Abstract
Purpose
To develop an accelerated, free-breathing, non-contrast, ECG-triggered, thoracic magnetic resonance angiography (NC-MRA) pulse sequence capable of achieving high spatial resolution at clinically acceptable scan time and test whether it produces clinically acceptable image quality in patients with suspected aortic disease.
Methods
We modified a “coronary” MRA pulse sequence to use a stack-of-stars k-space sampling pattern and combined it with GRASP reconstruction to enable self-navigation of respiratory motion and high data acceleration. The performance of the proposed NC-MRA was evaluated in 13 patients, where clinical standard contrast-enhanced MRA (CE-MRA) was used as control. For visual analysis, two readers graded the conspicuity of vessel lumen, artifacts, and noise level on a 5-point scale (overall score index = sum of three scores). The aortic diameters were measured at seven standardized locations. The mean visual scores, inter-observer variability, and vessel diameters were compared using appropriate statistical tests.
Results
The overall mean visual score index (12.1 ± 1.7 for CE-MRA vs. 12.1 ± 1.0 for NC-MRA) scores were not significantly different (p>0.16). The two readers’ scores were significantly different for CE-MRA (p = 0.01) but not for NC-MRA (p = 0.21). The mean vessel diameters were not significantly different, except at the proximal aortic arch (p < 0.03). The mean diameters were strongly correlated (R2 ≥ 0.96) and in good agreement (absolute mean difference ≤ 0.01 cm and 95% confidence interval ≤ 0.62cm).
Conclusion
This study shows that the proposed NC-MRA produces clinically acceptable image quality in patients at high spatial resolution (1.5 mm × 1.5 mm ×1.5 mm) and clinically acceptable scan time (~6 min).
Keywords: compressed sensing (CS), GRASP, non-contrast MRA, aortic disease, MRI
Introduction
Thoracic aortic aneurysms (TAAs), while rare, can lead to life-threatening conditions; including dissection and/or rupture. TAAs are usually caused by high blood pressure, injury or inherited connective tissue disorders such as Marfan’s and Ehlers-Danlos syndromes. TAAs were responsible for approximately 17,000 deaths in the United States (US) in 2009 (1). Given the poor prognosis, longitudinal screening with imaging is recommended to determine when surgical correction may be necessary (e.g., aortic diameter > 5.5 cm in adults (2)).
Contrast-enhanced magnetic resonance angiography (CE-MRA) is routinely used for evaluation of TAAs. The reliance on gadolinium-based contrast agent (GBCA) for CE-MRA has several drawbacks: (a) GBCA administration in patients with renal insufficiency has been associated with nephrogenic systemic fibrosis (NSF)(3), (b) recent evidence suggests that GBCA deposits in the brain after repeated exposure (4), (c) GBCA administration increases the cost of MRI, and (d) repeat scanning during the same MRI session is infeasible due to the residual contrast agent. Thus, there is a need for advancement of MRA methods that do not require GBCA.
The aforementioned drawbacks associated with CE-MRA have led to a renewed interest in non-contrast MRA pulse sequences (5,6). For non-contrast thoracic MRA, the most promising approach is derived from ECG-triggered, coronary MRA, which uses a combination of T2 preparation (7), fat suppression, balanced steady-state free precession (b-SSFP) readout (8), and navigator gating (9). Several studies have evaluated the performance of this pulse sequence for thoracic MRA in patients (10–12). Despite its early promise (10–12), the clinical translation of non-contrast thoracic MRA has been limited by inadequate spatial resolution and long scan time. For instance, averaging the imaging parameters from these three previous studies (10–12), the mean spatial resolution is 1.6 mm × 1.6 mm × 2.9 mm, and the mean scan time is 9:10 min. High spatial resolution is critically important for effective monitoring of aortic size, since a sizable subset of Marfan’s syndrome patients (“fast growers”) exhibit growth rates on the order of 0.15 cm/year for men and 0.18 cm/year for women (13). Thus, there is a need to further accelerate non-contrast thoracic MRA for achieving higher spatial resolution and shorter scan time.
The introduction of compressed sensing (CS)(14) to MRI has significantly accelerated cardiovascular MRI, including for NC-MRA (15,16). In this study, we sought to tailor a prior investigational reconstruction framework called Golden-angle Radial Sparse Parallel (GRASP) for non-contrast thoracic MRA (17). The two possible k-space patterns for GRASP reconstruction include 3D radial and stack-of-stars, with each having relative advantages and disadvantages. The relative advantages of 3D radial include isotropic spatial resolution and excellent motion properties for all three spatial encoding directions. The relative advantages of stack-of-stars include better k-space sampling efficiency for the partition (kz) encoding direction, especially when an organ of interest such as the thoracic aorta can be sampled with a thin slab, and insensitivity to off-resonance and gradient trajectory delays along kz. For this study, we elected to use a stack-of-stars k-space sampling pattern to image the thoracic aorta in an oblique sagittal orientation with a relatively thin slab thickness. The purposes of this study were to develop a non-contrast, ECG-triggered, thoracic MRA pulse sequence capable of achieving high resolution (1.5 mm × 1.5 mm × 1.5 mm) at clinically acceptable scan time (~5 min) and evaluate whether it produces clinically acceptable image quality in patients with suspected aortic disease.
Methods
Patients
We evaluated the performance of the proposed ECG-triggered, NC-MRA pulse sequence in 13 patients (10 men and 3 women, mean age = 56.2 ± 11.5 years) who underwent clinically indicated thoracic CE-MRA with a standard dose (0.15 – 0.2 mmol/kg, depending on kidney function) of gadobutrol (Gadavist, Bayer HealthCare Pharmaceuticals, Whippany, USA). This study was conducted in accordance with protocols approved by our institutional review board and was Health Insurance Portability and Accountability Act (HIPAA) compliant; all subjects provided written informed consent in writing. See Supporting Information Table S1 for relevant clinical profiles.
MRI Hardware
MRI examinations were conducted on two 1.5T whole-body MRI scanners (MAGNETOM Avanto, Siemens Healthcare, Erlangen, Germany) equipped with a gradient system capable of achieving a maximum gradient strength of 45 mT/m and a maximum slew rate of 200 T/m/s. The radio-frequency excitation was performed with the body coil, and the signal reception was made with the body matrix and spine coil arrays (~15-18 elements).
