Abstract
Introduction
Early-life exposure to antibiotics (ABX) has been linked to increases in asthma severity and prevalence in both children and laboratory animals. We explored the immunologic mechanisms behind this association using a mouse model of house dust mite (HDM)-induced asthma and early-life ABX exposure.
Methods
Mice were exposed to three short courses of ABX following weaning and experimental asthma was thereafter induced. Airway cell counts and differentials; serum IgE; pulmonary function; lung histopathology; pulmonary regulatory T cells (Tregs); and the fecal microbiome were characterized following ABX exposure and induction of experimental asthma.
Results
Asthma severity was increased in mice exposed to ABX, including: airway eosinophilia, airway hyper-reactivity, serum HDM-specific IgE, and lung histopathology. ABX treatment led to sharp reduction in fecal microbiome diversity, including the loss of pro-regulatory organisms such as Lachnospira. Pulmonary Tregs were reduced with ABX treatment, and this reduction was directly proportional to diminished microbiome diversity.
Conclusion
Intermittent exposure to ABX early in life worsened the severity of experimental asthma and reduced pulmonary Tregs, the latter change correlated with decreased microbiome diversity. These data may suggest targets for immunologic or probiotic therapy to counteract the harmful effects of childhood ABX.
Introduction
Asthma is an inflammatory disease of the airways which manifests as obstructive respiratory symptoms including cough, dyspnea, and wheeze(1). While multiple subtypes of asthma exist(2), perhaps the most widely recognized is allergic asthma, wherein the airway inflammation arises from an inappropriate immune response to otherwise innocuous environmental allergens(1). In this subtype, there is an imbalance favoring pro-inflammatory responses (e.g. Th2) as opposed to pro-regulatory responses (e.g. regulatory T or B cells)(3). The prevalence of the disease has nearly doubled since the 1980s, today affecting nearly 10% of children(4). The sharp increase in prevalence–termed the “asthma epidemic”(5)–carries significant costs, as asthma is the third leading cause of childhood hospitalization(6). Many factors have been proposed to explain the asthma epidemic, one of which is the role of early childhood exposure to antibiotics (ABX) and their effect on the host microbiome. Over 20 clinical studies have shown that ABX use, particularly in the first year of life, significantly increases the odds of developing asthma later in childhood(3,7,8).
Unfortunately, our recognition of this association has advanced much further than our understanding of the mechanisms behind it. Dissecting such mechanisms is complex in humans, leading multiple groups to explore the influence of antibiotics on experimental asthma in mice. These studies have been unified in their message: ABX exposure worsens experimental asthma and alters the immune response predisposing toward asthma development(9–13).
However, comparisons between these studies and human asthma and ABX have important limitations. Several investigations have focused on ABX exposure prior to birth until later in life, a rare treatment regimen(10,12). Others have exposed mice to ABX for prolonged periods of at least four weeks(11,13), much longer than a typical childhood antibiotic course(14). These reports have provided important insights into how antibiotic exposure may influence the developing immune system and susceptibility to asthma, but establishment of a more clinically-relevant model of early childhood antibiotic exposure would be of great benefit in determining which factors underlie the increased susceptibility to asthma seen in children. In the present study, we hypothesized that exposing laboratory mice to ABX in a manner similar to that of human children (i.e. brief, but repeated, exposures) would increase the severity of house dust mite (HDM)-induced allergic airway disease (AAD) later in life.
Methods
Animal Subjects
C57BL/6J nursing mothers with three week old female pups were purchased from the Jackson Laboratory (Bar Harbor, ME). Upon receipt, pups were weaned and ABX treatment initiated. Mice were housed in plastic cages with corncob bedding at UConn Health. Animal rooms were maintained at 22-24°C with a 12-hour light/dark cycle. Chow and water were supplied ad libitum. Work was performed under UConn Health Institutional Animal Care and Use Committee protocol #100983-0717.
House Dust Mite (HDM) Model
The HDM model of AAD was developed by Bracken et al (15). Briefly, mice were lightly anesthetized with vaporous isoflurane and intranasally (i.n.) instilled with 50 μL of phosphate-buffered saline (PBS) containing 25 μg lyophilized HDM extract (equal parts Dermatophagoides pteronyssinus and D. farinae, Greer Laboratories, Lenoir, NC). HDM was administered beginning at six weeks of age for five consecutive days followed by two days of rest for five consecutive weeks (Figure 1A). Control groups received equal volumes of i.n. PBS in a time-matched manner. All mice were sacrificed 72 hours following final HDM challenge.
Figure 1. Early-Life Antibiotic Exposure Increases the Severity of Experimental Asthma.
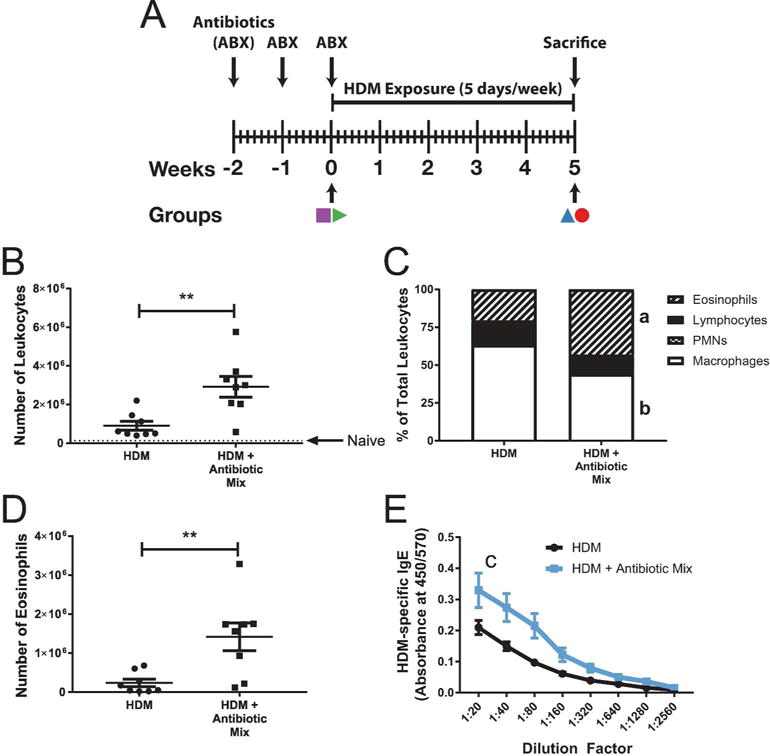
Experimental schematic is shown in (A), with colored symbols corresponding to groups in Figure 4. At sacrifice, broncho-alveolar lavage (BAL) was performed, total leukocytes counted (B), cellular differentials determined (C), and total eosinophils determined (D). Typical naïve BAL cell count is represented by a dotted line at 130,000 in (B). Serum taken at sacrifice was assessed for HDM-specific IgE using ELISA (E). Values in (B), (D), and (E) represent mean ± the SEM while data in (C) represent the mean. n=8 per group. ** p < 0.01- a: p < 0.01, HDM vs HDM + Antibiotic Mix eosinophil percent- b: p < 0.05, HDM vs HDM + Antibiotic Mix macrophage percent- c: p < 0.05, HDM vs HDM + Antibiotic Mix AUC.
