Abstract
Purpose
We sought to assess whether machine learning-based classification approaches can improve the classification of pancreatic tumor models relative to more simplistic analysis methods, using T1 relaxation, Chemical Exchange Saturation Transfer (CEST), and dynamic contrast-enhanced (DCE) MRI.
Methods
The T1 relaxation time constants, % CEST at five saturation frequencies, and vascular permeability constants from DCE MRI were measured from Hs 766T, MIA PaCa-2 and SU.86.86 pancreatic tumor models. We used each of these measurements as predictors for machine learning classifier algorithms. We also used principal component analysis (PCA) to reduce the dimensionality of entire CEST spectra and DCE signal evolutions, which were then analyzed using classification methods.
Results
The T1 relaxation time constants, % CEST amplitudes at specific saturation frequencies, and the relative Ktrans and kep values from DCE MRI could not classify all three tumor types. PCA was used to analyze entire CEST spectra, which predicted the correct tumor model with 87.5% accuracy. However, the AUC from DCE signal evolutions could classify each tumor type. PCA was used to analyze the entire CEST spectrum and DCE signal evolutions, which predicted the correct tumor model with 87.5% and 85.1% accuracy, respectively.
Conclusions
Machine learning applied to the entire CEST spectrum improved the classification of the three tumor models, relative to classifications that used % CEST values at single saturation frequencies. A similar improvement was not attained with machine learning applied to T1 relaxation times or DCE signal evolutions, relative to more simplistic analysis methods.
Keywords: PCA, CEST MRI, DCE MRI, machine learning, pancreatic cancer imaging
Introduction
Many MRI studies with small animal models of human cancers, or studies with patients who have cancer, reduce the information available from multiple images to provide a single value that represents the tumor (1–5). For example, a series of T1-weighted MR images with different relaxation times (6,7) or flip angles (8,9) can be used to measure a single T1 relaxation time constant; a temporal series of Dynamic Contrast Enhancement (DCE) MR images can be used to measure a single transport rate constant (3,10,11); and a series of Chemical Exchange Saturation Transfer (CEST) MR images can be used to measure a single % CEST value with saturation applied at a specific MR frequency (5,12). While this approach has advantages by producing a concise descriptor that is often simple to interpret, additional information contained in the images is discarded for the sake of this simplicity.
Principal component analysis (PCA), a type of machine learning, is a potential tool to address the complex dimensionality of MRI data (13–15). PCA can fundamentally reconstruct sparse data into linear combinations of elements that explain the variance of the data by systematically grouping the best predictors into principal components. Therefore, combining PCA with machine learning-based classification may improve the analyses of information-rich MRI data, including images acquired with a range of parameters such as multiple TR times for T1 MRI, multiple saturation frequencies for CEST MRI, and multiple time points for DCE MRI.
We sought to test the ability of machine learning to improve the analysis of MRI studies with small animal tumor models. More specifically, we developed tumor models of Hs 766T, Mia PaCa-2, and Su.86.86 pancreatic cancer that are known to have high, medium, and low hypoxia levels (16,17). We hypothesized that T1 MRI at 7T magnetic field strength would be insufficient to differentiate these tumor models, because T1 relaxation times are similar for most tissues at this high field strength (18). We also hypothesized that the different hypoxia levels would lead to measurable differences with DCE MRI, because hypoxia promotes angiogenesis that leads to increased vascular permeability (19). The ability to detect differences between these tumor models with CEST MRI was unknown, potentially providing an intermediate test case relative to T1 MRI and DCE MRI.
Methods
Tumor Models
All animals were cared for in compliance with protocols approved by the Institutional Animal Care and Use Committee of the University of Arizona. HS 766T, MIA PaCa-2, and SU.86.86 human pancreatic ductal adenocarcinoma cells were tripsinized, rinsed once in PBS, and suspended in 50% Matrigel™ (BD Biosciences, Franklin Lakes, NJ, USA) and 50% PBS. Thirty female SCID mice (Jackson Laboratory, Bar Harbor, ME, USA) were approximately 20 g and 6 to 8 weeks old. Three groups of 10 mice were injected subcutaneously in the right flank with 0.1 mL of saline that contained 10×106 HS 766T cells, MIA PaCa-2 cells, or SU.86.86 cells, respectively. Tumor volume measurements were performed using a caliper every two days, calculated as π/6 × (short axis)2 × (long axis). Tumors grew to ~300 mm3 before initiating MRI studies. Prior to the MRI scan, each mouse was anesthetized with 1.5–2.5% isoflurane in O2 carrier gas, and a 27 G catheter was placed in the tail vein. Physiologic respiration rate was monitored and core body temperature was maintained at 37.0 ± 0.2 °C using warm air controlled by a temperature feedback system (SA Instruments, New York, USA). MRI studies were performed with a 7T Biospec MRI instrument and 72 mm quadrature transceiver coil (Bruker Biospin, Inc., Billerica, MA). After the MRI scan, mice were allowed to recover for one day before being imaged a second and final time.
In vivo T1 MRI
We performed T2-weighted spin-echo MRI to localize the flank tumor, which required 1.5 minutes to acquire (Figure 1a). The T1 relaxation time of each flank tumor was measured using a Rapid Acquisition with Relaxation Enhancement (RARE) acquisition sequence and the following parameters: TR = 150, 300, 350, 500, 700, 900, 1200, 2000, 3000, and 6000 ms; 9.1 ms TE; 1 average; RARE factor = 2; linear encoding order; 1.0 mm slice thickness, for 3 slices centered in the tumor; 3.0 cm2 FOV; 128 × 128 matrix; and 0.23 mm2 in-plane spatial resolution (Figure 1c). The total scan time was 12.1 min. The signal profiles from a region of interest (ROI) that represented the tumor were fit with a monoexponential function without a constant offset using least squares curve fitting to estimate T1 and M0.