Stack-of-Stars k-space Sampling with Variable Density for GRASP Reconstruction
We adopted a GRASP framework (17) and enforced sparsity (18) along the respiratory dimension, in order to accomplish two goals: (a) self-navigation of respiratory motion and (b) higher acceleration of data acquisition than otherwise. We elected to use a stack-of-stars k-space sampling pattern with variable density (19) along the kz direction and leveraged the stack-of-stars’ intrinsic ability to efficiently sample a highly anisotropic field of view (FOV). Specifically, we designed a protocol with 80 partitions to sample a 120 mm FOV along the kz direction for thoracic aorta imaging. As shown in Figure 1, we elected to acquire a partial stack of rays (48 out of 80, 7 rays beyond the center of k-space, readout duration = 168 ms) along kz for each heartbeat in order to achieve a good balance between spatial resolution, slab thickness, and readout duration per shot (< 200 ms). Immediately prior to readout for each heart beat, we sampled the first two rays for self-navigation of motion, as previously described (20). The two navigator rays were oriented with in-plane rotation = 0° and 90° corresponding to the head-to-foot and oblique anterior-to-posterior directions, respectively. We elected to include two navigator rays to account for an operator error, but only the navigator ray oriented along the head-to-foot direction was used for respiratory motion tracking. For more details on the k-space sampling and ordering, see Appendix.
Figure 1.
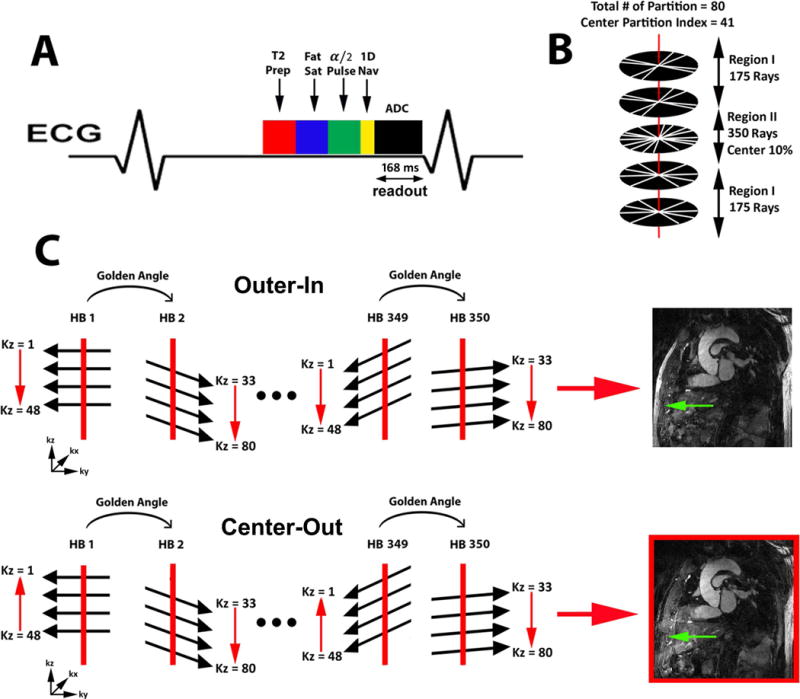
A) A schematic diagram including T2, fat suppression, and α/2 preparation RF pulses prior to b-SSFP readout. Two rays per heart beat are acquired along the head-to-foot and oblique anterior-to-posterior directions to track motion. B) The inner 10% of central partitions (labeled as region II) were sampled twice as densely as compared to the outer 90% partitions (labeled as region I). C) Outer-in and center-out kz orderings were compared to determine which ordering produces better fat suppression. The center-out kz ordering was found to produce superior fat suppression (green arrow) than the outer-in kz ordering as shown.
Imaging Parameters
The relevant imaging parameters for ECG-triggered, NC-MRA are as follows: image acquisition matrix size = 192 × 192 × 80, FOV = 288 mm × 288 mm × 120 mm, spatial resolution = 1.5 mm × 1.5 mm × 1.5 mm, receiver bandwidth = 745 Hz/pixel, flip angle = 80-90°, TE/TR = 1.8/3.5 ms, T2 preparation time = 50 ms, fat saturation, α/2 preparation pulse, total number of rays for the outer 90% along kz = 175, total number of rays for the inner 10% along kz = 350, 50 rays (48 for data + 2 for navigator) acquired per heartbeat, readout duration per shot = 168 ms, and scan time = 350 heart beats. As per clinical practice, we used the steady-state, inversion-recovery CE-MRA pulse sequence with navigator gating (21). This reference pulse sequence had the following imaging parameters: matrix size = 256 × 220 × 56-104 (depending on patient size), FOV = 384 mm × 326 mm × 84 -156 mm (depending on patient size), spatial resolution = 1.5 mm × 1.5 mm × 1.5 mm, Cartesian k-space sampling with GRAPPA (22) acceleration factor = 1.8, receiver bandwidth = 930 Hz/pixel, flip angle = 70°, TE/TR = 1.46/3.3 ms, and inversion time (TI) = 260 ms, k-space lines acquired per shot = 24, readout duration per shot = 79.2 ms. Note, both NC-MRA and CE-MRA were acquired in an oblique sagittal plane.
Image Reconstruction
We downloaded the GRASP reconstruction code from the Center for Advanced Imaging Innovation and Research website (http://cai2r.net/resources/software/grasp-matlab-code) and modified it to further reduce the reconstruction time. Specifically, the code was modified to shift a non-Cartesian k-space data set onto a Cartesian grid in one pre-processing step, as previously described (23). This modification enabled us to use more efficient FFT over NUFFT during the iterative CS step and achieve faster reconstruction. The GRASP reconstruction was performed off-line on a workstation equipped with MATLAB (R2014, The Mathworks Inc, Natick, MA, USA) running on Windows 7 (Microsoft Corporation, Redmond, WA, USA).