Broncho-Alveolar Lavage (BAL), Blood, and Tissue (Lymph Node and Lung) Collection
To obtain BAL fluid and cells from mice, were lavaged in situ at time of sacrifice with 5 mL of sterile saline in 1 mL aliquots and BAL cells pelleted at 600xg for ten minutes at 4°C. Hilar (lung-draining, mediastinal) lymph nodes (HLN) were removed, mechanically disrupted into single-cell suspensions, and filtered through a 100 μm Nytex screen (ELKO Filtering Co, Miami, FL). For histopathologic assessment, lavaged half lungs (right lung) were removed and fixed with 4% buffered formalin for 72 hours.
Manual total nucleated cell counts were obtained using a hemocytometer with nigrosin dye exclusion as a measure of viability. Cytospin preparations of BAL fluid were stained with May- Grünwald and Giemsa for differential cell analysis via light microscopy.
To isolate serum, blood was collected from mice through cardiac puncture at the time of sacrifice and allowed to clot for 30 minutes. Clotted blood was spun down at 1600xg for ten minutes and serum removed and stored at -80°C until needed for analysis.
Flow Cytometry
Cells isolated from BAL fluid and HLN were stained with the following antibodies: CD3epsilon (1:200, Tonbo Biosciences, San Diego, CA; Clone 145-2C11; Cat #35-0031-U100); CD4 (1:200, eBioscience, San Diego, CA; Clone GK1.5; Cat #47-0041-82); CD8α (1:200, Tonbo Biosciences; Clone 53-6.7; Cat #80-0081-U100); CD19 (1:200, Tonbo Biosciences; Clone 1D3; Cat #35-0193-U500); and CD45R/B220 (1:200, Tonbo Biosciences; Clone RA3-6B2; Cat #75-0452-U025). Samples were stained as previously described with minor modifications(15). Cells were washed in PBS containing 0.2% bovine serum albumin and 0.1% sodium azide. Aliquots containing 105-107 cells were incubated with anti-mouse CD16/CD32 (1:200, Fc Shield, Tonbo Biosciences; Cat #70-0161-U500) for 15 minutes. Cells were then incubated with AlexaFluor350 as a viability marker (1:500, Invitrogen/Thermo-Fisher Scientific, Waltham, MA; Cat #A10168) and surface antibodies for 30 minutes at 4°C. For staining of Foxp3, cells stained with surface markers were fixed and permeablized with Foxp3/Transcription Factor Staining Kit (Tonbo Biosciences; Cat #TNB-0607-KIT) according to the manufacturer’s instructions and stained with anti-Foxp3 (1:150, Tonbo Biosciences; Cat #20-5773-U100) for 30 minutes at 4°C. Samples were run with corresponding isotype controls on a BD LSR II (Becton Dickinson, Franklin Lakes, NJ) and analyzed using FlowJo (Tree Star Software, Ashland, OR).
Tissue Histology and Scoring
Formalin-fixed, paraffin-embedded lungs were sectioned and stained with hematoxylin and eosin (H&E, for gross pathology) and periodic acid-Schiff with hematoxylin counterstain (PAS-H, for mucus production). Sections from all four lobes of the right lung were evaluated in their entirety and representative images taken. Pathologic scoring was performed as described previously(15). Five blinded reviewers simultaneously graded the same visual fields on each slide using a multiheaded microscope. For inflammation scores, 0 corresponded to no detectable inflammation; 1 to mild peribronchiolar/perivascular cuffing with inflammatory cells; 2 to significant peribronchiolar/perivascular clustering; and 3 to significant clustering and airway remodeling (e.g. smooth muscle hypertrophy and hyperplasia). For mucus scores, 0 corresponded to no visible mucus; 1 to occasional and punctate mucus staining in the airways; 2 to presence of ring-like mucus structures in less than 10% of airways; and 3 to presence of ring-like mucus structures in greater than 10% of airways. Half scores were permitted.
Serum Immunoglobulin Assessment
Serum HDM-specific IgE ELISA was performed as described previously(15). 96-well Nunc MaxiSorp flat-bottom plates (Thermo-Fisher Scientific, Waltham, MA) were coated with 10 ug/mL HDM in sodium bicarbonate buffer (pH 9.5) for 48 hours at 4°C. Following coating, plates came to room temperature for 15 minutes and were washed six times with 0.05% PBS/Tween 20 (PBST) and blocked for one hour at 37°C with BD OptEIA Assay Diluent (BD Biosciences, San Diego, CA; Cat #555213). After washes, serum samples were added in two-fold serial dilutions (range 1:20 to 1:2560) and incubated at room temperature for 1.5 hours.
Plates were washed eight times with PBST and labeled with Biotin-SP-conjugated goat anti- mouse IgE (Southern Biotech, Birmingham, AL; Cat #1110-08) diluted 1:5000 in blocking buffer for one hour at room temperature. Plates were then washed six times and incubated with streptavidin HRP (BD Biosciences, San Jose, CA; Cat #554066) diluted 1:1000 in blocking buffer at room temperature for thirty minutes. Plates were washed seven times with PBST and incubated with 3,3',5,5'-tetramethylbenzidine (TMB, KPL, Gaithersburg, MD; Cat #50-76-01) for 20 minutes in the dark at room temperature. The reaction was stopped using 1 M phosphoric acid at equal volume to TMB and the plates were read at dual 450nm-570nm wavelengths using an iMark Microplate Reader (Bio-Rad Laboratories, Hercules, CA).
Airway Hyper-Reactivity (AHR) Assessment
AHR was assessed as previously described with modification(15). Total responses to increasing doses (0-300 mg/mL) of acetyl-β-methacholine chloride (Sigma-Aldrich, St. Louis, MO; Cat #A2251) were measured by a flexiVent FX1 mechanical ventilator (SCIREQ Scientific Respiratory Equipment, Montreal, QC, Canada). Mice were anesthetized using urethane (Sigma-Aldrich; Cat #U2500) administered in saline by intraperitoneal (i.p.) injection at 0.8 g/kg - 5.0 g/kg body weight. Once insensitive to toe pinch, respiratory muscles were paralyzed using 500 ng/kg i.p. pancuronium bromide (Sigma-Aldrich; Cat #P1918) and mice were immediately placed under mechanical ventilation. Airway reactivity was determined by assessing forced oscillatory mechanics every ten seconds for four minutes following each methacholine challenge. Electrocardiogram measurements were monitored to ensure viability for the duration of mechanical ventilation.
