine leukemia virus (BLV), avian leukosis virus subgroup J (ALV-J), and foamy viruses (FVs) encode miRNAs within their genomes (Kincaid et al., 2012, 2014; Whisnant et al., 2014; Yao et al., 2014). In this work, we assayed MMTV for expression of viral miRNAs in both cell culture based and in vivo models via high-throughput small RNA sequencing. We did not detect small RNA reads consistent with virally encoded miRNAs. In the library generated from infected HC11 mammary cells, the most abundant reads mapping to the viral reference sequence were shorter RNAs (~16-19 nt) that mapped to the primer binding site (Figure 1). These RNAs are most likely derived from host-encoded transfer RNAs (tRNAs) since reverse transcription is primed by host tRNAs. It was recently reported that tRNA-derived small RNAs of this size class could inhibit the retrotransposition of endogenous retroviruses (Schorn et al., 2017). Whether these small RNAs play a role in exogenous retrovirus infections remains unknown. Although we also observed tRNA-related sequences to a lesser extent in the mammary tumor samples, the most abundant cluster of reads in this context was in the pol gene region (Figure 2). However, these sequences are heterogeneous in start position and length and longer than typical miRNA sequences, which may be due to the activity of an internal promoter (Miller et al., 1992). MMTV RNA expression in lactating mammary glands is high, making it unlikely that our assays missed viral miRNAs due to lack of sensitivity (Figure S1) (Mok et al., 1992 and 2012). Finally, our inability to detect MMTV-encoded miRNAs is not due to RNA degradation since specifically processed miRNAs with defined termini of host origin were readily detected. We conclude that MMTV does not encode miRNAs.
In contrast to the absence of MMTV-encoded miRNAs, we detected 12 host miRNAs expressed at significantly different levels in MMTV-infected mammary tumors compared to normal lactating mammary glands (Table 1). Most of these small RNAs are upregulated, and several belong to the oncogenic miR-17-92 (oncomiR) gene cluster, which frequently has increased expression in human hematological malignancies and solid cancers (Mogilyansky and Rigoutsos, 2013; van Haaften and Agami, 2010; Volinia et al., 2006). Members of the miR-17-92 cluster are known to be a signature of human breast cancer (reviewed in (Mogilyansky and Rigoutsos, 2013)). The precise mechanism of how this cluster is induced (whether directly or via activation of c-myc, N-myc, etc.) and whether viral factors are involved in this process remain to be elucidated.
Conversely, miR-10b-5p, a “metastamiR” correlated with the cancer stem cell (CSC) phenotype (Bahena-Ocampo et al., 2016), was one of the few miRNAs significantly downregulated in MMTV-induced tumors. Unlike miR-19a-3p, miR-20a-5p, miR-17a-5p, miR-183a-5p, miR-93-5p, and miR-429-3p, which were upregulated in all infected lactating and mammary tumor tissues, decreased miR-10b-5p levels were only observed in the tumor tissue (Figure 3). This observation may explain why many MMTV-induced tumors are not metastatic.
Our failure to observe large changes in host miRNAs in our in vitro cell culture model likely reflects lower levels of infection. HC11 cells are undifferentiated mammary epithelial cells, and MMTV levels do not reach sufficiently high levels to give insertional mutagenesis and transformed foci under typical culture conditions, although MMTV envelope protein has been reported to transform epithelial cells in 3-dimensional mammary cultures (Katz et al., 2005). This observation highlights the importance of assaying viral and host miRNA interactions in vivo (Figure 1E, Supp. Data File 1). These findings demonstrate that MMTV infection in vivo promotes changes in levels of select cancer-associated miRNAs, some of which precede the appearance of tumors.