As shown in Figure 2, in the pre-processing step in polar coordinates, both the default (non-motion-resolved) and motion-resolved stack-of-stars k-space data sets were converted to the corresponding image sets in Cartesian space using Non-Uniform Fast Fourier Transform (NUFFT)(24) with a Kaiser Bessel kernel with alpha = 23.4 and 2D sector width of 10 pixels. The k-space sampling mask in polar coordinates was converted to the corresponding k-space sampling operator (included in variable F in the equation shown in Fig. 2) in Cartesian space using a nearest neighbor interpolation gridding algorithm. In the subsequent pre-processing step in Cartesian coordinates, the coil sensitivities (variable S in the equation shown in Fig. 2) were intrinsically calculated using the non-motion-resolved image and processing that image further as previously described (25,26). The motion-resolved image in Cartesian space was transformed to k-space using FFT and then multiplied by the k-space sampling operator to produce variable y in the equation shown in Fig. 2. Finally, the zero-filled, multi-coil, motion-resolved images (variable × in the equation shown in Fig. 2) were reconstructed using SENSE along with the coil sensitivities.
Figure 2.
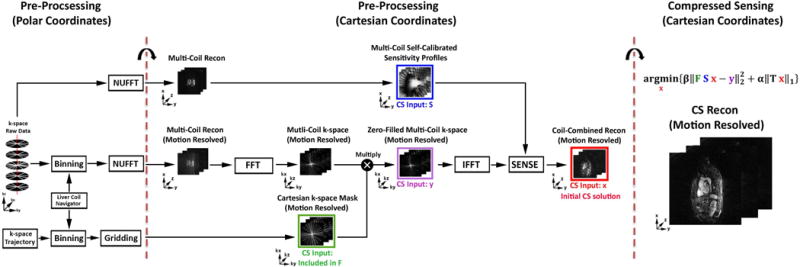
A schematic of the GRASP reconstruction pipeline. Using the navigator ray oriented along the head-to-foot direction to extract the respiratory motion (see Supporting Information Figure S1 for more details), each of six bins were populated with the same number of k-space data by adapting the bin width. In the pre-processing step in polar coordinates, both the non-motion-resolved and motion-resolved stack-of-stars k-space data sets were converted to the corresponding image sets in Cartesian space using NUFFT. The k-space sampling mask in polar coordinates was converted to the corresponding k-space sampling operator (included variable in F) Cartesian space using gridding. In the subsequent pre-processing step in Cartesian coordinates, the coil sensitivities (variable S) were calibrated intrinsically from the non-motion-resolved images as shown. The motion-resolved image in Cartesian space was transformed to k-space using FFT and then multiplied by the k-space sampling operator to produce variable y as shown. Next, the zero-filled, multi-coil, motion-resolved images (variable x) were reconstructed using SENSE along with coil sensitivities as shown. In the CS reconstruction step, we used the finite difference operator as the sparsifying transform along the respiratory dimension and nonlinear conjugate gradient with back-tracking line search as the optimization algorithm with 22 iterations. F represents undersampled FFT, S represents coil sensitivities, and × represents image to be reconstructed, y represents the k-space data, T represents the finite difference operator, α represents the normalized regularization weight, and β represents the normalized fidelity weight.
For estimating the diaphragm motion in the pre-processing step, we performed 1D FFT of navigator rays (head-to-foot direction) and then performed principal component analysis of this x-t signal to extract the respiratory motion. The contribution of cardiac motion was minimized by ECG-triggering the acquisition to late diastole. To make it easier to isolate the respiratory signal, we manually selected a single coil that sampled the liver. We elected to use the second principal component because our preliminary analysis showed that it reflects respiratory motion better than the first principal component. Next, we populated each of six bins with the same number of k-space data by adapting the bin width, as previously described (17). We elected to use six respiratory phases based on a preliminary analysis of training data for balancing between scan time and motion resolution. For additional details on respiratory motion tracking, see Supporting Information Figure S1 and Supporting Information Video S1.
As shown in Figure 2, in the CS portion of the GRASP reconstruction pipeline, we used the finite difference operator as the sparsifying transform (i.e., total variation (TV) as the regularization) along the respiratory dimension and nonlinear conjugate gradient with back-tracking line search as the optimization algorithm with 22 iterations. The normalized regularization weight (α) was 0.00075, and the normalized fidelity weight (β) was 0.002. These weights were determined empirically based on a visual inspection of training data sets, as previously described (28).
As shown in Figure 3, in the post-processing step, the remaining aliasing artifacts were further suppressed using a block matching filter as described by Dabov et al (29). This step was deemed necessary in our preliminary analysis because the CS reconstruction with finite difference is inadequate to fully remove aliasing artifacts with only six respiratory phases. A 3D block matching filter was applied to each individual 2D plane with a single iteration using a gradient descent with a step size of 0.385. These parameters were derived empirically based on a visual inspection of training data.
Figure 3.
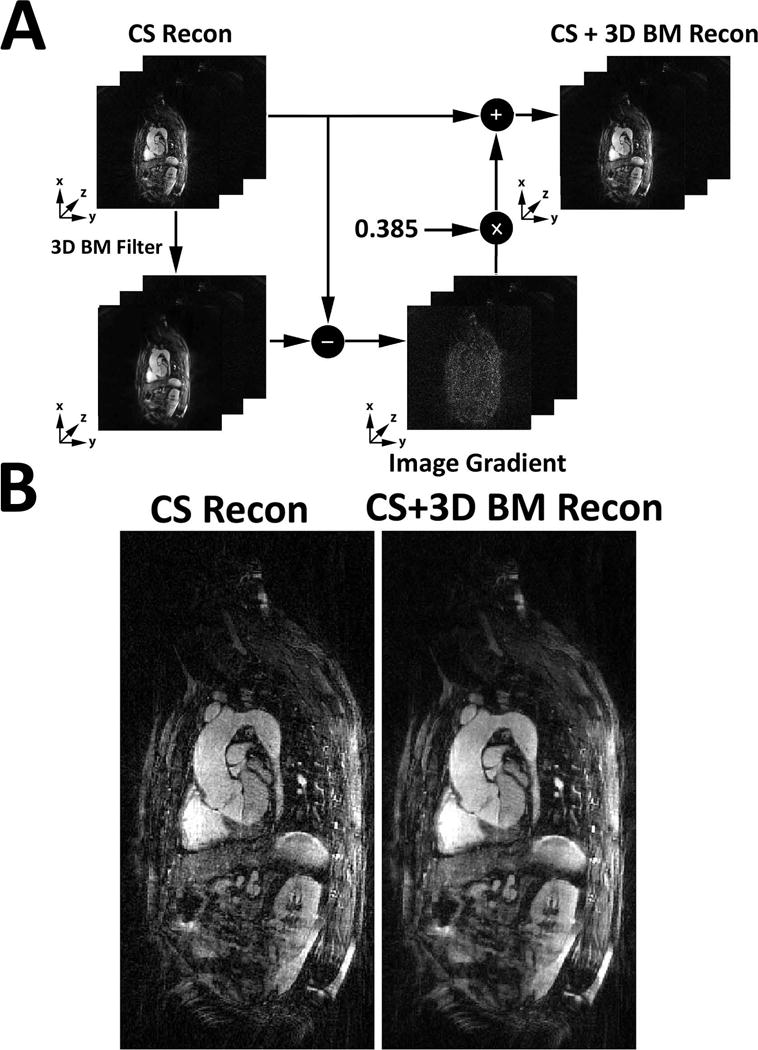
A) A schematic illustrating how a 3D block matching filter was used to suppress image artifacts remaining after the CS reconstruction. A 3D block matching filter was applied to each 2D plane individually with a single iteration using a gradient descent with a step size of 0.385. B) A comparison between NC-MRA images before and after 3D block matching filtering.
