Abstract
Drought and salinity are the major factors that limit the faba bean (Vicia faba L.) production worldwide. The aim of this study is to identify the water stress differentially expressed genes (DEGs) through the root transcriptome analyses of the drought-tolerant Hassawi 2 genotype at vegetative and flowering stages. A total of 624.8 M high-quality Illumina reads were generated and assembled into 198,155 all-unigenes with a mean length of 738 bp and an N50 length of 1347 bp. Among all-unigenes, 78,262 were assigned to non-redundant (Nr), 66,254 to nucleotide (Nt), 54,034 to KEGG, and 43,913 to gene ontology (GO) annotations. A total of 36,834 and 35,510 unigenes were differentially expressed at the vegetative and flowering stages of Hassawi 2 under drought stress, respectively. The majority of unigenes were down-regulated at both developmental stages. However, the number of genes up-regulated (15,366) at the flowering stage exceeded the number of those up-regulated (14,097) at the vegetative stage, and the number of genes down-regulated (20,144) at the flowering stage was smaller than the number of those down-regulated (22,737) at the vegetative stage. The drought stress-responsive differentially expressed unigenes coded for various regulatory proteins, including protein kinases and phosphatases, transcription factors and plant hormones and functional proteins including enzymes for osmoprotectant, detoxification and transporters were differentially expressed, most of which were largely up-regulated. Moreover, a substantial proportion of the DEGs identified in this study were novel, most exhibited a significant change in their expression levels under water stress, making them an unexploited resource that might control specific responses to drought stress in the faba bean. Finally, qRT-PCR results were found almost consistent with the results of next-generation sequencing. Our data will help in understanding the drought tolerance mechanisms in plants and will provide resources for functional genomics.
Electronic supplementary material
The online version of this article (10.1007/s13205-018-1518-2) contains supplementary material, which is available to authorized users.
Keywords: Faba bean, Drought, Transcriptome, Hassawi2, Differentially expressed genes, Vegetative and flowering stages
Introduction
The faba bean (Vicia faba L.) is one of the most important grain legumes in the world. Because of its multiple uses, high nutritional value, and ability to grow over a wide range of climatic and soil conditions, it is suitable for sustainable agriculture in many marginal areas (Nadal et al. 2003). The crop contributes to human nutrition via its high protein and nutrient contents (Aykroyd et al. 1982), and the faba bean seeds are considered an important source of nutrients, protein, energy, and dietary fiber for both humans and animals (Haciesferogullari et al. 2003). Furthermore, owing to its symbiotic N2-fixation in association with Rhizobium bacteria, the faba bean is utilized as a green manure, especially in organic farming. It provides vegetable proteins to animal and poultry feeds. Although the faba bean is less consumed as human food in Western countries, it is considered a main source of cheap protein and energy for many people in Africa, Asia, and Latin America, where meat is expensive. It is becoming increasingly important in Saudi diets and is consumed fresh and as dry seeds (Ahmed and Shehata 1992; Alghamdi 2003). The high lysine content of its seeds has encouraged its use as a protein supplement for cereals (El-Fiel et al. 2002). Despite its importance, the faba bean faces drought stress throughout its lifetime because of the water shortage due to low rain fall, largely in the central part of Saudi Arabia. Increasing drought tolerance and crop water-use efficiency has always been a concern because of the high demand for water and improved environmental quality by the human population (Hatfield et al. 2001). Recent genetic and breeding improvements in the faba bean have resulted in plants with several important traits such as resistance to environmental stresses, high yield, high protein content, and lack of major anti-nutritional factors (tannins, vicine-convicine) (Hoshikawa 1991; Bond and Duc 1993; Duranti and Gius 1997). Some genotypes of the faba bean, especially Hassawi 2, have evolved mechanisms that exhibit tolerance to various abiotic stresses such as drought (Ammar et al. 2014). These tolerant genotypes are, therefore, considered excellent genetic resources for stress tolerance for low-yield genotypes. Therefore, some initiatives would certainly help to develop the faba bean genotypes that produce better yields in water-limited environments.
The genome size of faba bean (2n = 2x = 12 chromosomes) is 13,000 Mb (Cubero 1974), which is larger than that of other legume crops such as soybean (~ 1200 Mb), pea (~ 4000 Mb), or Medicago (~ 450 Mb) (Pearce et al. 1996). Some recent development in the faba bean genomics, such as expressed sequence tags (ESTs) (Gong et al. 2010, 2011; Kaur et al. 2012; Suresh et al. 2015) or genome survey sequences (GSSs) (Yang et al. 2012a), have been reported for marker development, particularly for simple sequence repeats (SSRs) and single-nucleotide polymorphisms (SNPs). The improvement of genetic map resolution with SNP and SSR markers have also been described previously (Satovic et al. 2013; Kaur et al. 2014; El-Rodeny et al. 2014; Webb et al. 2016). Ammar et al. (2016) identified some limited number of potential drought-responsive genes in the drought-tolerant Hassawi 2 variety of faba bean, using a suppressive subtraction hybridization approach (SSH). However, no information is available on the genome-wide response of the faba bean to drought stress.
In recent years, the sequencing technology has undergone several fundamental changes. The development of the next-generation high-throughput DNA sequencing (NGS) platforms has deeply improved the efficiency and speed of gene discovery (Ansorge 2009; Asmann et al. 2009; Hershkovitz et al. 2013; Mortazavi et al. 2008; Zhang and Dolan 2010). The present study aims to study the transcriptomic profile of the faba bean drought-tolerant genotype Hassawi 2 at the vegetative and flowering stages under drought stress using the RNA-seq next-generation sequence technology. In this study, the paired-end sequencing technology was used to characterize the root transcriptomes under drought stress to create a sequencing resource. Subsequently, the gene expression profiles were analyzed during the vegetative and flowering stages under drought stress to obtain a list of candidate genes related to drought tolerance. Our study will enrich the understanding of drought tolerance mechanisms in the faba bean with possible candidate genes for various cellular processes and allow further trait manipulation.
Materials and methods
Plant materials and stress treatments
Hassawi 2, the local faba bean variety, is a genotype well-adapted to drought (Ammar et al. 2014). The Hassawi 2 seeds were grown under the normal and drought stress conditions in a controlled growth chamber at the College of Food and Agriculture Sciences, King Saud University. The seeds were grown in the pots filled with sandy soil. The fully developed seedlings were separated in four batches, two controlled (one for vegetative and one for flowering) and two stressed (one for vegetative and one for flowering), with ten pots/batch and one plant /pot. At the seedling stage, one batch of plants was exposed to stress using 15% PEG6000 (− 1.65 MPa). The stress was monitored daily using an osmometer 800 CL (SLAMED, Germany). When the plants were about to reach wilting point, the samples were collected from the control and drought-stressed plants. The remaining plants were grown under the normal conditions till flowering, when the second batch of samples was stressed as described and collected. All these collected samples from roots at the vegetative and flowering stages at both normal and stressed plants were immediately dipped in liquid N2 until RNA isolation. The roots of the plants were harvested in at least three biological replicates from each drought-treated and well-watered (control) plant. The pooled samples of each treatment were used for RNA extraction. The control plants at the vegetative and flowering stages were labeled as VCR-H2 and FCR-H2, respectively, while the drought-stressed plants at the vegetative and flowering stage were labeled as VSR-H2 and FSR-H2, respectively.
RNA isolation, RNA-seq library preparation and sequencing
Total RNA was extracted from 100 mg of each control and drought-stressed root tissue at different developmental stages using total RNA extraction kit (SV Total RNA isolation system, Promega, USA) incorporating on-column digestion of any DNA with RNase-free DNase I, according to the manufacturer’s instructions. The integrity of RNA samples (quality and quantity) was assessed using the Nanodrop spectrophotometer (NanoDrop Technologies) and Agilent Bioanalyzer 2100 (Agilent Technologies, Singapore). Only those RNA samples with A260/A280 ratios ranging from 1.9 to 2.1, A260/A230 ratios ≥ 2, and RNA integrity values > 7 were used in the subsequent experiments. The total RNA from these samples were submitted to Beijing Genome Institute (Hong Kong) for sequencing on the Illumina HiSeq 4000 platform to generate 2 × 150-bp long paired-end (PE) reads for each sample. Briefly, after extracting the total RNA and treating it with DNase I, the oligo (dT) primers were used to isolate mRNA. The isolated mRNA was fragmented into short sequences which were used as template for cDNA synthesis using random hexamer primer. After end repair and single nucleotide A (adenine) addition, short fragments are ligated to adapters. The suitable fragments were selected for the PCR amplification and paired-end cDNA libraries were constructed. The Agilent 2100 Bioanaylzer and ABI StepOnePlus Real-Time PCR System were used for the quantification and qualification of the sample library. Finally, the paired-end cDNA library was sequenced using Illumina HiSeq 4000 sequencing platform.
Filtering of sequencing reads and de novo assembly
Raw FASTQ files obtained from the sequencer were checked for quality parameters of the sequences using Phred quality score. The raw reads were filtered, and high-quality clean read data were obtained by removing adaptor sequences, removing reads containing more than 5% ambiguous bases (undetermined bases) and low-quality reads containing more than 10% bases with a Q value ≤ 20. After filtering, the remaining reads were called “clean reads” and stored in FASTQ (Cock et al. 2010) format.
Trinity (Haas et al. 2013) (v2.0.6 http://sourceforge.net/projects/trinityrnaseq/files, parameters:--min_contig_length 150 --CPU 8 --min_kmer_cov 3 --min_glue 3 --bfly_opts ‘-V 5 --edgethr = 0.1 --stderr’) was used for de novo assembly with clean reads and then TGICL (v2.0.6, parameters: -l 40 -c 10 -v 25 -O ‘-repeat_stringency 0.95 -minmatch 35 -minscore 35’) was used to cluster transcripts to unigenes. The unigenes from each sample were assembled for the unique all-unigene sequences using the TGICL software (Pertea et al. 2003). The unigenes were divided to two classes—clusters and singletons. The clusters have a prefix CL with the cluster IDs behind them (A single cluster contains several unigenes with more than 70% similarity among them). The singletons have a prefix unigene.
Unigene functional annotation
All unigene sequences were aligned against NCBI non-redundant nucleotide (Nt) (ftp://ftp.ncbi.nlm.nih.gov/blast/db), NCBI non-redundant protein (Nr) (ftp://ftp.ncbi.nlm.nih.gov/blast/db), Swiss-Prot protein (http://ftp.ebi.ac.uk/pub/databases/swissprot), the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway (http://www.genome.jp/kegg), and Eukaryotic Orthologous Groups (KOG) (http://www.ncbi.nlm.nih.gov/COG) databases. Green plant database was used as reference. To retrieve Gene Ontology (GO) terms regarding biological process, molecular function and cellular component (Ashburner et al. 2000) descriptions, the resulting Nr hits against Nr database were analyzed by Blast2GO software (Conesa et al. 2005) (version 2.5.0; http://www.blast2go.org/), and InterProScan5 (version v5.11-51.0) to get the InterPro annotation.
