Abstract
Background:
Gradient index (GRIN) lenses can be used to image deep brain regions otherwise inaccessible via standard optical imaging methods. Brain tissue aspiration before GRIN lens implantation is a widely adopted approach. However, typical brain tissue aspiration methods still rely on a handheld vacuum needle, which is subjective to human error and low reproducibility. Therefore, a high-precision automated surgical instrument for brain tissue aspiration is desirable.
New Method:
We developed a robotic surgical instrument that utilizes robotic control of a needle connected to a vacuum pump to aspirate brain tissue. The system was based on a commercial stereotaxic instrument, and the additional parts can be purchased off-the-shelf or Computer Numerical Control (CNC) machined. A MATLAB-based user- friendly graphical user interface (GUI) was developed to control the instrument.
Results:
We demonstrated the GRIN lens implantation procedure in the dorsal striatum utilizing our proposed surgical instrument and confirmed the surgical results by microscope after the implantation.
Compare with Existing Method(s):
Compared to the traditional handheld method, the automatic tissue aspiration can be performed by interacting with GUI. The instrument was designed specifically for microendoscope implantation, but it can also be easily adapted for robotic craniotomy. This robotic surgical instrument can minimize human error, reduce training time, and greatly increase surgical precision.
Conclusions:
Our robotic surgical instrument is an ideal solution for brain tissue aspiration prior to GRIN lens implantation. It will be useful for neuroscientists performing in vivo deep brain imaging using miniature microscope or two-photon microscope combined with microendoscopes.
Keywords: GRIN lens implantation, MATLAB, automated surgical instrument, calcium imaging, miniature microscope, tissue aspiration
1. Introduction
Recent advances in miniature epi-fluorescence microscope (Barbera et al., 2016; Cai et al., 2016; Ghosh et al., 2011) enable recording of calcium activities of hundreds of neurons simultaneously in freely behaving animals, allowing the use of well-established freely moving behavior procedures rather than head-fixed animals. Via the implanted gradient index (GRIN) lens (Jung et al., 2004), neural calcium activities in deep brain regions such as the hippocampus (Ziv et al., 2013), hypothalamus (Betley et al., 2015; Jennings et al., 2015), and striatum (Barbera et al., 2016) can be recorded. GRIN lenses with different diameters and lengths were used for different brain regions. On the one hand, for ultra-deep regions such as the hypothalamus, smaller diameter (e.g., 0.5 mm) GRIN lenses were often used to reduce damages to brain tissue. GRIN lenses were slowly inserted towards the target brain region (Betley et al., 2015; Resendez et al., 2016). On the other hand, for medium-depth cortical and subcortical brain regions such as medial prefrontal cortex, hippocampus and dorsal striatum, larger diameter GRIN lenses were used to obtain a larger field of view and to record a greater number of neurons. Under such circumstances, brain tissue above the target brain region was often aspirated by using a blunt needle attached to a vacuum pump before the GRIN lens implantation (Resendez et al., 2016). To date, the neuroscience community still relies on handheld vacuum needles for brain tissue aspiration, which can inadvertently introduce human errors and low reproducibility. Consequently, this will lead to higher inter-subject variability. Although there are commercially available automated stereotaxic instruments (Pak et al., 2015) and open-source robotic instruments for craniotomy (Coffey et al., 2013; Ghanbari et al., 2018; Pak et al., 2015), these instruments cannot be readily adapted for tissue aspiration purposes.
Here we present an open-source robotic surgical instrument for brain tissue aspiration that greatly facilitates GRIN lens implantation. The main hardware of this instrument can be easily modified from commercially available stereotaxic instruments with a few Computer Numerical Control (CNC) machined parts. It is well-accepted that the aspiration should be performed layer by layer until slowly reaching the desired brain region (Resendez et al., 2016). Based on this principle, we developed user-friendly open-source software with a graphical user interface (GUI) that allows the experimenter to define the surgical parameters by interacting with the software interface on a personal computer (PC). This open-source robotic surgical instrument is an ideal device specifically designed for rodent brain tissue aspiration, the use of which can greatly increase the surgical precision and minimize training time for novice.