Visual Analysis of MRA
Two radiologists (PJA & AMS) graded the image quality of both NC-MRA and CE-MRA sets. In total, 26 MRA sets were randomized and de-identified for display on a 3D workstation (LEONARDO, Siemens). Prior to visual evaluation, the two readers were given training data sets with poor to excellent quality to calibrate their scores together. Following this training session, each reader was blinded to image acquisition type, the other reader, and clinical history for independent evaluation. Each reader graded the images on a 5-point Likert scale for three categories: conspicuity of vessel lumen (1 = nondiagnostic, 2 = poor, 3 = clinically acceptable, 4 = good, 5 = excellent), artifact (1 = nondiagnostic, 2 = poor, 3 = clinically acceptable, 4 = mild, 5 = minimal), noise (1 = nondiagnostic, 2 = severe, 3 = clinically acceptable, 4 = mild, 5 = minimal). An overall visual score index was defined as the sum of three scores (ranging from 3 to 15).
Aortic Diameter Measurements
Another reader (HH) measured two diameters (longest and its orthogonal) at seven standardized locations established by guidelines (2)(1: aortic sinuses of valsalva; 2: sinotubular junction; 3: mid ascending aorta; 4: proximal aortic arch; 5: mid aortic arch; 6: proximal descending thoracic aorta; 7: mid descending aorta) on a 3D workstation (LEONARDO, Siemens). Anatomic landmarks were used to reformat the MRA and find appropriate views.
Statistical Analysis
A Kolmogorov-Smirnov test was performed to test the null hypothesis that each variable is normally distributed at the 5% significance level. A paired t-test was performed for normal distribution, whereas the Wilcoxon signed rank test was performed for non-normal distribution. Assuming normal distribution, the inter-observer variability in the overall visual score index was inferred using a paired t-test, and the mean vessel diameters were compared using a paired t-test. For statistical analysis between MRA methods, the two readers’ scores were averaged. Assuming non-normal distribution, the individual visual scores were compared using the Wilcoxon signed rank test. The Bland-Altman and linear regression analyses were conducted on vessel diameters to determine their agreement and association, respectively. A p value < 0.05 was considered significant for all statistical tests.
Results
The mean scan time for NC-MRA (5:55 ± 0:48 min) was significantly (p < 0.05) lower than that of CE-MRA (6:56 ± 2:10 min). The mean offline image reconstruction time for NC-MRA was 4 hr and 41 min and 13 s (pre-processing steps = 29 min and 17 s, CS step = 4 hr 10 min, post-processing step = 1 min 34 s). Figure 4 shows representative thin reformatted maximum-intensity-projection (MIP)s of two patients. As shown in Figure 4, one of the patients exhibited a ductus diverticulum (red arrow) which is clearly visible in both CE-MRA and NC-MRA MIPs.
Figure 4.
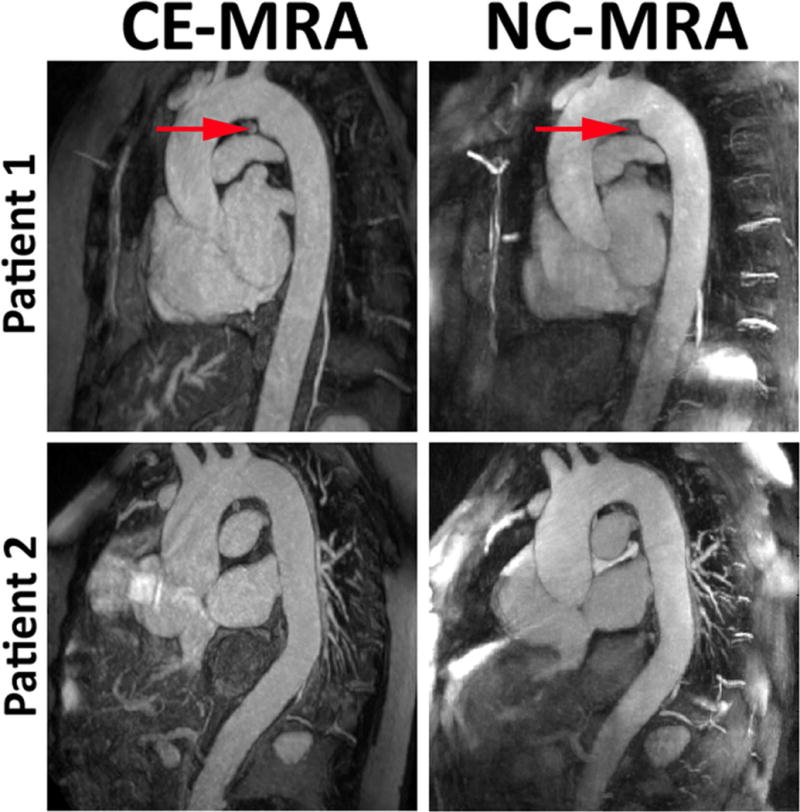
Representative thin reformatted MIPs of two patients CE-MRA (left) and NC-MRA (right). Patient 1 exhibited a ductus diverticulum (red arrow) which is clearly visible in both the CE-MRA and NC-MRA.