Figure 1.
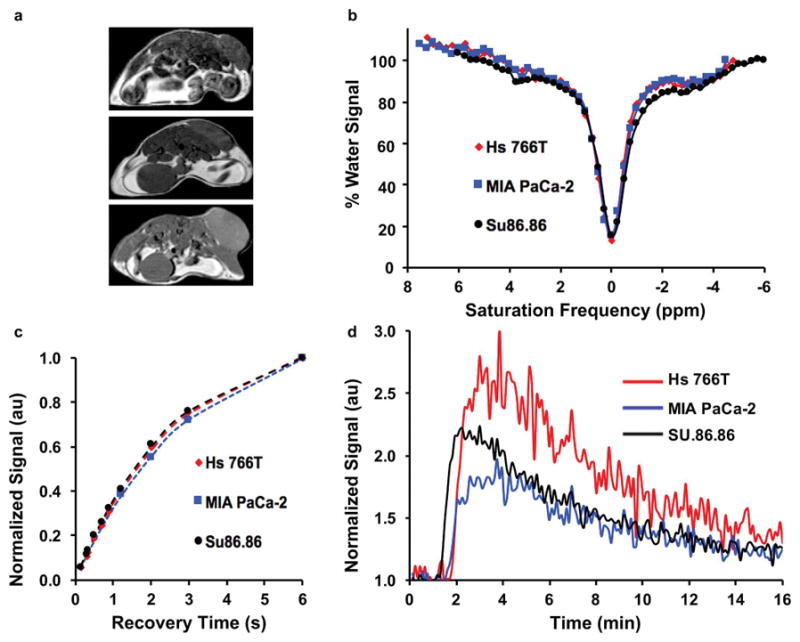
Experimental work flow for in vivo mouse studies. (a) Examples of T2-weighted anatomical images of the Hs 766T (top), MIA PaCa-2 (middle), and SU.86.86 (bottom) tumor models. (b) Examples of CEST spectra for the three tumor models. (c) Examples of T1-weighted recovery time curves for the three tumor models. (d) Examples of DCE CNR evolutions for the three tumor models.
In vivo CEST MRI
Endogenous CEST MRI studies were performed with a respiration-gated CEST-FISP MRI acquisition protocol using a 3.7 ms TR; 1.7 ms TE; 1 average; 15º excitation pulse angle; centric encoding order; unbalanced “FID” mode; 1.0 mm slice thickness; 1 slice; 3.0 cm2 FOV; 128 × 128 matrix; 0.23 mm2 in-plane spatial resolution; 516 ms acquisition time (20,21). A saturation period was applied prior to each FISP acquisition, with ten 600 ms continuous wave radio frequency pulses (totaling 6.0 s of saturation time), at 1.0 μT saturation power with no additional spoiling and fat saturation pulses. Selective saturation was applied at 49 frequencies ranging from −4.5 to 7.5 ppm in 0.25 ppm increments, which required a total of 8.1 min. During the initial optimization of the endogenous CEST MRI acquisition protocol, the Hs 766T tumor model imaged on the first time point (day 1) were imaged with different parameters, using a 5.6 ms TR; 2.8 ms TE; 30º excitation pulse angle; 4.0 cm2 FOV; 128 × 128 matrix; 0.32 mm2 in-plane spatial resolution; 500 ms scan time; a saturation period of 3.0 s; and selective saturation at 101 frequencies ranging from −6 to 6 ppm in 0.12 ppm increments, which required a total time of 6.8 min. CEST spectra from the ROI of the tumor were fit with a sum of six Lorentzian line shapes to measure % CEST at 3.5, 3.0, 2.0, −1.6, and −3.5 ppm, and to account for direct saturation of water at 0 ppm (Figure 1b) (22).
In vivo DCE MRI
A series of dynamic images were acquired using a spoiled gradient-echo MRI protocol (23) using a 50 ms TR; 8.1 ms TE; 1 average; 3.0 cm2 FOV; 0.23 mm2 in-plane spatial resolution; matrix =128 × 128; 1.0 mm slice thickness, for a single slice centered on the tumor (Figure 1d). Each image was acquired in 6.4 s, and repeated 150 times for a total acquisition time of 16 min. An initial set of baseline images was acquired for 30 s prior to intravenous injection of 50 μL of 100 mM MultiHance (Bracco Diagnostics Inc., Milan, Italy), which corresponds to a dose of 0.25 mmolKg−1. DCE MRI signal of the tumor ROI was first normalized to the average baseline signal before injection of contrast agent, then divided by the standard deviation of noise (measured from a region of the image that represented air) to obtain a contrast-to-noise ratio (CNR), and then thresholded at ≥ 2√2 CNR to ensure that only real contrast was analyzed (24). The area under the curve (AUC) was calculated from injection to 2 minutes, from injection to 3 minutes, and from injection to other values beyond 3 minutes. The DCE MRI results were also analyzed with the Linear Reference Region Model to estimate the relative Ktrans and kep values for each tumor (25).