2. Methods
2.1. Overview
The surgical instrument robotically controls a vacuum needle to aspirate brain tissue above the target region. To facilitate adaptation across different labs, we designed the instrument based on a commercial stereotaxic frame, with additional parts purchased off-shelf or CNC machined. The total cost of the modification is under $1000.
2.2. Hardware and electronics
The surgical instrument (Fig. 1A) was designed based on a commercial small animal stereotaxic instrument (Model 963, Kopf). The assembly procedure can be described as the following (Fig.1B,C): (1) Remove the adjustment knobs of all the three axes from the stereotaxic manipulators. (2) Mount the adapters on the manipulators and tighten with #4–40 screws that are taken from step 1. There are two types of adapters, one for the lateral-medial manipulator and the dorsal-ventral manipulator, and the other for the anterior-posterior manipulator according to the different hole positions. (3) Use the shaft coupler to connect the shaft of the manipulator to the shaft of the stepper motor, tighten the set screw on the shaft coupler, then connect the adapter with the stepper motor using M3 socket-head screws. The adapters and shaft couplers were CNC-machined (Millitnow.com). The shaft couplers can also be purchased online (https://www.aliexpress.com/item/5–8mm-Motor-shaft-Rigid-Steel-coupling-8mm-A-Diameter-5mm-B-Diameter-16mm-OD-22m/1566611975.html). The schematics, assembly photos, Solidworks/stl models, and parts list can be found at our Github repository (https://github.com/liang-bo/AutoStereota). A 30-gauge blunt needle was connected to the vacuum pump (standard laboratory vacuum, 40 ~ 50 psi) through a Medi-Vac suction canister (Cardinal Health, OH). The syringe holding the needle was parallelly mounted onto the dorsal-ventral manipulator. This needle, carried by the robotic manipulators, was used to slowly aspirate brain tissue via a predefined trajectory in the software.
Fig.1. Surgical instrument system design.
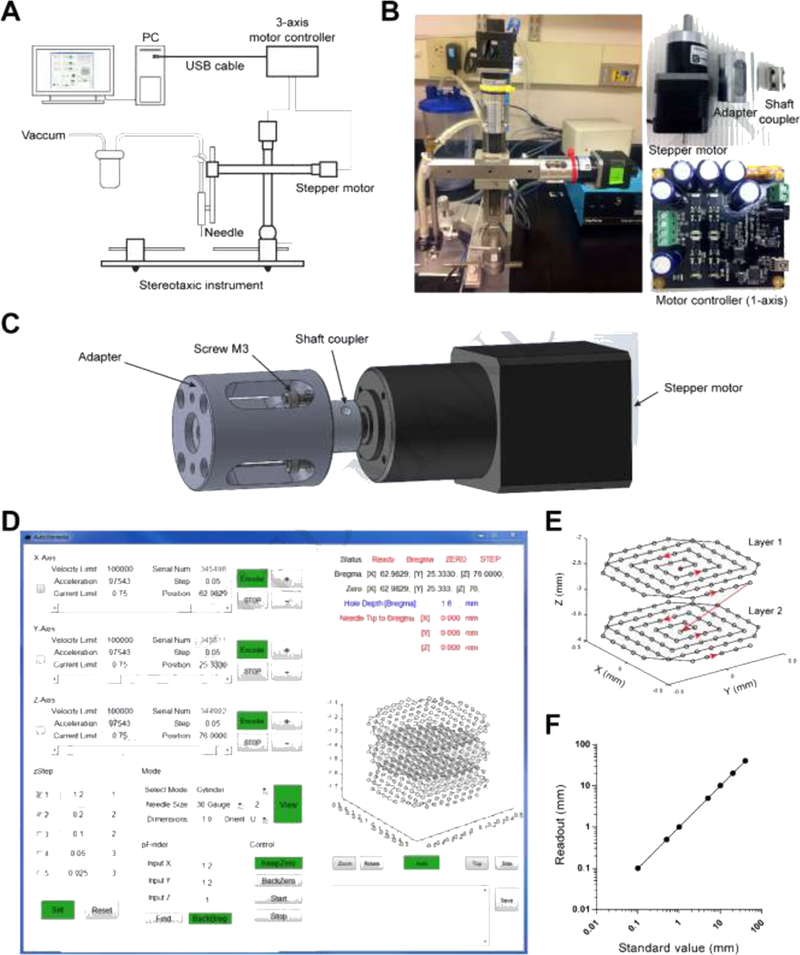
(A) Schematic design of the system. (B) Photos of the actual system (left), stepper motor, adapter and shaft coupler (top right), and stepper motor controller (bottom right). (C) 3D model of the assembly of the stepper motor, adapter and shaft coupler. (D) Graphical user interface. (E) Representative two-layer trajectory of the needle tip coordinates during tissue aspiration. Red dot indicates the start point and the red arrows show the direction in which the needle tip should be moving. (F) Calibration of the stepper motor driving the stereotaxic manipulator: the standard value of movement set in the software versus the actual readout from the manipulator vernier scales.