The overall visual score index was not significantly different (p = 0.94) between CE-MRA (12.1 ± 1.7) and NC-MRA (12.1 ± 1.0). The two readers’ overall visual score index was significantly different for CE-MRA (p = 0.01) but not for NC-MRA (p = 0.21). The median vessel conspicuity (4.5; range 3 - 5 for CE-MRA vs. 4; range 3 - 5 for NC-MRA), artifact (4; range 2 -4.5) for CE-MRA vs. 4; range 3 - 4.5 for NC-MRA), and noise (4; range 3 - 4.5 for CE-MRA vs. 4; range 3.5 - 5 for NC-MRA) scores were not significantly different (p > 0.16) between CE-MRA and NC-MRA, and NC-MRA was deemed clinically acceptable to good for each category (i.e., scores ≥ 3.0).
As summarized in Supporting Information Table S2 and Supporting Information S3, the mean vessel diameters were not significantly different, except at the proximal aortic arch (p < 0.03). Figure 5 shows the scatter plots showing the results from the Bland-Altman and linear regression analyses. These results show that the vessel diameters are strongly correlated (R2 ≥ 0.96) and in good agreement (absolute mean difference ≤ 0.01 cm and 95% confidence interval ≤ 0.62 cm).
Figure 5.
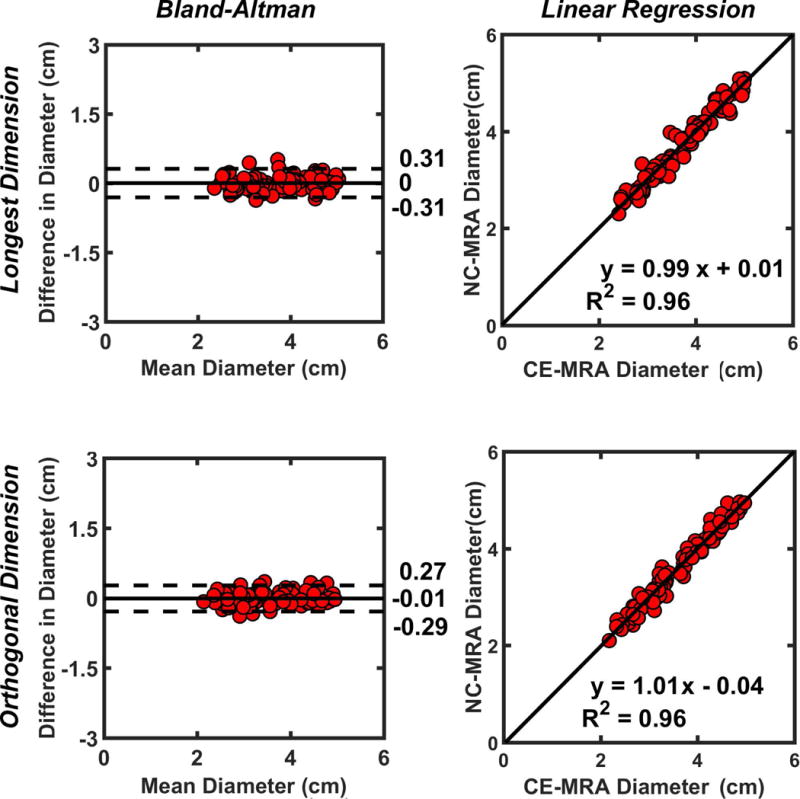
The scatter plots (91 points, 13 patients) derived from the Bland-Altman (left column) and linear regression (right column) analyses on the longest (top row) and orthogonal (bottom row) diameters at 7 locations along the aorta.
Discussion
This study describes the development and evaluation of an accelerated, free-breathing, non-contrast, ECG-triggered, thoracic MRA pulse sequence with stack-of-stars k-space sampling and GRASP reconstruction. The proposed pulse sequence produces clinically acceptable to good image quality at 1.5 mm × 1.5 mm × 1.5 mm spatial resolution and 6 min scan time. The overall visual score index was not significantly different between CE-MRA and NC-MRA. Between CE-MRA and NC-MRA, the vessel diameters were not significantly different, except at the proximal aortic arch (p < 0.03), and the vessel diameters were strongly correlated and in good agreement.
This study has several important implications. First, a rapid NC-MRA may serve as a viable alternative test for patients with poor renal function and/or intravenous access and for reducing the cost and exposure to GBCA. Second, an advancement of NC-MRA is particularly noteworthy considering the lack of viable intravascular contrast agents in the market. Gadofosveset trisodium was recently withdrawn as a product by its manufacturer due to poor sales. Ferumoxytol is not approved as a contrast agent for MRI, especially since the FDA has issued a Boxed Warning. Third, an advantage of NC-MRA over CE-MRA is that the scan can be repeated as needed to achieve clinically acceptable image quality. Fourth, the proposed NC-MRA pulse sequence produces higher spatial resolution (1.5 mm × 1.5 mm × 1.5 mm) and lower scan time (5 min 55 s) than prior works using parallel imaging (10–12), which as a group averaged spatial resolution of 1.6 mm × 1.6 mm × 2.9 mm and scan time of 9:10 min. This corresponds to a 55% reduction in the voxel size and a 33% reduction in scan time. Fifth, the proposed NC-MRA does not rely on an operator’s expertise to properly prescribe navigator gating and achieves 100% gating efficiency, and is thereby capable of achieving shorter and more consistent scan times than pulse sequences using conventional navigator gating.