Analysis Methods
All analyses of T1, CEST, and DCE MRI results were completed with MATLAB 2017b (MathWorks, Inc., Natick, MA, USA). Boxplots were constructed for average T1 relaxation time, % CEST at specific saturation frequencies, and AUC of DCE MRI for each tumor model using RStudio (RStudio, Inc., Boston, MA, USA) and Rattle (Togaware, Canberra, Australia) (Figure 2). These boxplots showed the median of each tumor model as a thick horizontal line; means as asterisks; 95% confidence intervals of the median as notches; 25% and 75% quartiles as thin horizontal lines; open circles as data points that are beyond the interquartile ranges, and vertical dashed lines as the range. Groups were considered to be different when a Wilcoxon Rank-Sum analysis showed p ≤ 0.05 between groups.
Figure 2.
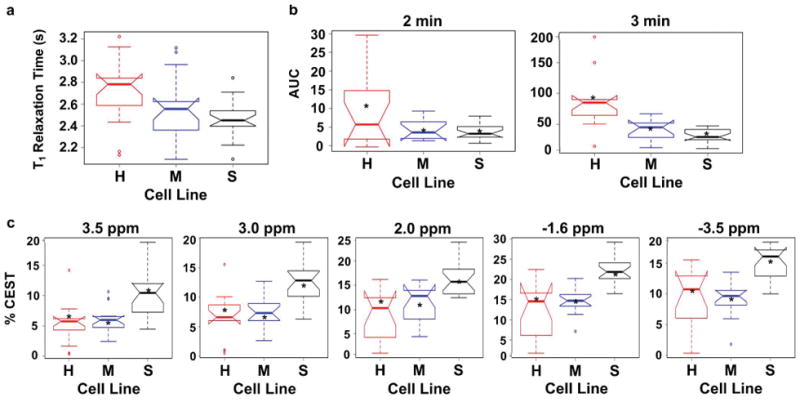
Distributions of MRI results in notched boxplot representations. H = Hs 766T tumor model; M = MIA PaCa-2 tumor model; S = SU.86.86 tumor model. (a) The distributions of T1 relaxation times for H, M, and S tumor models. The Area under the Curve (AUC) from injection to 2 minutes (left), and from injection to 3 minutes (right), for H, M, and S tumor models. (c) The distributions of % CEST signals at saturation frequencies of 3.5, 3.0, 2.0, −1.6, and −3.5 ppm for H, M, and S tumor models.
Four analysis methods, including four variations of the support vector machine (SVM) analysis method (Supporting Information Table S1), were used to build tumor classification models with the T1 relaxation times (Figure 1c, 3; Supporting Information Figure S1, S2), CEST spectra (Figure 1b, 4; Supporting Information Figure S3, S4), and DCE CNR evolutions (Figure 1d, 5; Supporting Information Figure S5, S6) (13–15). To train the analysis method, a 30-fold cross validation method was used to prevent over-fitting the model. The performance of each model was measured using the AUC of the receiver operating curve (ROC). In addition, the predictive classification models were assessed to evaluate true positive rates verses false negative rates, and positive predictive values versus false discovery rates (true positive and false positive rates for classifications). The k-Nearest Neighbor classification of CEST spectra was performed with 5 PCA components; a one-vs-one multiclass method; 3 neighbors; cosine distance metric; equal distance weight. Training required 5.5 seconds and the prediction speed was ~71 observations per second. The Gaussian SVM classifications of DCE CNR evolutions were performed with 10 PCA components; a one-vs-one multiclass method; 1 box constraint; 1.0 and 2.3 kernel scales, for fine and medium SVM methods. Training required 7.4 seconds and the prediction speed was ~52 observations per second.
Figure 3.
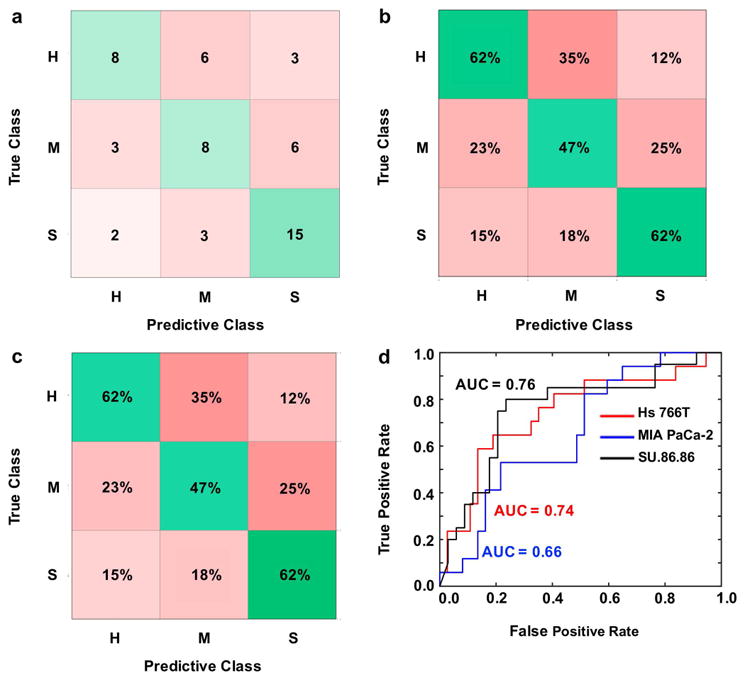
Error matrix representations after machine learning classification with a Fine Gaussian Support Vector Machine (FG SVM) algorithm using T1 relaxation times as predictors. H = Hs 766T tumor model; M = MIA PaCa-2 tumor model; S = SU.86.86 tumor model. (a) Number of correct (green) and incorrect (red) predicted observations. (b) Positive predictive value rates (green) and false discovery rates (red). (c) True positive rates (green) and false negative rates (red). (d) Area under the curve (AUC) for the receiver operator characteristic (ROC) curves represents classifier algorithm prediction accuracy.