Antibiotic (ABX) Treatment
The ABX treatment schedule was adapted from Nobel et al(16). Mice were exposed to Antibiotic Mix (ABX Mix), a combination of ampicillin (Sigma-Aldrich; Cat #A0166), metronidazole (Sigma-Aldrich; Cat #M1547), neomycin (Sigma-Aldrich; Cat #N6386), and vancomycin (Sigma-Aldrich; Cat #V1130), each at 1 g/L, in the drinking water or control (untreated) water. ABX-treated water was sterile filtered prior to placement in mouse cages and provided to mice ad libitum. Control water was treated identically to ABX-containing water (including sterile filtering) save for addition of ABX. ABX exposure occurred for three four-day periods (Thursday to Sunday), with a three-day rest between treatments where all animals were supplied untreated water (Figure 1A). Treatment began immediately after weaning (three weeks old). Following the third ABX treatment (six weeks old), mice were administered HDM for five weeks according to the model described earlier. For one experiment, two groups of animals (naïve, who received untreated water- and ABX-treated, who received ABX-treated water) were sacrificed following ABX-treated or untreated water exposure (Supplemental Figure S1A).
Microbiome Sampling and DNA Extraction
The mouse fecal microbiome was sampled by collecting fresh fecal pellets upon defecation using sterile, RNA-/DNA-/RNAse-/DNAse-free microcentrifuge tubes. Collected pellets were immediately placed at -80°C for storage. DNA was extracted using a PowerSoil DNA Isolation Kit (MoBio Laboratories, Carlsbad, CA; Cat #12888-100) according to the manufacturer’s protocol with one modification. At step five (bacterial cell disruption), fecal samples were placed in PowerSoil bead tubes and bead-beaten for five minutes using a single-speed (3450 oscillations/min) Mini-Beadbeater-16 (Biospec Products, Bartlesville, OK). Extracted DNA was quantified using a Qubit dsDNA Broad Range Assay Kit (ThermoFisher Scientific; Cat #Q32853) and a Qubit 2.0 fluorometer (ThermoFisher Scientific; Cat #Q32866).
16S rRNA Amplification and Sequencing
Amplicon library preparation, cleaning, and dilution were performed by the Microbial Analysis, Resources, and Services (MARS) Facility at UConn. The V4 region of the 16s rRNA gene was amplified in triplicate using the dual barcoded primers (515F and 806R) of Caporaso et al (8 basepair Golay-barcoded indices on the 3’ end)(17) and Kozich et al (8 basepair on the 5’ end)(18). Amplification was performed using 30 ng of DNA and AccuPrime Taq DNA Polymerase (Thermo-Fisher Scientific; Cat #12342010). The PCR reaction was as follows: 95°C for 3.5 minutes; 30 cycles of 30 sec at 95.0°C, 30 sec at 50.0°C and 90 sec at 72.0°C; and finally 72.0°C for 10 minutes. PCR products were pooled, quantified, and visualized using QIAxcel DNA Fast Analysis (Qiagen, Hilden, Germany). PCR products were normalized based on the concentration of DNA from 250-400 bp and pooled using the QIAgility liquid handling robot (Qiagen). Pooled PCR products were cleaned using the Gene Read Size Selection kit (Qiagen; Cat #180514) according to the manufacturer’s protocol. Samples were sequenced on an Illumina MiSeq using v3 chemistry (Illumina, San Diego, CA) according to the manufacturer’s instructions. PhiX (5-20%) was added to the run to increase sample complexity. All sequence read data are available in the Sequence Read Archive (BioProject ID PRJNA401190).
Analysis of 16S rRNA Sequence Data
Sequence data were analyzed using QIIME 1.9.1(19). The analysis pipeline was derived from Nelson et al(20). Samples were demultiplexed and paired-ends joined with a Phred score cut-off of 19. Joined reads were filtered to be between 245 and 260 bp. Chimeric sequences were removed using UCHIME(21) and operational taxonomic units (OTUs) picked at 97% similarity using USEARCH6(22) in an open-reference manner against the GreenGenes May 2013 release(23). Results were subsampled to 10,000 sequences. For depiction of taxonomic tables, only organisms with an abundance of greater than 1% in more than one group or timepoint were represented as distinct bars. All other organisms are represented by the “Other” category. Supplementary File S1 contains the analysis script used. Supplementary File S2 contains the QIIME parameters used for analysis.
Statistical Analysis
Statistical comparisons between two groups were made using the non-parametric Mann-Whitney U test. Comparisons between three or more groups were made using a two-way analysis of variance (ANOVA). Multiple-comparisons between individual groups were made using Sidak’s multiple comparisons test. Comparisons of serum immunoglobulin levels and AHR were made by comparing the area under the curve (AUC). Tests were performed using Prism 7 (GraphPad Software, La Jolla, CA). Measures of within-sample (alpha) and between-sample (beta) microbe diversity, including Simpson’s diversity index(24), were made using QIIME software. PCoA of unweighted UniFrac distances(25) was performed using QIIME. Biomarker and effect size assessment for taxa were made using LDA Effect Size (LEfSe)(26). LEfSe performs linear discriminant analysis (LDA) to estimate effect sizes of biomarkers (i.e. microbial OTUs) and represents these as LDA scores, where a higher score indicates a stronger association between that biomarker and the variable of interest. For all tests, a threshold of p < 0.05 was used to determine statistical significance.
Results
Antibiotic exposure increased the severity of HDM-induced AAD
Mice developed profound airway leukocytosis (Figure 1B) following HDM exposure, in agreement with previous results(15), and HDM + Antibiotic Mix animals exhibited three-fold higher BAL leukocytosis (Figure 1B) as compared to HDM alone (29.0 × 105 vs 9.0 × 105 cells, p < 0.01). Leukocyte counts for both groups were several-fold higher than the level seen in naïve mice (dotted line in Figure 1B). Naïve mice and mice treated with ABX but not exposed to HDM had similar BAL leukocyte counts (Supplemental Figure S1B).
Exposure to HDM resulted in airway eosinophilia (Figure 1C), with over 20% of BAL leukocytes being eosinophils in HDM-exposed animals. HDM + Antibiotic Mix animals had two-fold higher relative eosinophilia as compared to HDM animals (43% vs 20%, p < 0.01). Similarly, total eosinophil numbers (Figure 1D) were over five-fold higher in HDM + Antibiotic Mix animals as compared to HDM exposed animals (14.0 × 105 vs 2.4 × 105 eosinophils, p < 0.01). Naïve mice and mice treated with ABX but not exposed to HDM did not demonstrate airways eosinophilia, and both were not significantly different from each other (Supplemental Figure S1C-D).
