Abstract
Introduction
Keratoconus (KC) is a complex, genetically heterogeneous, multifactorial degenerative disorder that is accompanied by corneal ectasia which usually progresses asymmetrically. With an incidence of approximately 1 per 2000 and 2 cases per 100,000 population presenting annually, KC follows an autosomal recessive or dominant pattern of inheritance and is, apparently, associated with genes that interact with environmental, genetic, and/or other factors. This is an important consideration in refractive surgery in the case of familial KC, given the association of KC with other genetic disorders and the imbalance between dizygotic twins. The present review attempts to identify the genetic loci contributing to the different KC clinical presentations and relate them to the common genetically determined comorbidities associated with KC.
Methods
The PubMed, MEDLINE, Google Scholar, and GeneCards databases were screened for KC-related articles published in English between January 2006 and November 2017. Keyword combinations of “keratoconus,” “risk factor(s),” “genetics,” “genes,” “genetic association(s),” and “cornea” were used. In total, 217 articles were retrieved and analyzed, with greater weight placed on the more recent literature. Further bibliographic research based on the 217 articles revealed another 124 relevant articles that were included in this review. Using the reviewed literature, an attempt was made to correlate genes and genetic risk factors with KC characteristics and genetically related comorbidities associated with KC based on genome-wide association studies, family-based linkage analysis, and candidate-gene approaches.
Results
An association matrix between known KC-related genes and KC symptoms and/or clinical signs together with an association matrix between identified KC genes and genetically related KC comorbidities/syndromes were constructed.
Conclusion
Twenty-four genes were identified as potential contributors to KC and 49 KC-related comorbidities/syndromes were found. More than 85% of the known KC-related genes are involved in glaucoma, Down syndrome, connective tissue disorders, endothelial dystrophy, posterior polymorphous corneal dystrophy, and cataract.
Keywords: Keratoconus comorbidities, Keratoconus genes, Keratoconus risk factors
Introduction
Keratoconus (KC) is a relatively common [1, 2] bilateral [1, 3–21] corneal disease that is accompanied by corneal ectasia [1, 3, 4, 6, 9, 10, 13–19, 21–46] which usually progresses asymmetrically [1, 5, 9, 11–13, 15, 17–19, 30, 31, 33, 38, 39, 45]. Its main clinical manifestation is thinning and protrusion of the cornea [1, 2, 4–6, 10, 13–17, 19–22, 25–34, 36–44, 47–58], which assumes a conical shape [1, 2, 4, 13, 15–19, 27, 29–31, 33, 34, 36–40, 42, 44, 47, 48, 51, 55, 56, 58, 59]. These characteristics, even in the absence of clinically manifest KC, are also major risk factors for the condition [60, 61]; depending on the stage of the disease, KC presents with a variety of clinical manifestations and combinations of these characteristics/clinical signs [15, 55, 62]. KC is characterized by refractive errors that include myopia and irregular astigmatism [1, 3–8, 10, 13, 15–22, 25–28, 31, 32, 38–40, 42, 44–46, 50, 53, 57, 58, 63, 64], vision distortion, sensitivity to light, and multiple images [13, 31, 50, 65]. It is accompanied by a loss of visual acuity [1, 3–6, 13, 15–19, 22, 31, 35, 38–40, 44–47, 58, 66, 67] because of the distortions of the corneal curvature [1, 3–8, 10, 17–19, 21, 22, 31, 32, 38–42, 45, 46, 53, 58, 63, 64], which compromise its role in vision by distorting the refraction of light and its transmission onto the retina [1]. Corneal changes in KC also include acute corneal edema and scar formation [7, 8, 42, 45, 46], as in rare cases keratoconus presents with a central dense corneal stromal edema (hydrops) with linear oblique Descemet’s tears (ruptures in Descemet’s membrane), followed by corneal edema and scarring [68]. Finally, KC is associated with abnormal enzymatic activity within the cornea [38, 69].
Given that KC is an insidious and irreversible disease [70], it is vital to diagnose it as soon as possible [8, 19], and to treat each patient on an individual basis because of the unique nature of the disease [8, 19].
The term “ectasia” has been broadly used by ophthalmologists, vision scientists, and optometrists to characterize different conditions that affect the shape of the cornea. Keratoconus, pellucid marginal degeneration (PMD), keratoglobus, and post-refractive surgery progressive corneal ectasia are ectatic diseases, while keratoconus and PMD are different clinical presentations of the same basic disease process. In contrast, Terrien marginal degeneration, dellen, rheumatoid/autoimmune melts, and other similar conditions are corneal thinning disorders. Against this background, ectasia progression is considered to correspond to at least two of the following three changes: an increase in the steepness of the anterior corneal surface; an increase in the steepness of the posterior corneal surface; and a thinning or thickening of the cornea upon transversing it from the periphery to the thinnest point [71]. This means that there is an average difference of 75 μm in the central corneal thickness (CCT) between keratoconus patients and normal controls [34].
Method
The present review is based on a search of the PubMed, MEDLINE, Google Scholar, and GeneCards databases for articles related to KC. The keywords used were “keratoconus,” “risk factor(s),” “genetics,” “genes,” “genetic association(s),” and “cornea,” as well as all relevant/meaningful combinations of those terms. The search focused on articles written in English from January 2006 until November 2017. A total of 217 articles were identified and reviewed, and both the text and references in each paper were analyzed. The analysis revealed an additional 124 relevant articles, which were also reviewed. The review is based on previously conducted studies and does not contain any studies with human participants or animals performed by any of the authors. The aim of the review is to summarize current research into the genetics of KC. It also represents an attempt to correlate genes and genetic risk factors with KC characteristics and genetically related KC comorbidities based on genome-wide association studies, family-based linkage analysis, and candidate-gene approaches.
Demographics
The incidence of KC is approximately 1 per 2000 [1, 2, 4, 10, 32, 49, 51–53], while the annual incidence of KC is estimated at 2 per 100,000 [1, 13, 15, 18]. However, estimates of its incidence in the general population vary widely, from 1/500 to 1/2000 per year [1, 6, 29, 31, 33–35, 39]. The prevalence of KC is estimated as between 8.8 and 54.5 per 100,000 [1, 13, 15, 18, 29, 59] and between 50 and 230 per 100,000 depending on ethnicity [25, 27, 35, 40, 56, 58, 72] and the diagnostic criteria used [1, 4, 53]. Its prevalence may rise further as the use of new technologies permits better and earlier diagnosis [2, 73].
The role of ethnicity is very important; there is a sixfold increase in the incidence of KC in Asians compared with Caucasians [74, 75]; its incidence is 25/100,000 among Asians [72] but only 3,3/100,000 in Caucasians [72], while differences have been reported in the central corneal thinning (CCT) distribution between Asian and Caucasian patients [76]. Apart from the differences between Asian and Caucasian patients, the incidence of KC is greater in Indians than in Chinese or other ethnic groups [27, 75, 77]. In the UK, the prevalence of KC in Asian (Indian, Pakistani, and Bangladeshi) subjects is 4.4–7.5 times greater than that in white Caucasians, in line with the high prevalence of KC in India [72, 75, 78] and the higher prevalence and incidence of keratoconus in Asian populations in comparison to Caucasians [72, 79]. Finally, KC appears earlier and is more severe in Chinese patients [80], requiring corneal grafting at an earlier age [81].
Among various other ethnicities, the prevalence of KC ranges from 0.3 per 100,000 in Russia [78, 82] to 2300 per 100,000 in Central India (0.0003–2.3%) [53, 78]. A relatively high prevalence has been reported in Minnesota, USA (0.054%) [2, 78] and in Jerusalem [78, 83]. Although the incidence of KC in Saudi Arabia is 20/100,000 (0.02%), its incidence and severity in specific regions of Saudi Arabia (e.g., Asir Province) are high, with an early onset and more rapid progress to the severe disease stage at a young age [78, 84]. Finally, worldwide estimates range from 1/100,000 in the United Kingdom to 2/100,000 in Minnesota (USA), 2.2/100,000 in Finland, 2.5/100,000 in Holland, and 50/100,000 in New Zealand [78]. Apparently, locations with a lot of sunshine and hot weather such as India [53, 78] and the Middle East [78, 84] tend to be associated with a higher prevalence of keratoconus than locations with cooler climates and less sunshine, such as Finland [78, 85], Denmark [51, 78], Minnesota [2, 78], Japan [78, 86], and Russia [78, 82].