This study includes several limitations worth mentioning. First, we did not compare the proposed NC-MRA method with prior NC-MRA methods using parallel imaging, because that would have added an extra 9-10 min to a clinical MRI, which would significantly delay the clinical workflow. Second, this study did not compare the proposed NC-MRA to ECG-gated, breath-hold CE-MRA with a bolus chase (30), because our clinical practice prefers time-resolved MRA during first-passage and steady-state, inversion-recovery CE-MRA with navigator gating immediately after contrast agent administration. The reason why a steady-state, inversion-recovery CE-MRA with navigator gating is preferred over a breath-hold CE-MRA is that the former is capable of achieving higher spatial resolution and more extensive spatial coverage than the latter and is less operator dependent. Another study is warranted to compare the proposed NC-MRA to ECG-gated, breath-hold CE-MRA in patients. Third, this study did not compare the proposed stack-of-stars k-space sampling pattern with other investigational k-space sampling patterns proposed for NC-MRA, because they are not commercially available. A future collaborative study is warranted to compare the performance of different investigational NC-MRA methods. Fourth, the image reconstruction time was considerably long on a standalone computer. One approach to reduce the reconstruction time is to employ hardware (GPU) and software solutions [e.g., Split Bregman (31) and software coil compression (32)]. This can be implemented with an engineering approach by systematically testing for reconstruction fidelity along each step in improving computational efficiency (Split Bregman, coil compression). Specific benchmarks one may use to evaluate success include normalized root mean square error less than 5% and structural similarity index (33) greater than 0.95, where 1 is a perfect score. Fifth, this study included only 13 patients and did not include diagnosis of aortic disease. Future clinical studies including a larger patient cohort are warranted to further evaluate the clinical utility of NC-MRA. Sixth, the proposed stack-of-stars k-space sampling pattern with overlapping kz stacks achieved 10% oversampling at the center relative to its periphery. This design was driven by our desire to achieve a good balance between spatial resolution, slab thickness, and readout duration per shot (< 200 ms). Our preliminary analysis on training data showed that oversampling the center improves CS performance. However, we did not perform an exhaustive search to identify an optimal level of oversampling the center of k-space. One approach to accomplish that goal is to acquire a training data set at Nyquist sampling rate and retrospectively downsample and reconstruct to determine an optimal pattern per acceleration rate. Unfortunately, this would be difficult to accomplish in a clinical environment, because the scan time would be considerably long and it is nearly impossible to match critical conditions such as magnetization weighting, eddy currents, motion between the accelerated and non-accelerated acquisitions. Seventh, we manually selected a single coil sampling the liver to make it easier to isolate the respiratory motion from the navigator signal. One approach to automate this step is to use an approach described by Zhang et. al (34).
In summary, this study describes an accelerated, free-breathing, non-contrast, ECG-triggered, thoracic MRA pulse sequence with stack-of-stars k-space sampling and GRASP reconstruction. The proposed NC-MRA pulse sequence produces clinically acceptable to good image quality at high spatial resolution (1.5 mm × 1.5 mm × 1.5 mm) and clinically acceptable scan time (6 min). The overall visual score index was not significantly different between NC-MRA and CE-MRA, and the mean vessel diameters were strongly correlated and in good agreement. Future work includes a study aimed at evaluating the diagnostic accuracy of the proposed NC-MRA pulse sequence in patients with aortic diseases with respect to clinical standard CE-MRA methods.
Supplementary Material
Supporting Video S1. Movie display of respiratory motion of the images shown in Supporting Information Figure S1.
Table S1. The Pertinent clinical profiles of the 13 patients enrolled in this study.
Table S2: The longest diameters at seven standardized locations. Values represent mean ± standard deviation. Location 1: aortic sinuses of valsalva; location 2: sinotubular junction; location 3: mid ascending aorta; location 4: proximal aortic arch; location 5: mid aortic arch; location 6: proximal descending thoracic aorta; location 7: mid descending aorta; location. *P < 0.05 was considered statistically significant.
Table S3: The “orthogonal” diameters at seven standardized locations. Values represent mean ± standard deviation. Location 1: aortic sinuses of valsalva; location 2: sinotubular junction; location 3: mid ascending aorta; location 4: proximal aortic arch; location 5: mid aortic arch; location 6: proximal descending thoracic aorta; location 7: mid descending aorta. *P < 0.05 was considered statistically significant.
Figure S1. A schematic illustrating how the diaphragmatic motion is extracted from the navigator ray oriented along the head-to-foot direction. In step 1, we performed 1D FFT to generate an x-t image (left column, row 1). In step 2, we performed PCA and selected the second principal component, superimposed as yellow line (left column, row 2). In step 3, we populated each of six bins with the same number of k-space data by adapting the bin width. Performing NUFFT on motion-resolved k-space data produced aliased images with six respiratory states (right column). For cross reference, see Supporting Video S1 for movie display of six respiratory states of images. Note, aliasing artifacts are suppressed later with CS reconstruction.
Acknowledgments
The authors thank funding support from the National Institutes of Health (R01HL116895, R01HL138578, R21EB024315, R21AG055954).
Grant Support: This work was supported in part by funding from the National Institutes of Health (R01HL116895, R01HL138578, R21EB024315, R21AG055954)
Appendix: Optimization of k-space Sampling and Ordering
As shown in Figure 1, we elected to acquire a partial stack of rays (48 out of 80, 7 rays beyond the center of k-space, readout duration = 168 ms) along kz for each heartbeat in order to achieve a good balance between spatial resolution, slab thickness, and readout duration per shot (< 200 ms). Note, the rays are rotated with golden angle ratio (35) over the heartbeat dimension. This sampling scheme features three advantages: (a) the inner 10% of the center of k-space is twice as dense as the periphery which results in a better initial solution for CS, (b) eddy currents are minimized by acquiring identical stacks along kz, which is important for artifact reduction in b-SSFP readout (36–38), and (c) the degree of incoherence of aliasing artifacts is increased by rotating the rays with golden angle ratio, which is important for CS reconstruction performance. For this sampling pattern, we explored two different k-space ordering strategies along the kz direction (a) outer-in and (b) center-out. The center-out ordering is expected to produce better fat suppression, because in this trajectory the center of k-space is sampled by the 8th ray after the fat suppression pulse. In contrast, in the outer-in trajectory, the center of k-space is sampled by the 41st ray after the fat saturation pulse. Figure 1C shows the preliminary results which highlight this difference.