We used Transdecoder (v3.0.1https://transdecoder.github.io) to identify the candidate gene-coding region. The longest ORF (open reading frame) was extracted, and then the Pfam protein homologous sequences were searched using the SwissProt blast tool, and Hmmscan was used to predict the gene coding regions.
Unigene expression
The expression level from the four libraries (control and stress treatment, vegetative and flowering phase) were determined by mapping the clean reads of each library to the unigene-all using the Bowtie2 software [(Langmead and Salzberg 2012) (website:http://bowtie-bio.sourceforge.net/Bowtie2/index.shtml)]. The fragments per kilobase of transcripts per million fragments mapped (FPKM) values were obtained using RESM [(Li and Dewey 2011) (website: http://deweylab.biostat.wisc.edu/RSEM)].
Differential gene expression analysis
The differentially expressed genes (DEGs) were detected with the PossionDis algorithm, which is based on the poisson distribution process as described by (Audic and Claverie 1997). The hierarchical clustering of DEGs was performed using Pheatmap, a function of R. For clustering more than two groups, the intersection and union test was applied to the DEGs. The false discovery rate (FDR) was used to determine the threshold of P-value in multiple tests and analyses. FDR < = 0.001 and foldChange ≥ 2.00 were used as the threshold values to determine the significance of differences in gene expression.
Gene ontology and pathway analysis of DEGs
With the GO and KEGG annotation result, we classified the DEGs according to the official classification and performed the GO and pathway functional enrichment using Phyper, a function of R. The formula used for calculating the p-value in the hypergeometric test was:
where N is the number of all genes with GO annotation or KEGG annotation, n is the number of DEGs genes in N, M is the number of all genes that are annotated to certain GO terms or specific pathways, m is the number of DEGs in M. Subsequently, we calculated the FDR for each p value; in general, the terms in which FDR was ≤ 0.001 were defined as significantly enriched.
Identification of SSRs
MISA (MIcroSAtellite v1.0;http://pgrc.ipk-gatersleben.de/misa/) was used for SSR identification in the unigenes. The search criteria in the MISA script included more than 11 mono-nucleotide repeats, five di-nucleotides repeats, four tri- and quad-nucleotide repeats, and three penta- and hexa-nucleotide repeats.
Unigene transcription factors prediction
A transcription factor (TF) is a protein that binds to the specific DNA sequences, thereby controlling the rate of transcription of the genetic information from DNA to messenger RNA. The getorf algorithm (version EMBOSS:6.6.0.0;http://genome.csdb.cn/cgi-bin/emboss/help/getorf) was used to find the ORF of each unigene. Subsequently, the ORFs were aligned to the TF domains (form PlntfDB) using hmmsearch (version 3.0; http://hmmer.org), and TFs were identified according to the regulations described here [form PlantfDB (http://plntfdb.bio.uni-potsdam.de/v3.0/)].
qRT-PCR validation
To validate the results of the RNA-Seq experiments, a total of eight potential candidate DEGs related to drought stress were chosen for qRT-PCR. Total RNA was isolated from the root tissues of the three replicates of the control and drought-stressed samples using total RNA extraction kit (SV Total RNA isolation system, Promega, USA) according to manufacturer’s protocol. The first strand cDNA synthesis was performed using the ProtoScript First Strand Synthesis Kit (New England BioLabs) using an oligo (dT) primer and 5 µg of total RNA according to the manufacturer’s protocol. The quantitative reaction was performed using the SYBR Green PCR master mix (Applied Biosystems). Each reaction consisted of 1 µL of cDNA (100 ng), 10 µL of 2X SYBR Green PCR master mix (Applied Biosystems), 0.7 µL each of the forward and reverse primers (10 µM) and sufficient quantity of deionized water to make a final volume of 20 µL. The PCR amplification was performed under the following conditions: 50 °C for 2 min, initial denaturation at 95 °C for 10 min, followed by 40 cycles at 95 °C for 10 s, 57 °C for 15 s, and 60 °C for I min. All primers were designed using batch primers3 v1.0 software (https://probes.pw.usda.gov/batchprimer3). The relative expression levels of the potential genes were calculated using the 2−△△CT method (Livak and Schmittgen 2001). The relative mRNA levels were quantified with respect to the reference gene “Actin.” To compare the RNA-Seq and qRT-PCR data, the fold change values in the gene-expression levels of the drought-stressed samples were compared relative to the control samples used for the RNA-Seq and qRT-PCR analyses.
Results
RNA-Sequencing and transcriptomic analysis
We used the faba bean genotype Hassawi 2, which has been previously characterized for drought tolerance, for transcriptomic analysis (Ammar et al. 2014). Hassawi 2 revealed dominance of drought tolerance characters included high chlorophyll content; accumulation of compatible solutes, and maintenance of tissue water, efficiency of photosystem-II and grain weight during drought stress and this helped to classify this genotype to be drought tolerance (Ammar et al. 2014).
In this study, four cDNA libraries (as described in methodology section) were sequenced by Illumina deep-sequencing using the Illumina HiSeq 4000 platform. The summaries of transcriptome sequencing and assembly are summarized in Table 1. The clean reads Q20 (%) comprised more than 95% of the total reads in all libraries, thus suggesting high-quality sequencing. The four cDNA libraries yielded a total of 675.51 M of sequences raw data with the total clean reads yielding 624.8 M of reads.
Table 1.
Statistics of transcriptome sequencing
| Library | VCR-2 | VSR-H2 | FCR-H2 | FSR-H2 |
|---|---|---|---|---|
| Raw reads | 189,411,266 (189.41 M) | 143,468,264 (143.47 M) | 188,651,532 (188.65 M) | 153,980,020 (153.98 M) |
| Clean reads | 174,859,804 (174.86 M) | 132,495,282 (132.5 M) | 173,152,596 (173.15 M) | 144,290,892 (144.29 M) |
| Total clean bases (Gb) | 26.23 | 19.87 | 25.97 | 21.64 |
| Clean reads Q20 (%) | 96.79 | 96.78 | 96.44 | 96.64 |
| Clean reads Q30 (%) | 93.34 | 93.26 | 92.72 | 93.07 |
| Clean reads ratio (%) | 92.32 | 92.35 | 91.78 | 93.71 |
The clean reads of each library were separately de novo assembled at high stringency levels; the results are summarized in Table 2. The clean-read assembly resulted in 140,904, 76,160, 78,003, and 74,810 unigenes for VCR-H2, VSR-H2, FCR-H2, and FSR-H2, respectively. A total of 198,155 unigenes were obtained from the de novo assembled unigenes obtained from each library with an N50 length of 1347 bp and a mean length of 738 bp.
Table 2.
Statistics of transcriptome assembly
| Library | VCR-H2 | VSR-H2 | FCR-H2 | FSR-H2 | All-unigene |
|---|---|---|---|---|---|
| Total number | 140,904 | 76,160 | 78,003 | 74,810 | 198,155 |
| Total length | 94,088,842 | 59,085,461 | 70,239,709 | 61,040,418 | 146,377,199 |
| Mean length bp | 667 | 775 | 900 | 815 | 738 |
| N50 | 1136 | 1275 | 1529 | 1325 | 1347 |
| N70 | 571 | 753 | 928 | 809 | 702 |
| N90 | 254 | 291 | 343 | 318 | 261 |
| GC (%) | 40.16 | 40.13 | 39.27 | 39.29 | 39.78 |
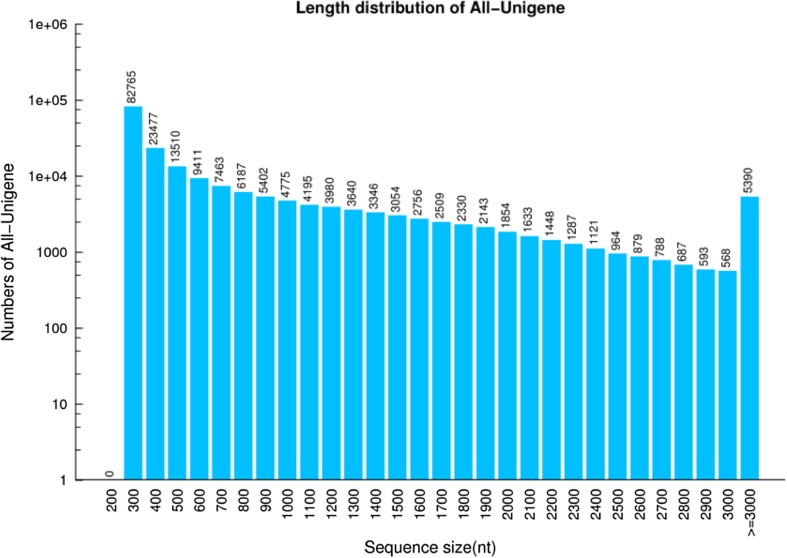
The size distribution of all-unigenes is shown (Fig. 1). The majority of the transcripts were 300–400 bp in length (82,765), followed by those that were 400–500 bp in length (23,477). The longest transcripts were > 3000 bp in length and contained 5390 unigenes.
Fig. 1.
Size distribution of sequences in all-unigenes
The transcriptome raw data sequences of the four libraries of the faba bean under control and drought stress conditions have been deposited in the NCBI under SRA (sequence reads archives) under the following accession numbers: SRX3182040, SRX3182041, SRX3182044, and SRX3182045.
Unigene functional annotation
The sequence similarity search of the 198 155 assembled all-unigenes was performed against the public databases, including the NCBI non-redundant protein (Nr) database, NCBI non-redundant nucleotide sequence (Nt) database, Gene Ontology (GO) database, Swiss-Prot, Interpro, Kyoto Encyclopedia of Genes and Genomes (KEGG), and Cluster of Orthologous Groups of proteins (COG). The annotation summary is shown in Table 3. Of the 198,155 all-unigenes, 66,254 (33.44%) shared significant homology with the NCBI Nt database and 78,262 (39.50%) unigenes with the NCBI Nr database. Similarly, 44,011 (22.21%) unigenes were annotated in Swiss-Prot, 56,535 (28.53%) unigenes in KOG, 54,034 (27.27%) unigenes in KEGG, 55,281 (27.90%) unigenes in Interpro, and 43,913 (22.16%) unigenes in the GO database (Table 3). In total, 95,844 (48.37%) annotated unigenes were obtained. The details of the functional annotation of all unigenes with seven functional databases are presented in Supplementary Table S1.
Table 3.
Summary of functional annotation of all-unigenes with different functional databases
| Categories | Number | Percentage (%) |
|---|---|---|
| All-unigenes | 198,155 | 100 |
| Nr | 78,262 | 39.50 |
| Nt | 66,254 | 33.44 |
| Swissprot | 44,011 | 22.21 |
| KEGG | 54,034 | 27.27 |
| KOG | 56,535 | 28.53 |
| Interpro | 55,281 | 27.90 |
| GO | 43,913 | 22.16 |
| Intersection | 20,557 | 10.37 |
| All annotated unignenes | 95,844 | 48.37 |
For the KOG and GO annotation, the functional distribution of the annotated unigenes was analyzed as shown as Fig S1 and Fig S2, respectively. All unigenes were compared against the KOG database to predict and classify their putative functions. Overall, 56,535 (28.53%) unigenes showed nonredundant hits assigned to the KOG classifications. The KOG-annotated putative proteins were functionally classified into 25 molecular families, such defence mechanisms, cytoskeleton, and different kinds of metabolism. Several categories (molecular families) were quite interesting, for instance, signal transduction (6140 unigenes); transcription (3619 unigenes); secondary metabolites biosynthesis, transport, and catabolisim (1947 unigenes); posttranslational modification, protein turnover, and chaperones (5740 unigenes); inorganic ion transport and metabolism (1361 unigenes); energy production and conservation (2492 unigenes); defence mechansims (939 unigenes); carbohydrate transport and metabolism (3274 unigenes); and amino acid transport and metabolism (2147 unigenes).