Gross and Histological Pathology
Frequent stimulation and/or damage to the corneal epithelium contribute to the pathogenesis of KC [87]. Several types of collagen are reduced in KC epithelium and stroma. It has been postulated that altered expression and/or activity of lysyl oxidase, which plays a critical role in the biogenesis of connective tissue, weakens covalent bonds between the collagen and elastin fibrils and leads to biomechanical deterioration of the cornea [43, 88]. This potentially accounts for abnormal microstructures detected in the keratoconus cone that result in slippage, folding, and even rupture of the anterior lamellae and to decreasing biomechanical strength during the progression of keratoconus. Furthermore, these alterations may encourage keratocyte activation and its conversion into fibroblasts, and even myofibroblasts [43, 88].
Inflammatory and apoptotic pathways are associated with KC [38, 44, 89, 90], and the resulting imbalance between the enhanced apoptosis and the proliferation of the corneal epithelial cells [91] is thought to play a vital role in the thinning of the cornea [38, 92]. Histologically, 75% of the normal corneal stroma thickness is lost [93], resulting in hypoplasia [6] and extensive corneal distortion characterized by severe decreases in the amount and distribution of corneal collagen fibers [6, 43, 54, 58]. The collagen fibrils appear morphologically normal but there is fragmentation of the epithelial basement membrane. There are membrane anomalies of the keratocytes, fibrillation, and disintegrations of Bowman’s membrane, while electron microscopy reveals degenerative changes in the basal epithelial cells [14, 88].
Another feature is Vogt’s striae, which appear as fine vertical (rarely horizontal) whitish lines in the deep stroma and Descemet’s membrane that can be observed in a slit-lamp examination; they disappear on the application of gentle pressure [1, 19, 25, 26, 56, 94, 95]. Other signs are anterior stromal scars, epithelial nebulae, and increased visibility of corneal nerves [1, 4, 6, 11, 12, 19, 42, 46, 53, 96, 97]. In addition, iron deposits are frequently observed on the basal layers of the corneal epithelium, together with breaks in the continuity of Bowman’s layer [1, 5, 6, 19, 96, 97].
In the more severe cases, acute stromal edema may develop, which causes breaks in Descemet’s membrane and leads to aqueous leakage (hydrops) [19, 56, 94] and, ultimately, corneal scarring [19, 21, 27, 42, 45, 46, 62].
A V-shaped conformation of the lower lid when the patient’s gaze is directed downwards, which is due to corneal ectasia (Munson’s sign), can contribute to the diagnosis of KC [1, 6, 24, 26]. Other clinical signs that can contribute to the diagnosis of KC are Rizzuti’s sign and Charleaux’s sign [1, 26]. Also, the scissoring of the retinoscopic reflex with a fully dilated pupil can contribute to the diagnosis of keratoconus [26, 98, 99].
Natural History
Although the onset of KC is a controversial topic, most studies agree that KC appears in the teens and early twenties and progresses until the fourth decade of life [1, 6–8, 16, 18, 19, 27, 29, 31, 35, 44, 47, 48, 56, 100–103], and that it occurs in all ethnicities [6, 10, 30, 40, 74, 104–106] without a male or female predominance [6, 30, 31, 104–106]. Other studies, however, place its onset at an average age of 39.3 years [59], while others estimate the male/female ratio at 1.7 [1]. Finally, differences between populations in the biometric parameters of the eyes are attributable to environmental and interethnic factors [107, 108].
Heritability
KC is a complex, genetically heterogeneous, multifactorial degenerative disorder [1, 109–114] with a sporadic distribution [1, 2, 104, 109, 115–117]. Reports indicate that proportions ranging from 5–10% [2, 31, 39, 104] to 8–10% [1, 29] or 14% [59], and even up to 23% of KC patients have a family history of KC. It follows an autosomal recessive or dominant pattern of inheritance [1, 35, 39, 104, 118–122]. In the autosomal dominant pattern, there are many phenotypes with incomplete penetrance [39, 123]. In other words, there appears to be a genetic predisposition for KC [39, 122, 124–126], and the genes associated with KC interact with environmental, genetic, and/or other factors [1, 6, 24, 39, 49, 67, 70, 99, 109, 120, 127–129]. This underscores the potential role of genetic factors in KC pathogenesis [72, 95, 125, 128, 130], which is an important consideration in refractive surgery to treat familial KC [130, 131], given the association of KC with other genetic disorders and the imbalance between dizygotic twins [59, 120, 127, 130, 132]. Indeed, the prevalence of KC is 3.34% in first-degree relatives of KC patients (i.e., 15–67 times higher than that for the general population [125]), while its concordance in monozygotic twins adds strong support to the notion of a genetic basis for KC [1, 124, 125, 133–135].
Another heritability-related issue is that of increased corneal curvature. The heritability of this trait has been reported to range between 60 and 95% [136–140]. The Beaver Dam Eye Study (which included 715 individuals in 185 families) estimated it at 95% [137], whereas the Danish Twin Registry estimated it at 90% based on a study of 114 pairs of twins [140]. It is apparent that an increasing corneal curvature is associated with various factors aside from heredity, including age, gender, height, ethnic background, geographical area, and environmental conditions [120, 124, 136, 141–146], and is influenced by both lifestyle and genetic factors [63, 108, 141].
Associations of KC with Different Comorbidities
The genetic etiology of KC is far from being fully clarified, and the pathophysiological processes involved are largely unknown [24, 147–150]. Multifactorial interactions of KC with environmental and genetic factors [1, 4, 6, 31, 56, 57, 104, 121, 123–125, 128, 132, 133, 148, 151–162] have already been mentioned. The pathophysiology of keratoconus is likely to include the following components: genetic disorder, biochemical disorder, biomechanical disorder, and environmental disorder [29].
KC has been associated with spring conjunctivitis [163], contact lens use, atopic disease, UV light, and eye rubbing [1, 31, 33, 104, 128, 151, 154, 156–160], while it can be triggered by trauma [164]. KC is also associated with several diseases, especially those belonging to the atopic diathesis. Atopic disorders affect up to one-third of the population in developed countries. They affect several epithelia, including the skin (atopic dermatitis, AD), the respiratory tract (asthma, allergic rhinitis), and the eye (allergic conjunctivitis). Atopic diseases, especially AD, which is typically the first clinical manifestation of atopy, have been associated with KC in several uncontrolled studies with conflicting data according to the type of patient recruitment [29]. KC is also associated with a constellation of genetic syndromes, including Down syndrome [32, 165–168], Marfan syndrome [169], osteogenesis imperfecta [170], GAPO syndrome [171], Ehlers–Danlos syndrome and its subtypes [56, 57, 172, 173], Noonan syndrome, pigmentary retinopathy [174], Leber congenital amaurosis (LCA) [57, 127, 175–178], Apert syndrome, mitral valve prolapse [1, 4, 104, 115, 124, 128, 132, 133, 148, 151–153, 155, 161, 165–167, 179, 180], congenital hip dysplasia [181], Rieger syndrome [182], focal dermal hypoplasia [183], Crouzon syndrome [184], floppy eyelid syndrome [185], monosomy X (Turner syndrome), Bardet–Biedl syndrome, nail-patella syndrome, ichthyosis, neurofibromatosis, xeroderma pigmentosum, collagenosis, neurocutaneous angiomatosis, pseudoxanthoma elasticum, retinitis pigmentosa [104, 124, 174], atopy, vernal keratoconjunctivitis [127, 165, 166, 176–180], and other noninflammatory connective tissue disorders such as joint hypermobility [1, 56, 167]. Finally, other disorders associated with KC are cataracts, Avellino corneal dystrophy, and granular corneal dystrophy, although there is a school of thought which insists that KC is not a feature of any specific syndrome but mostly an isolated ocular disorder [147, 148, 186, 187].