Footnotes
This work will be presented in part at the 2018 ISMRM conference
References
- 1.Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Borden WB, Bravata DM, Dai S, Ford ES, Fox CS, Franco S, Fullerton HJ, Gillespie C, Hailpern SM, Heit JA, Howard VJ, Huffman MD, Kissela BM, Kittner SJ, Lackland DT, Lichtman JH, Lisabeth LD, Magid D, Marcus GM, Marelli A, Matchar DB, McGuire DK, Mohler ER, Moy CS, Mussolino ME, Nichol G, Paynter NP, Schreiner PJ, Sorlie PD, Stein J, Turan TN, Virani SS, Wong ND, Woo D, Turner MB. Heart disease and stroke statistics–2013 update: a report from the American Heart Association. Circulation. 2013;127(1):e6–e245. doi: 10.1161/CIR.0b013e31828124ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hiratzka LF, Bakris GL, Beckman JA, Bersin RM, Carr VF, Casey DE. 2010 ACCF/AHA/AATS/ACR/ASA/SCA/SCAI/SIR/STS/SVM guidelines for the diagnosis and management of patients with thoracic aortic disease A report of the American College of Cardiology Foundation/American Heart Association task force on practice guidelines, American Association for Thoracic Surgery, American College of Radiology American Stroke Association, Society of Cardiovascular Anesthesiologists, Society for Cardiovascular Angiography and Interventions, Society of Interventional Radiology, Society of Thoracic Surgeons, and Society for Vascular Medicine. J Am Coll Cardiol. 2010;55 doi: 10.1016/j.jacc.2010.02.015. [DOI] [PubMed] [Google Scholar]
- 3.Broome DR, Girguis MS, Baron PW, Cottrell AC, Kjellin I, Kirk GA. Gadodiamide-associated nephrogenic systemic fibrosis: why radiologists should be concerned. American Journal of Roentgenology. 2007;188(2):586–592. doi: 10.2214/ajr.06.1094. [DOI] [PubMed] [Google Scholar]
- 4.Olchowy C, Cebulski K, Łasecki M, Chaber R, Olchowy A, Kałwak K, Zaleska-Dorobisz U. The presence of the gadolinium-based contrast agent depositions in the brain and symptoms of gadolinium neurotoxicity-A systematic review. PloS one. 2017;12(2):e0171704. doi: 10.1371/journal.pone.0171704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wheaton AJ, Miyazaki M. Non-contrast enhanced MR angiography: physical principles. J Magn Reson Imaging. 2012;36(2):286–304. doi: 10.1002/jmri.23641. [DOI] [PubMed] [Google Scholar]
- 6.Miyazaki M, Lee VS. Nonenhanced MR angiography. Radiology. 2008;248(1):20–43. doi: 10.1148/radiol.2481071497. [DOI] [PubMed] [Google Scholar]
- 7.Brittain JH, Hu BS, Wright GA, Meyer CH, Macovski A, Nishimura DG. Coronary angiography with magnetization-prepared T2 contrast. Magn Reson Med. 1995;33(5):689–696. doi: 10.1002/mrm.1910330515. [DOI] [PubMed] [Google Scholar]
- 8.Weber OM, Martin AJ, Higgins CB. Whole-heart steady-state free precession coronary artery magnetic resonance angiography. Magn Reson Med. 2003;50(6):1223–1228. doi: 10.1002/mrm.10653. [DOI] [PubMed] [Google Scholar]
- 9.Muller MF, Fleisch M, Kroeker R, Chatterjee T, Meier B, Vock P. Proximal coronary artery stenosis: three-dimensional MRI with fat saturation and navigator echo. J Magn Reson Imaging. 1997;7(4):644–651. doi: 10.1002/jmri.1880070406. [DOI] [PubMed] [Google Scholar]
- 10.Srichai MB, Kim S, Axel L, Babb J, Hecht EM. Non-gadolinium-enhanced 3-dimensional magnetic resonance angiography for the evaluation of thoracic aortic disease: a preliminary experience. Tex Heart Inst J. 2010;37(1):58–65. [PMC free article] [PubMed] [Google Scholar]
- 11.Francois CJ, Tuite D, Deshpande V, Jerecic R, Weale P, Carr JC. Unenhanced MR angiography of the thoracic aorta: initial clinical evaluation. AJR Am J Roentgenol. 2008;190(4):902–906. doi: 10.2214/AJR.07.2997. [DOI] [PubMed] [Google Scholar]
- 12.Amano Y, Takahama K, Kumita S. Non-contrast-enhanced MR angiography of the thoracic aorta using cardiac and navigator-gated magnetization-prepared three-dimensional steady-state free precession. J Magn Reson Imaging. 2008;27(3):504–509. doi: 10.1002/jmri.21256. [DOI] [PubMed] [Google Scholar]
- 13.Meijboom LJ, Timmermans J, Zwinderman AH, Engelfriet PM, Mulder BJ. Aortic root growth in men and women with the Marfan’s syndrome. Am J Cardiol. 2005;96(10):1441–1444. doi: 10.1016/j.amjcard.2005.06.094. [DOI] [PubMed] [Google Scholar]
- 14.Lustig M, Donoho D, Pauly JM. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magn Reson Med. 2007;58(6):1182–1195. doi: 10.1002/mrm.21391. [DOI] [PubMed] [Google Scholar]
- 15.Nam S, Akcakaya M, Basha T, Stehning C, Manning WJ, Tarokh V, Nezafat R. Compressed sensing reconstruction for whole-heart imaging with 3D radial trajectories: a graphics processing unit implementation. Magn Reson Med. 2013;69(1):91–102. doi: 10.1002/mrm.24234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piccini D, Feng L, Bonanno G, Coppo S, Yerly J, Lim RP, Schwitter J, Sodickson DK, Otazo R, Stuber M. Four-dimensional respiratory motion-resolved whole heart coronary MR angiography. Magn Reson Med. 2017;77(4):1473–1484. doi: 10.1002/mrm.26221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng L, Grimm R, Block KT, Chandarana H, Kim S, Xu J, Axel L, Sodickson DK, Otazo R. Golden-angle radial sparse parallel MRI: combination of compressed sensing, parallel imaging, and golden-angle radial sampling for fast and flexible dynamic volumetric MRI. Magn Reson Med. 2014;72(3):707–717. doi: 10.1002/mrm.24980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otazo R, Kim D, Axel L, Sodickson DK. Combination of compressed sensing and parallel imaging for highly accelerated first-pass cardiac perfusion MRI. Magn Reson Med. 2010;64(3):767–776. doi: 10.1002/mrm.22463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhat H, Yang Q, Zuehlsdorff S, Li K, Li D. Contrast-enhanced whole-heart coronary magnetic resonance angiography at 3 T with radial EPI. Magn Reson Med. 2011;66(1):82–91. doi: 10.1002/mrm.22781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng L, Axel L, Chandarana H, Block KT, Sodickson DK, Otazo R. XD-GRASP: Golden-angle radial MRI with reconstruction of extra motion-state dimensions using compressed sensing. Magn Reson Med. 2016;75(2):775–788. doi: 10.1002/mrm.25665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galizia MS, Febbo JA, Popescu AR, Bi X, Collins J, Markl M, Edelman RR, Carr J. Steady state imaging of the thoracic vasculature using inversion recovery FLASH and SSFP with a blood pool contrast agent. J Cardiovasc Magn Reson. 2012;14(Suppl 1):P51. [Google Scholar]