Serum HDM-specific IgE is elevated in mice treated with antibiotics early in life
A hallmark of allergic asthma and AAD is the appearance of allergen-specific IgE in the serum. In agreement with our previous results, serum HDM-specific IgE appeared following HDM exposure (in naïve animals, it was virtually undetectable(15)) (Figure 1E). However, HDM + Antibiotic Mix animals had significantly higher levels of HDM-specific IgE than HDM animals (0.95 vs 0.50 mean AUC, p < 0.05).
Antibiotic exposure increased the severity of tissue inflammation in the lungs
HDM exposure led to evidence of perivascular and peribronchial inflammation in the lungs (Figure 2A-B), particularly clusters of inflammatory cells surrounding the airways and vasculature and airway smooth muscle (ASM) hyperplasia and hypertrophy around the small airways. Blinded comparison of the inflammation and ASM remodeling revealed that HDM + Antibiotic Mix mice harbored more severe histopathologic changes than HDM mice (2.5 vs 1.5 median score, p < 0.01, Figure 2E). Pathology scores for HDM mice were similar to those noted in our previous description of the model(15). Evaluation of mucus production (Figure 2C-D) showed no significant differences in mucus scores (Figure 2F).
Figure 2. Antibiotic Exposure Is Associated with Increased Disease Pathology.
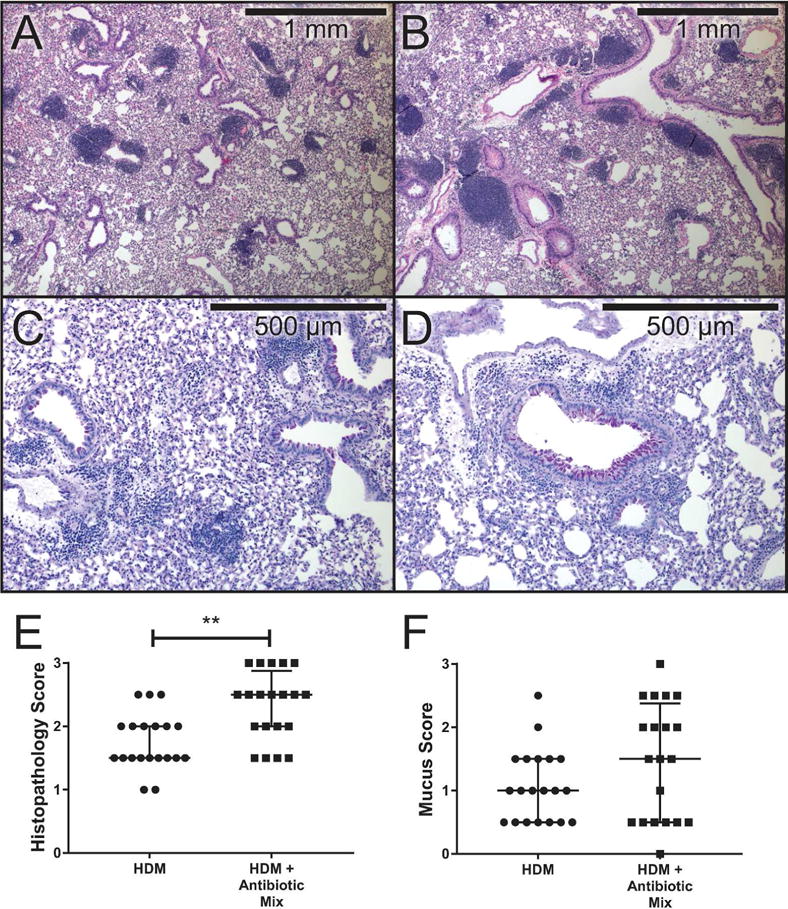
Formalin-fixed tissue sections were stained with hematoxylin and eosin (top row, A & B, 4X) or periodic acid- Schiff with hematoxylin counterstain (bottom row, C & D, 10X). Inflammation scores (E) and mucus scores (F) were determined in a blinded manner on a severity scale of 0-3 by five independent graders. Data represent mean ± interquartile ranges. n=3 slides per group. ** p < 0.01.
Airway hyper-reactivity (AHR) was significantly elevated in antibiotic-treated mice
Exposure to HDM induces ASM remodeling that results in AHR and bronchoconstriction in response to allergens and agents such as the non-selective muscarinic receptor agonist methacholine(15). Experimental assessment of respiratory function using the flexiVent system revealed that HDM + Antibiotic Mix animals had significantly higher increases in total respiratory system resistance (Rrs) in response to methacholine challenge as compared to HDM animals (5.1 vs 3.8 mean AUC, p < 0.05, Figure 3).
Figure 3. Antibiotic Exposure Significantly Increases Airway Hyper-Reactivity.
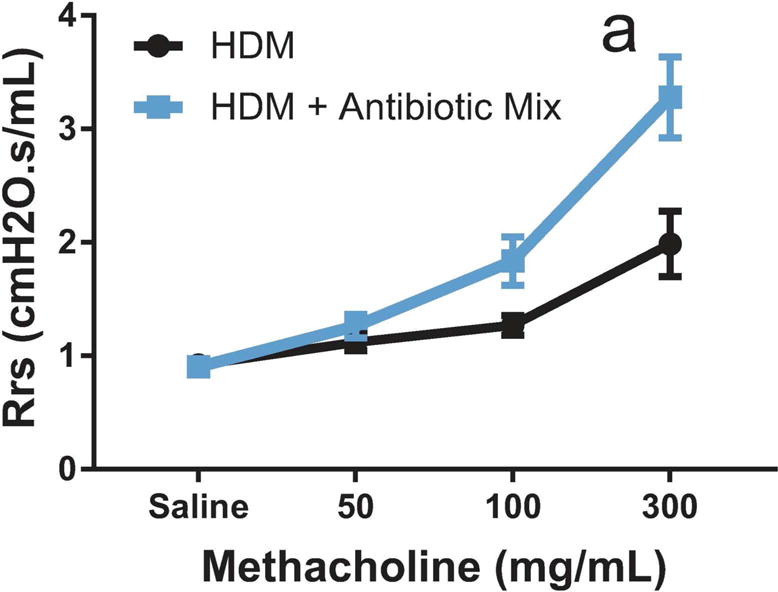
Total respiratory system resistance (Rrs) was measured using the flexiVent system following increasing doses of aerosolized methacholine. Rrs was compared using Area Under the Curve (AUC). Data represent mean ± SEM. n=4 for HDM, n=9 for HDM + Antibiotic Mix. a: p < 0.05, HDM vs HDM + Antibiotic Mix AUC.
Antibiotic exposure reduced the diversity of the fecal microbiome
Antibiotic exposure produced marked alterations in the fecal microbial community (Figure 4). The composition of the community shifted dramatically (Figure 4A). Antibiotic Mix treatment, as compared to No Treatment, saw greatly diminished phylum Bacteroidetes, particularly order Bacteroidales, and shifted the composition of phylum Firmicutes from primarily class Clostridia, particularly members of order Clostridiales, to primarily members of genus Turcibacter and genus Enterococcus.