KC therefore can be divided into three broad categories: (a) KC associated with genetic disorders such as Down syndrome, neurofibromatosis, LCA, Turner syndrome, nail-patella syndrome, etc.; (b) KC associated with eye rubbing, mitral valve prolapse, atopy, contact lens use, LCA, and a positive family history; and (c) isolated KC without any associations [188–190].
The Role of Genetic Factors in KC
Although the genetic etiology of diseases such as age-related macular degeneration and pseudoexfoliative glaucoma is almost fully known [191], much about heritable blinding diseases such as primary open-angle glaucoma, KC, and myopia is still unknown [191]. Observations regarding KC heritability and the association of KC with a multitude of genetic syndromes have prompted a shift of focus to the genetic factors involved in KC [95, 128]. A host of genetic epidemiological data, segregation analyses, and gene mapping studies have indicated that genetic factors play a significant role [95, 122, 124–126, 128], and many gene loci and chromosomal regions associated with familial KC have been mapped by linkage analysis [104, 109, 114, 117–119, 121, 192–196], especially in Caucasians, Australians, and Chinese [51]. However, no specific genes have been identified in these regions as yet [118, 121, 192–196], and reports on differentially expressed genes in KC patients are scarce [197–200].
CCT meta-analysis of a sample of more than 20,000 individuals suggested that there are 27 CCT-associated loci [201]. Eleven SNPs showed nominal associations with KC, while 6 had significant associations after correcting for multiple testing [70]. These 11 CCT-associated loci were located 100 kb upstream [202, 203] of the ZNF469 gene (zinc finger 469 gene; MIM 612078)—the gene most strongly associated with CCT [76, 201, 204–209]. The SNPs rs12447690 and rs9938149 [201] upstream of the ZNF469 gene were also identified in Australian, UK, Croatian, Scottish, Indian, Malay, Caucasian, and Latino populations [203, 205, 208], a finding that suggests the involvement of these SNPs in CCT variation. Studies from Australia and the UK revealed two genome-wide significant signals at 16q24.2 and 13q14.1 in intragenic regions that influence the genes ZNF469 and FOX01, respectively [210]. However, sequencing analysis of the gene ZNF469 in patients with KC and high myopia showed no significant variants [211].
Genome-wide association studies (GWASs) and other studies have also associated KC with more than 60 genes/loci, although the roles of these genes/loci are inconclusive, as the reported associations were inconsistent across different study cohorts. Among them are the genes HGF (Hepatocyte Growth Factor, which has well-established effects on epithelial cells), LOX (a lysyl oxidase whose copper-dependent amine oxidase activity functions in the crosslinking of collagens and plays a significant role in collagen chain trimerization), FOXO1 (a gene of the forkhead family of transcription factors, the specific function of which is yet to be determined, but which participates in the cytokine signaling pathways in the immune system), and FNDC3B (Fibronectin Type III Domain Containing 3B). GWASs have also associated KC with more than 150 polymorphisms [70, 201, 212–214]. It should be noted, however, that this number is inflated, and a significant association with KC has only been proven for some of them [70]. Thus, GWAS identified 7 genes/loci, including the LOX, FOXO1, HGF, and FNDC3B genes and the 2q21.3, 3p26, and 19q13.3 loci [70]. Again, these associations differ from study to study, so their roles remain uncertain [201, 212–214]. Moreover, the genetic association profiles of sporadic and familial keratoconus could be different [70]. For example, GWASs of KC report that rs3735520, located upstream of the HGF gene, is associated with KC in American and Australian populations [213, 215]. In addition, a relation between HGF and refractive error has been reported in Caucasian and Han Chinese populations [216]. Other studies of KC families from Australia report an association between the LOX variants (rs10519694 and rs2956540) located at 5q23.2 and KC [196, 214]. Furthermore, there are reports that rs3735520 in the promoter of HGF and rs2956540 in LOX are associated with KC in the European population [35, 217]. In contrast, a recent meta-analysis showed that there is no significant association between KC and SNPs in ten reportedly associated genes/loci, including IL1A, IL1B, BIRC8, BHLHB2, LRRN1, KIF26B, VSX1, PPP3CA, 3q26.2, and 12p13.3 [201].
Another focus of investigations of genetic contributions to KC is their role in KC progression. This is a very complex subject because of the genetic heterogeneity of KC [195]; many of these studies focus on different chromosomal loci, have moderate sample sizes, and have difficulties in localizing regions through linkage [195]. Despite the lack of sufficient information for selecting KC susceptibility genes [212], GWASs are a very effective means for investigating KC progression [218, 219]. Two other GWASs, the Singapore Malay Eye Study (SiMES) and the Singapore Indian Eye Study (SINDI), reported two genetic regions (RXRA/COL5A1 and COL8A2) that are associated with central corneal thinning in Asians. RXRA (Retinoid X Receptor Alpha) is a protein coding gene with a well-established association with keratoconus; it participates in the apoptotic pathways in synovial fibroblasts and in organelle biogenesis and maintenance. COL5A1 encodes an alpha chain and appears to regulate the assembly of heterotypic fibers composed of both type I and type V collagen. COL8A2 encodes the alpha 2 chain of type VIII collagen; this protein is a major component of the basement membrane of the corneal endothelium and forms homo- or heterotrimers with alpha 1 (VIII)-type collagens. Defects in this gene are associated with Fuchs endothelial corneal dystrophy and posterior polymorphous corneal dystrophy (PPCD) type 2 [220]. The SiMES and SINDI studies also agreed with the results for Europeans regarding the role of ZNF469 [206] (which encodes a zinc-finger protein); it may function as a transcription factor or extranuclear regulator factor for the synthesis and/or organization of collagen fibers. Mutations in this gene cause brittle cornea syndrome.
Another GWAS, which used microarray analysis of scraped-off epithelial tissue to compare gene expression in normal corneal epithelium with that from KC patients, identified 15 keratoconus-associated SNPs from 13 loci [197–199, 221]. Other reports focus on the vital role of apoptosis in KC corneal thinning [38, 44, 200] or the associations between different SNPs and KC [24, 95, 166, 188, 201, 213, 222–230]. Meta-analysis revealed that there is an association between KC and the SNP rs4954218 located near RAB3GAP1, which encodes the catalytic subunit of the RAB3GAP enzyme (i.e., the RAB3GTPase-activating protein); this enzyme controls the exocytosis of neurotransmitters and hormones and the RAB3 cycle [231]. Alterations in RAB3GAP1 are related to the autosomal recessive Martsolf syndrome and the Warburg Micro syndrome, which are combined with ocular and neurodevelopmental dysfunctions [221, 232].
SNPs and Loci Identification
Apart from the contributions of GWASs to known KC-related gene identification, substantial contributions to the clarification of the genetics of KC have arisen from the identification of relevant SNPs and loci. Five loci with six independent SNPs associated with KC have been reported, of which two have been confirmed in relation to KC risk in an independent cohort of patients of European origin [233]: rs1324183, upstream of the MPDZ gene (Multiple PDZ Domain Crumbs Cell Polarity Complex Component, which plays a role in nonsyndromic autosomal recessive 2 hydrocephalus and in congenital communicating hydrocephalus), and rs9938149, upstream of the ZNF469 gene. It is not known if other SNPs among those found in European patients are relevant to the SNPs found in Asians [76].