Figure 4.
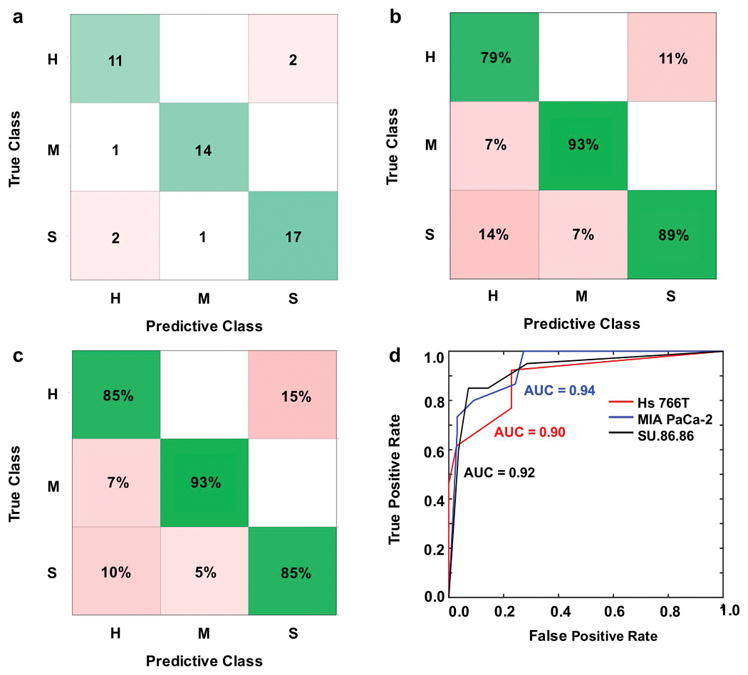
Error matrix representations after machine learning classification with a k-Nearest Neighbor algorithm using principal components from entire CEST spectra as predictors. H = Hs 766T tumor model; M = MIA PaCa-2 tumor model; S = SU.86.86 tumor model. (a) Number of correct (green) and incorrect (red) predicted observations. (b) Positive predictive value rates (green) and false discovery rates (red). (c) True positive rates (green) and false negative rates (red). (d) Area under the curve (AUC) for the receiver operator characteristic (ROC) curves represents classifier algorithm prediction accuracy.
Figure 5.
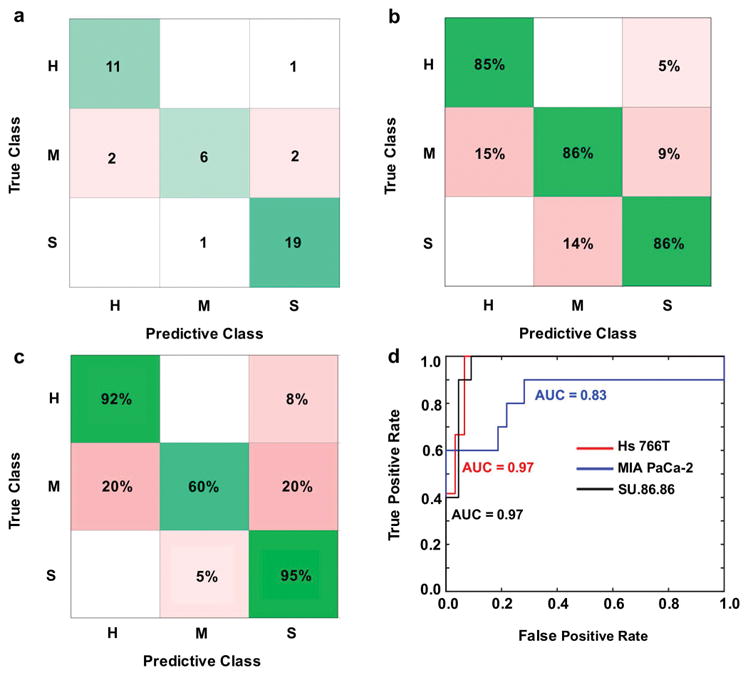
Error matrix representations after machine learning classification with a Medium Gaussian Support Vector Machine (MG SVM) algorithm using principal components from entire DCE pharmacokinetic curves as predictors. H = Hs 766T tumor model; M = MIA PaCa-2 tumor model; S = SU.86.86 tumor model. (a) Number of correct (green) and incorrect (red) predicted observations. (b) Positive predictive value rates (green) and false discovery rates (red). (c) True positive rates (green) and false negative rates (red). (d) Area under the curve (AUC) for the receiver operator characteristic (ROC) curves represents classifier algorithm prediction accuracy.
Results
Twenty eight were successfully imaged at two time points. One mouse from the Hs 766T group and one mouse from the MIA PaCa-2 group expired after their first imaging scan. The T1-weighted MRI signals of the tumor ROIs were very similar for each mouse model, indicating similar T1 relaxation times for each model (Figure 1c). CEST spectra of the tumor ROIs showed a decrease in water signal 3.5, 3.0, and 2.0 ppm, which corresponds to CEST arising from endogenous amides, amines, and hydroxyls that have been observed in previous studies (Figure 1b) (5). A decrease in signal was also observed at −1.6 and −3.5 ppm from the relayed NOE from aliphatic protons on lipids and proteins, respectively, as previously reported (26–29). These decreases in water signals in the CEST spectra were different for each tumor model. The CNR evolution of the DCE MRI studies also showed differences between tumor models (Figure 1d).