Figure 4. Antibiotic Exposure Reduces the Diversity of the Fecal Microbiome.
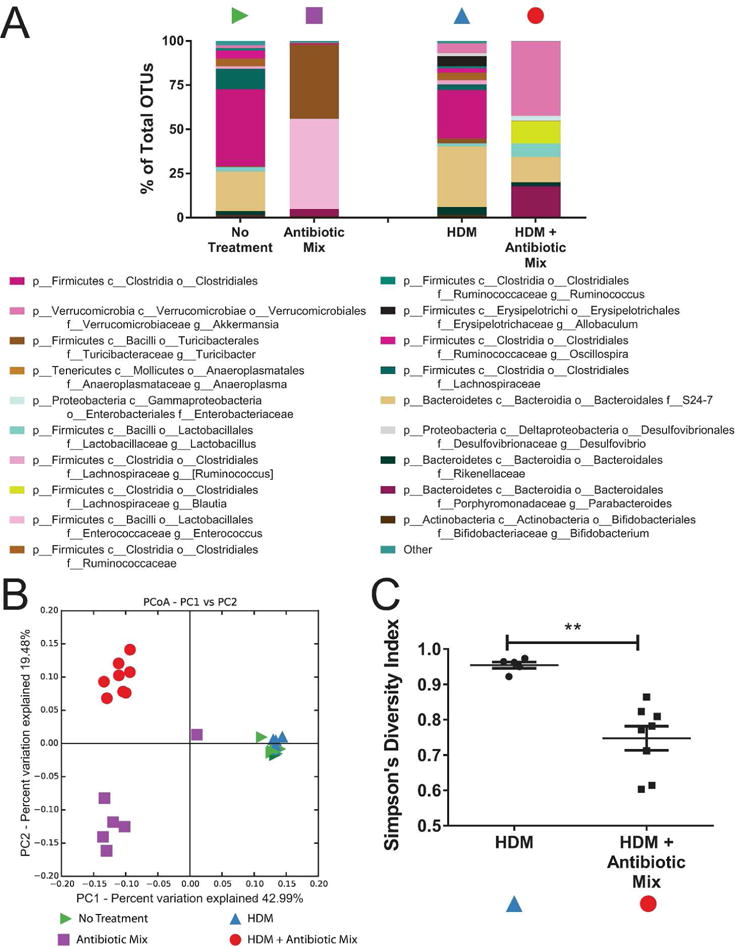
QIIME was used to analyze 16S sequence data and pick operational taxonomic units (OTUs). Symbols in (A-C) refer to those in Figure 1A. OTUs are represented as bar graphs of the average percent of total organisms represented by each OTU in each group (A). In (A), p__ refers to phylum, c__ to class, o__ to order, f__ to family, and g__ to genus. OTUs lacking a lower classification refer to a single OTU classifiable only to the lowest taxonomic level noted. Principal coordinate analysis (PCoA) of the unweighted UniFrac distances was performed on samples before and after HDM exposure (B). In (B), the closer two points are to each other, the more similar they are to each other. Variance among samples is represented by the X and Y axes, where the percentage noted represents the proportion of all variance represented on that axis. The higher the number, the greater the dissimilarity a given distance along that axis represents. Simpson’s Diversity Index for the fecal microbiome following HDM exposure was calculated (C). n=8 for No Treatment and HDM + Antibiotic Mix, n=5 for HDM, n=6 for Antibiotic Mix. ** p < 0.01.
Following HDM exposure, animals not treated with antibiotics (denoted HDM) had similar community composition to No Treatment animals. However, in Antibiotic Mix mice, five weeks of HDM treatment (HDM + Antibiotic Mix) witnessed notable changes in the microbial community, including recovery of many “lost” organisms. However, this recovery was not exact. Organisms of phylum Bacteroidetes, lost with ABX exposure, reappeared, but nearly half of these consisted of genus Parabacteroides, an organism almost absent from either No Treatment or HDM animals. Nearly half of the organisms present were of genus Akkermansia, an organism making up less than 6% of the microbiome in both No Treatment and HDM animals.
Principle coordinate analysis (PCoA) of the unweighted UniFrac distances confirmed these changes (Figure 4B). No Treatment and HDM animals were virtually indistinguishable by analysis of PC1 and PC2 (which together represented over 50% of the variance among the samples). Both differed considerably from either Antibiotic Mix or HDM + Antibiotic Mix animals along both PC1 (nearly 43% of the variance) and PC2 (over 19% of the variance). Antibiotic Mix and HDM + Antibiotic Mix saw appreciable difference along PC2 only.
In addition to assessment of the microbiome by examining community composition (Figure 4A), many metrics exist to quantify the overall diversity of the microbiome. Assessment of the alpha diversity (within-sample diversity) revealed that the Simpson diversity index was a third higher for HDM as compared to HDM + Antibiotic Mix (0.954 vs 0.747, p < 0.01, Figure 4C).
To separate the effects of antibiotic exposure from HDM exposure on the composition of the microbiome, LEfSe was utilized to identify taxonomic biomarkers linked to antibiotic treatment (Figure 5A). The identified taxa lost with antibiotic treatment (represented by rightward bars) matched those described in Figure 4 and included several, such as Lachnospiraceae (3rd row) and Clostridium (7th row), with known immunologic activity. Those taxa which increased in proportion with antibiotic treatment (represented by leftward bars) included organisms, such as genus Enterococcus (last row), with known pathogenic properties. Further analysis to isolate those organisms whose effects were due to HDM and not ABX (Figure 5B) revealed several organisms, including those with pro-regulatory properties such as genus Blautia (4th row), more represented in HDM-treated animals (leftward bars). However, the absence or near-absence of these organisms in the HDM + Antibiotic Mix exposed animals (Figure 4B) suggested this effect was likely driven solely by the non-ABX treated, HDM-exposed animals.
Figure 5. Antibiotic Exposure Results in Proportional Changes in Multiple Taxa.