Specific CCT SNPs associated with KC are rs4894535 (FNDC3B), rs1324183 (MPDZ-NF1B), rs1536482 (RXRA-COL5A1), rs7044529 (COL5A1), rs2721051 (FOXO1), and rs9938149 (BANP-ZNF469) [201]. Furthermore, linkage analysis has shown that there are six chromosomal loci for isolated KC [201, 212]: 2p24, 3p14–q13, 5q14.3–q21.1, 13q32, 16q22.3–q23.1, and 20q12. However, no other disease-related mutation was identified for these loci [53, 118, 148, 192–194]. Of those, the chromosomal regions 2p24, 3p14–q13, 5q14.3–q21.1, and 16q22.3–q23.1 were mapped by genome-wide scans of various resolutions in family studies, while the linkage between KC and locus 13q32 [148] was identified by linkage analysis. Also, heterozygous nucleotide substitutions were identified in c.2262A > C (exonic region of DOCK9), in c.2377-132A > C (intronic region of IPO5 which codes for a protein of the important beta family involved in nucleocytoplasmic transport), and in c.1053 + 29G > C (STK24), showing 100% segregation with the affected phenotype (in an Ecuadorian family) [148]. It should be noted that the STK24 gene encodes a serine/threonine protein kinase which is cleaved into two chains by caspases; the N-terminal fragment (MST3/N) translocates to the nucleus and promotes programmed cell death. Moreover, a Gln754His substitution (because of a variant c.2262A > C in DOCK9) might negatively affect the VSX1 (Visual System Homeobox 1)-coded protein function and structure [233]. Meta-analysis studies, therefore, have identified 8 SNPs in 6 genes/loci which are thought to be significant genetic markers for KC in whites, including FNDC3B rs4894535, BANP-ZNF469 rs9938149, RXRA-COL5A1 rs1536482, FOXO1 rs2721051, COL4A4 rs2228557 and rs2229813, and IMMP2L rs214884 and rs757219 [70]. They have also identified 10 genes/loci with suggestive evidence of associations with keratoconus [70].
Regarding 3p14–q13 and 5q14–q21 mentioned above, and 15q22–q24 (which is also involved in congenital cataract), a link has been established with autosomal dominant forms of KC [119, 195]. Moreover, the 2p24 locus has been linked with KC in families from Europe and the West Indies and the locus 16q22–q23 has been linked to KC in families from Finland [118, 192, 194, 196]. It should be noted that there are no references to these regions in other populations [117].
Also, affected-only linkage analyses related regions 4q31, 5q31, 9q34, 12p12, 14p11, 17q24, and 20q12 to KC [31, 188, 193, 194, 222, 234]. Of these, locus 9q34 has been linked with KC in families from Spain and locus 20q12 with KC in families from Tasmania. In addition, a copy number variation (deletion) of 5q31 has been reported in a family with autosomal dominant KC as well as in KC patients with other ocular and developmental abnormalities [235].
In another study, a large four-generation Caucasian pedigree was identified and the responsible gene was subsequently mapped to a novel 8.2 MB (megabases or million base pairs) genomic region located at 5q14.3–q21.1 that contains more than 50 known or predicted genes [35, 196]. Further genotyping of tightly spaced SNPs in the linkage region by the same group resulted in the narrowing down of the region to approximately 5mb (96mb–100mb) [35, 121], though this locus still needs confirmation. The 5q31.1–q35.3 linkage region overlaps with two of the other loci on 5q associated with KC, namely with 5q31 in Caucasian/Hispanic populations and 5q32–q33 in Southern Italian populations, supporting the possibility that the 5q31.1–q35.3 locus might actually be linked with KC [119, 236]. Similarly, SPARC (Secreted Protein Acidic and Cysteine Rich gene), which encodes a cysteine-rich acidic matrix-associated protein involved in extracellular matrix (ECM) synthesis [237] and the promotion of changes to cell shape) and LOX, which are located on 5q31.3–q32 and 5q23.2, respectively, are indicated as candidate genes for KC (linkage in familiar KC) [35, 121, 196].
Specific Genes Involved in Keratoconus
Several genes from the chromosomal regions COL6A1 (which encodes the alpha 1 subunit of type VI collagen), SOD1 (which encodes a soluble cytoplasmic protein which converts superoxide radicals to molecular oxygen and hydrogen peroxide), and COL8A1 (which encodes one of the two alpha chains of type VIII collagen, which is a major component of the basement membrane of the corneal endothelium) have been excluded as KC causal agents. Also, only a few KC patients have been reported to carry VSX1 mutations [35, 188, 193, 194, 222, 224]. VSX1 mutations cause PPCD and keratoconus (e.g., the SNP rs6050307 in a Han Chinese population) [238]. However, studies into the role of VSX1 have yielded conflicting results [149, 163, 176, 186, 188, 239–241], while there are no definitive or consistent findings about the roles of most genes [104, 118, 119, 148, 166, 180, 188, 193, 199, 200, 242, 243].
Other genes that are associated with KC are DOCK9 (Dedicator of Cytokinesis) and MIR184 (microRNA 184) [188, 228, 229, 239]. MIR184 regulates the VEGF and Akt signaling pathways, and it can inhibit corneal angiogenesis; mutations in the seed region of MIR184 cause familial keratoconus with cataract, which is known as EDICT syndrome.
The VSX1 Gene
Despite the often conflicting results of studies focusing on the role of VSX1 in KC, there are many studies that report specifically on the role of VSX1 in the pathogenesis of KC [117, 121, 148–150, 153, 166, 186, 188, 195, 226, 228, 239–241, 244–254], as well as studies focusing on both the VSX1 and SOD1 genes [166, 188, 222, 239–241, 255], which are highly conserved across many species [188–190]. Studies that failed to associate VSX1 with KC [105, 148, 149, 166, 167, 186, 188, 190, 212, 241, 244, 245, 247, 250–252, 254, 256] most likely did so because of ethnic variation, the low frequency of changes, and the multifactorial and polygenic character of KC [167]. The VSX1 gene plays a role in corneal wound repair in the corneal stroma. This involves the development of myofibroblasts by corneal stromal cells, and is evidenced by the increased expression of VSX1 during the process of wound healing and abnormal stromal repair [257].
The VSX1 gene is located within chromosome 20p11–q11 [166] and encodes a protein acting as a homeodomain transcription factor which is responsible for cone opsin expression during early ocular development and the differentiation of cells in craniofacial development [149, 249, 258–260]. Moreover, it is associated with the core of the locus region that controls the red-green visual pigment gene cluster [166]. It has also been observed that there are six transcripts and five variants which encode a truncated protein, while two of them retain the DNA-binding domain [261]. The five exons of VSX1 encode a protein consisting of 365 amino acids with a homeobox DNA-binding domain, a ceh-10 domain, and a Chx10/VSX1 domain [166, 249, 262]. The mRNA from the VSX1 gene is detected in embryonic craniofacial tissue, in the inner layer of the retina, and in the cornea [249, 257, 258]. Mutations in VSX1 trigger developmental abnormalities in retinal cells, craniofacial tissues, sella turcica, and in corneal endothelium [253].
There are 5 exons in the VSX1 gene which span 6.2 kb of the coding sequence [247, 249, 258]. Other reports mention that the exons of VSX1 span 6.7 kb of DNA on chromosome 20 with two main transcripts; transcript 1 (NM_014588) encodes protein isoform A (NP_199452.1) and transcript 2 (NM_199425) encodes protein isoform B (NP_955457) [252, 261]. Two more exons of VSX1 produce four previously unknown VSX1 transcripts [241, 249, 258, 261]. Therefore, VSX1 spans 10.65 kb of genomic DNA and has 7 exons producing 6 transcripts (via different splicing patterns). According to Genbank, these are NM_199425 (or DQ854808), NM_014588 (or DQ854807), DQ854809, DQ854810, DQ854811, and DQ854812 [241, 249, 258, 261]. Exons 2, 3, and 4 of VSX1 apparently have an increased probability of being mutated [244]. Even so, not all VSX1 mutations are causally connected to KC. Attempts to associate Q195H in VSX1 with KC were unsuccessful [153].