- 22.Griswold MA, Jakob PM, Heidemann RM, Nittka M, Jellus V, Wang J, Kiefer B, Haase A. Generalized autocalibrating partially parallel acquisitions (GRAPPA) Magn Reson Med. 2002;47(6):1202–1210. doi: 10.1002/mrm.10171. [DOI] [PubMed] [Google Scholar]
- 23.Benkert T, Tian Y, Huang C, DiBella EVR, Chandarana H, Feng L. Optimization and validation of accelerated golden-angle radial sparse MRI reconstruction with self-calibrating GRAPPA operator gridding. Magn Reson Med. 2018;80(1):286–293. doi: 10.1002/mrm.27030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fessler JA. On NUFFT-based gridding for non-Cartesian MRI. J Magn Reson. 2007;188(2):191–195. doi: 10.1016/j.jmr.2007.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Griswold MAWD, Heidemann RM, Haase A, Jakob PM. The use of an adaptive reconstruction for array coil sensitivity mapping and intensity normalization In Proceedings of the 10th Annual Meeting of. Vol. 2002. ISMRM; Honolulu, Hawaii, USA: p. 2410. [Google Scholar]
- 26.Walsh DO, Gmitro AF, Marcellin MW. Adaptive reconstruction of phased array MR imagery. Magn Reson Med. 2000;43(5):682–690. doi: 10.1002/(sici)1522-2594(200005)43:5<682::aid-mrm10>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 27.Pang J, Sharif B, Fan Z, Bi X, Arsanjani R, Berman DS, Li D. ECG and navigator-free four-dimensional whole-heart coronary MRA for simultaneous visualization of cardiac anatomy and function. Magn Reson Med. 2014;72(5):1208–1217. doi: 10.1002/mrm.25450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng L, Srichai MB, Lim RP, Harrison A, King W, Adluru G, Dibella EV, Sodickson DK, Otazo R, Kim D. Highly accelerated real-time cardiac cine MRI using k-t SPARSE-SENSE. Magn Reson Med. 2013;70(1):64–74. doi: 10.1002/mrm.24440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dabov K, Foi A, Katkovnik V, Egiazarian K. Image denoising by sparse 3-D transform-domain collaborative filtering. IEEE Trans Image Process. 2007;16(8):2080–2095. doi: 10.1109/tip.2007.901238. [DOI] [PubMed] [Google Scholar]
- 30.Krinsky GA, Rofsky NM, DeCorato DR, Weinreb JC, Earls JP, Flyer MA, Galloway AC, Colvin SB. Thoracic aorta: comparison of gadolinium-enhanced three-dimensional MR angiography with conventional MR imaging. Radiology. 1997;202(1):183–193. doi: 10.1148/radiology.202.1.8988210. [DOI] [PubMed] [Google Scholar]
- 31.Goldstein T, Osher S. The Split Bregman Method for L1-Regularized Problems. Siam Journal on Imaging Sciences. 2009;2(2):323–343. [Google Scholar]
- 32.Adluru G, DiBella E. Compression2: compressed sensing with compressed coil arrays. J Cardiovasc Magn Reson. 2012;14(Suppl 1):P242. [Google Scholar]
- 33.Wang Z, Bovik AC, Sheikh HR, Simoncelli EP. Image quality assessment: from error visibility to structural similarity. IEEE Trans Image Process. 2004;13(4):600–612. doi: 10.1109/tip.2003.819861. [DOI] [PubMed] [Google Scholar]
- 34.Zhang T, Cheng JY, Chen Y, Nishimura DG, Pauly JM, Vasanawala SS. Robust self-navigated body MRI using dense coil arrays. Magn Reson Med. 2016;76(1):197–205. doi: 10.1002/mrm.25858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winkelmann S, Schaeffter T, Koehler T, Eggers H, Doessel O. An optimal radial profile order based on the Golden Ratio for time-resolved MRI. IEEE Trans Med Imaging. 2007;26(1):68–76. doi: 10.1109/TMI.2006.885337. [DOI] [PubMed] [Google Scholar]
- 36.Bieri O, Markl M, Scheffler K. Analysis and compensation of eddy currents in balanced SSFP. Magn Reson Med. 2005;54(1):129–137. doi: 10.1002/mrm.20527. [DOI] [PubMed] [Google Scholar]
- 37.Wundrak S, Paul J, Ulrici J, Hell E, Rasche V. A Small Surrogate for the Golden Angle in Time-Resolved Radial MRI Based on Generalized Fibonacci Sequences. IEEE Trans Med Imaging. 2015;34(6):1262–1269. doi: 10.1109/TMI.2014.2382572. [DOI] [PubMed] [Google Scholar]
- 38.Schaeffter T, Weiss S, Eggers H, Rasche V. Projection reconstruction balanced fast field echo for interactive real-time cardiac imaging. Magn Reson Med. 2001;46(6):1238–1241. doi: 10.1002/mrm.1322. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Video S1. Movie display of respiratory motion of the images shown in Supporting Information Figure S1.
Table S1. The Pertinent clinical profiles of the 13 patients enrolled in this study.
Table S2: The longest diameters at seven standardized locations. Values represent mean ± standard deviation. Location 1: aortic sinuses of valsalva; location 2: sinotubular junction; location 3: mid ascending aorta; location 4: proximal aortic arch; location 5: mid aortic arch; location 6: proximal descending thoracic aorta; location 7: mid descending aorta; location. *P < 0.05 was considered statistically significant.
Table S3: The “orthogonal” diameters at seven standardized locations. Values represent mean ± standard deviation. Location 1: aortic sinuses of valsalva; location 2: sinotubular junction; location 3: mid ascending aorta; location 4: proximal aortic arch; location 5: mid aortic arch; location 6: proximal descending thoracic aorta; location 7: mid descending aorta. *P < 0.05 was considered statistically significant.
Figure S1. A schematic illustrating how the diaphragmatic motion is extracted from the navigator ray oriented along the head-to-foot direction. In step 1, we performed 1D FFT to generate an x-t image (left column, row 1). In step 2, we performed PCA and selected the second principal component, superimposed as yellow line (left column, row 2). In step 3, we populated each of six bins with the same number of k-space data by adapting the bin width. Performing NUFFT on motion-resolved k-space data produced aliased images with six respiratory states (right column). For cross reference, see Supporting Video S1 for movie display of six respiratory states of images. Note, aliasing artifacts are suppressed later with CS reconstruction.