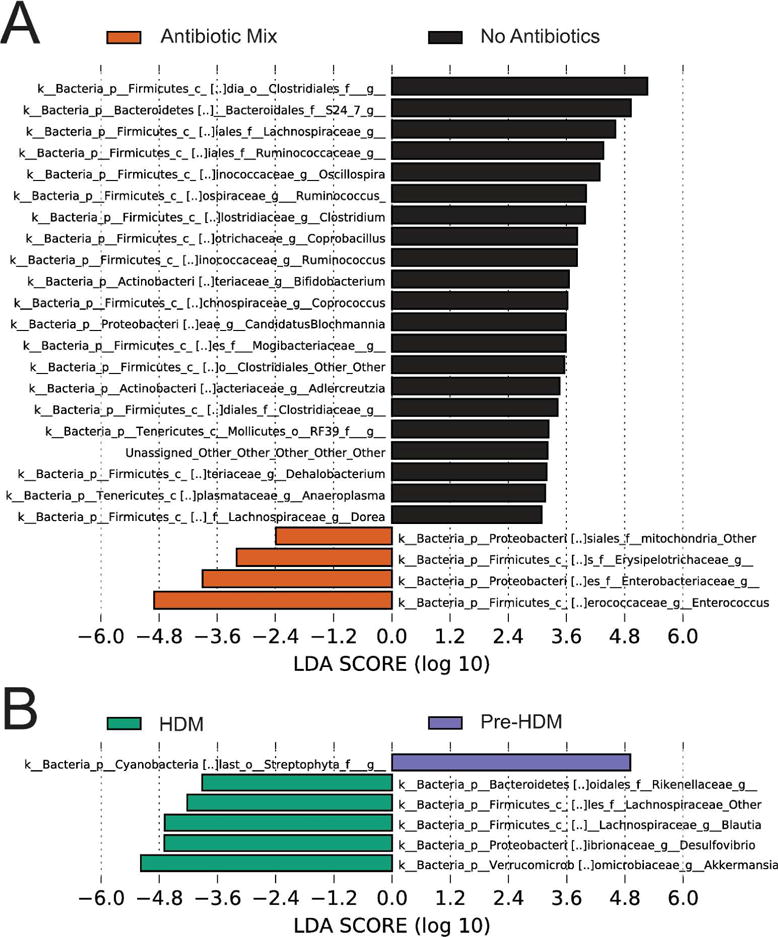
Linear Discriminant Analysis with Effect Size (LEfSe) was performed on the taxonomy table of abundances for all taxa based on (A) antibiotic treatment as the class and HDM exposure as the subclass or (B) HDM exposure as the class and antibiotic exposure as the subclass. In (A) bars represent taxa associated with antibiotic treatment only (left) or no antibiotic treatment only (right). In (B) bars represent taxa associated with HDM exposure only (left) or not associated with HDM exposure only (right). LDA scores represent the strength of the association between an identified taxa and the variable (HDM, antibiotics) in question. n=13 for No Antibiotics and n=16 for Antibiotic Mix. n=14 for HDM and n=15 for Pre-HDM.
Decreased diversity of the fecal microbiome directly correlates with the proportion of Tregs in the HLN
Our laboratory(15,27) and others(28,29) have shown that Foxp3+ regulatory T Cells (Tregs) play a critical role in the control of asthma and allergic disease, including orchestration of disease suppression and the development of tolerance to both inhaled ovalbumin(27,30) and HDM(15) from within the in the lung-draining, or hilar, lymph node (HLN). Assessing the frequency of Foxp3+ Tregs in the HLN revealed that HDM + Antibiotic Mix mice had significantly lower proportions of Tregs as a percent of total CD4+ T Cells than HDM mice (8.9% vs 11.1%, vs p < 0.05, Figure 6A). Plotting the Simpson diversity index against the proportion of Foxp3+ Tregs (Figure 6B) for both HDM and HDM + Antibiotic Mix mice revealed that there was a positive correlation between microbiome diversity and the proportion of Tregs (Spearman r=0.7109, p < 0.01). As diversity increased, so too did Treg proportions. While limited by sample size, assessment of the effect of antibiotic exposure alone on Tregs did not demonstrate a difference with ABX treatment in the absence of HDM exposure (Supplemental Figure S2).
Figure 6. Fecal Microbiome Diversity Directly Correlates with Regulatory T Cell Proportions.
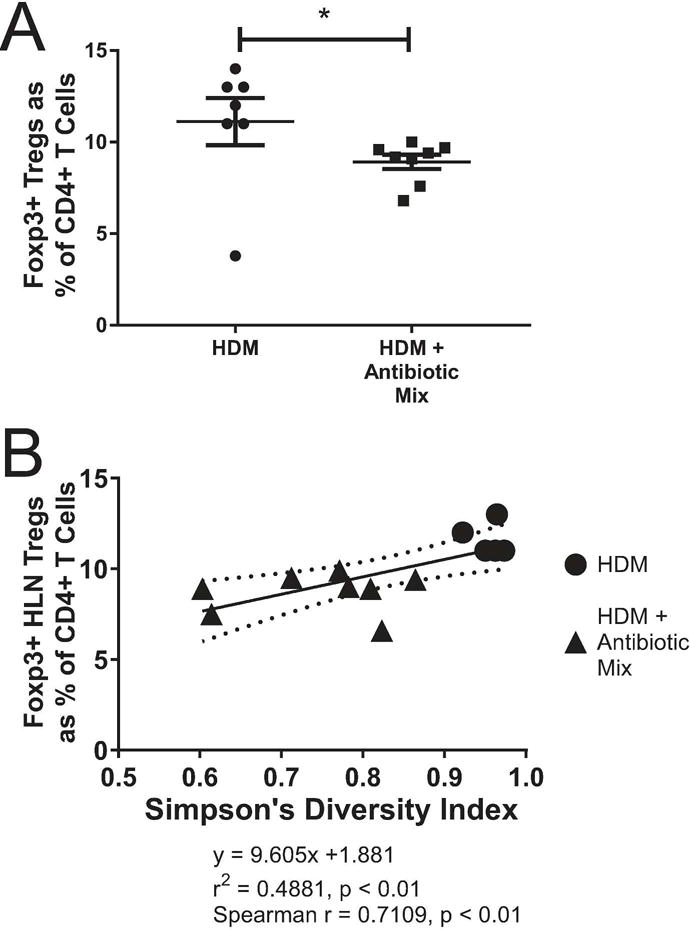
At sacrifice, Lung-Draining (Hilar, Mediastinal) Lymph Nodes (HLNs) were removed and processed to identify CD3+CD4+Foxp3+ regulatory T cells (Tregs) (A). Simpson’s Diversity Index was plotted against the proportion of all CD4+ T Cells in the HLN that are Foxp3+ Tregs (B). The Spearman r coefficient of correlation was calculated. The solid line is the linear regression line with the equation shown below the graph. The curved, dotted lines represent the 95% confidence band for the linear regression line. Error bars represent mean ± SEM. For (A), n=7 for HDM and n=8 HDM + Antibiotic Mix. For (B), n=5 for HDM and n=8 for HDM + Antibiotic Mix. Not all animals from (A) were able to be assessed for (B) due to poor DNA extraction and/or 16S gene amplification. * p < 0.05.
Discussion
In the present study, we demonstrated that brief, repeated exposure to ABX following weaning significantly worsened the severity of subsequent experimental asthma, including airway leukocytosis, serum IgE levels, and airway hyper-reactivity. ABX administration dramatically reduced the diversity of the fecal microbiome, with differences persisting even following five weeks of HDM administration. Tregs, a critical cell population for moderation of the immune response, were reduced in the lung-draining lymph node following ABX administration, and this reduction correlated directly with microbiome diversity. The present study expands previous reports which have identified increased severity of experimental asthma following long-term ABX(11,13) or pre- and post-natal ABX treatment(10,12). While we are not the first to explore alternatives to commonly-used ABX treatment regimens in laboratory mice–Martin Blaser’s group has shown that similar ABX treatment can produce lasting metabolic changes(16)–the present study provides strong evidence that such ABX exposure also induces lasting alterations in immune function.