Boxplots of T1 relaxation times for each tumor model showed that the Hs 766T vs. MIA PaCa-2 models, and the MIA PaCa-2 vs. SU.86.86 models, were not statistically different (Figure 2a). However, the Hs 766T vs. SU.86.86 models had statistically different T1 relaxation times. Boxplot analyses of CEST results showed that the Hs 766T vs. MIA PaCa-2 models were not statistically different at each saturation frequency (Figure 2c). The Hs 766T vs. SU.86.86 models, and the MIA PaCa-2 vs. SU.86.86 models were statistically different at each saturation frequency. Therefore, T1 relaxation times and CEST at single saturation frequencies could not differentiate all tumor models.
Quantitative pharmacokinetics analysis of the DCE MRI results using the Linear Reference Region Model produced values of relative Ktrans and kep. However, a comparison of these parameters showed a lack of statistical significance between all tumor models (Supporting Information). A boxplot analysis of DCE CNR evolution also showed a lack of statistical significance between all tumor models when results were analyzed for the first two minutes after injection (Figure 2b, left). However, all three tumor models had a statistically significant difference when results were analyzed for the first three minutes after injection (Figure 2b, right) (p < 0.002 for Hs 766T and MIA PaCa-2; p < 0.01 for MIA PaCa-2 and SU.86.86; and p < 0.001 for Hs 766T and SU.86.86). Notably, each tumor model showed statistically significant differences when DCE CNR evolutions were analyzed for >3 minutes after injection. These results showed that an adequately long 3-minute DCE acquisition was needed to differentiate the tumor models, but a very long DCE MRI acquisition beyond 3 minutes was unnecessary. Moreover, these results showed that a simple AUC analysis of DCE MRI results could differentiate all tumor models.
When T1 relaxation times were used as predictors, the Gaussian SVM with a fine kernel scale had the best performance among the classification models tested (Supporting Table S1). However, even this best-performing model was only able to classify 75% of the SU.86.86 tumors, and classified a very poor 47% of MIA PaCa-2 and Hs 766T tumors (Figure 3c; Supporting Information Figure S1, S2). The AUC of the ROCs ranged from 0.66 to 0.76 for the tumor models, indicating a relatively poor classification based on T1 relaxation time (Figure 3d; Supporting Information Figure S1, S2). Therefore, machine learning (Figure 3; Supporting Information Figure S1, S2) did not improve the ability to classify each tumor type relative to analyses of the values and distributions of T1 relaxation times (Figure 2a).
CEST spectra (Figure 1b) provided 101 predictors (corresponding to each saturation frequency) for classifying tumor models. Principal component analysis (PCA) using a k-Nearest Neighbors algorithm produced the best classification of the models tested, with 85–93% correct classifications for the three tumor types (Figure 4c; Supporting Information Figure S3, S4; Supporting Information Table S1). PCA reduced the CEST spectra to 5 components, where the first two components explained 75% and 21% of the classification. These components resulted in excellent AUC of the ROCs with true positive rates ≥ 0.90 for each tumor classification (Figure 4d; Supporting Information Figure S3, S4; Supporting Information Table S1). Therefore, machine learning (Figure 4; Supporting Information Figure S3, S4) improved the ability to classify each tumor type relative to analyses of CEST at each saturation frequency (Figure 2c).
DCE CNR evolutions (Figure 1d) provided 150 predictors (corresponding to each image repetition). A Gaussian SVM with medium kernel scale produced the best classification among the tested models, with 92–95% correct classifications for Hs 766T and SU.86.86 tumor models, and 60% correct classifications for the MIA PACa-2 tumor model (Figure 5c; Supporting Information Figure S5, S6; Supporting Information Table S1). PCA reduced the DCE results to 3 components, where the first component explained 99% of the classification. These components resulted in excellent AUC of the ROCs with true positive rates of 0.97 for Hs 766T and SU.86.86 tumor models, and 0.83 for the MIA PACa-2 tumor model (Figure 5d; Supporting Information Figure S5, S6; Supporting Information Table S1). Therefore, machine learning (Figure 5; Supporting Information Figure S5, S6) had the same ability to classify each tumor type relative to more traditional DCE MRI analyses of the AUC for at least 3 minutes after injection of agent (Figure 2c).
Discussion
Our results showed that machine learning improved the classification of tumor models based on entire CEST spectra, relative to classifications based on a single saturation frequency. For comparison, machine learning failed to improve classifications using T1 relaxation times, and did not improve on classification of tumor models using more simplistic analyses of DCE MRI AUC in our studies. This lack of classification based on T1 was anticipated due to similar T1 relaxation times of tissues at 7T magnetic field strength. The classification of all three tumor models with the AUC from DCE MRI was also anticipated, because the tumor models differ in levels of hypoxia that leads to differences in angiogenesis and vascular permeability. The classification of tumor models when CEST MRI was analyzed with machine learning was unexpected. Also, machine learning has previously been applied to assess results involving T1 relaxation rates and DCE MRI, but machine learning has only recently been applied to analyze CEST MRI.