The GO classification is used for annotating and analyzing the functional categorization of the annotated genes. A total of 43,913 unigenes (22.16%) were assigned to the GO ontologies based on their sequence similarity with the previously known functions. The annotated unigenes were also classified into the GO categories for biological processes, cellular components, and molecular functions; indicating high accuracy of annotation. The majority of unigenes were assigned to more than one GO term; therefore, the total number of GO terms was higher than the total number of the unigenes with GO assignments. This GO classification is a source of the diverse set of putative biological functions. Among biological processes, the most abundant function was related to the metabolic processes (23,415 unigenes), followed by the cellular processes (21,602 unigenes), single organism processes (13,023 unigenes), biological regulation (5802 unigenes), regulation of biological processes (4960), responses to stimuli (4477), and signaling (1648 unigenes). In the cellular component category, the most abundant unigenes were present in the cell (17 884 unigenes), membrane (14 423 unigenes), and cell parts (17 746 unigenes). In the molecular function category, the catalytic activity was the most abundant subcategory (22 764 unigenes), followed by binding (21 040 unigenes). Among the interesting groups in the molecular function category were the transporter activity (2631 unigenes), nucleic acid-binding transcription factor activity (782 unigenes), and antioxidant activity (293 unigenes). These classes, undoubtedly, are involved in the stress tolerance mechanisms.
The KEGG analysis revealed that the 54 034 KEGG-annotated unigenes were classified into five different functional groups (Fig S3). Of these, 37 267 unigenes were classified into the category of metabolism, including the subcategories of biosynthesis of other secondary metabolites (2012 unigenes), genetic information processing (14 786 unigenes), and environmental information processing (3706). In the secondary metabolism category, many important subcategories comprised of phenylpropanoid biosynthesis [1211 (2.24%) unigenes], carotenoid biosynthesis [258 (0.48%) unigenes], terpenoid backbone biosynthesis [361 (0.67%) unigenes], flavonoid biosynthesis [236 (0.44%) unigenes], zeatin biosynthesis [76 (0.14%) unigenes], and flavone and flavonol biosynthesis [65 (0.12%) unigenes].
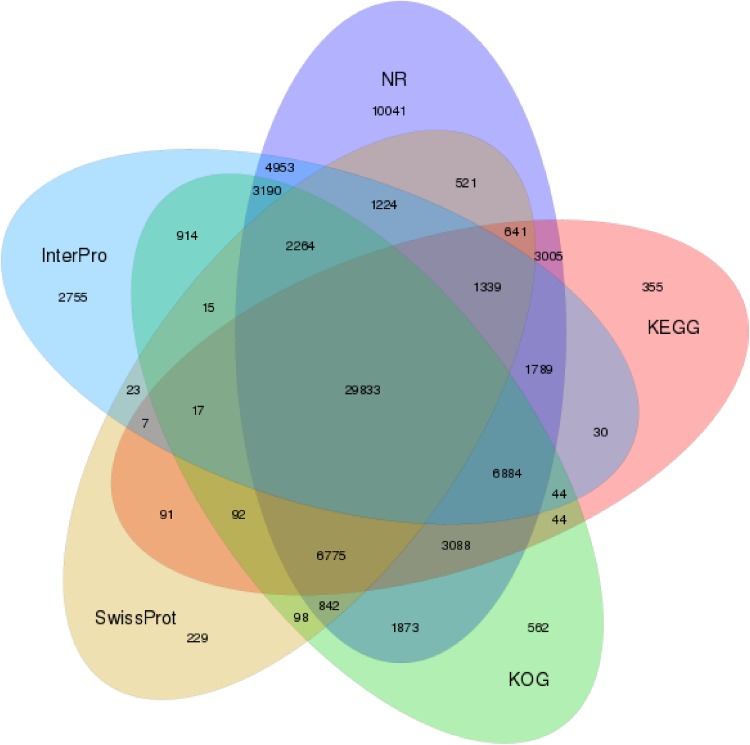
The Venn diagrams between the NR, KOG, KEGG, Swissprot, and Interpro annotations are shown in Fig. 2. The Venn diagrams classified some genes in a unique manner. Venn diagrams had the classified 29,833 unigenes in common. On the other hand, the NR database had 10,041 unique transcripts, KEGG had 355, InterPro had 2755, KOG had 582, and SwissPort had 229 transcripts. These differences might be attributed to the various algorithms used in each tool.
Fig. 2.
Venn diagram between NR, COG, KEGG, Swissprot and Interpro. The numbers of unique and shared unigenes are shown
The annotated percentages were obtained with the Nr annotation; the distribution of annotated species was analyzed as shown in Table S2. Approximately 41.2% of the annotated sequences were similar to the model legume Medicago truncatula, 23.25% were similar to Cicer arietinum, 4.92% were similar to Glycine max, 3.24% were similar to Pisum sativum, and 27.38% were similar to other organisms. These results showed that more than 80% of the annotated sequences showed high similarity with the legume crops, while approximately 20% showed similarities with other organisms.
After functional annotation, we selected the segment of unigenes that best mapped to functional databases as its gene coding sequence (CDS). For the unigenes that unannotated, ESTScan was used to predict CDS. A total of 66,557 unigenes (42.83% of all-unigenes) were predicted as cds showing N50 length 1215 with maximum and minimum length of 16,194 and 297 bp, respectively. Distribution of CDS length is shown in Fig S4.
All-unigene SSR detection
SSRs are one of the most commonly used marker systems consisting of varying numbers of the tandemly repeated di-, tri-, or tetranucleotide DNA motifs. After assembly, the SSR-containing unigenes were identified in all-unigenes. The SSR size summary is shown in Table S3. A total of 18,327 potential SSRs were identified in 15,361 unigenes (Fig. 3). The largest category of the SSR markers was the trinucleotide repeats (6927), followed by the dinucleotide (5594) and mono-nucleotide (4359) repeats. The other types of SSRs, such as tetra- (288), penta- (407), and hexa-(752) nucleotide motifs, were also detected.
Fig. 3.
SSR size distribution. X axis represents the type of SSR. Y axis represents the number of SSR
Differential gene expression
The expression levels of the unigenes from the four samples were determined using the FPKM method (fragments per kb per million fragments). To quantify the gene expression, the total reads of each library were aligned to the de novo assembled all-unigenes. The summary of the mapped reads is shown in Table 4. For gene expression quantification, the expression level of each unigene in all samples was determined, and the FPKM range of all unigenes in each sample is shown in Table 4. The large number of unigenes in each sample showed the FPKM value greater than 10. A total of 11,022 unigenes for VCR-H2, 11,025 unigenes for VSR-H2, 10,354 unigenes for FCR-H2, 8444 unigenes for FSR-H2 have shown FPKM > 10. Similarly, the majority of unigenes in each sample showed the FPKM value between 1 and 10 and less than 1.
Table 4.
Summary of sample mapping results and unigene FPKM range in the samples
| VCR-H2 | VSR-H2 | FCR-H2 | FSR-H2 | |
|---|---|---|---|---|
| Mapping type | # of reads | # of reads | # of reads | # of reads |
| Total reads | 168,919,008 | 132,495,282 | 173,152,596 | 144,290,892 |
| Total mapped reads (%) | 119,515,634 (70.75%) | 109,307,056 (82.5%) | 138,427,148 (79.95%) | 116,911,796 (81.03%) |
| Perfect match (%) | 65,678,862 (38.88%) | 60,722,868 (45.83%) | 78,248,375 (45.19%) | 68,117,391 (47.21%) |
| Mismatch (%) | 53,836,772 (31.87%) | 48,584,188 (36.67%) | 60,178,773 (34.75%) | 48,794,405 (33.82%) |
| Unique match (%) | 69,672,500 (41.25%) | 63,678,416 (48.06%) | 79,407,602 (45.86%) | 69,740,706 (48.33%) |
| Multi-position match (%) | 49,843,134 (29.51%) | 45,628,640 (34.44%) | 59,019,546 (34.09%) | 47,171,090 (32.69%) |
| Total unmapped reads (%) | 49,403,374 (29.25%) | 23,188,226 (17.5%) | 34,725,448 (20.05) | 27,379,096 (18.97%) |
| FPKM > = 10 | 11,022 | 11,025 | 10,354 | 84,44 |
| FPKM 1–10 | 39,737 | 34,068 | 24,760 | 27,604 |
| FPKM < = 1 | 65,024 | 23,983 | 44,046 | 33,347 |
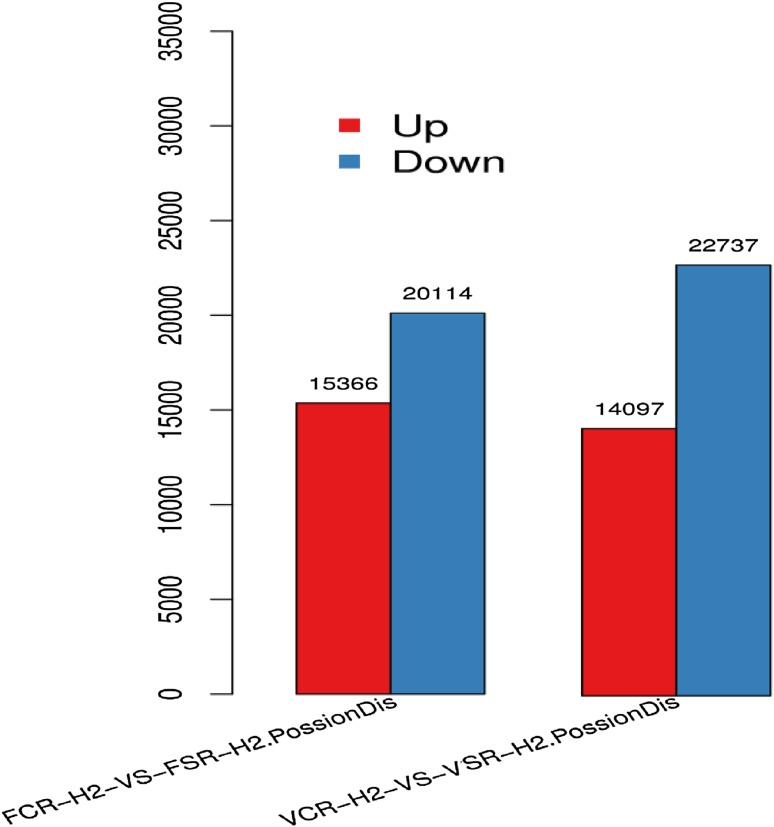
The drought-responsive DEGs were identified in root samples at the vegetative and flowering or reproductive stages by performing an expression analysis between VCR-H2 and VSR-H2 and between FCR-H2 and FSR-H2 to unravel the similarities and differences between the control and drought samples and to further analyze their biological functions. For this, firstly, the unigene expression level for each gene was calculated and normalized to the RPKM value. With unigene expression result, the DEGs between these two combinations were identified. The distribution of these genes is shown in Fig. 4. The majority of unigenes were down-regulated at both developmental stages. The differential gene expression analysis at the vegetative stage (VCR-H2 vs. VSR-H2) revealed a total of 36,834 differentially expressed unigenes. Among them, 14,097 unigenes were up-regulated and 22,737 unigenes were down-regulated. Moreover, at the flowering stage, among the 35,510, differentially expressed unigenes identified in the control and drought-stressed samples (FCR-H2 vs. FSR-H2), a large majority (20,114) of the unigenes were down-regulated, while only 15,366 unigenes were up-regulated. However, it was found that at the flowering stage, the number of up-regulated DEGs exceeded the number of up-regulated DEGs at the vegetative stage, and the number of down-regulated DEGs was smaller than that of the down-regulated genes at the vegetative stage. Among differentially expressed genes, 14,262 unigenes were found common in both development stages.