The stepper motors (3326_0, Phidgets, Canada) used here were NEMA-17 bipolar stepper motors with 14:1 gearbox. This has a 0.13° step angle and 30 kg cm torque at low speeds. For Kopf Model 963, the travel distance is 1mm per 360-degrees. Thus, the resolution of each step is approximately 0.36 μm, which is sufficiently precise for tissue aspiration. Three stepper motors were controlled by three USB-based motor controllers (1067_0, Phidgets, Canada) via MATLAB-based (Mathworks, MA) GUI software (See next section). It is worth noting that each stepper motor controller has a unique serial number that allows the master PC to identify and control the device. A 12V/2.5A power adapter was used to power the motor controller to drive the stepper motor.
2.3. Software
The Phidgets APIs in C were used to develop the software. To use the C APIs in MATLAB, the C header file was first compiled and then loaded by the MATLAB function “loadlibrary”. The software GUI (Fig.1D) consists of three main panels: (1) the motor parameter setting panel, labelled with x-axis, y-axis and z-axis representing the lateral-medial, anterior-posterior, and dorsal-ventral manipulator, respectively. This panel includes the basic setting of the motor parameters for all three axes such as peak speed, acceleration, serial number, step and current position. For each axis, buttons labelled with “+” and “-” were designed to adjust the arm position. The edit box labelled “Position” was designed to set the current position of each manipulator. (2) The surgery setting and control panel, labelled with “pFinder”, “zStep”, “Mode” and “Control”. This panel is related to the surgical procedure, and the functions include: (a) Find the surgical zero coordinates based on the bregma position (pFinder). (b) The setting of the sequence of steps (or depths) of layers to be aspirated by the vacuum needle (zStep). The left edit boxes are the input of the step value in mm and the right edit boxes are the repeat times of each step. These steps are additive. (c) Generating the aspiration coordinates for different types of microendoscopic implantation (Mode), i.g., GRIN lens and micro-prism. For GRIN lenses, the overall coordinates form a cylinder shape matching the size of the GRIN lens (also see next section for detailed settings). Aspiration was performed in a layer-by-layer fashion (Fig.1E). Vertically, for each step in one layer, the needle tip was first lowered from the zero-coordinate to the pre-defined z-coordinate and then returned to the zero-coordinate. This maintains the level of artificial cerebral spinal fluid (aCSF) in order to keep the physiological environment for the brain tissue (also see next section). Horizontally, within one layer, the needle tip scans with an aspiral pattern starting from the center. The spiral pattern is not strictly necessary but it can simplify the positioning of the GRIN lens. The aspiration sampling frequency can also be adjusted in this panel according to the gauge size of the aspiration needle. For the micro-prism, the overall coordinates form a shape matching the size of the micro-prism. Horizontally the needle tip moves along a line-by-line pattern. (d) The control panel with functions to keep zero-coordinates, and start, pause or resume a surgical procedure (Control). (3) The real-time trajectory monitoring panel (right panel). This panel is to display the real-time parameters and coodinates of the needle tip to remind the experimenter the status of the surgery. The middle part of this panel shows a 3-dimentional representation of aspiration coordinates for the 1-mm GRIN lens implantation. These three layers correspond to the steps set in the zStep panel. The bottom of this panel is the log panel which allows the experimenter to record the current parameters to a text file such as bregma coordinates during the surgery. The MATLAB source code and packaged installer (for Windows 64-bit OS, it is not necessary to install MATLAB, but must install the free shared library MATLAB Runtime) can be found at our Github repository (https://github.com/liang-bo/AutoStereota).