Our preliminary study provides a foundation for additional research that investigates the utility of machine learning for CEST MRI analyses. Our studies only evaluated T1 MRI, DCE MRI, and CEST MRI results that were expected to have low, high, and unknown potential to classify each tumor model, respectively. Future studies may include other MRI methods, including T2* MRI, diffusion-based MRI, or hyperpolarized MR spectroscopic imaging, and may also include multiparametric assessments with machine learning. Our studies only evaluated 3 tumor models that differed in levels of hypoxia and vasculature, while future studies can test differences in tumor models with other tumor volumes (because tumor size may affect the levels of hypoxia and vascular permeability), differences in other tumor models, or changes in tumor models before and after treatment. Furthermore, our study only tested a limited number of mice, and performing imaging studies with a larger number of mice can provide opportunities to further investigate other machine learning methods to analyze results. Yet our preliminary studies contribute to growing evidence that machine learning provides advantages for more comprehensively analyzing multifaceted MRI data such as CEST spectra.
Conclusions
Machine learning improved the classification of the three tumor models based on CEST MRI results. A similar improvement was not attained with machine learning based on T1 relaxation times. Tumor classification was successfully accomplished with DCE MRI using a simple AUC analysis, and machine learning did not further improve this classification. Therefore, machine learning is a useful analysis methodology for evaluating CEST spectra.
Supplementary Material
Supporting Information Figure S1. Error matrix representations after machine learning classification with a Linear Discriminant Analysis (LDA) algorithm using T1 relaxation times as predictors. H = Hs 766T tumor model; M = MIA PaCa-2 tumor model; S = SU.86.86 tumor model. (a) Number of correct (green) and incorrect (red) predicted observations. (b) Positive predictive value rates (green) and false discovery rates (red). (c) True positive rates (green) and false negative rates (red). (d) Area under the curve (AUC) for the receiver operator characteristic (ROC) curves represents classifier algorithm prediction accuracy.
Supporting Information Figure S2. Error matrix representations after machine learning classification with a Subspace Discriminant (SD) algorithm using T1 relaxation times as predictors. H = Hs 766T tumor model; M = MIA PaCa-2 tumor model; S = SU.86.86 tumor model. (a) Number of correct (green) and incorrect (red) predicted observations. (b) Positive predictive value rates (green) and false discovery rates (red). (c) True positive rates (green) and false negative rates (red). (d) Area under the curve (AUC) for the receiver operator characteristic (ROC) curves represents classifier algorithm prediction accuracy.
Supporting Information Figure S3. Error matrix representations after machine learning classification with a Subspace Discriminant (SD) algorithm using principal components from whole CEST spectra as predictors. H = Hs 766T tumor model; M = MIA PaCa-2 tumor model; S = SU.86.86 tumor model. (a) Number of correct (green) and incorrect (red) predicted observations. (b) Positive predictive value rates (green) and false discovery rates (red). (c) true positive rates (green) and false negative rates (red). (d) area under the curve (AUC) for the receiver operator characteristic (ROC) curves represents classifier algorithm prediction accuracy.
Supporting Information Figure S4. Error matrix representations after machine learning classification with a Quadratic Support Vector Machine (Q SVM) using principal components from whole CEST spectra as predictors. H = Hs 766T tumor model; M = MIA PaCa-2 tumor model; S = SU.86.86 tumor model. (a) Number of correct (green) and incorrect (red) predicted observations. (b) Positive predictive value rates (green) and false discovery rates (red). (c) True positive rates (green) and false negative rates (red). (d) Area under the curve (AUC) for the receiver operator characteristic (ROC) curves represents classifier algorithm prediction accuracy.
Supporting Information Figure S5. Error matrix representations after machine learning classification with a Linear Support Vector Machine (L SVM) algorithm using principal components from whole DCE pharmacokinetic curves as predictors. H = Hs 766T tumor model; M = MIA PaCa-2 tumor model; S = SU.86.86 tumor model. (a) Number of correct (green) and incorrect (red) predicted observations. (b) Positive predictive value rates (green) and false discovery rates (red). (c) True positive rates (green) and false negative rates (red). (d) Area under the curve (AUC) for the receiver operator characteristic (ROC) curves represents classifier algorithm prediction accuracy.
Supporting Information Figure S6. Error matrix representations after machine learning classification with a Linear Discriminant Analysis (LDA) algorithm using principal components from whole DCE pharmacokinetic curves as predictors. H = Hs 766T tumor model; M = MIA PaCa-2 tumor model; S = SU.86.86 tumor model. (a) Number of correct (green) and incorrect (red) predicted observations. (b) Positive predictive value rates (green) and false discovery rates (red). (c) True positive rates (green) and false negative rates (red). (d) Area under the curve (AUC) for the receiver operator characteristic (ROC) curves represents classifier algorithm prediction accuracy.
Supporting Information Table S1. Error matrix values for prediction models using T1, CEST, and DCE MRI. The data highlighted in red corresponds to Figs. 3–5, respectively. H = Hs 766T tumor model; M= MIA PaCa-2 tumor model; S = SU.86.86 tumor model.
Acknowledgments
This work was supported by NIH grants R01CA169774, P30CA023074 and P50CA95060, and by Institutional Research Grant number 128749-IRG-16-124-37-IRG from the American Cancer Society.