Variants of VSX1 such as G160D, P247R, L17P, G160V, N151S, D144E, H244R, L159M, and R166W are possibly associated with KC [153, 166, 248, 258]. For example, the G160D variant is pathogenic while P247R is not pathogenic [153, 166, 248, 258]; however, P247R seems to co-segregate with KC [188]. There is also confusion about the role of the D144E mutation in KC, as some studies mention that it is pathogenic whereas others do not [188, 248, 251, 252]. In addition, the variants H244R, L159M, and R166W are not considered sufficient to cause KC, although they have been identified in KC patients [240]. Also, the roles of mutations of H244R, L159M, and R166W in VSX1 in KC [166], as well as the role of the mutation of D144E, have been disputed [153, 186, 240, 245, 248, 251, 252, 261]. Finally, in the Korean population, two other variants, G160V and N151S, were identified but have not been demonstrated in other populations [239].
Other VSX1 variants identified in KC patients are c.432CAG (Asp144Glu), c.475T4A (Leu159Met), c.496C4T(Arg166Trp), and c.731A4G(His244Arg) [166]. Three of these, i.e., Asp144Glu, Leu159Met and Arg166Trp, as well as Pro247Arg and Gly160Asp (also in the VSX1 gene), are possibly associated with two corneal dystrophies, KC and PPCD [166, 220]. Other mutations in VSX1 such as Leu17Pro [188], the missense mutations Asn151Ser and Gly160Val, and one intragenic polymorphism are associated with KC only [239]. In addition, susceptibility to KC is evident in patients with LCA and mutations in CRB1 (which encodes a protein that localizes to the inner segment of mammalian photoreceptors), CRX (which encodes a photoreceptor-specific transcription factor that participates in the differentiation of photoreceptor cells and is essential for normal cone and rod function), and AIPL1 (which is expressed in photoreceptors and the pineal gland; it encodes aryl-hydrocarbon-interacting protein-like 1 and is involved in nuclear transport activity) [175, 178]. Note that, apart from LCA, N mutations are also associated with a severe form of retinitis pigmentosa (RP12); CRX mutations are associated with photoreceptor degeneration, LCA type III, and autosomal dominant cone-rod dystrophy 2; while AIPL1 mutations are responsible for 20% of recessive LCA.
VSX1 and PPCD
The VSX1 gene is associated with the pathogenesis of endothelial corneal dystrophy, Fuchs, and PPCD [166, 188–190, 220, 246, 254, 263–266]. PPCD is a typically bilateral hereditary corneal dystrophy characterized by abnormalities in Descemet’s membrane and the corneal endothelium, with a primarily asymmetric clinical presentation [220, 267]. PPCD is, reportedly, associated with KC [155, 220, 268, 269]; it is genotypically heterogeneous [270, 271], with one-third of the PPCD cases associated with mutations in the PPCD3 locus, in the AIPL1gene (which encodes a zinc finger transcription factor; mutations of it are associated with posterior polymorphous corneal dystrophy-3 and late-onset Fuchs endothelial corneal dystrophy) [270, 271]. PPCD is also affected by the COL4A3 gene (which encodes the collagen type IV alpha 3 chain) expression in the cornea. Similarly, mutations in COL8A2 are associated with PPCD and Fuchs [220, 266, 270, 271]. Finally, the chromosome 20p11–q11 is associated with PPCD1 (cytogenetic location 20p11.23), while VSX1 gene mutations (genetic locus VSX1; MIM#605020 and genetic locus KTCN1; MIM#148300) are involved in PPCD1 and KC, respectively [166, 220, 239, 248].
The SOD1 Gene
The SOD1 gene has a role in the pathogenesis of KC [147, 150, 166, 222], albeit without a definite association [186, 272, 273]; SOD1 mutations have also been implicated in amyotrophic lateral sclerosis [274, 275]. The SOD1 enzyme binds zinc and copper ions and destroys free superoxide radicals, thus protecting the cell from damage [147, 276, 277]. It is of note that the distribution of superoxide dismutase isoenzymes differs between the healthy cornea and the corneas of KC patients [278].
SOD1 is located on chromosome 21q22.11 and encodes superoxide dismutase enzymes [222, 274, 275]. A SOD1 variant with a seven-base deletion in intron 2 (IVS2 + 50del7 bp) is associated with KC [147, 222]; mRNA analysis showed the presence of two additional transcript splice variants coding for proteins lacking the active site of the SOD1 enzyme [222], while a deletion has been identified within intron 2 close to the 5′ splice junction of SOD1 in 3 KC patients [186, 222, 272, 273]. In addition, the skipping of SOD1 exon 2 or SOD1 exon 2 + 3 results in weak protein expression or no expression at all, since it diminishes the enzyme levels and activities [154, 186, 222, 272, 273]. It is probable that mutations on chromosome 21 may be related to the effects of oxidative stress on the cornea, while trisomy 21 is associated with an increased risk of KC [150, 255, 278].
The SOD Isoenzymes and ROS
Oxygen tension, light exposure, and high metabolic activity contribute to the production of reactive oxygen species (ROS) [277], while the human eye is particularly vulnerable to oxidative stress [277]. There are three superoxide dismutase (SOD) isoenzymes (SOD1, SOD2, and SOD3). They are compartmentalized in the mitochondrial matrix, the cytosol, and the extracellular space, and they trigger the generation of hydrogen peroxide by catalyzing the dismutation of the superoxide radicals [277]. In KC patients, these antioxidant enzymes are altered [110, 222, 278, 279], resulting in an increase in the byproducts of the nitric oxide and lipid peroxidation pathways [111, 280]. Furthermore, increased levels of reactive oxygen species (ROS) result in a decrease in mitochondrial membrane potential [281], apoptosis of corneal fibroblasts, and oxidative damage that leads to the upregulation of altered proteins, degradation of enzymes, cellular dysfunction, and DNA damage [93, 282–284].
The ZNF 469 Gene
ZNF469 is a two-exon gene that codes a 413 kDa protein consisting of 3,925 amino acid residues [285]. It is detected in the human cornea as well as in various tissues [210]. ZNF469 shows a 30% sequence similarity to the helical parts of COL1A1 (encoding the pro-alpha1 chains of type I collagen, which is abundant in the cornea), COL1A2 (encoding the pro-alpha2 chains of type I collagen, also abundant in the cornea), and COL4A1 (which encodes a type IV collagen alpha protein, an integral component of basement membranes). The COL1A1, COL1A2, and COL4A1 genes are highly expressed in the cornea [31, 210].
There are five classical C2H2 zinc finger domains (ZNFs) in the C-terminus; these are the most important parts of the ZNF469 protein. ZNFs work as sequence-specific DNA-binding motifs that regulate some specific transcription processes [207]. Although the role of ZNF469 is not well established, evidence shows that it regulates the development and maintenance of the ECM [286]. It is also possible that ZNF469 is a transcription factor or an extranuclear regulator for the synthesis and organization of the corneal collagen fibers in humans [114, 207, 286]. This organization of the collagen fibers occurs in conjunction with the gene PRDM5 (which encodes a transcription factor of the PR-domain protein family); heterozygous mutations of PRDM5 have been linked with mildly reduced CCT (480–505 μm), KC, and blue sclera [286].
Brittle Cornea Syndrome (BCS)
BCS (also known as brittle cornea syndrome 1) can lead to blindness. It is a tissue disorder characterized by thinning and fragility of the cornea that can develop even after minor trauma, and is associated with homozygous mutations in ZNF469 [120]. It is an autosomal recessive generalized connective tissue disorder associated with extreme corneal thinning to between 220 and 450 μm, leading to a high risk of corneal rupture [286, 287]. Other common features of BCS include deafness, leading to combined sensory deprivation [287] and joint hypermobility, among other features of connective tissue disorders, including scoliosis [287, 288].