The stepper motor has a step resolution of 0.36 μm. It also allows for fractional step of 1/16. The manipulators have a precision of 10 μm. Therefore the precision of the system would be limited by the precision of the manipulators. We validated the performance of the stepper motors by comparing the standard value of movement set in the software with the actural readout from the manipulator vernier scales (Fig.1F, R2 = 1 with linear regression).
3. Results
All experiments were conducted in accordance with the guidelines of Institutional Animal Care and Use Committee, the Intramural Research Program, National Institute on Drug Abuse, National Institutes of Health. Both wild type (C57BL/6J) mice and a transgenic D1-Cre mouse (FK150 line, C57BL/6J congenic, Gensat) were used in the experiments. To image GCaMP6s fluorescence in dorsal striatum (DS), we injected AAV1.CAG.Flex.GCaMP6s.WPRE.SV40 (University of Pennsylvania Vector Core) into the dorsal striatum of the D1-Cre mouse. For the experiments comparing the performance of the automated surgical instrument and handheld method, we injected AAV1.hSyn.Cre-GFP.WPRE.hGH into the dorsal striatum of the wildtype mice. The AAV viruses were injected using the stereotactic coordinates A/P: −0.93 mm, M/L: +1.8 mm, D/V: −3.46 mm, with 30 angle shift to caudate.
Figure 2A shows the GRIN lens implantation procedure in the dorsal striatum utilizing the automated surgical instrument. The GRIN lens implantation was performed 1 week after the viral injection. In this study, ~4-mm long GRIN lenses (GRINTECH) with 1-mm diameter were used. First, the mice were anaesthetized with 100 mg/kg ketamine and 15 mg/kg xylazine. A 1.1 mm-diameter craniotomy was made at the coordinates. The needle tip was then located to the bregma and the “Breg” button in the “pFinder” panel was pressed to record the bregma coordinates. Second, the coordinates for GRIN lens implantation were entered in the software “pFinder” panel, and the needle tip moved over to the centroid of the craniotomy, ready for the aspiration.The z-axis input is typically 0.3 mm above the bregma to allow enough clearance for the needle. The “KeepZero” button in the Control panel was pressed at this point to keep the zero-coordinates. Third, the vacuum pump (standard laboratory vacuum, ~ 40–50 psi) was turned on and the artificial cerebral spinal fluid (aCSF) flush was started. Pre-warmed aCSF was administered by 1- meter-gravity difference via a 30-gauge needle (Fig.2A1). Aspiration was performed very slowly, layer by layer until reaching the desired brain region (Fig.1E). The steps of the aspiration can be set in the “zStep” panel. The first step is typically 0.5 mm, taking into account that the needle tip is 0.3 mm above the bregma and the actual aspiration depth is 0.2 mm. The aspiration depth was gradually increased by 0.2 mm per layer until the desired brain region was close, then the z-step was set to 0.1 mm per layer. When all the settings were configured properly in the “zStep” setting, the “Start” button in the “Control” panel was pressed to start the tissue aspiration. The 30-gauge needle would follow the predefined pattern to aspirate brain tissue. Bleeding from vessel rupture during aspiration was expected during the aspiration process. Continue aCSF irrigation and tissue aspiration until all steps were completed. A stereomicroscope (Zeiss Stemi 2000, with 0.4× objective) together with an LED fiber optic illuminator (LED-50W, AmScope) were used to monitor the tissue aspiration. For the dorsal striatum (D/V: −1.75mm), the brain tissue was visible through the stereomicroscope. Thus, after the removal of the corpus callosum, the access to the striatum could be visually confirmed by its relatively darker color compared to the corpus callosum. Once aspiration was completed (needle tip reached target z-coordinate), aCSF irrigation was continued for 5 minutes to let small, ruptured vessels to seal. Subsequently, a single-layer aspiration was performed to clean the remaining debris during which the aspiration depth was set 0.2 mm above the surgical site. This cleaning step was repeated until no visible blood was aspirated. Finally, the needle was removed and aCSF irrigation was stopped. The disinfected GRIN lens (15 min immersion in 70% alcohol followed by saline immersion until placed) was gently dropped and seated onto the apirated brain surface. To ensure the bottom of the GRIN lens was in contact with the brain tissue at the bottom of the tissue well, a soft, sterilized paper towel was used to gently press the GRIN lens. Excess aCSF around the GRIN lens was removed using the sterilized paper towel. Melted agarose (1% in water, melted in microwave, and kept in a 42-degree water bath) was applied onto the skull around the GRIN lens to seal the gap between the skull and the GRIN lens. Excess agarose was removed using a micro blade after the gel hardened. The skull was cleaned again with saline or hydrogen peroxide using cotton swabs to remove any tissue debris. A layer of dental cement (Metabond, S380, Parkell) was applied around the GRIN lens and over the exposed skull to secure the GRIN lens. Dental cement was cured for 5 minutes. A second layer of dental cement mixed with carbon powder was applied to reduce the scattered light reflection during in vivo imaging. At this point, the mouse was be placed under microscope (Zeiss Axio examiner Z1 with 10× 0.5NA objective and Fs 38HE filter set) to confirm targeting of GRIN lens to the desired brain region by the fluorescence of GCaMP6s (Fig.2B). A cap made out of PCR tube bottom was glued on the skull to cover the GRIN lens, protecting the lens from potential damages during recovery phase.