Several clinical studies have linked early-life microbiome diversity, both gut (31) and environmental(32), to subsequent protection from asthma and allergy development, although others have found no connection(33). One important difference which may explain this discrepancy is the age of children assessed, as the latter report examined children at ages two or three(33), while the other two studies evaluated asthma at age seven(31,32). Others have noted that day care is associated with increased wheeze in the toddler years but reduced asthma by age six(34), and at least one study has linked daycare attendance with increased fecal microbiome diversity(35), suggesting that the true effect of increased early-life microbiome diversity may manifest as early disease but later protection.
Our finding that organisms of family Lachnospiraceae were reduced with ABX exposure matches clinical data linking reduction in these organisms to asthma development in children(33). These organisms have been linked to Treg induction in mice, as members of the family Lachnospiraceae fall within the Clostridial clusters known to induce Tregs through fermentation of dietary fiber and production of short-chain fatty acids(36). Their loss may be one of the factors behind the decrease in Tregs observed in our model.
One interesting observation is that the microbiome appeared to shift most dramatically during HDM administration for those animals exposed to ABX. Perhaps the most likely explanation is that these changes reflect post-ABX changes independent of HDM. Studies of the human and laboratory animal microbiome have shown that ABX rapidly induce microbiome changes but that the microbial community does not remain fixed once antibiotics are withdrawn(37). It is plausible that the same is happening to the animals in the present study. A second possibility, and one not necessarily mutually-exclusive with the first, is that HDM itself is influencing the microbiome to a greater degree in ABX-treated mice. It may be that the inflammation induced by HDM in the lungs leads to systemic alterations in mucosal and epithelial barriers, including in the gut, and that the ABX-depleted microbiome is simply more susceptible to these influences due to its lower diversity and likely lower bacterial burden. Additionally, it is likely that some of the inhaled HDM is eventually expelled from the lung by the action of the pulmonary mucociliary system and swallowed, where the HDM, which contains proteinases and microbial products which may potentially impact the gut microbiome, could conceivably cause greater microbiome changes in the disrupted gut microbiome of ABX-treated mice than those not treated with ABX.
The present study may also provide insight into the struggle to translate experimental benefits of probiotics into human therapies. Despite abundant experimental evidence of asthma protection from probiotics (reviewed in (3)), multiple clinical trials have failed to demonstrate such benefit in humans(38). As our data suggest overall diversity is associated with Tregs, it may be that administration of one or even a few organisms is insufficient in humans to provide meaningful protection. More comprehensive therapy, such as fecal microbiome transplantation (FMT), may be required to overcome the loss of diversity seen with ABX administration. Indeed, some have suggested that childhood ABX courses should be accompanied by FMT in pill form to counteract the disruption of the microbiome(39).
One limitation of the present study is the choice of the ABX cocktail. While based in part on another group’s cocktail of five ABX(11), the fact remains that the present study’s combination is unlikely to be utilized clinically. However, it does provide a means to assess the effect of significant antibiotic disturbance of the microbiome without resorting to germ free mice and the many complications therein. An important avenue for future investigation would be to determine if a subset of this cocktail was sufficient to increase disease severity (e.g. vancomycin alone or neomycin and metronidazole together). While the composition is not ideal, the use of a series of three short doses rather than the typical prenatal dosing or weeks or more of postnatal dosing is closer to clinical reality. It will be important to complement existing work evaluating one or multiple ABX prior to birth or for an extended period following birth with future studies evaluating treatment regimens in the pulsed manner of the present study. However, results identified in asthma should be applied with caution to other diseases, as there is some evidence that modulation of disease severity by ABX can depend as much on the disease as the ABX(10,40).
A final limitation of the present study concerns the source of the immunologic disturbances seen with ABX therapy. While we have shown that the microbiome is disrupted by ABX therapy, the present study does not conclusively establish that the immunologic changes seen are exclusively derived from disruption of the microbiome. Some ABX, with macrolides being perhaps the classic example, have been recognized to have immunomodulatory properties (41). It is conceivable that the immunologic disturbances we have reported arise as much from the impact of ABX on the host as on the microbiome, although our conclusion is suggested by the body of evidence showing that disruption of the microbiome specifically disturbs the host immune system. A more conclusive assessment could be made using fecal microbiota transplantation to counter the impact of antibiotics, and such work should be the subject of future studies which build on the present one.
To conclude, the present study describes a model of brief, early-life ABX exposure on the subsequent severity of experimental asthma. Exposure to ABX increased the severity of all parameters of experimental asthma, an increase accompanied by a loss of Tregs, known allergic inflammatory modulators, which correlated with a loss of diversity of the fecal microbiome. These data suggest a mechanism for clinical studies that have linked early-life microbiome diversity with subsequent asthma development in children.
Supplementary Material
Acknowledgments
The authors thank Jacqui Benjamino, Susan Janton, and Michael Nelson of the Graf Lab (UConn Storrs) for feedback and assistance. The authors thank David Benson (UConn Storrs) for use of his laboratory space for experimentation.
Statement of Financial Support
This work was supported by National Institutes of Health grants R01-AI43573 (RST and CMS), F30-HL122018 (SJB), and F30 HL-126324 (AJA).
Footnotes
Author Disclosures: The authors declare no conflicts of interest related to this study.