There are two types of BCS. BCS type 1 (OMIM#229200) features all of the signs mentioned above and is a result of homozygous mutations in the ZNF469 gene [210]. As previously mentioned, the gene was originally mapped to chromosome 16q24 [210], while five homozygous mutations of the ZNF469 have been reported: a homozygous frameshift mutation (c.9527delG) resulting in a premature termination codon (p.Gln3178ArgfsX23) [210]; p.Gln1392X [210]; p.Phe717SerfsX14; p.Gln1757X [286]; and the homozygous missense mutation p.Cys3339Tyr [288]. BCS type 2 is an autosomal recessive condition (like BCS type 1) results from mutations in the PRMD5 gene [286]. The wide variety of mutations in ZNF469 have been reported to be linked to an increased risk of isolated KC and BCS type 1 in patients of different ethnicities (23% of patients with KC in New Zealand, 12.5% of the European KC patients from three different cohorts and 50% of Maori or Polynesian KC patients). Among the KC patients in Polynesian populations, 23% were reported to have a rare missense mutation in ZNF469 [203].
The TGFβ Pathway
The TGFβ pathway (transforming growth factor-β pathway) alters the modulation of the ECM, thus contributing to the pathogenesis of KC [289, 290]. TGFβ consists of three isoforms: TGFβ1, TGFβ2, and TGFβ3, and binds to TGFβ receptors (which also present three different isoforms) [291]. TGFβ1 (transforming growth factor beta 1) plays a role in myofibroblast transformation and proliferation, wound healing, keratocyte activation, chemotaxis, and corneal dystrophies [292]. TGFβ2 is increased in the aqueous humor of KC patients, although no elevation in TGFβ2 levels was observed in immunofluorescence studies [291].
The TGFBI Gene
Another gene that may be associated with KC is TGFBI (Transforming Growth Factor β-Induced), which encodes the protein βig-h3 that binds to type I, II, and IV collagens and plays a role in cell–collagen interactions [224]. βig-h3 is involved in the development of corneal stroma, contributing to the movement, cell adhesion, and interaction with the ECM [293]. This protein is decreased in the ECM and epithelium of KC patients, implying the involvement of the TGFBI gene in KC [293]. The role of the TGFBI gene in KC was elucidated by observing that the c.1603G4T mutation located in exon 12 of TGFBI led to the Gly535Ter substitution at the protein level in a Chinese patient with sporadic KC [224]. This result has not been replicated, and an association between TGFBI and KC is yet to be established [235].
Expression of cytokine nuclear factor κβ (NF-κβ), anti-inflammatory marker transforming growth factor β (TGF-β), interleukin 6 (IL-6), and the proinflammatory marker tumor necrosis factor α (TNF-α) increases in the corneas of KC patients [294]. TGF-β is associated with corneal dystrophies [295, 296], while there is aberrant TGF-β signaling in KC [297]. The TGF-β ligand binds to TGF-βR1, which then dimerizes with TGF-βR2 and stimulates the phosphorylation of SMAD2/3 [298]. It is then translocated to the nucleus and the transcription of genes targeted by TGF-β is activated [298]. TGF-β signaling is negatively regulated by SMAD6 and SMAD7 [298, 299]. The role of SMAD 6 and SMAD7 is to compete for the binding of receptor-regulated SMAD3, to bind histone deacetylases and inhibit the transcription of TGF-β-responsive genes, and to promote the recruitment of ubiquitin E3 ligases that cause the dissolution of TGF-β receptors [298, 299]. TGF-β1, TGF-β2, and TGF-β3 are the three isoforms of TGF-β that modulate the expression of the matrix metalloproteinase, terminal differentiation to the myofibroblast, and ECM remodeling [300, 301]. In addition, TGF-β1 and TGF-β2 contribute to the stimulation of a profibrotic response after injury [302, 303], while TGF-β3 has an antifibrotic role [304, 305]. Finally, TGF-β3 can stimulate human KC cells to assemble normal stroma [304].
The Role of Long Noncoding RNAs in the Pathophysiology of Keratoconus
Long noncoding RNAs (lncRNAs), i.e., RNAs that are at least 200 nucleotides long but do not code for proteins, participate in the complex pathophysiology of KC [306], and there are several lncRNAs that contribute to KC pathogenesis [307]. Recent surveys have accounted for more than 200,000 lncRNA transcripts in humans [308]. These are believed to be powerful regulators of transcriptomes [307], even though their function is not clear [308]. lncRNAs have been found to regulate gene expression in both pathologic and physiologic situations at the transcriptional and post-transcriptional levels [307], contributing to the cellular processes of transcription, translation, protein localization, splicing, imprinting, stem cell pluripotency, cellular structure integrity, migration, oxidative stress response, wound healing, the cell cycle, and apoptosis [309]. lncRNAs are also linked to a variety of human diseases such as cancer [306].
Microarray analyses of RNAs isolated from KC-affected epithelium and keratocytes [307] and RNA-Seq were recently used to study the expression of lncRNAs in KC and to determine the differential expression of RNAs in KC patients and normal subjects [307], as well as to assess the potential roles of lncRNAs in lncRNA–RNA duplexes [307]. These studies highlighted the role of the AQP5, lnc-WNT4-2:1, lnc-ALDH3A2-2:1, SFRP1, and CTGF genes and the WNT, TGF-β and PI3K/AKT pathways in KC [307]. In addition, many coding and noncoding RNAs that potentially contribute to KC have been identified by RNA-Seq-based analyses [307].
The lncRNA–RNA duplexes affect the metabolism of the more than 50,000 transcripts that exist [306]. Bioinformatics analysis yielded 870 lncRNAs, some of which potentially affect genes putatively associated with KC [306]. There are also genes that are differentially expressed under oxidative stress. For example, the expression of lnc-ALDH3A2-2:1 increases more than threefold under oxidative stress. The lnc-ALDH3A2-2:1 sequence overlaps with the last exon of the ALDH3A1 gene and may alter protein levels without affecting mRNA [307].
The WNT signaling pathway is essential for normal corneal development, and variants of the WNT7B and WNT10A genes have been associated with CCT and KC risk [307], while dysregulation of WNT signaling in the corneal epithelium in KC was highlighted by a recent RNA-Seq-based study [307]. Also, the expression of lnc-WNT4-2:1 (a sense transcript for its overlap with exon 5 of the WNT4 gene) is increased in KC patients [307]. Finally, significant changes in WNT4 mRNA of KC corneas apparently eliminate the role of lnc-WNT4-2:1 in regulating the WNT signaling pathway [307].
Micrornas and the Role of miRNA 184 in Keratoconus
Gene expression is also regulated by microRNAs (miRNAs), which are single-stranded noncoding RNAs [310]. miRNAs, which mediate mRNA degradation and the suppression of translation, consist of 19–25 nucleotides and bind to the 3′ untranslated regions (UTRs) of mRNAs [229, 311]. Many miRNA mutations can cause disease [114, 312, 313], making them possible therapeutic targets [229, 311, 314, 315]. In addition, miRNAs target the mRNAs from many genes that regulate the abundance of proteins in some organs and tissues [229]. The miRNA that is primarily expressed in the cornea and the lens is miRNA 184 [229, 311, 314, 316], which is localized in the endothelium, the basal and the immediate suprabasal cells of the epithelium of the cornea, but not in the limbus nor the conjunctival epithelia [229, 311, 314, 316]. miRNA 184 competitively inhibits the binding of miR205 to its mRNA [230, 313] so that miR205 cannot encode integrin beta 4 (ITGB4) and inositol polyphosphate like 1 (INPPL1), which it generally can [229, 311].