Fig.2. Surgical instrument calibration and representative surgical procedure.
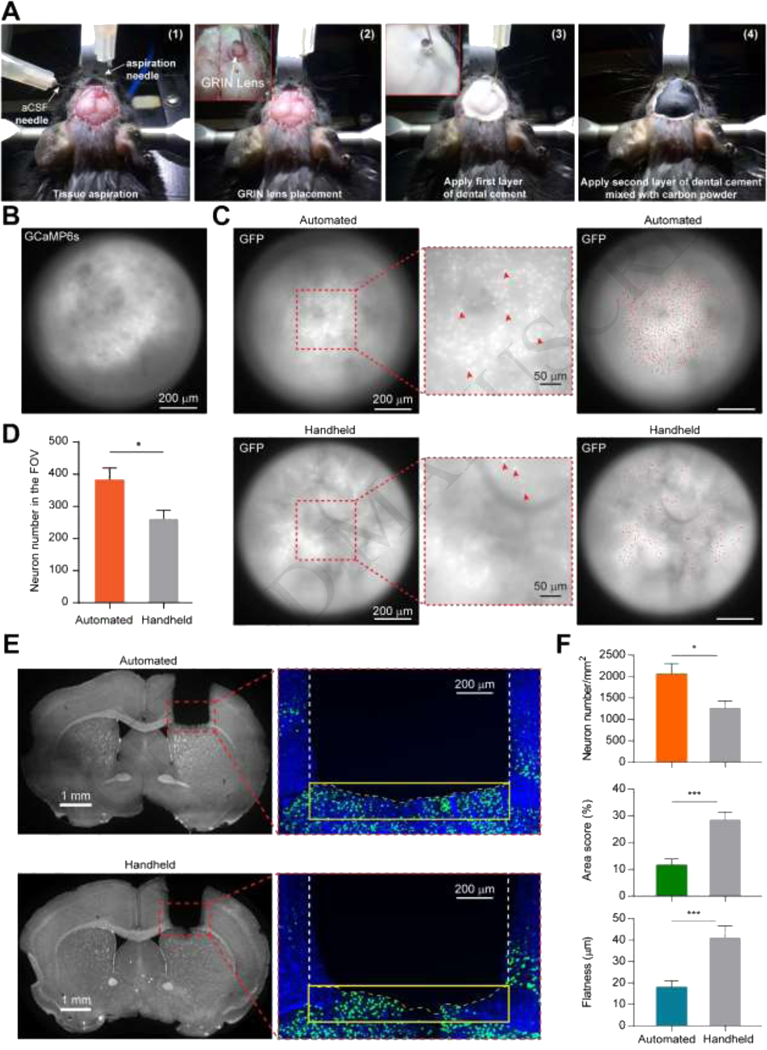
(A) Photo illustration of the surgical procedure. (A-1): Tissue aspiration; (A-2): GRIN lens placement; (A-3): First thin layer of dental cement application; (A-4): Application of second layer of dental cement mixed with carbon powder to reduce the scattered light reflection during in vivo imaging. (B) Image of the dorsal striatum from Zeiss microscope to confirm the result of implantation of the GRIN lens. (C) Representative fluorescence images of the GFP-expressing neurons. Top left and bottom left panels show the fluorescence images of GFP-expressing neurons in the dorsal striatum right after GRIN lens implantation using automated instrument and handheld manner, respectively. Red arrows in the middle insets indicate the representative GFP-expressing neurons. Top right (automated) and bottom right (handheld) panels show GFP fluorescence images overlaid with all the identified neurons (red dots) in the field of view. (D) Number of identified GFP-expressing neurons in the field of view for both automated and handheld groups. n = 10 mice, automated group; n = 10 mice, handheld group; * p<0.05, Mann-Whitney test. (E) Representative brain images after GRIN lens implantation. Left panels: representative brain images with DAPI immunostaining from automated and handheld groups, respectively. Right panels: representative images around the aspiration surfaces from automated and handheld groups, respectively. Yellow box indicates the underlying area within 200 μm range of the aspiration surface. Yellow dashed line indicates the contour of the aspiration surface. (F) Quantifications of the neuron density (left), the amount of damage (middle) and the aspirated tissue surface flatness (right) for automated and handheld groups. * p<0.05, *** p<0.001, Mann-Whitney test. Automated group: n = 9 slices from 9 mice; handheld group: n = 8 slices from 8 mice. Data are represented as mean ± s.e.m.
To show that our proposed automated surgical instrument can decrease human error and improves reproducibility, we performed experiments to compare the performance of the tissue aspiration using the automated surgical instrument as shown above (n = 10 wild type mice, automated group) and the handheld manner (n = 10 wild type mice, handheld control group). For the handheld control group, the tissue was first slowly aspirated by hand. When the aspiration was close to the target region, the needle was then attached to the manual manipulator. The last layer of aspiration was performed by manually controlling the vacuum needle through the manipulator.