Fig. 4.
Differentially expressed genes in drought vs. control at vegetative (VCR-H2 vs. VSR-H2) and flowering stage (FCR-H2 vs. FSR-H2). Numbers of up-regulated and down-regulated genes were summarized
Analysis of DEGs
The list of important significantly up- and down-regulated DEGs (> 8 log2 fold change and <-10 log2 fold change) at the vegetative stage (VCR-H2 vs. VSR-H2) is presented in Table 5 and the list of DEGs with functional annotation is presented in Table S4. Some of the up-regulated DEGs belonged to the LRR receptor-like kinase (CL13710.Contig2_All), pseudo response regulator 37 (CL1026.Contig10_All), ethylene response factor (CL5278.Contig4_All), mitogen-activated protein kinase kinase kinase (CL19595.Contig15_All), and vacuolar H+-ATPase (Unigene64029_All). Similarly, the important down-regulated unigenes included the argininosuccinate synthase (CL10254.Contig3_All), putative histone deacetylase (CL1072.Contig1_All), phospholipid-transporting ATPase (CL2724.Contig2_All), trehalose-6-phosphate synthase domain protein (Unigene127707_All), and alpha-mannosidase (CL253.Contig8_All).
Table 5.
List of significantly differentially expressed genes in VCR-H2 vs. VSR-H2
| GeneID | Length | log2FoldChange | FDR | Description |
|---|---|---|---|---|
| CL13710.Contig2_All | 441 | 10.90087 | 3.18E−57 | LRR receptor-like kinase |
| CL1026.Contig10_All | 3122 | 10.66622 | 0 | Pseudo response regulator 37 |
| CL5278.Contig4_All | 1073 | 10.15861 | 6.36E−109 | Ethylene response factor |
| Unigene28218_All | 1151 | 9.726218 | 1.58E−87 | Glycoside hydrolase family 18 protein |
| CL19595.Contig15_All | 830 | 9.575539 | 2.12E−53 | Mitogen-activated protein kinase kinase kinase |
| Unigene71188_All | 217 | 9.283088 | 2.44E−05 | ABC-transporter B family protein |
| CL5915.Contig6_All | 224 | 8.882643 | 0.0002 | Aldehyde dehydrogenase family 7 member |
| CL6578.Contig2_All | 3299 | 8.78136 | 3.91E−143 | Putative respiratory burst oxidase-like protein C |
| Unigene64029_All | 253 | 8.717676 | 4.94E−05 | Vacuolar H+-ATPase |
| Unigene62821_All | 228 | 8.584963 | 0.000802 | Calcium-dependent protein kinase |
| CL10254.Contig3_All | 1672 | − 12.3565 | 0 | Argininosuccinate synthase |
| CL1072.Contig1_All | 1243 | − 12.0481 | 0 | Putative histone deacetylase |
| CL2724.Contig2_All | 1836 | − 11.8607 | 0 | Phospholipid-transporting ATPase |
| CL5262.Contig5_All | 658 | − 11.5022 | 0 | Dormancy-associated protein |
| Unigene24541_All | 840 | − 11.3393 | 2.67E−180 | Vicia faba heat shock protein |
| Unigene127707_All | 3001 | − 11.2354 | 0 | Trehalose-6-phosphate synthase domain protein |
| CL9947.Contig5_All | 2008 | − 11.0063 | 0 | CBL-interacting serine/threonine-protein kinase 8-like |
| CL253.Contig8_All | 3579 | − 11.0021 | 0 | Alpha-mannosidase |
| CL1512.Contig5_All | 5632 | − 10.9894 | 0 | Eukaryotic translation initiation factor 4G |
| CL18210.Contig2_All | 1644 | − 10.8186 | 2.30E−270 | Protein disulfide-isomerase |
The list of the up- and down-regulated DEGs at the flowering stage (FCR-H2 vs. FSR-H2) that showed > 1 log2 (fold change) are presented in Table S5 and the list of highly upregulated (> 9 log2foldchange) DEGs is presented in Table 6. Among these, some of the important up-regulated DEGs belonged to the phospholipid glutathione peroxidase (CL14906.Contig1_All), gibberellin-regulated family protein (Unigene58316_All), auxin-induced protein 6B-like (Unigene49384_All), LRR receptor-like kinase (Unigene46583_All), and transcription factor bHLH18 (Unigene53927_All). The list of highly down-regulated (<-10 log2foldchange) DEGs is presented in Table 6. Among these, some of the important down-regulated DEGs belonged to glutathione S-transferase, amino-terminal domain protein (Unigene12892_All), extensin-2 (CL19713.Contig1_All), ubiquitin-conjugating enzyme E2 (CL3497.Contig3_All), peroxidase family protein (Unigene20216_All), and aquaporin (major intrinsic protein family) (CL2349.Contig1_All).
Table 6.
List of significantly differentially expressed genes in FCL-H2 vs. FSL-H2
| GeneID | Length | log2FoldChange | FDR | Description |
|---|---|---|---|---|
| Unigene46583_All | 2403 | 11.01053 | 0 | LRR receptor-like kinase |
| CL14906.Contig1_All | 614 | 10.78709 | 2.74E−98 | Phospholipid glutathione peroxidase |
| Unigene58316_All | 597 | 10.63027 | 5.01E−85 | Gibberellin-regulated family protein |
| Unigene18741_All | 911 | 10.54045 | 0 | Oxygen-evolving enhancer protein |
| Unigene49384_All | 526 | 10.46557 | 5.89E−64 | Auxin-induced protein 6B-like |
| CL2907.Contig6_All | 1811 | 10.45018 | 9.51E−277 | ABC-transporter D family protein |
| CL15820.Contig1_All | 1942 | 9.839204 | 8.05E−193 | Serine/threonine kinase family protein |
| Unigene12779_All | 457 | 9.717676 | 4.76E−31 | Lysine histidine transporter 1-like |
| CL5203.Contig1_All | 3179 | 9.625709 | 1.38E−284 | Solute carrier family 40 member |
| Unigene53927_All | 840 | 9.543032 | 1.40E−61 | Transcription factor bHLH18 |
| Unigene12892_All | 1197 | − 14.028 | 0 | Glutathione S-transferase, amino-terminal domain protein |
| CL19713.Contig1_All | 1115 | − 13.2406 | 0 | Extensin-2 |
| CL3497.Contig3_All | 4321 | − 12.6991 | 0 | Ubiquitin-conjugating enzyme E2 |
| Unigene20216_All | 1275 | − 12.6425 | 0 | Peroxidase family protein |
| CL2349.Contig1_All | 1802 | − 12.2656 | 0 | Aquaporin (major intrinsic protein family) |
| Unigene4635_All | 1949 | − 11.6188 | 0 | Type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2 |
| Unigene8676_All | 2067 | − 11.617 | 0 | Squalene synthase |
| Unigene7377_All | 2376 | − 11.6036 | 0 | Heat shock cognate 70 kDa-like protein |
| Unigene31019_All | 1313 | − 11.5083 | 0 | 1-Aminocyclopropane-1-carboxylate oxidase |
| CL16169.Contig2_All | 2036 | − 11.4998 | 0 | Beta-fructofuranosidase |
In addition, the potential drought-responsive regulatory genes were detected based on the criteria of log2 fold change > 1 for up-regulation and log2 fold change <-1 for down-regulation at FDR < 0.001 in the VCR-H2 vs. VSR-H2 and FCR-H2 vs. FSR-H2 combinations as previously identified and validated in plants. These genes are further detailed in the Supplementary Tables S6, S7, S8, and S9. These DEGs included the transcriptional factors (i.e., BHLH, NAC, MYP, and WRKY), protein kinases and receptors (e.g., calcium-dependent protein kinase, mitogen-activated protein kinase kinases kinase, LRR receptor-like kinase, receptor-like kinase, and histidine kinase), phosphatases, (protein phosphatase 2C and serine/threonine-protein phosphatase 2A), and genes related to different hormones (e.g., abscisic acid aldehyde oxidase, beta-carotene hydroxylase, and molybdenum cofactor sulfurase, zeaxanthin epoxidase, abscisic acid receptor), ethylene (1-aminocyclopropane-1-carboxylate oxidase, 1-aminocyclopropane-1-carboxylate synthase, ethylene insensitive 3, ethylene response factor, EIN3-binding F-box protein 1), gibberellic acid (Ent-kaurenoic acid oxidase, gibberellin 2-beta-dioxygenase), jasmonic acid (lipoxygenase, jasmonate O-methyltransferase), auxin (indole-3-pyruvate monooxygenase, auxin transporter-like protein, auxin efflux carrier component, auxin response factor), cytokinin (adenylate isopentenyltransferase, cytokinin dehydrogenase), and salicylic acid (salicylic acid-binding protein).