The observation that MMTV infection induces host miRNAs suggests that these RNAs provide an advantage for virus replication. Infection with other viruses is known to alter host miRNAs, and some of these changes have been proposed to be advantageous for viral replication (Amaral et al, 2017 (HIV-1); Sun et al., 2016 (HIV-1); Bruce and Alcorn, 2011 (RSV); Lagos et al. 2010 (KSHV); Harden et al., 2017a and b (HPV); Marthaler et al., 2017 (HPV); Melar-New and Laimins, 2010 (HPV); Gunasekharan et al, 2013 and 2016 (HPV); Linnstaedt et al., 2010 (EBV); Skalsky and Cullen, 2010 (EBV); Wang et al., 2017 (HCMV); Saldaña et al., 2017 (Zika)). MMTV is transmitted to the virgin mammary gland during puberty by infected B and T lymphocytes (Golovkina et al., 1992 and 1998; Held et al., 1993). Although MMTV expression is relatively low in virgin mammary tissues, inactivation of the Cux1 transcriptional repressor in late pregnancy and activation of the MMTV promoter and transcriptional activators by lactogenic hormones allows high levels of viral RNA during milk-borne transmission of the virus (Zhu et al., 2004; Maitra et al., 2006). Increased viral production for milk-borne transmission is stimulated by cell proliferation to allow increased numbers of infected cells as well as proviral access to the nucleus for integration. Although the mechanism for MMTV induction of host miRNAs remains to be determined, our findings are consistent with viral subversion of host miRNAs to promote infection and/or transmission of the virus.
This study demonstrates that MMTV infection does not lead to expression of viral miRNAs, yet alters the levels of select host miRNAs, several of which have been implicated in human breast cancer (Mogilyansky and Rigoutsos, 2013; van Haaften and Agami, 2010; Volinia et al., 2006). This observation lends further support to the use of the MMTV-induced mammary tumors as a relevant model system for human breast cancer. As alterations of several of these miRNAs precedes detectable tumor formation, these results are consistent with a possible role of infection-induced host miRNAs in promoting MMTV-associated tumors. Future studies will address whether viral perturbation of host miRNA levels contributes directly to MMTV-mediated tumorigenesis.
Supplementary Material
Supp. Data File 1: Identification and quantification of microRNAs by high-throughput small RNA-seq. This file includes the differential expression analysis and the raw outputs from miRDeep2 and mirUtils.
Highlights.
The MMTV genome does not encode viral miRNAs.
MMTV infection alters host miRNA levels, including the oncogenic miR-17-92 cluster.
Host miRNA changes following MMTV infection occur prior to tumor formation.
MMTV-induced changes to host miRNAs may help drive tumor initiation/maintenance.
Acknowledgements
This study was supported in part by funds to FM from the Higher Education Commission, Pakistan, Sheikh Hamdan Award for Medical Sciences (MRG 39), the United Arab Emirates University Start-up (31M122), and NRF (31M101) grants, and National Institutes of Health grant R01 CA167053 to JPD. Work performed by RPK and CSS was supported by a Burroughs Wellcome Investigators in Pathogenesis Award and a grant from the Cancer Prevention and Research Institute of Texas (RP140842) to CSS.
Footnotes
Conflict of Interest
None of the authors declare any financial or personal interest that could have influenced the work reported in this study.
Supplemental Data
Supplemental material is available for this article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altuvia Y, Landgraf P, Lithwick G, Elefant N, Pfeffer S, Aravin A, Brownstein MJ, Tuschl T, Margalit H, 2005. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res 33, 2697–2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral AJ, Andrade J, Foxall RB, Matoso P, Matos AM, Soares RS, Rocha C, Ramos CG, Tendeiro R, Serra-Caetano A, Guerra-Assunção JA, Santa-Marta M, Gonçalves J, Gama-Carvalho M, Sousa AE 2017. miRNA profiling of human naive CD4 T cells links miR-34c-5p to cell activation and HIV replication. EMBO J 36, 346–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahena-Ocampo I, Espinosa M, Ceballos-Cancino G, Lizarraga F, Campos-Arroyo D, Schwarz A, Garcia-Lopez P, Maldonado V, Melendez-Zajgla J, 2016. miR-10b expression in breast cancer stem cells supports self-renewal through negative PTEN regulation and sustained AKT activation. EMBO Rep 17,1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP, 2009. MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP, 2004. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 116, 281–297. [DOI] [PubMed] [Google Scholar]