We conducted both in vivo and histological evaluations to quantitatively compare the performances of these two methods. After the GRIN lens implantation, we took fluorescence images through GRIN lenses using a Zeiss Axio microscope while the mice were under anesthesia (Fig.2C). The numbers of the GFP-expressing neurons were counted for both automated and handheld groups (Fig.2D). We found that there were significantly greater numbers of neurons identified in the automated group than in the handheld group (automated group: 383±36, n = 10 mice; handheld group: 262±27, n = 10 mice; p = 0.01, Mann-Whitney test).
For histological evaluation, mice were anesthetized with an overdose of ketamine (150 mg/kg) and xylazine (22.5 mg/kg), perfused with phosphate buffer solution (PBS) and then followed by a fixation buffer containing 4% paraformaldehyde (PFA) in PBS. Mice brains were dissected and post-fixed by 4% PFA in PBS overnight at 4°C. Fixed mice brains were sectioned via a Vibratome (LEICA VT1000S) and 30 μm coronal brain slices were collected. Immunostaining of DAPI, a cell marker, was performed following a protocol for free-floating brain slicing (Zhang et al., 2016). Fluorescent images were obtained through an Olympus VS120 scanner.
The histological evaluation was quantified by three metrics (Fig.2E, F): (1) the GFP-expressing neuronal density in the underlying tissue within 200 μm range of the aspirated tissue surface; (2) the percentage of damaged tissue within the underlying 200 μm range of the aspirated tissue surface; and (3) the flatness of the aspirated tissue surface. We found that the neuron density of the automated group was significantly higher than that of the handheld group (p = 0.0164, Mann Whitney test; automated group: 2059±241 per mm2, n = 9 slices from 9 mice; handheld group: 1269±153 per mm2, n = 8 slices from 8 mice). We also discovered that the automated surgical instrument caused significantly less amount of damage to the underlying tissue than the handheld method (p = 0.001, Mann Whitney test; automated group: 11.63% ± 2.29%, n = 9 slices from 9 mice; handheld group: 28.59% ± 2.72%, n = 8 slices from 8 mice). The area score was defined as the percentage of the damaged area compared with the whole evaluated area. The tissue boundary was defined by the fluorescence of the DAPI immunostaining. We extracted the edges of the stained tissue surfaces and calculated the flatness of the tissue surfaces, which was quantified by the root mean square (RMS) of the surface coordinates. We found that the tissue surface flatness of the automated group was significantly higher than that of the of handheld group (p = 0.001, Mann Whitney test; automated group: 18.03±2.99, n = 9 slices from 9 mice; handheld group: 41.15±5.46, n = 8 slices from 8 mice).