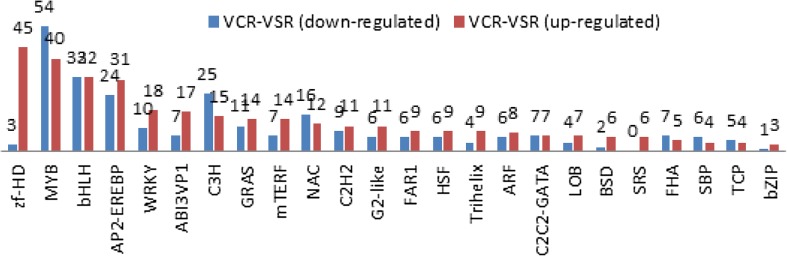
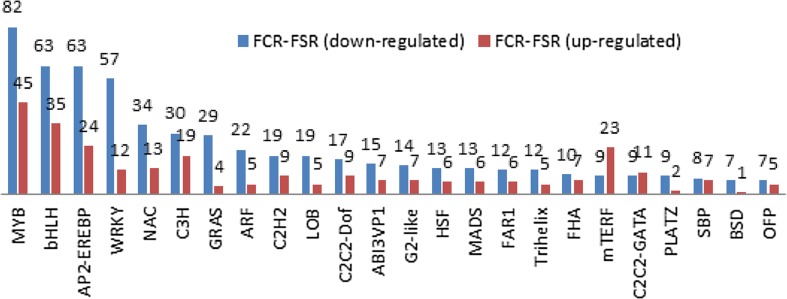
At the vegetative stage, a total of 578 differentially expressed drought-responsive TFs were identified, among which 311 were up-regulated and 267 were down-regulated (Table S8). However, at the flowering stage, a total of 987 differentially expressed drought-responsive TFs were identified, among which 338 were up-regulated and 649 were down-regulated (Table S9). The differentially expressed TFs belonging to the MYB family were the most abundant, followed by the bHLH- and AP2-EREBP-family TFs during different vegetative and flowering stages under drought stress. A considerable number of the DEGs identified in the majority of TFs in the VCR-H2 and VSR-H2 samples were found up-regulated (Fig. 5). The AP2-EREBP-, zf-HD-, and WRKY-family TFs were largely up-regulated, while the MYP- and NAC-family TFs were largely down-regulated. However, at the flowering stage, a large number of differentially expressed TFs were largely down-regulated (Fig. 6), suggesting different patterns of regulation at different growth stages. The identification of such a large number of regulatory genes indicates that plants use diverse signaling mediators and complex pathways to control drought stress.
Fig. 5.
Differentially expressed genes for transcription factor families in VCR-H2 vs. VSR-H2
Fig. 6.
Differentially expressed genes for transcription factor families in FCR-H2 vs. FSR-H2
In this study, we identified different DEGs encoding different functional proteins in the VCR-H2 vs. VSR-H2 and FCR-H2 vs. FSR-H2 combinations (Tables S10 and S11). During drought conditions, the reactive oxygen species (ROS) induce oxidative damage. To combat this damage, plants have evolved several enzymatic compounds that help maintain the redox homeostasis by detoxifying these ROS. A large number of DEGs encoding different enzymes were identified: glutathione S transferases, superoxide dismutase, glutathione peroxidase, respiratory burst oxidase, catalase, ascorbate peroxidases, glutathione reductase and non-enzymatic machinery, including metallothionein. Some of these were largely down-regulated and some were largely up-regulated. In addition, a large number of DEGs were annotated in the channels and transporters category. Among those, the chloride channel, guard-cell S-type anion channel, different types of aquaporin, ABC-transporter protein family, multidrug resistance protein, ABC-transporter protein, betaine aldehyde dehydrogenase, delta-1-pyrroline-5-carboxylate synthetase, and trehalose-phosphate synthase were identified, and the majority of these were largely up-regulated. The late embryogenesis-abundant (LEA) genes are commonly induced during drought stress; thus, we identified the LEA gene, which is related to dehydrin and group 3 late embryogenesis-abundant proteins. Moreover, drought-induced protein, dehydration-induced protein and heat shock protein were also differentially expressed in both vegetative and flowering stages under drought stress.
Functional annotation of DEGs
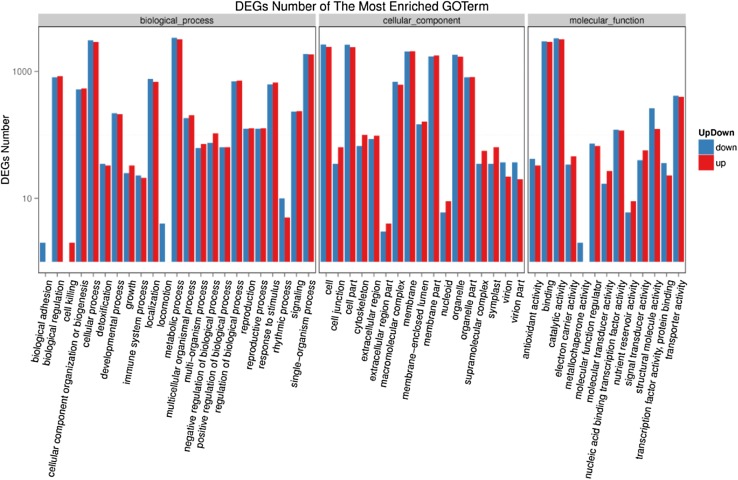
To understand the functions of DEGs, we mapped all genes to the terms in the GO database, looking for the significantly enriched GO terms. Figure 7 summarizes the most significant BP, CC, and MF ontology terms that resulted from the VCR-H2 and VSR-H2 samples. The most significant GO terms for the biological processes were biological regulation, cellular process, and metabolic process, response to stimulus, signaling, and detoxification. Furthermore,a comparatively higher number of up-regulated genes were found in a subcategory of the main biological process category, especially in the “biological regulation,” “signaling,” and “response to stimulus” terms. In the cellular process category, a large number of DEGs were up-regulated in 11 GO terms. However, more DEGS were down-regulated in 6 GO terms (Fig. 7). According to the molecular function, the DEGs represented a high proportion the GO terms including binding, catalytic activity, nucleic acid-binding TF activity, structural molecule activity, transporter activity, and antioxidant activity. However, in this category maximum GO terms were down-regulated, except for the “electron carrier activity,” “molecular transducer activity,” “signal transducer activity,” and “nutrient reservoir activity,” which have comparatively up-regulated DEGs. Overall, the most stress-related GO terms in the categories of biological processes, cellular components, and molecular function were largely up-regulated.
Fig. 7.
GO function enrichment analysis of VCR-H2 VS VSR-H2. X axis represents GO term. Y axis represents number of DEGs
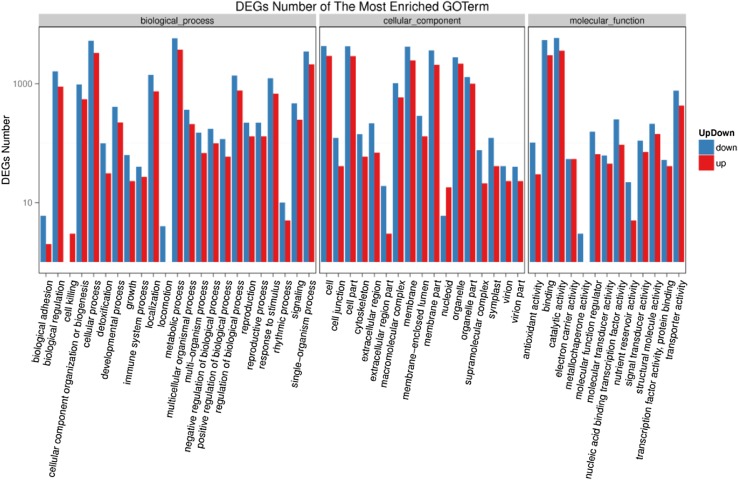
Similarly, Fig. 8 summarizes the most significant BP, CC, and MF ontology terms that resulted from the FCR-H2 and FSR-H2 samples. Almost similar GO terms were enriched in the FCR-H2 vs. FSR-H2 samples as described for the VCR-H2 vs. VSR-H2 samples for the three main categories (biological process, molecular function, and cellular component) (Fig. 8) Overall, the majority of DEGs in these GO terms in the categories of biological processes, cellular components, and molecular function were comparatively down-regulated.
Fig. 8.
GO function enrichment analysis of FCR-H2 vs. FSR-H2. X axis represents GO term. Y axis represents number of DEGs
KEGG pathways enrichment
To identify biochemical pathways, among the 36,834 DEGs of the VCR-H2 vs. VSR-H2 combination, only (15 055) unigenes were mapped to 136 KEGG pathways, which were classified into five main categories. The complete set of matched pathways in the VCR-H2 vs. VSR-H2 combination is summarized in Supplementary Table S12. Most genes were annotated with “metabolic function” (10 771 unigenes), followed by “genetic information processing” (3980 unigenes), “environmental information processing” (1013 unigenes), “cellular processes” (815 unigenes), and “organismal systems” (680 unigenes). However, the highest number of unigene pathways associated with the “cellular process” category were endocytosis [ko04144, 511 (3.39%)] and peroxisome [ko04146, 190 (1.26%)]. The highest number of unigenes associated with the “genetic information processing” category were spliceosome [ko03040, 516 (3.43%)], and protein processing in endoplasmic reticulum [ko04141, 676 (4.49%)]. The only four pathways associated with the “environmental information processing” category were plant hormone signal transduction [ko04075, 403 (2.68%)], plant MAPK signaling pathway [ko04016, 385 (2.56%)], ABC transporters [ko02010, 258 (1.71%)], and phosphatidylinositol signaling system [ko04070, 132 (0.88%)]. The plant hormone signal transduction was the largest complex comprising several plant hormone, including ABA (abscisic acid), cytokinins, auxin, gibberellins (GA), ethylene (ET), jasmonic acid (JA), and brassinosteroid (BR).The highest number of unigene representation pathways in the “metabolism” category were biosynthesis of secondary metabolites [ko01110, 2006 (13.32%)], biosynthesis of amino acids [ko01230, 553 (3.67%) unigenes], and carbon metabolism [ko01200, 454 (3.02%)]. The two pathways associated with the “organismal systems” (environmental adaptation) category were plant-pathogen interaction [ko04626, 578 (3.84%)] and plant circadian rhythm [ko04712, 102 (0.68%)].
Similarly, among the 35,510 DEGs for the combination FCR-H2 vs. FSR-H2, 19,035 unigenes were annotated to 136 KEGG pathways, which were classified into six main categories. The complete set of matched pathways in the FCR-H2 vs. FSR-H2 combination is summarized in Supplementary Table S13. Most genes were annotated with “metabolic function” (13 745 unigenes), followed by “genetic information processing” (4360 unigenes), “environmental information processing” (1406 unigenes), “cellular processes” (1030 unigenes), and “organismal systems” (791 unigenes). However, the highest number of unigene pathways associated with the “cellular process” category were endocytosis [ko04144, 673 (3.54%)] and peroxisome [ko04146, 223 (1.17%)]. The highest number of unigene pathways associated with the “genetic information processing” category were spliceosome [ko03040, 541 (2.84%)] and protein processing in endoplasmic reticulum [ko04141, 709 (3.72%)]. The only four pathways associated with the “environmental information processing” category were plant hormone signal transduction [ko04075, 575 (3.02%)], plant MAPK signaling pathway [ko04016, 538 (2.83%)], ABC transporters [ko02010, 300 (1.58%)], and phosphatidylinositol signaling system [ko04070, 191 (1%)]. The highest number of unigene representation pathways associated with the “metabolism” category were biosynthesis of secondary metabolites (ko01110, 3707), biosynthesis of amino acids [ko01230, 2717 (14.27%)], and carbon metabolism [ko01200, 566 (2.97%)]. The two pathways associated with organismal systems category (environmental adaptation) were plant–pathogen interaction [ko04626, 633 (3.33%)] and plant circadian rhythm [ko04712, 158 (0.83%)].
qRT-PCR validation of DEGs from RNA-Seq
To confirm the correctness and reproducibility of the Illumina RNA-Seq results, eight candidate DEGs were selected, and their expression levels were compared with the results obtained from the qRT-PCR analysis. The list of the primers specific for these genes is presented in Table S14. The expression profiles of the five up-regulated genes, including the Unigene62821_All (Calcium-dependent protein kinase), CL5915.Contig1_All (Aldehyde dehydrogenase family 7 member), CL5019.Contig4_All (Aquaporin PIP2-7), Unigene14261_All (late embryogenesis-abundant protein), CL12207.Contig3_All (unkown),, three down-regulated genes include CL18827.Contig2_All (unknown), CL6858.Contig1_All (Serine/threonine-protein phosphatase PP2A), and Unigene76735_All (RR receptor-like kinase family protein). The expression patterns obtained from the qRT-PCR assay were more or less consistent with those obtained from the Illumina sequencing except one down-regulated gene which is slightly up-regulated in qPCR (Table S14). Therefore, these qRT-PCR data support the high confidence of the RNA-Seq data.