- Beutner U, Kraus E, Kitamura D, Rajewsky K, Huber BT, 1994. B cells are essential for murine mammary tumor virus transmission, but not for presentation of endogenous superantigens. J. Exp. Med 179, 1457–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhadra S, Lozano MM, Dudley JP, 2005. Conversion of mouse mammary tumor virus to a lymphomagenic virus. J. Virol 79, 12592–12596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittner JJ, 1936. Some possible effects of nursing on the mammary gland tumor incidence in mice. Science 84, 162–162. [DOI] [PubMed] [Google Scholar]
- Bruce SR, Alcorn JL 2011. "RSV Induced Changes in miRNA Expression in Lung" in Human Respiratory Syncytial Virus Infection, Resch Dr. Bernhard (Ed.), ISBN: 978-953-307-718-5, InTech, Available from: http://www.intechopen.com/books/human-respiratory-syncytial-virus-infection/rsv-induced-changes-inmirna-expression-in-lung. [Google Scholar]
- Callahan R, Drohan W, Gallahan D, D’hoostelaere L, Potter M, 1982. Novel class of mouse mammary tumor virus-related DNA sequences found in all species of Mus, including mice lacking the virus proviral genome. Proc. Natl. Acad. Sci 79, 4113–4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan R, Smith GH, 2000. MMTV-induced mammary tumorigenesis: gene discovery, progression to malignancy and cellular pathways. Oncogene 19, 992. [DOI] [PubMed] [Google Scholar]
- Courreges MC, Burzyn D, Nepomnaschy I, Piazzon I, Ross SR 2007. Critical role of dendritic cells in mouse mammary tumor virus in vivo infection. J. Virol 81, 3769–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David M, Dzamba M, Lister D, Ilie L, Brudno M, 2011. SHRiMP2: sensitive yet practical short read mapping. Bioinformatics 27, 1011–1012. [DOI] [PubMed] [Google Scholar]
- Di Leva G, Garofalo M, Croce CM, 2014. MicroRNAs in cancer. Annu. Rev. Pathol. Mech. Dis 9, 287–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley JP, Golovkina TV, Ross SR, 2016. Lessons learned from mouse mammary tumor virus in animal models. ILAR J 57, 12–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzuris JL, Zhu W, Kapkov D, Golovkina TV, Ross SR, 1999. Expression of mouse mammary tumor virus envelope protein does not prevent superinfection in vivo or in vitro. Virology 263, 418–426. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ, 2006. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer 6, 259–269. [DOI] [PubMed] [Google Scholar]
- Finke D, Acha-Orbea H, 2001. Differential migration of in vivo primed B and T lymphocytes to lymphoid and non-lymphoid organs. Eur. J. Immunol. 31, 2603–2611. [DOI] [PubMed] [Google Scholar]
- Friedländer MR, Mackowiak SD, Li N, Chen W, Rajewsky N, 2011. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res 40, 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillet NA, Hamaidia M, De Brogniez A, Gutierrez G, Renotte N, Reichert M, Trono K, Willems L, 2016. Bovine leukemia virus small noncoding RNAs are functional elements that regulate replication and contribute to oncogenesis in vivo. PLoS Pathog. 12, e1005588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovkina TV, Chervonsky AV, Dudley JP, Ross SR 1992. Transgenic mouse mammary tumor virus superantigen expression prevents viral infection. Cell 69, 637–645. [DOI] [PubMed] [Google Scholar]
- Golovkina TV, Dudley JP, Ross SR, 1998. B and T cells are required for mouse mammary tumor virus spread within the mammary gland. J. Immunol. 161, 2375–2382. [PubMed] [Google Scholar]
- Gottwein E, Mukherjee N, Sachse C, Frenzel C, Majoros WH, Chi J-TA, Braich R, Manoharan M, Soutschek J, Ohler U, Cullen BR, 2007. A viral microRNA functions as an orthologue of cellular miR-155. Nature 450, 1096–1099. doi: 10.1038/nature05992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, Van Dongen S, Bateman A, Enright AJ, 2006. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34, D140–D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundhoff A, Sullivan CS, 2011. Virus-encoded microRNAs. Virology 411, 325–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunasekharan V, Laimins LA 2013. Human papillomaviruses modulate microRNA 145 expression to directly control genome amplification. J. Virol. 87, 6037–43. doi: 10.1128/JVI.00153-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunasekharan VK, Li Y, Andrade J, Laimins LA 2016. Post-Transcriptional Regulation of KLF4 by High-Risk Human Papillomaviruses Is Necessary for the Differentiation-Dependent Viral Life Cycle. PLoS Pathog. 2016 July 7;12(7):e1005747. doi: 10.1371/journal.ppat.1005747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harden ME, Prasad N, Griffiths A, Munger K 2017a. Modulation of microRNA-mRNA Target Pairs by Human Papillomavirus 16 Oncoproteins. MBio 8(1). pii: e02170–16. doi: 10.1128/mBio.02170-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harden ME, Munger K 2017b. Human papillomavirus 16 E6 and E7 oncoprotein expression alters microRNA expression in extracellular vesicles. Virology 508, 63–69. doi: 10.1016/j.virol.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hede K, 2010. MicroRNAs as Onco-miRs, drivers of cancer. Oxford University Press. [DOI] [PubMed] [Google Scholar]