There are numerous miRNAs in specific chromosomal loci that are related to KC [223, 317]. Their target genes, MBNL (which encodes a C3H-type zinc finger protein that modulates alternative splicing of pre-mRNAs) and ZIC5 (which encodes a member of the ZIC family of C2H2-type zinc finger proteins that acts as a transcriptional repressor), are located on 13q32. MBNL2 (which encodes a C3H-type zinc finger protein that modulates alternative splicing of pre-miRNAs) is targeted by miRNA 548ab and miR5688, while ZIC5 is targeted by miR568 [223, 317]. The chromosome locus of miRNA 548 (which was identified in a white family from western Europe) is 8q13.1–q21.11. Finally, SMAD2 is also considered a target for miR568 [117, 297].
Another characteristic mutation that is found in KC patients is in the 5.5 Mb linkage region of miR184 on 15q25.1 (MIRN184 (MIM 613146)) [229]. This heterozygous mutation c.57 C > U was found in a Northern Irish family who had anterior polar cataract and KC [229]. Also, two KC patients were found to have two heterozygous mutations in the region of MIR184 (+3A > G and +8C > A) [291, 318].
KC mutations are associated with the region within the EDICT interval [119, 319]. EDICT is an autosomal dominant syndrome characterized by congenital cataract, KC with stromal thinning, iris hypoplasia, and endothelial dystrophy [291, 318]. Both the EDICT syndrome and KC are, in some families, caused by the same mutations (c.57C > T) [291, 318]. However, although the mutations in miR184 cause congenital cataract, there are differences in the changes they inflict on the cornea. The same mutation has been found in a family from Galicia in Spain whose main symptoms were severe KC, congenital cataract, and nonectatic corneal thinning [310]. Therefore, mutations in miR184 are associated with KC regardless of the presence or absence of other lens and corneal defects.
Mitochondrial DNA
Mitochondria play an important role in the cell cycle, cell signaling, differentiation, death, and growth [320], and they are a significant endogenous source of reactive oxygen species (ROS) [321]. Mitochondria carry their own genome; mitochondrial DNA (mtDNA) is inherited maternally and has a characteristic absence of recombination and a high mutation rate. Certain mtDNA polymorphisms may predispose to certain diseases [322]. Such diseases that are focused on the eye include Leber hereditary optic neuropathy (LHON) [323], type 2 diabetes [324] and Wolfram syndrome [325], nonarteritic ischemic optic neuropathy [326], chronic progressive external ophthalmoplegia, and pigmentary retinopathy [327]. Mitochondrial haplogroups H and R have been linked to an increased risk of developing KC in Saudi patients [328].
The involvement of mitochondrial abnormalities in KC is well documented [281, 329, 330]. Compared to control subjects, KC patients have significantly lower levels of leukocyte mtDNA [331]. KC corneas also have a lower mtDNA-to-nDNA ratio, as well as a higher level of mtDNA damage when compared to normal corneas [330]. The mitochondria in KC corneal tissues appear swollen under transmission electron microscopy [329].
Conclusions
The present review focused on the genetic basis of KC and its associations with different comorbidities. Data came primarily from genome-wide association studies, SNP studies, and genetic loci identification. Emphasis was placed on the most definitively implicated genes involved in KC: the VSX1 gene, which is involved in (among other conditions) posterior polymorphous corneal dystrophy; the SOD1 gene, which determines the effects of reactive oxygen species; the ZNF 469 gene, which is also involved in brittle cornea syndrome; the TGF8 pathway, which is involved in the regulation of the extracellular matrix composition; the TGFI gene, which plays a role in cell–collagen interactions; and the roles of microRNAs (especially miRNA 184), mitochondrial DNA, and reactive oxygen species in KC.
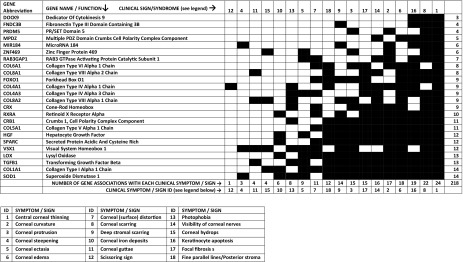
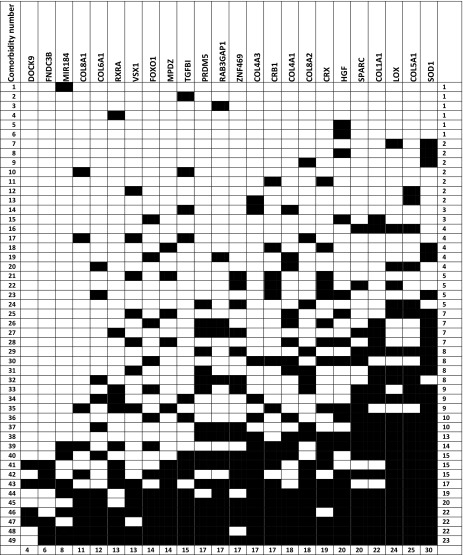
A total of 18 different KC symptoms and clinical signs were identified, documented, and cross-referenced to 24 different genes/genetic loci; each of the symptoms was associated with between 3 and 14 identified KC genes, as presented in Table 1. In addition, 49 diseases/syndromes that involve at least some of the KC-implicated genes were also identified and cross-referenced to the 24 identified KC genes, and each of those 49 diseases/syndromes was associated with between 1 and 23 identified KC genes, as presented in Tables 2 and 3.
Table 1.
Correlation matrix between specific genes implicated in keratoconus and clinical symptoms/signs
Symptoms/signs are represented according to the number of genes they correspond to, in an ascending order from top to bottom. The correspondence between symptoms/signs and their ID in this table is presented in the columns to the left of this legent
Table 2.
Correlation matrix between genes implicated in keratoconus and different keratoconus-related comorbidities
The comorbidities have been tabulated from top to bottom according to the number of genes that are implicated in both the comorbidity and Keratoconus. The numbers on the left column correspond to the different Keratoconus related comorbidities as they are listed in Table 3
Table 3.
List of keratoconus-related comorbidities
| ID# | Disease/syndrome |
|---|---|
| 1 | EDICT syndrome |
| 2 | Granular corneal dystrophy (GCD) |
| 3 | Martsolf syndrome |
| 4 | Xeroderma pigmentosum |
| 5 | Noonan syndrome |
| 6 | Vernal keratoconjunctivitis |
| 7 | Pseudoexfoliative glaucoma |
| 8 | Spring conjunctivitis |
| 9 | Atopy |
| 10 | Avellino corneal dystrophy |
| 11 | Bardet–Biedl syndrome |
| 12 | Turner syndrome (monosomy X) |
| 13 | Nail-patella syndrome |
| 14 | Ichthyosis |
| 15 | Neurofibromatosis |
| 16 | Osteogenesis imperfecta |
| 17 | Fuchs endothelial corneal dystrophy (FECD) |
| 18 | Leber hereditary optic neuropathy (LHON) |
| 19 | Warburg Micro syndrome |
| 20 | Floppy eyelid syndrome |
| 21 | Leber congenital amaurosis (LCA) |
| 22 | Pseudoxanthoma elasticum |
| 23 | Chronic progressive external ophthalmoplegia |
| 24 | Ehlers–Danlos syndrome |
| 25 | Nonsyndromic autosomal recessive 2 hydrocephalus |
| 26 | Rieger syndrome |
| 27 | Atopic disease |
| 28 | Congenital communicating hydrocephalus |
| 29 | Mitral valve prolapse |
| 30 | Pigmentary retinopathy |
| 31 | Marfan syndrome |
| 32 | Joint hypermobility |
| 33 | BCS (brittle cornea syndrome) |
| 34 | Focal dermal hypoplasia |
| 35 | Retinitis pigmentosa |
| 36 | Rheumatoid arthritis |
| 37 | Scoliosis |
| 38 | Congenital hip dysplasia |
| 39 | Age-related macular degeneration |
| 40 | Iris hypoplasia |
| 41 | Noninflammatory connective tissue disorders |
| 42 | Primary open-angle glaucoma |
| 43 | Eye rubbing |
| 44 | Cataract |
| 45 | Posterior polymorphous corneal dystrophy (PPCD) |
| 46 | Endothelial dystrophy |
| 47 | Connective tissue disorders |
| 48 | Down syndrome |
| 49 | Glaucoma |
Numbers in the left column correspond to the numbering (comorbidity identification number) in the left column of Table 2
The expectation underscoring the present review is that it will promote due vigilance by ophthalmologists to prevent KC, and will encourage medical practitioners to refer KC-related comorbidities to ophthalmologists so that they may prevent the further development of KC.