Discussion
The objective of this study was to generate the root transcriptome of the drought-tolerant faba bean genotype Hassawi 2 for its wide-ranging characterization with the final aim of detecting the potential drought stress-responsive genes. The root is the first organ to be exposed to and to deal with drought stress and the evaluation of plant responses to drought stress is often based on the evaluation of the environment around root. In the current study, we performed an in-depth sequencing analysis of root transcriptome of the faba bean genotype Hassawi 2 in response to drought stress because it is tolerant to drought and very little is known about its genome.
For this purpose, four selected cDNA libraries were sequenced using the Illumina HiSeq 4000. A total of 675.51 M raw reads were obtained from the four selected libraries and assembled by the De Novo Assembler. The total clean reads in each library were far more than those reported by Ocaña et al. (2015), who generated 33,023,160 reads from three libraries. A total of 198,155 all-unigenes from four libraries were obtained after de novo assembly in our experiment. The complete unigene set (198,155 all-unigenes) was functionally annotated using BlastN against NCBI non-redundant (Nr) nucleotide, BLASTx against NCBI non-redundant (Nr) protein, Swiss-Prot protein, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway, and Cluster of Orthologous Groups (COG) databases. A total of 95,844 (48.37%) annotative genes were obtained, and approximately 41.2% of the annotated sequences were found similar to the model legume Medicago truncatula, and 23.25% were similar to Cicer arietinum. In addition, a large number of unigenes were not annotated, accounting for more than 50%, suggesting these unigenes were novel and may be unique to faba bean. Furthermore this is a first report of faba bean transcriptome sequencing under drought stress. The functional annotation with GO terms showed that the most abundant GO function in the biological and molecular function category indicated that a large range of activities related to plant growth, development, and stress tolerance mechanisms in the faba bean. This type of distribution in the faba bean unigenes was more or less similar to that previously reported in arabidopsis and lentil transcriptomes (Gan et al. 2011; Kaur et al. 2011).
In this study, the important molecular families of the KOG-annotated putative proteins were found related to signal transduction, inorganic ion transport and metabolism, and defence mechansims. The similar COG classification has been reported previously for Hevea brasiliensis transcriptome (Li et al. 2012). The KEGG analysis can help us further understand the specific processes, gene functions, and gene interactions at a transcriptome level. In our study, 54 034 unigenes were mapped to 135 predicted metabolic pathways through the KEGG database. The largest category was metabolic pathways and biosynthesis of secondary metabolites.
The SSR-based marker systems have been popular for the population genetic analyses and genetic mapping studies (Luikart et al. 2003). However, only a few SSRs were available for the faba bean until recently (Pozarkova et al. 2002). Therefore, the genetic diversity studies have been performed mainly with the random amplification of polymorphic DNA or amplified fragment length polymorphism techniques, which are less effective than the sequence-based co-segregation markers (Zeid et al. 2009). In our study, a total of 18,327 potential SSRs were identified and the majority SSRs showed trinucleotide repeats, followed the dinucleotide and mono-nucleotide repeats. The large number of SSRs identified in present study, thus, offers a cost-effective way to further develop the functional markers for marker-assisting breeding purposes. These SSRs will also allow fingerprinting for taxonomic and phylogenetic comparisons in a wide range of closely related organisms.
In this study, the differential expression of the drought-treated samples at the vegetative (VCR-H2 vs. VSR-H2) and flowering stages (FCR-H2 vs. FSR-H2) identified the DEGs effectively across a wide range of transcript abundance with a fold change log2 ratio > 1 or < − 1 and an FDR (of < 0.001). Based on this criteria, a total of 36,834 (14,097 up-regulated and 22,737 down-regulated) and 35,510 genes (15,366 up-regulated and 20,114 down-regulated) were found to be differentially expressed in the VCR-H2 vs. VSR-H2 and FCR-H2 vs. FSR-H2 samples, respectively. In both stages, a large number of unigenes were down-regulated; however, at the flowering stage, the number of up-regulated genes was more than that of the up-regulated genes at the vegetative stage, suggesting the presence of different expression patterns in the root tissues. Based on annotation, the DEGs identified under the drought stress condition were closely related with the plant stress functions, such as stress tolerance (i.e., sodium hydrogen exchanger, sodium calcium exchanger, universal stress protein, stress protein, calcineurin B-like protein, dehydration-induced protein, drought-induced protein, heat shock protein, LEA and protein dehydrin), detoxification signaling (i.e., glutathione S transferases, superoxide dismutase, glutathione peroxidase, respiratory burst oxidase, catalase, ascorbate peroxidases, glutathione reductase), energy production and conversion (i.e., ATP synthase beta subunit, ATP synthase delta subunit, ATP-binding protein, ATP citrate synthase, vacuolar ATP synthase subunit b, vacuolar-type H+ATPase, and vacuolar proton ATPase b subunit), signal transduction (mitogen-activated protein kinase kinase Kinase, SNF1-related protein kinase, LRR receptor-like kinase, calmodulin and calcium calmodulin-dependent protein kinase), and inorganic ion transport (Na+/H+antiporter, inorganic pyrophosphatase, transmembrane protein, plasma membrane H+-ATPase) (Dorothea and Ramanjulu 2005; Shinozaki and Yamaguchi-Shinozaki 2007) (Supplementary Table S4, Table S5, Table S6, Table S7, Table S8, Table S9, Table S10 and S11). These putative functional unigenes identified in the present study can provide clues for further investigations and valuable information for inferring the putative function of novel genes. However, detailed studies are needed to understand their precise roles and functions.
Generally, plants exhibit a large number of responses at the biological, molecular, and cellular levels under drought stress and can be broadly identified by the GO and KEGG enrichment analyses. In the present study, several drought-related biological processes were enriched in both GO and KEGG analyses. A large number of GO terms were identified in the root transcriptomes of the VCR-H2 vs. VSR-H2 and FCR-H2 vs. FSR-H2 samples, and most of them were related to abiotic stresses, e.g., binding, catalytic activity, nucleic acid-binding TF activity, transporter activity, signal transducer activity and antioxidant activity (Figs. 7, 8).
In addition to the GO analysis, a KEGG pathway enrichment analysis is an alternative approach to categorize gene functions with an emphasis on biochemical pathways. According to the KEGG analysis result, several unigenes were mapped onto the predicted metabolic pathways. Some important drought stress-related pathways represented in all combinations studied were “plant hormone signal transduction,” “flavonoid biosynthesis,” “oxidative phosphorylation,” “fatty acid biosynthesis,” “ABC transporters,” and “biosynthesis of other secondary metabolites”. The KEGG pathway analysis is a very useful tool for the prediction of potential/putative genes and their functions. These pathways were well documented to play roles in the acclimatization of plants under stress conditions. The enrichment of the stress-related GO terms and KEGG pathways among the DEGs is indicative of the improved drought-stress management in the VCR-H2 vs. VSR-H2 and FCR-H2 vs. FSR-H2 samples.
Before the occurrence of any transcriptional change in response to dehydration-related stresses, such as drought and salinity, plants first sense the signal from water deficit in the soil and then elicit their signal transduction cascades following the activation of their effector genes to accordingly adjust their metabolisms in response to the stress conditions. The perception of stress stimuli occurs through the G-protein coupled receptors (GPCR), inositol polyphosphates, or receptor-like kinases (RLKs) (Xiong et al. 2002). In this study, we identified a large number of RLK-encoding DEGs, which were largely up-regulated in the VCL-H2 vs. VSL-H2 samples; however, a substantial number of DEGs also showed down-regulation in the FCL-H2 vs. FSL-H2 comparisons (Supplementary Table S6 and Table S7).
Furthermore, we identified a large number of DEGs that participate in plant hormone regulation (especially, ABA ethylene, auxin) (Supplementary Table S6 and Table S7). In this context, ABA is a very important drought signal-sensing molecule. ABA is synthesized in roots and leaves, and its early response to water deficit in root is well-known (Schachtman and Goodger 2008). In this study, a 9-cis-epoxycarotenoid dioxygenase -encoding DEGs were identified under drought stress. These DEGs were induced in roots during water deficit, thus enhancing the synthesis of ABA. Further, we identified a large number of DEGs that encode protein phosphatases PP2C. They are the major ABA regulators, which were reported to be up-regulated in several previous studies on drought stress (Yang et al. 2015; Magalhães et al. 2016; Iovieno et al. 2016). Besides, a large number of DEGs encoding calcium-dependent protein kinase, mitogen-activated protein kinase kinases kinase, histidine kinases, and SNF1-related protein kinase were differentially regulated (Supplementary Table S6 and Table S7). A potential protein phosphatase 2C and serine/threonine-protein phosphatase 2A were also differently regulated. The phosphorylated kinases play an important role in response to drought stress by initiating phosphorylation cascades (Singh and Laxmi 2015).
Over the last years, several TF-encoding genes identified in response to various biotic/abiotic stresses in roots stimulate downstream target genes to increase stress tolerance (Shinozaki et al. 2003). Majority large number of TF DEGs annotated to different transcription factors families, including BHLH, AP2-EREBP, NAC, MYB, and WRKY were identified (Supplementary Table S8 and Table S9). A large array of transcription factors (TFs) involved in abscisic acid (ABA)-dependent and -independent pathways were found to be involved in water-deficit stress. In this study, the members of the NAC, MYB, bZIP, WRKY, and AP2/EREBP families were found to be differentially regulated under drought-stress conditions. The NAC TFs also played vital roles in stress responses (Bianchi et al. 2015) and showed up and down-regulation, predominantly at both developmental stages. MYB is a large family of TFs that is involved in response to the ABA-mediated drought stress in Arabidopsis (Abe et al. 2003), cold and drought stress in transgenic apples (Pasquali et al. 2008) and rice (Yang et al. 2012b).
Plants adapt to osmotic stress conditions by activating various mechanisms of maintaining cell membrane structure, cell turgor pressure, and water status (Delauney and Verma 1993; Seki et al. 2007). In this regard, the accumulation of molecules that function as osmotically active solutes (e.g., proline, glycine, betaine, and trehalose) during drought stress play a critical role in maintaining cellular functions as well as protein and enzyme activities (Burg and Ferraris 2008). Therefore, different enzymes involved in the metabolism of these osmolytes have also been differentially expressed in this study (Supplementary Tables S10 and S11).