- Held W, Shakhov AN, Izui S, Waanders GA, Scarpellino L, MacDonald HR, Acha-Orbea H, 1993. Superantigen-reactive CD4+ T cells are required to stimulate B cells after infection with mouse mammary tumor virus. J. Exp. Med 177, 359–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S, Okumoto M, Iwai M, Haga S, Mori N, Miyashita N, Moriwaki K, Hilgers J, Sarkar NH, 1994. Distribution of mouse mammary tumor virus in Asian wild mice. J. Virol. 68, 3437–3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz E, Lareef MH, Rassa JC, Grande SM, King LB, Russo J, Ross SR, Monroe JG, 2005. MMTV Env encodes an ITAM responsible for transformation of mammary epithelial cells in three-dimensional culture. J Exp Med 201, 431–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC 2009. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol 10, 126–139. [DOI] [PubMed] [Google Scholar]
- Kincaid RP, Burke JM, Sullivan CS, 2012. RNA virus microRNA that mimics a B-cell oncomiR. Proc. Natl. Acad. Sci 109, 3077–3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kincaid RP, Chen Y, Cox JE, Rethwilm A, Sullivan CS, 2014. Noncanonical microRNA (miRNA) biogenesis gives rise to retroviral mimics of lymphoproliferative and immunosuppressive host miRNAs. MBio 5, e00074–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kincaid RP, Sullivan CS, 2012. Virus-encoded microRNAs: an overview and a look to the future. PLoS Pathog 8, e1003018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos D, Pollara G, Henderson S, Gratrix F, Fabani M, Milne RS, Gotch F, Boshoff C 2010. miR-132 regulates antiviral innate immunity through suppression of the p300 transcriptional co-activator. Nat Cell Biol 12, 513–9. doi: 10.1038/ncb2054. [DOI] [PubMed] [Google Scholar]
- Linnstaedt SD, Gottwein E, Skalsky RL, Luftig MA, Cullen BR, 2010. Virally induced cellular microRNA miR-155 plays a key role in B-cell immortalization by Epstein-Barr virus. J. Virol. 84, 11670–11678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M, 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17, 10–12. [Google Scholar]
- Maitra U, Seo J, Lozano MM, Dudley JP 2006. Differentiation-induced cleavage of Cutl1/CDP generates a novel dominant-negative isoform that regulates mammary gene expression. Mol. Cell Biol 26, 7466–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marthaler AM, Podgorska M, Feld P, Fingerle A, Knerr-Rupp K, Grässer F, et al. 2017. Identification of C/EBPα as a novel target of the HPV8 E6 protein regulating miR-203 in human keratinocytes. PLoS Pathog 13(6): e1006406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure LV, Lin Y-T, Sullivan CS, 2011. Detection of viral microRNAs by northern blot analysis Methods Mol. Biol Clifton NJ: 721, 153–171. [DOI] [PubMed] [Google Scholar]
- Melar-New M, Laimins LA 2010. Human papillomaviruses modulate expression of microRNA 203 upon epithelial differentiation to control levels of p63 proteins. J Virol. 84, 5212–21. doi: 10.1128/JVI.00078-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CL, Garner R, Paetkau V 1992. An activation-dependent, T-lymphocyte-specific transcriptional activator in the mouse mammary tumor virus env gene. Mol Cell Biol 12, 3262–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogilyansky E, Rigoutsos I, 2013. The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ 20, 1603–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok E, Golovkina TV, Ross SR, 1992. A mouse mammary tumor virus mammary gland enhancer confers tissue-specific but not lactation-dependent expression in transgenic mice. J. Virol. 66, 7529–7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustafa F, Lozano M, Dudley JP, 2000, C3H mouse mammary tumor virus superantigen function requires a splice donor site in the envelope gene. J. Virol. 