Together, these histological evaluation results suggest that tissue aspiration using our automated instrument is more precisely controlled than the handheld method. The automated tissue aspiration generates a uniform surface and causes less amount of damage to the neighboring tissue, and thus is beneficial to endoscope imaging.
4. Discussion
In this manuscript, we reported a robotic surgical instrument that allows us to aspirate brain tissue in an automated manner. This instrument was controlled by GUI-based software, which provided high precision surgical outcomes.
The ability to record cellular-level neural calcium activities holds great potential for unraveling neural encoding mechanisms for behavior control. Using GRIN lenses to access deep brain regions is an important approach for deep brain calcium activity recording. A reliable surgical instrument for GRIN lens implantation and related procedures is necessary. Based on the literature and our experiences (Barbera et al., 2016; Jung et al., 2004; Pinto and Dan, 2015; Ziv et al., 2013), brain tissue aspiration performed in a slow and layer-by-layer fashion was highly recommended for brain region of medium depth such as the prefrontal cortex, the hippocampus and the striatum with a large diameter GRIN lens (≥1mm). Our proposed robotic surgical instrument is specifically customized for this purpose and will serve as a highly suitable tool. With the growing popularity of the miniScope and GRIN lens imaging toolset, our open-source surgical solution will also be highly beneficial to the neuroscience community.
Open-source automated tools for biological experiments have become increasingly desirable to enhance experimental accuracy and efficiency (Ardesch et al., 2017; Bolanos et al., 2017; Coffey et al., 2013; Giovannucci et al., 2017; Pak et al., 2015). It can also reduce training time and unnecessary manpower. Recently, high precision automated surgical stereotaxic tools for craniotomies have been developed (Coffey et al., 2013; Ghanbari et al., 2018; Pak et al., 2015). These tools provided low-cost and easy-to-adopt solutions for a variety of surgeries such as cranial window/hole drilling, skull thinning. It is possible to implement similar trajectory or pattern on these systems to have similar functions as we proposed in this study. However, in addition to the trajectory and speed control, our study provides a comprehensive solution for brain tissue aspiration including software, hardware and surgical protocol, which can be easy used by neuroscientists who perform endoscopic calcium imaging experiments, even without programing experiences. Our customized user-friendly software was specialized for GRIN lens implantation experiments, which will fulfill the unmet needs for a sophisticated tool for brain tissue aspiration prior to GRIN lens implantation. Besides GRIN lens implantation, our instrument can also be used for micro-prism implantation or cranial window generation for the striatum or hippocampus (Andermann et al., 2013; Howe and Dombeck, 2016). We selected MATLAB as the programming platform because MATLAB as a high-level language allows us to prototype in a quick and efficient way. MATLAB satisfies the majority of requirements regarding the control performance, such as response time. However, if one wants to improve performance, the stepper motor controller also provides APIs of several other popular languages such as Java, Python, C# and so on.
5. Conclusion
The robotic surgical instrument presented here is a practical solution for brain tissue aspiration prior to GRIN lens implantation. It can be useful for neuroscientists who are performing in vivo deep brain imaging using miniscope or two-photon microscope combined with microendoscopes.
Highlights:
Automated surgical instrument for brain tissue aspiration
Simply modified from commercial stereotaxic instrument
Open-source user friendly GUI software in MATLAB
Gradient index lens or micro-prism implantation for in vivo calcium imaging
Minimize human error, reduce training time, and increase surgical precision
Acknowledgements
The authors would like to thank the valuable suggestions from Drs. Wen Zhang, Sarah Hawes, and Rajtarun Madangopal. The project was supported by National Institute on Drug Abuse Intramural Research Program, NIH.
Footnotes
Competing financial interests
The authors declare no competing financial interests.