Owing to its osmoprotective function, proline is considered an important component of drought tolerance (Szabados and Savoure 2010). In this study, two enzymes, Δ1 -pyrroline-5-carboxylate synthetase (P5CS1), which is involved in proline biosynthesis, and proline dehydrogenase 1 (PDH1), which is involved in catabolism, were differentially expressed. The enzyme P5CS is rate-limiting for proline biosynthesis in higher plants (Delauney and Verma 1993). Another enzyme, Δ1-pyrroline-5-carboxylate reductase (P5CR), was found to be up-regulated and was considered to play extra role in the proline biosynthetic pathway (De Ronde et al. 2000). Another osmolyte, trehalose, is an important osmolyte and signaling molecule expressed under drought stress (Avonce et al. 2004; Garg et al. 2002). The important enzymes involved in trehalose synthesis are trehalose-phosphate synthases (TPS) and trehalose-phosphate phosphatases (TPP) (Schluepmann et al. 2004). The trehalose-6-phosphate synthase (TPS) gene was largely down-regulated.
Various abiotic stresses, especially drought, lead to the overproduction of ROS in plants, eventually resulting in oxidative stress (Gill and Tuteja 2010). The major ROS scavenging enzymes include ascorbate peroxidase (APX), superoxide dismutase (SOD), mono-dehydroascorbate reductase (MDAR), dehydroascorbate reductase, catalase (CAT), glutathione-S-transferase (GST), glutathione peroxidase (GPX), and glutathione reductase (GR). The roles of detoxification enzymes in cell protection have been well-known in other plants (Sappl et al. 2009; Das and Roychoudhury 2014). The balance between the activities of these enzymes is vital for determining the steady-state level of ROS, otherwise the overproduction of ROS would be detrimental to plants because they cause DNA damage, lipid oxidation, and programmed cell death (Das and Roychoudhury 2014). In this study, a large number of DEGs encoding these enzymes, which play an important role in the detoxification of ROS, were found to be differentially expressed (Supplementary Tables S10 and S11).
Finally, a large number of DEGs identified in this study belonged to the aquaporin protein family, whose members facilitate water uptake across cell membranes in maintaining cellular water homeostasis (Javot and Maurel 2002). In this study, a large number of aquaporin proteins families showed differential expression showing up and down-regulation (Supplementary Tables S10 and S11). These proteins have been reported to differentially accumulate in drought-resistant plants (Montalvo-Hernandez et al. 2008). In addition, the LEA proteins, which are important dehydration-protective proteins with their expression levels linked to drought tolerance (Wise and Tunnacliffe 2004), showed differential expression (Supplementary Tables S10 and S11).
Furthermore, a substantial proportion of the DEGs identified in this study were novel as they could not be assigned to any database (Supplementary Tables S4 and S5). However, many of these DEGs exhibited a considerable change in their expression levels throughout the drought stress period. Therefore, these novel DEGs might regulate specific responses to drought and other stresses in the faba bean. Further analysis of these transcripts will be helpful for the better understanding of the drought tolerance mechanisms in plants.
Conclusion
The present study contributes a non-redundant set of 198,155 all-unigenes identified from the four libraries of the drought-tolerant V. faba genotype, Hassawi 2 and provides a global view of the genes expressed during drought stress. Using the llumina HiSeq 4000 platform, a total of 675.51 M raw reads were obtained from four selected libraries. A large number of SSRs identified in the present study provide a cost-effective way to further develop the functional markers for marker-assisted breeding purposes. The differential expression analysis of the RNA-seq data of the control vs. drought stress at the vegetative and flowering stages identified a large number of the significantly differentially expressed genes. Furthermore, the transcripts identified under the specific categories of the regulatory and functional proteins represent a valuable resource for faba bean crop improvement by detecting several stress-response genes and the genes involved in important metabolic pathways. Furthermore, no RNA sequencing-mediated transcriptome analysis studies have been carried out thus far on the faba bean under drought stress. This makes this study a pioneer in candidate gene identification for drought tolerance at the transcriptome level. The identified candidate genes related to drought could be used for molecular breeding and/or biotechnological approaches in the drought-sensitive genotypes of the faba bean with the aim of improving its yield.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 2 (XLSX 26326 KB)
Acknowledgements
The authors of this project in number AT-35-128 are highly appreciated the encouragement and the assistances provided by the King Abdulaziz City for Science and Technology. Simultaneously, the research team would also express their appreciation to Department of Plant Production, College of Food and Agriculture Sciences, King Saud University for their facilities and cooperation during the project period.
Author contributions
S.S.A and M.A conceptualized the project, participated in the experimental design, and assisted in drafting of the manuscript. M.A.K. conducted the experiment and assisted in the data analysis and drafted the manuscript. Q.S. and L.H. analyzed the data. S.A, H.M.M and E.H.E contributed to drafting and reviewing the manuscript. All authors read and approved the final manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interests.
References
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell. 2003;15:63–78. doi: 10.1105/tpc.006130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed M, Shehata T. Hard-to-cook phenomenon in legumes. Food Rev Int. 1992;8(2):191–221. doi: 10.1080/87559129209540938. [DOI] [Google Scholar]
- Alghamdi S. Effects of Various Water Regimes on Productivity of Some Faba Bean (Vicia faba L.) Varieties under central region of Saudi Arabia. College of Agriculture, Agriculture Research Center, Saudi Arabia. Res Bull. 2003;95:522. [Google Scholar]
- Ammar MH, Anwar F, El-Harty EH, Migdadi HM, Abdelkhalik SM, Alghamdi SS. Physiological and yield responses of faba bean (Vicia faba L.) to drought stress. J Agron Crop Sci. 2014;201(4):280–287. doi: 10.1111/jac.12112. [DOI] [Google Scholar]
- Ammar MH, Khan MA, Migdadi HM, Samah MA, Alghamdi SS. Faba bean drought responsive gene identification and validation. Saudi J Biol Sci. 2016;24:80–89. doi: 10.1016/j.sjbs.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansorge WJ. Next-generation DNA sequencing techniques. Nat Biotechnol. 2009;25:195–203. doi: 10.1016/j.nbt.2008.12.009. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, et al. Gene ontology, tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asmann YW, Klee EW, Thompson EA, Perez EA, Middha S, Oberg AL, et al. 3′ tag digital gene expression profiling of human brain and universal reference RNA using illumina genome analyzer. BMC Genom. 2009;10:531. doi: 10.1186/1471-2164-10-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res. 1997;7(10):986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- Avonce N, Leyman B, Mascorro-Gallardo JO, Van DP, Thevelein JM, Iturriaga G. The Arabidopsis trehalose-6-P synthase AtTPS1 gene is a regulator of glucose, abscisic acid, and stress signaling. Plant Physiol. 2004;136:3649–3659. doi: 10.1104/pp.104.052084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aykroyd WR, Doughty J, Walker A. Legumes in human nutrition. Food and Agricultural Organization of the United Nations (FAO), Food and Nutrition Paper. Rome: FAO; 1982. [PubMed] [Google Scholar]
- Bianchi VJ, Rubio M, Trainotti L, Verde I, Bonghi C, Martínez GP. Prunus transcription factors, breeding perspectives. Front Plant Sci. 2015;6:443. doi: 10.3389/fpls.2015.00443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond DA, Duc G (1993) Plant breeding as a means of reducing antinutritional factors in grain legumes. In, Recent advances of research in antinutritional factors in legume seeds (p. 379–396). EAAP Publication (70). Presented at 2. International workshop on antinutritional factors (ANFs) in legume seeds, Wageningen, NLD (1993-12-01–1993-12-03). Wageningen, NLD, Wageningen Academic Publishers
- Burg MB, Ferraris JD. Intracellular organic osmolytes, Function and regulation. J Biol Chem. 2008;283:7309–7313. doi: 10.1074/jbc.R700042200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock P, Fields CJ, Goto N, Heuer ML, Rice PM. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 2010;38:1767–1771. doi: 10.1093/nar/gkp1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO, a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Cubero JI. On the evolution of Vicia faba L. Theor Appl Genet. 1974;45:4751. doi: 10.1007/BF00283475. [DOI] [PubMed] [Google Scholar]
- Das K, Roychoudhury A. Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front Environ Sci. 2014;2:53. doi: 10.3389/fenvs.2014.00053. [DOI] [Google Scholar]
- De Ronde JA, Spreeth MH, Cress WA. Effect of antisense L-∆1-pyrroline-5-carboxylate reductase transgenic soybean plants subjected to osmotic and drought stress. Plant Growth Regul. 2000;32:13–26. doi: 10.1023/A:1006338911617. [DOI] [Google Scholar]
- Delauney AJ, Verma DP. Proline biosynthesis and osmoregulation. The plant j. 1993;4:215–223. doi: 10.1046/j.1365-313X.1993.04020215.x. [DOI] [Google Scholar]
- Dorothea B, Ramanjulu S. Drought and Salt Tolerance in Plants. Cr Rev Plant Sci. 2005;24:(1):23–58. doi: 10.1080/07352680590910410. [DOI] [Google Scholar]
- Duranti M, Gius C. Legume seeds, Protein content and nutritional value. Field Crops Res. 1997;53:31–45. doi: 10.1016/S0378-4290(97)00021-X. [DOI] [Google Scholar]
- El-Fiel WE, El-Tiany AH, El-Sheikh EAE. Effect of nutritional status of faba bean (Vicia faba L.) on protein solubility profiles. Food Chemist. 2002;76:219–223. doi: 10.1016/S0308-8146(00)00314-9. [DOI] [Google Scholar]
- El-Rodeny W, Kimura M, Hirakawa H, Sabah A, Shirasawa K, Sato S, Tabata S, Sasamoto S, Watanabe A, Kawashima K, et al. Development of EST-SSR markers and construction of a linkage map in faba bean (Vicia faba) Breed Sci. 2014;64:252–263. doi: 10.1270/jsbbs.64.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan X, Stegle O, Behr J, Steffen JG, Drewe P, Hildebr KL, Lyngsoe R, Schultheis SJ, Osborne EJ, Sreedharan VT, et al. Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature. 2011;477:419–423. doi: 10.1038/nature10414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg AK, Kim JK, Owens TG, Ranwala AP, Choi YD, Kochian LV, Wu RJ. Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses. PNAS. 2002;99:15898–15903. doi: 10.1073/pnas.252637799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SS, Tuteja N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem. 2010;48:909–930. doi: 10.1016/j.plaphy.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Gong YM, Xu SC, Mao WH, Hu QZ, Zhang GW, Ding J, Li ZY. Generation and characterization of 11 novel EST-derived microsatellites from Vicia faba (fabaceae) Am J Bot. 2010;97:e69–e71. doi: 10.3732/ajb.1000166. [DOI] [PubMed] [Google Scholar]