74, 9431–9440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekarsky Y, Croce CM, 2010. Is miR-29 an oncogene or tumor suppressor in CLL? Oncotarget 1, 224–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdy A, Case L, Duvall M, Overstrom-Coleman M, Monnier N, Chervonsky A, Golovkina T, 2003. Unique resistance of I/LnJ mice to a retrovirus is due to sustained interferon gamma-dependent production of virus-neutralizing antibodies. J Exp Med 197, 233–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross SR, 2010. Mouse mammary tumor virus molecular biology and oncogenesis. Viruses 2, 2000–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldaña MA, Etebari K, Hart CE, Widen SG, Wood TG, Thangamani S, Asgari S, Hughes GL. 2017. Zika virus alters the microRNA expression profile and elicits an RNAi response in Aedes aegypti mosquitoes. PLoS Negl Trop Dis 2017 July 17;11(7):e0005760. doi: 10.1371/journal.pntd.0005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorn AJ, Gutbrod MJ, LeBlanc C, Martienssen R, 2017. LTR-Retrotransposon Control by tRNA-Derived Small RNAs. Cell 170, 61–71. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, Lightfoot S, Menzel W, Granzow M, Ragg T, 2006. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol January 31;7:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackleford GM, Varmus HE, 1988. Construction of a clonable, infectious, and tumorigenic mouse mammary tumor virus provirus and a derivative genetic vector. Proc. Natl. Acad. Sci 85, 9655–9659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalsky RL, Cullen BR, 2010. Viruses, microRNAs, and host interactions. Annu. Rev. Microbiol 64, 123–141. doi:10.1146/annurev.micro.112408.134243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalsky RL, Samols MA, Plaisance KB, Boss IW, Riva A, Lopez MC, Baker HV, Renne R, 2007. Kaposi’s sarcoma-associated herpesvirus encodes an ortholog of miR-155. J. Virol. 81, 12836–12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart THM, Sage RD, Stewart AFR, Cameron DW, 2000. Breast cancer incidence highest in the range of one species of house mouse, Mus domesticus. Br. J. Cancer 82, 446–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B, Yang R, Mallardo M 2016. Roles of microRNAs in HIV-1 Replication and Latency. Microrna. 5, 120–123. [DOI] [PubMed] [Google Scholar]
- Tremblay PJ, Pothier F, Hoang T, Tremblay G, Brownstein S, Liszauer A, Jolicoeur P, 1989. Transgenic mice carrying the mouse mammary tumor virus ras fusion gene: distinct effects in various tissues. Mol. Cell. Biol 9, 854–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Haaften G, Agami R, 2010. Tumorigenicity of the miR-17-92 cluster distilled. Genes Dev 24, 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM, 2006. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA 103, 2257–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhao P, Qian D, Hu M, Zhang L, Shi H, Wang B 2017. MicroRNA-613 is downregulated in HCMV-positive glioblastoma and inhibits tumour progression by targeting arginase-2. Tumour Biol 2017 July;39(7):1010428317712512. doi: 10.1177/1010428317712512. [DOI] [PubMed] [Google Scholar]
- Whisnant AW, Kehl T, Bao Q, Materniak M, Kuzmak J, Löchelt M, Cullen BR, 2014. Identification of novel, highly expressed retroviral microRNAs in cells infected by bovine foamy virus. J. Virol. 88, 4679–4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Smith LP, Nair V, Watson M, 2014. An avian retrovirus uses canonical expression and processing mechanisms to generate viral microRNA. J. Virol. 88, 2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitels LR, Acharya A, Shi G, Chivukula D, Chivukula RR, Anandam JL, Abdelnaby AA, Balch GC, Mansour JC, Yopp AC, 2014. Tumor suppression by miR-26 overrides potential oncogenic activity in intestinal tumorigenesis. Genes Dev 28, 2585–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Yao Y, Xu H, Lambeth L, Smith LP, Kgosana L, Wang X, Nair V, 2009. A functional microRNA-155 ortholog encoded by the oncogenic Marek’s disease virus. J. Virol. 83, 489–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Maitra U, Johnston D, Lozano M, Dudley JP, 2004. The homeodomain protein CDP regulates mammary-specific gene transcription and tumorigenesis. Mol. Cell. Biol 24, 4810–4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Yi M, Kim CH, Deng C, Li Y, Medina D, Stephens RM, Green JE 2011. Integrated miRNA and mRNA expression profiling of mouse mammary tumor models identifies miRNA signatures associated with mammary tumor lineage. Genome Biol 12, R77. doi: 10.1186/gb-2011-12-8-r77. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supp. Data File 1: Identification and quantification of microRNAs by high-throughput small RNA-seq. This file includes the differential expression analysis and the raw outputs from miRDeep2 and mirUtils.
Data Availability Statement
RNA-seq data has been deposited through NCBI GEO (accession no GSE101333).