Supplementary data
None.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andermann ML, Gilfoy NB, Goldey GJ, Sachdev RN, Wolfel M, McCormick DA, Reid RC, Levene MJ. Chronic cellular imaging of entire cortical columns in awake mice using microprisms. Neuron, 2013; 80: 900–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardesch DJ, Balbi M, Murphy TH. Automated touch sensing in the mouse tapered beam test using Raspberry Pi. J Neurosci Methods, 2017; 291: 221–6. [DOI] [PubMed] [Google Scholar]
- Barbera G, Liang B, Zhang L, Gerfen CR, Culurciello E, Chen R, Li Y, Lin DT. Spatially Compact Neural Clusters in the Dorsal Striatum Encode Locomotion Relevant Information. Neuron, 2016; 92: 202–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betley JN, Xu S, Cao ZFH, Gong R, Magnus CJ, Yu Y, Sternson SM. Neurons for hunger and thirst transmit a negative-valence teaching signal. Nature, 2015; 521: 180–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolanos F, LeDue JM, Murphy TH. Cost effective raspberry pi-based radio frequency identification tagging of mice suitable for automated in vivo imaging. J Neurosci Methods,2017; 276: 79–83. [DOI] [PubMed] [Google Scholar]
- Cai DJ, Aharoni D, Shuman T, Shobe J, Biane J, Song W, Wei B, Veshkini M, La-Vu M, Lou J, Flores SE, Kim I, Sano Y, Zhou M, Baumgaertel K, Lavi A, Kamata M, Tuszynski M, Mayford M, Golshani P, Silva AJ. A shared neural ensemble links distinct contextual memories encoded close in time. Nature, 2016; 534: 115–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey KR, Barker DJ, Ma S, West MO. Building an open-source robotic stereotaxic instrument. J Vis Exp, 2013: e51006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanbari L, Rynes M, Hu J, Sousa Shulman D, Johnson G, Laroque M, Shull G, Kodandaramaiah SB. Principles of Computer Numerical Controlled Machining Applied to Cranial Microsurgery. bioRxiv, 2018. [Google Scholar]
- Ghosh KK, Burns LD, Cocker ED, Nimmerjahn A, Ziv Y, Gamal AE, Schnitzer MJ. Miniaturized integration of a fluorescence microscope. Nat Methods, 2011; 8: 871–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannucci A, Pnevmatikakis EA, Deverett B, Pereira T, Fondriest J, Brady MJ, Wang SS, Abbas W, Pares P, Masip D. Automated gesture tracking in head-fixed mice J Neurosci Methods, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe MW, Dombeck DA. Rapid signalling in distinct dopaminergic axons during locomotion and reward. Nature, 2016; 535: 505–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings JH, Ung RL, Resendez SL, Stamatakis AM, Taylor JG, Huang J, Veleta K, Kantak PA, Aita M, Shilling-Scrivo K, Ramakrishnan C, Deisseroth K, Otte S, Stuber GD. Visualizing hypothalamic network dynamics for appetitive and consummatory behaviors. Cell, 2015; 160: 516–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JC, Mehta AD, Aksay E, Stepnoski R, Schnitzer MJ. In vivo mammalian brain imaging using one- and two-photon fluorescence microendoscopy. J Neurophysiol, 2004; 92: 3121–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak N, Siegle JH, Kinney JP, Denman DJ, Blanche TJ, Boyden ES. Closed-loop, ultraprecise, automated craniotomies. J Neurophysiol, 2015; 113: 3943–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto L, Dan Y. Cell-Type-Specific Activity in Prefrontal Cortex during Goal-Directed Behavior. Neuron, 2015; 87: 437–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resendez SL, Jennings JH, Ung RL, Namboodiri VM, Zhou ZC, Otis JM, Nomura H, McHenry JA, Kosyk O, Stuber GD. Visualization of cortical, subcortical and deep brain neural circuit dynamics during naturalistic mammalian behavior with head-mounted microscopes and chronically implanted lenses. Nat Protoc, 2016; 11: 566–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Zhang L, Liang B, Schroeder D, Zhang ZW, Cox GA, Li Y, Lin DT. Hyperactive somatostatin interneurons contribute to excitotoxicity in neurodegenerative disorders. Nat Neurosci, 2016; 19: 557–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv Y, Burns LD, Cocker ED, Hamel EO, Ghosh KK, Kitch LJ, El Gamal A, Schnitzer MJ. Long term dynamics of CA1 hippocampal place codes. Nat Neurosci, 2013; 16: 264–6. [DOI] [PMC free article] [PubMed] [Google Scholar]