References
- 1.O’Connor JPB, Aboagye EO, Adams JE, et al. Imaging biomarker roadmap for cancer studies. Nature Rev Clin Oncology. 2017;14:169–86. doi: 10.1038/nrclinonc.2016.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abramson RG, Arlinghaus L, Dula A, et al. MRI biomarkers in oncology clinical trials. Magn Reson Imaging Clin N Am. 2016;24:11–29. doi: 10.1016/j.mric.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tofts PS, Brix G, Buckley DL, et al. Estimating kinetic parameters from dynamic contrast-enhanced T1-weighted MRI of a diffusible tracer: standardized quantities and symbols. J Magn Reson Imaging. 1999;10:223–232. doi: 10.1002/(sici)1522-2586(199909)10:3<223::aid-jmri2>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 4.Jafar MM, Parsai A, Miquel ME. Diffusion-weighted magnetic resonance imaging in cancer: Reported apparent diffusion coefficients, in-vitro and in-vivo reproducibility. World J Radiol. 2016;8:21–49. doi: 10.4329/wjr.v8.i1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones KM, Pollard AC, Pagel MD. Clinical applications of chemical exchange saturation transfer (CEST) MRI. J Magn Reson Imaging. 2018;47:11–27. doi: 10.1002/jmri.25838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crawley AP, Henkelman RM. A comparison of one-shot and recovery methods in T1 imaging. Magn Reson Med. 1988;7:23–34. doi: 10.1002/mrm.1910070104. [DOI] [PubMed] [Google Scholar]
- 7.Haase A. Snapshot FLASH MRI. Applications to T1, T2, and cheicla shift imaging. Magn Res Med. 1990;13:77–89. doi: 10.1002/mrm.1910130109. [DOI] [PubMed] [Google Scholar]
- 8.Haase A, Frahm J, Matthaei D, Hanicke W, Merboldt KD. FLASH imaging: Rapid NMR imaging using low flip-angle pulses. J Magn Reson. 1986;67:258–266. doi: 10.1016/j.jmr.2011.09.021. [DOI] [PubMed] [Google Scholar]
- 9.Wang HZ, Riederer SJ, Lee JN. Optimizing the precision in T1 relaxation estimation using limited flip angles. Magn Reson Med. 1987;5:399–416. doi: 10.1002/mrm.1910050502. [DOI] [PubMed] [Google Scholar]
- 10.Jackson A, Buckley D, Parker GJM. Dynamic contrast-enhanced magnetic resonance imaging in oncology. Berlin: Springer; 2005. [Google Scholar]
- 11.Yankeelov TE, Gore JC. Dynamic Contrast Enhanced Magnetic Resonance Imaging in Oncology: Theory, Data Acquisition, Analysis, and Examples. Curr Med Imaging Rev. 2007;3:91–107. doi: 10.2174/157340507780619179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu G, Song X, Chan KWY, McMahon MT. Nuts and bolts of chemical exchange saturation transfer MRI. NMR Biomed. 2013;26:810–828. doi: 10.1002/nbm.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sajda P. Machine learning for detection and diagnosis of disease. Annu Rev Biomed Eng. 2006;8:537–565. doi: 10.1146/annurev.bioeng.8.061505.095802. [DOI] [PubMed] [Google Scholar]
- 14.Wernick MN, Yang Y, Brankov JG, Yourganov G, Strother SC. Machine learning in medical imaging. IEEE Signal Process Mag. 2010;27:25–38. doi: 10.1109/MSP.2010.936730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeGrandchamp JB, Cárdenas-Rodríguez J. A machine-learning approach to measuring tumor pH using MRI. The University of Arizona; 2017. pp. 1–13. [Google Scholar]
- 16.Wojtkowiak JW, Cornnell HC, Matsumoto S, et al. Pyruvate sensitizes pancreatic tumors to hypoxia-activated prodrug TH-302. Cancer and Metabolism. 2015;3:1–13. doi: 10.1186/s40170-014-0026-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deer EL. Phenotype and genotype of pancreatic cancer cell lines. Pancreas. 2010;39:425–435. doi: 10.1097/MPA.0b013e3181c15963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van de Ven RCG, Hogers B, van den Maagdenberg AMJM, et al. T1 relaxation in in vivo mouse brain at ultra-high field. Magn Reson Med. 2007;58:390–395. doi: 10.1002/mrm.21313. [DOI] [PubMed] [Google Scholar]
- 19.Gillies RJ, Gatenby RA. Hypoxia and adaptive landscapes in the evolution of carcinogenesis. Cancer Met Rev. 2007;26:311–317. doi: 10.1007/s10555-007-9065-z. [DOI] [PubMed] [Google Scholar]
- 20.Shah T, Lu L, Dell KM, Pagel MD, Griswold MA, Flask CA. CEST-FISP: a novel technique for rapid chemical exchange saturation transfer MRI at 7T. Magn Reson Med. 2011;65:432–437. doi: 10.1002/mrm.22637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones KM, Randtke EA, Howison CM, Pagel MD. Respiration gating and Bloch fitting improve pH measurements with acidoCEST MRI in an ovarian orthotopic tumor model. Proc SPIE. 2016;9788:978815. doi: 10.1117/12.2216418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu G, Li Y, Sheth VR, Pagel MD. Imaging in vivo extracellular pH with a single paramagnetic chemical exchange saturation transfer magnetic resonance imaging contrast agent. Mol Imaging. 2012;11:47–57. [PMC free article] [PubMed] [Google Scholar]