References
- 1.Fanta CH. Asthma. N Engl J Med. 2009;360:1002–14. doi: 10.1056/NEJMra0804579. [DOI] [PubMed] [Google Scholar]
- 2.Lötvall J, Akdis CA, Bacharier LB, et al. Asthma endotypes: a new approach to classification of disease entities within the asthma syndrome. J Allergy Clin Immunol. 2011;127:355–60. doi: 10.1016/j.jaci.2010.11.037. [DOI] [PubMed] [Google Scholar]
- 3.Adami AJ, Bracken SJ. Breathing Better Through Bugs: Asthma and the Microbiome. Yale J Biol Med. 2016;89:309–24. [PMC free article] [PubMed] [Google Scholar]
- 4.Sears MR. Trends in the prevalence of asthma. Chest. 2014;145:219–25. doi: 10.1378/chest.13-2059. [DOI] [PubMed] [Google Scholar]
- 5.Eder W, Ege MJ, von Mutius E. The asthma epidemic. N Engl J Med. 2006;355:2226–35. doi: 10.1056/NEJMra054308. [DOI] [PubMed] [Google Scholar]
- 6.National Center for Health Statistics. Health, United States, 2014: With Special Feature on Adults Aged 55–64 [Internet] Hyattsville, MD: Centers for Disease Control; 2015. Available from: http://www.cdc.gov/nchs/hus.htm. [PubMed] [Google Scholar]
- 7.Yamamoto-Hanada K, Yang L, Narita M, Saito H, Ohya Y. Influence of antibiotic use in early childhood on asthma and allergic diseases at age 5. Ann Allergy Asthma Immunol. 2017;119:54–8. doi: 10.1016/j.anai.2017.05.013. [DOI] [PubMed] [Google Scholar]
- 8.Walker WA. The importance of appropriate initial bacterial colonization of the intestine in newborn, child, and adult health. Pediatr Res. 2017;82:387–95. doi: 10.1038/pr.2017.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noverr MC, Noggle RM, Toews GB, Huffnagle GB. Role of antibiotics and fungal microbiota in driving pulmonary allergic responses. Infect Immun. 2004;72:4996–5003. doi: 10.1128/IAI.72.9.4996-5003.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Russell SL, Gold MJ, Hartmann M, et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 2012;13:440–7. doi: 10.1038/embor.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hill DA, Siracusa MC, Abt MC, et al. Commensal bacteria-derived signals regulate basophil hematopoiesis and allergic inflammation. Nat Med. 2012;18:538–46. doi: 10.1038/nm.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Russell SL, Gold MJ, Willing BP, Thorson L, McNagny KM, Finlay BB. Perinatal antibiotic treatment affects murine microbiota, immune responses and allergic asthma. Gut Microbes. 2013;4:158–64. doi: 10.4161/gmic.23567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zaiss MM, Rapin A, Lebon L, et al. The Intestinal Microbiota Contributes to the Ability of Helminths to Modulate Allergic Inflammation. Immunity. 2015;43:998–1010. doi: 10.1016/j.immuni.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Little P, Gould C, Williamson I, Moore M, Warner G, Dunleavey J. Pragmatic randomised controlled trial of two prescribing strategies for childhood acute otitis media. BMJ. 2001;322:336–42. doi: 10.1136/bmj.322.7282.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bracken SJ, Adami AJ, Szczepanek SM, et al. Long-Term Exposure to House Dust Mite Leads to the Suppression of Allergic Airway Disease Despite Persistent Lung Inflammation. Int Arch Allergy Immunol. 2015;166:243–58. doi: 10.1159/000381058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nobel YR, Cox LM, Kirigin FF, et al. Metabolic and metagenomic outcomes from early-life pulsed antibiotic treatment. Nat Commun. 2015;6:7486. doi: 10.1038/ncomms8486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caporaso JG, Lauber CL, Walters WA, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–4. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79:5112–20. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nelson MC, Morrison HG, Benjamino J, Grim SL, Graf J. Analysis, optimization and verification of Illumina-generated 16S rRNA gene amplicon surveys. PloS One. 2014;9:e94249. doi: 10.1371/journal.pone.0094249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 23.McDonald D, Price MN, Goodrich J, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–8. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simpson EH. Measurement of diversity. Nature. 1949;163:688. [Google Scholar]
- 25.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–35. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carson WF, 4th, Guernsey LA, Singh A, Vella AT, Schramm CM, Thrall RS. Accumulation of regulatory T cells in local draining lymph nodes of the lung correlates with spontaneous resolution of chronic asthma in a murine model. Int Arch Allergy Immunol. 2008;145:231–43. doi: 10.1159/000109292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taylor A, Verhagen J, Akdis CA, Akdis M. T regulatory cells in allergy and health: a question of allergen specificity and balance. Int Arch Allergy Immunol. 2004;135:73–82. doi: 10.1159/000080523. [DOI] [PubMed] [Google Scholar]
- 29.Robinson DS. Regulatory T cells and asthma. Clin Exp Allergy. 2009;39:1314–23. doi: 10.1111/j.1365-2222.2009.03301.x. [DOI] [PubMed] [Google Scholar]
- 30.Natarajan P, Singh A, McNamara JT, et al. Regulatory B cells from hilar lymph nodes of tolerant mice in a murine model of allergic airway disease are CD5+, express TGF-β, and co-localize with CD4+Foxp3+ T cells. Mucosal Immunol. 2012;5:691–701. doi: 10.1038/mi.2012.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abrahamsson TR, Jakobsson HE, Andersson AF, Björkstén B, Engstrand L, Jenmalm MC. Low gut microbiota diversity in early infancy precedes asthma at school age. Clin Exp Allergy. 2014;44:842–50. doi: 10.1111/cea.12253. [DOI] [PubMed] [Google Scholar]
- 32.Dannemiller KC, Mendell MJ, Macher JM, et al. Next-generation DNA sequencing reveals that low fungal diversity in house dust is associated with childhood asthma development. Indoor Air. 2014;24:236–47. doi: 10.1111/ina.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arrieta M-C, Stiemsma LT, Dimitriu PA, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. 2015;7:307ra152. doi: 10.1126/scitranslmed.aab2271. [DOI] [PubMed] [Google Scholar]
- 34.Ball TM, Castro-Rodriguez JA, Griffith KA, Holberg CJ, Martinez FD, Wright AL. Siblings, day-care attendance, and the risk of asthma and wheezing during childhood. N Engl J Med. 2000;343:538–43. doi: 10.1056/NEJM200008243430803. [DOI] [PubMed] [Google Scholar]
- 35.Thompson AL, Monteagudo-Mera A, Cadenas MB, Lampl ML, Azcarate-Peril MA. Milk-and solid-feeding practices and daycare attendance are associated with differences in bacterial diversity, predominant communities, and metabolic and immune function of the infant gut microbiome. Front Cell Infect Microbiol. 2015;5:3. doi: 10.3389/fcimb.2015.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Atarashi K, Tanoue T, Oshima K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–6. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 37.Koenig JE, Spor A, Scalfone N, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4578–85. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elazab N, Mendy A, Gasana J, Vieira ER, Quizon A, Forno E. Probiotic administration in early life, atopy, and asthma: a meta-analysis of clinical trials. Pediatrics. 2013;132:e666–676. doi: 10.1542/peds.2013-0246. [DOI] [PubMed] [Google Scholar]
- 39.Blaser MJ. Antibiotic use and its consequences for the normal microbiome. Science. 2016;352:544–5. doi: 10.1126/science.aad9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Russell SL, Gold MJ, Reynolds LA, et al. Perinatal antibiotic-induced shifts in gut microbiota have differential effects on inflammatory lung diseases. J Allergy Clin Immunol. 2015;135:100–9. doi: 10.1016/j.jaci.2014.06.027. [DOI] [PubMed] [Google Scholar]
- 41.Labro MT, Abdelghaffar H. Immunomodulation by macrolide antibiotics. J Chemother Florence Italy. 2001;13:3–8. doi: 10.1179/joc.2001.13.1.3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