- Gong YM, Xu SH, Mao WH, Li ZY, Hu QZ, Zhang GW, Ding J. Genetic diversity analysis of faba Bean (Vicia faba L.) based on EST-SSR markers. Agric Sci China. 2011;10(6):838–844. doi: 10.1016/S1671-2927(11)60069-2. [DOI] [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, et al. De novo transcript sequence reconstruction from RNA-Seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haciseferogullari H, Gezer I, Bahtiyarca Y, Menges HO. Determination of some chemical and physical properties of sakis faba bean (Vicia faba L. var. major) J Food Eng. 2003;60:475–479. doi: 10.1016/S0260-8774(03)00075-X. [DOI] [Google Scholar]
- Hatfield JL, Sauer TJ, Prueger JH. Managing soils to achieve greater water use efficiency, a review. Agron J. 2001;93:271–280. doi: 10.2134/agronj2001.932271x. [DOI] [Google Scholar]
- Hershkovitz V, et al. De-novo assembly and characterization of the transcriptome of Metschnikowia fructicola reveals differences in gene expression following interaction with Penicillium digitatum and grapefruit peel. BMC Genom. 2013;14:168. doi: 10.1186/1471-2164-14-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshikawa K. Significance of legume crops in improving the productivity and stability of cropping systems. In: Johansen C, Lee KK, Sahrawat KL, editors. Phosphorus nutrition of grain legumes in the semi-arid tropics. India: ICRISAT; 1991. pp. 173–181. [Google Scholar]
- Iovieno P, Punzo P, Guida G, Mistretta C, Van Oosten MJ, Nurcato R, et al. Transcriptomic changes drive physiological responses to progressive drought stress and rehydration in tomato. Front Plant Sci. 2016;7:371. doi: 10.3389/fpls.2016.00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iseli C, Jongeneel CV, Bucher P (1999) EST-Scan, a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proc Int Conf Intell Syst Mol Biol 138–148 [PubMed]
- Javot H, Maurel C. The role of aquaporins in root water uptake. Ann Bot. 2002;90:301–313. doi: 10.1093/aob/mcf199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur, et al. Transcriptome sequencing of lentil based on second-generation technology permits large-scale unigene assembly and SSR marker discovery. BMC Genom. 2011;12:265. doi: 10.1186/1471-2164-12-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S, Pembleton LW, Cogan NOI, Savin KW, Leonforte T, Paull J, Materne M, Forster JW. Transcriptome sequencing of field pea and faba bean for discovery and validation of SSR genetic markers. BMC Genome. 2012;13:104. doi: 10.1186/1471-2164-13-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S, Kimber RB, Cogan NO, Materne M, Forster JW, Paull JG. SNP discovery and high-density genetic mapping in faba bean (Vicia faba L.) permits identification of QTLs for ascochyta blight resistance. Plant Sci. 2014;217–218:47–55. doi: 10.1016/j.plantsci.2013.11.014. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM, accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011;4(12):323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li DJ, Deng Z, Qin B, Liu XH, Men ZH. De novo assembly and characterization of bark transcriptome using Illumina sequencing and development of EST-SSR markers in rubber tree (Hevea brasiliensis Muell. Arg.) BMC Genome. 2012;13:192. doi: 10.1186/1471-2164-13-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using realtime quantitative PCR and the 2(– Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Luikart G, England PR, Tallmon D, Jordan S, Taberlet P. The power and promise of population genomics, from genotyping to genome typing. Nat Rev Genet. 2003;4:981–994. doi: 10.1038/nrg1226. [DOI] [PubMed] [Google Scholar]
- Magalhães AP, Verde N, Reis F, Martins I, Costa D, LinoNeto T, et al. RNA-Seq and gene network analysis uncover activation of an ABA-dependent signalosome during the cork oak root response to drought. Front Plant Sci. 2016;6:1195. doi: 10.3389/fpls.2015.01195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montalvo-Hernandez L, Piedra-Ibarra E, Gomez-Silva L, LiraCarmona R, Acosta-Gallegos JA, Vazquez-Medrano J, Xoconostle-CazaRes B, Ruiz-Medrano R. Differential accumulation of mRNAs in drought-tolerant and susceptible common bean cultivars in response to water deficit. New Phytol. 2008;77:102–113. doi: 10.1111/j.1469-8137.2007.02247.x. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-SEq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Nadal S, Suso MJ, Moreno MT. Management of Vicia faba genetic resources changes associated to the selfing process in the major, equina and minor groups. Genet Res Crop Evol. 2003;50:183–192. doi: 10.1023/A:1022944017530. [DOI] [Google Scholar]
- Ocaña S, Seoane P, Bautista R, Palomino C, Claros GM, Torres AM, et al. Large-scale transcriptome analysis in faba bean (Vicia faba L) under Ascochyta fabae Infection. PLoS One. 2015;10(8):e0135143. doi: 10.1371/journal.pone.0135143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquali G, Biricolti S, Locatelli F, Baldoni E, Mattana M. Osmyb4 expression improves adaptive responses to drought and cold stress in transgenic apples. Plant Cell Rep. 2008;27:1677–1686. doi: 10.1007/s00299-008-0587-9. [DOI] [PubMed] [Google Scholar]
- Pearce SR, Harrison G, Li D, Heslop-Harrison J, Kumar A, Flavell AJ. The Ty1-copia group retrotransposons in Vicia species, copy number, sequence heterogeneity and chromosomal location. Mol Gen Genet. 1996;250:305–315. doi: 10.1007/BF02174388. [DOI] [PubMed] [Google Scholar]
- Pertea G, Huang X, Liang F, Antonescu V, Sultana R, Karamycheva S, Lee Y, White J, Cheung F, Parvizi B, Tsai J, Quackenbush J. TIGR Gene Indices clustering tools (TGI CL), a software system for fast clustering of large EST datasets. Bioinformatics. 2003;19(5):651–652. doi: 10.1093/bioinformatics/btg034. [DOI] [PubMed] [Google Scholar]
- Pozarkova D, Koblizkova A, Roman B, Torres AM, Lucretti S, Lysak M, Dolezel J, Macas J. Development and characterization of microsatellite markers from chromosome 1-specific DNA libraries of Vicia faba. Biol Plantarum. 2002;45:337–345. doi: 10.1023/A:1016253214182. [DOI] [Google Scholar]
- Sappl PG, Carroll AJ, Clifton R, Lister R, Whelan J, Harvey Millar A, et al. The Arabidopsis glutathione transferase gene family displays complex stress regulation and co-silencing multiple genes results in altered metabolic sensitivity to oxidative stress. Plant J. 2009;58:53–68. doi: 10.1111/j.1365-313X.2008.03761.x. [DOI] [PubMed] [Google Scholar]
- Satovic Z, Avila CM, Cruz-Izquierdo S, Díaz-Ruíz R, García-Ruíz GM, Palomino C, Gutiérrez N, Vitale S, Ocaña-Moral S, Gutiérrez MV, et al. A reference consensus genetic map for molecular markers and economically important traits in faba bean (Vicia faba L.) BMC Genome. 2013;14:932. doi: 10.1186/1471-2164-14-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachtman DP, Goodger JQ. Chemical root to shoot signaling under drought. Trends Plant Sci. 2008;13:281–287. doi: 10.1016/j.tplants.2008.04.003. [DOI] [PubMed] [Google Scholar]
- Schluepmann H, van Dijken A, Aghdasi M, Wobbes B, Paul M, Smeekens S. Trehalose mediated growth inhibition of Arabidopsis seedlings is due to trehalose-6-phosphate accumulation. Plant Physiol. 2004;135:879–890. doi: 10.1104/pp.104.039503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Umezawa T, Urano K, Shinozaki K. Regulatory metabolic networks in drought stress responses. Curr Opin Plant Biol. 2007;10:296–302. doi: 10.1016/j.pbi.2007.04.014. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Gene networks involved in drought stress response and tolerance. J Exp Bot. 2007;58:221–227. doi: 10.1093/jxb/erl164. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K, Seki M. Regulatory network of gene expression in the drought and cold stress responses. Curr Opin Plant Biol. 2003;6:410–417. doi: 10.1016/S1369-5266(03)00092-X. [DOI] [PubMed] [Google Scholar]
- Singh D, Laxmi A. Transcriptional regulation of drought response, a tortuous network of transcriptional factors. Front Plant Sci. 2015;6:895. doi: 10.3389/fpls.2015.00895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suresh S, Kim TS, Raveendar S, Cho JH, Yi JY, Lee MC, Lee SY, Baek HJ, Cho GT, Chung JW. Transcriptome characterization and large-scale identification of SSR/SNP markers in symbiotic nitrogen fixation crop faba bean (Vicia faba L.) Turk J Agric For. 2015;39:459–469. doi: 10.3906/tar-1409-3. [DOI] [Google Scholar]
- Szabados L, Savoure A. Proline, a multifunctional amino acid. Trends Plant Sci. 2010;15:89–97. doi: 10.1016/j.tplants.2009.11.009. [DOI] [PubMed] [Google Scholar]
- Webb A, Cottage A, Wood T, Khamassi K, Hobbs D, Gostkiewicz K, White M, Khazaei H, Ali M, Street D, et al. A SNP-based consensus genetic map for synteny-based trait targeting in faba bean (Vicia faba L.) Plant Biotechnol J. 2016;14:177–185. doi: 10.1111/pbi.12371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise MJ, Tunnacliffe A. POPP the question, what do LEA proteins do? Trends Plant Sci. 2004;9:13–17. doi: 10.1016/j.tplants.10.012. [DOI] [PubMed] [Google Scholar]
- Xiong L, Schumaker KS, Zhu JK. Cell signaling during cold, drought, and salt stress. Plant Cell. 2002;14:S165S183. doi: 10.1105/tpc.000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Bao SY, Ford R, Jia TJ, Guan JP, He YH, Sun XL, Jiang JY, Hao JJ, Zhang XY, et al. High-throughput novel microsatellite marker of faba bean via next generation sequencing. BMC Genome. 2012;13:602. doi: 10.1186/1471-2164-13-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Dai X, Zhang WH (2012b) A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J Exp Bot err 431. 10.1093/jxb/err431 [DOI] [PMC free article] [PubMed]
- Yang S, Ha D, Song Z, Yang G, Wang L, Su Y. RNA-Seq analysis of differentially expressed genes in rice under varied nitrogen supplies. Gene. 2015;555:305–317. doi: 10.1016/j.gene.2014.11.021. [DOI] [PubMed] [Google Scholar]
- Ye J, Fang L, Zheng H, Zhang Y, Chen J, et al. WEGO, a web tool for plotting GO annotations. Nucleic Acids Res. 2006;34:293–297. doi: 10.1093/nar/gkl031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye YY, Ding YF, Jiang Q, Wang FJ, Sun JW, Zhu C. The role of receptor-like protein kinases (RLKs) in abiotic stress response in plants. Plant Cell Rep. 2017;36:235–242. doi: 10.1007/s00299-016-2084-x. [DOI] [PubMed] [Google Scholar]
- Zeid M, Mitchell S, Link W, Carter M, Nawar A, Fulton T, Kresovich S. Simple sequence repeats (SSRs) in faba bean, new loci from Orobanche-resistant cultivar ‘Giza 402’. Plant Breed. 2009;128:149–155. doi: 10.1111/j.1439-0523.2008.01584.x. [DOI] [Google Scholar]
- Zhang W, Dolan ME. Impact of the 1000 genomes project on the next wave of pharmacogenomic discovery. Pharmacogenomics. 2010;11:249e256. doi: 10.2217/pgs.09.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 2 (XLSX 26326 KB)