- 23.Cárdenas-Rodríguez J, Li Y, Galons JP, Cornnell H, Gillies RJ, Pagel MD, Baker AF. Imaging biomarkers to monitor response to the hypoxia-activated prodrug TH-302 in the MiaPaCa2 flank xenograft model. Magn Reson Imaging. 2012;30:1002–1009. doi: 10.1016/j.mri.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu G, Ali M, Yoo B, Griswold MA, Tkach JA, Pagel MD. PARACEST MRI with improved temporal resolution. Magn Reson Med. 2009;61:399–408. doi: 10.1002/mrm.21863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cárdenas-Rodríguez J, Howison CM, Pagel MD. A linear algorithm of the reference region model for DCE-MRI is robust and relaxes requirements for temporal resolution. Magn Reson Imaging. 2013;31:497–507. doi: 10.1016/j.mri.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song X, Xu J, Xia S, et al. Multi-echo length and offset varied saturation (MeLOVARS) method for improved CEST imaging. Magn Reson Med. 2015;73:488–496. doi: 10.1002/mrm.25567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zu Z, Xu J, Li H, et al. Imaging amide proton transfer and nuclear overhauser enhancement using chemical exchange rotation transfer (CERT) Magn Reson Med. 2014;72:471–476. doi: 10.1002/mrm.24953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang XY, Wang F, Afzal A, et al. A new NOE-mediated MT signal at around -1. 6 ppm for detecting ischemic stroke in rat brain. Magn Reson Imaging. 2016;34:1100–1106. doi: 10.1016/j.mri.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zaiss M, Windhscuh J, Goerke S, et al. Downfield-NOE-Suppressed amide-CEST-MRI at 7 Tesla provides a unique contrast in human glioblastoma. Magn Reson Med. 2017;77:196–208. doi: 10.1002/mrm.26100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Figure S1. Error matrix representations after machine learning classification with a Linear Discriminant Analysis (LDA) algorithm using T1 relaxation times as predictors. H = Hs 766T tumor model; M = MIA PaCa-2 tumor model; S = SU.86.86 tumor model. (a) Number of correct (green) and incorrect (red) predicted observations. (b) Positive predictive value rates (green) and false discovery rates (red). (c) True positive rates (green) and false negative rates (red). (d) Area under the curve (AUC) for the receiver operator characteristic (ROC) curves represents classifier algorithm prediction accuracy.
Supporting Information Figure S2. Error matrix representations after machine learning classification with a Subspace Discriminant (SD) algorithm using T1 relaxation times as predictors. H = Hs 766T tumor model; M = MIA PaCa-2 tumor model; S = SU.86.86 tumor model. (a) Number of correct (green) and incorrect (red) predicted observations. (b) Positive predictive value rates (green) and false discovery rates (red). (c) True positive rates (green) and false negative rates (red). (d) Area under the curve (AUC) for the receiver operator characteristic (ROC) curves represents classifier algorithm prediction accuracy.
Supporting Information Figure S3. Error matrix representations after machine learning classification with a Subspace Discriminant (SD) algorithm using principal components from whole CEST spectra as predictors. H = Hs 766T tumor model; M = MIA PaCa-2 tumor model; S = SU.86.86 tumor model. (a) Number of correct (green) and incorrect (red) predicted observations. (b) Positive predictive value rates (green) and false discovery rates (red). (c) true positive rates (green) and false negative rates (red). (d) area under the curve (AUC) for the receiver operator characteristic (ROC) curves represents classifier algorithm prediction accuracy.
Supporting Information Figure S4. Error matrix representations after machine learning classification with a Quadratic Support Vector Machine (Q SVM) using principal components from whole CEST spectra as predictors. H = Hs 766T tumor model; M = MIA PaCa-2 tumor model; S = SU.86.86 tumor model. (a) Number of correct (green) and incorrect (red) predicted observations. (b) Positive predictive value rates (green) and false discovery rates (red). (c) True positive rates (green) and false negative rates (red). (d) Area under the curve (AUC) for the receiver operator characteristic (ROC) curves represents classifier algorithm prediction accuracy.
Supporting Information Figure S5. Error matrix representations after machine learning classification with a Linear Support Vector Machine (L SVM) algorithm using principal components from whole DCE pharmacokinetic curves as predictors. H = Hs 766T tumor model; M = MIA PaCa-2 tumor model; S = SU.86.86 tumor model. (a) Number of correct (green) and incorrect (red) predicted observations. (b) Positive predictive value rates (green) and false discovery rates (red). (c) True positive rates (green) and false negative rates (red). (d) Area under the curve (AUC) for the receiver operator characteristic (ROC) curves represents classifier algorithm prediction accuracy.
Supporting Information Figure S6. Error matrix representations after machine learning classification with a Linear Discriminant Analysis (LDA) algorithm using principal components from whole DCE pharmacokinetic curves as predictors. H = Hs 766T tumor model; M = MIA PaCa-2 tumor model; S = SU.86.86 tumor model. (a) Number of correct (green) and incorrect (red) predicted observations. (b) Positive predictive value rates (green) and false discovery rates (red). (c) True positive rates (green) and false negative rates (red). (d) Area under the curve (AUC) for the receiver operator characteristic (ROC) curves represents classifier algorithm prediction accuracy.
Supporting Information Table S1. Error matrix values for prediction models using T1, CEST, and DCE MRI. The data highlighted in red corresponds to Figs. 3–5, respectively. H = Hs 766T tumor model; M= MIA PaCa-2 tumor model; S = SU.86.86 tumor model.