Acknowledgements
Funding
No funding or sponsorship was received for this study or the publication of this article.
Writing Assistance
The authors thank Professor Harry Whitaker, retired from the Department of Psychological Science, Northern Michigan University, for his help with the English language editing of the present text.
Authorship
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published.
Disclosures
All authors (Eleftherios Loukovitis, Konstantinos Sfakianakis, Panagiota Syrmakesi, Eleni Tsotridou, Myrsini Orfanidou, Dimitra Rafailia Bakaloudi, Maria Stoila, Athina Kozei, Spyridon Koronis, Zachos Zachariadis, Paris Tranos, Nikos Kozeis, Miltos Balidis, Zisis Gatzioufas, Aliki Fiska, and George Anogeianakis) have nothing to disclose.
Compliance with Ethics Guidelines
This article does not contain any studies with human participants or animals performed by any of the authors.
Data Availability
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
Open Access
This article is distributed under the terms of the Creative Commons Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/), which permits any noncommercial use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Footnotes
Enhanced Digital Features
To view enhanced digital features for this article go to 10.6084/m9.figshare.7000493.
References
- 1.Rabinowitz YS. Keratoconus. Surv Ophthalmol. 1998;42(4):297–319. doi: 10.1016/s0039-6257(97)00119-7. [DOI] [PubMed] [Google Scholar]
- 2.Kennedy RH, Bourne WM, Dyer JA. A 48-year clinical and epidemiologic study of keratoconus. Am J Ophthalmol. 1986;101(3):267–273. doi: 10.1016/0002-9394(86)90817-2. [DOI] [PubMed] [Google Scholar]
- 3.Ertan A, Kamburoglu G. Intacs implantation using a femtosecond laser for management of keratoconus: comparison of 306 cases in different stages. J Cataract Refract Surg. 2008;34(9):1521–1526. doi: 10.1016/j.jcrs.2008.05.028. [DOI] [PubMed] [Google Scholar]
- 4.Krachmer JH, Feder RS, Belin MW. Keratoconus and related noninflammatory corneal thinning disorders. Surv Ophthalmol. 1984;28(4):293–322. doi: 10.1016/0039-6257(84)90094-8. [DOI] [PubMed] [Google Scholar]
- 5.Vazirani J, Basu S. Keratoconus: current perspectives. Clin Ophthalmol. 2013;7:2019–2030. doi: 10.2147/OPTH.S50119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Romero-Jiménez M, Santodomingo-Rubido J, Wolffsohn JS. Keratoconus: a review. Cont Lens Anterior Eye. 2010;33(4):157–166. doi: 10.1016/j.clae.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 7.Jhanji V, Sharma N, Vajpayee RB. Management of keratoconus: current scenario. Br J Ophthalmol. 2010;95(8):1044–1050. doi: 10.1136/bjo.2010.185868. [DOI] [PubMed] [Google Scholar]
- 8.Kumar NL, Rootman DS. Newer surgical techniques in the management of keratoconus. Int Ophthalmol Clin. 2010;50(3):77–88. doi: 10.1097/IIO.0b013e3181e2464e. [DOI] [PubMed] [Google Scholar]
- 9.Orucoglu F, Toker E. A novel scoring system for distinguishing keratoconus from normal eyes. Cont Lens Anterior Eye. 2016;39(5):369–374. doi: 10.1016/j.clae.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 10.Moreira LB, Bardal RA, Crisigiovanni LR. Contact lenses fitting after intracorneal ring segments implantation in keratoconus. Arq Bras Oftalmol. 2013;76(4):215–217. doi: 10.1590/s0004-27492013000400004. [DOI] [PubMed] [Google Scholar]
- 11.Zadnik K, Steger-May K, Fink BA, Joslin CE, Nichols JJ, Rosenstiel CE, et al. Between-eye asymmetry in keratoconus. Cornea. 2002;21(7):671–679. doi: 10.1097/00003226-200210000-00008. [DOI] [PubMed] [Google Scholar]
- 12.Nichols JJ, Steger-May K, Edrington TB, Zadnik K. The relation between disease asymmetry and severity in keratoconus. Br J Ophthalmol. 2004;88(6):788–791. doi: 10.1136/bjo.2003.034520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belin MW, Duncan J, Ambrosio R, Jr, et al. A new tomographic method of grading keratoconus: the ABCD grading system. Int J Keratoconus Ectatic Corneal Dis. 2015;4(3):85–93. [Google Scholar]
- 14.Cannon DJ, Foster CS. Collagen cross linking in keratoconus. Investig Ophthalmol Vis Sci. 1978;17:63–65. [PubMed] [Google Scholar]
- 15.Duncan JK, Belin MW, Borgstrom M. Assessing progression of keratoconus: novel tomographic determinants. Eye Vis Lond. 2016;11:3–6. doi: 10.1186/s40662-016-0038-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lema I, Sobrino T, Duran JA, et al. Subclinical keratoconus and inflammatory molecules from tears. Br J Ophthalmol. 2009;93(6):820–824. doi: 10.1136/bjo.2008.144253. [DOI] [PubMed] [Google Scholar]
- 17.Lopes BT, Lopes T, Ramos IC, Faria-Correia F, Luz A, Valbon BDF, et al. Correlation of topometric and topographic indices with visual acuity in patients with KCN. Int J Keratoconus Ectatic Corneal Dis. 2012;1(3):167–172. [Google Scholar]
- 18.Mahmoud AM, Nuñez MX, Blanco C, Koch DD, Wang L, Weikert MP, et al. Expanding the cone location and magnitude index to include corneal thickness and posterior surface information for the detection of keratoconus. Am J Ophthalmol. 2013;156(6):1102–1111. doi: 10.1016/j.ajo.2013.07.018. [DOI] [PubMed] [Google Scholar]
- 19.Mannion LS, Tromans C, O’Donnell C. Reduction in corneal volume with severity of keratoconus. Curr Eye Res. 2011;36(6):522–527. doi: 10.3109/02713683.2011.553306. [DOI] [PubMed] [Google Scholar]
- 20.Pramanik S, Musch D, Sutphin J, Farjo A. Extended long-term outcomes of penetrating keratoplasty for keratoconus. Ophthalmology. 2006;113(9):1633–1638. doi: 10.1016/j.ophtha.2006.02.058. [DOI] [PubMed] [Google Scholar]
- 21.Shah S, Laiquzzaman M, Bhoiwani R, et al. Assessment of the biomechanical properties of the cornea with the ocular response analyzer in normal and keratoconic eyes. Investig Ophthalmol Vis Sci. 2007;48(7):3026–3031. doi: 10.1167/iovs.04-0694. [DOI] [PubMed] [Google Scholar]
- 22.Wojcik K, Blasiak J, Szaflik J, Szaflik J. Role of biochemical factors in the pathogenesis of keratoconus. Acta Biochim Pol. 2014;1:55–62. [PubMed] [Google Scholar]
- 23.Ghosheh FR, Cremona FA, Rapuano CJ, Cohen EJ, Ayres BD, Hammersmith KM, et al. Trends in penetrating keratoplasty in the United States 1980–2005. Int Ophthalmol. 2008;28(3):147–153. doi: 10.1007/s10792-007-9177-z. [DOI] [PubMed] [Google Scholar]
- 24.Wheeler J, Hauser MA, Afshari NA, Allingham RR, Liu Y. The genetics of keratoconus: a review. Reprod Syst Sex Disord. 2012;(Suppl 6). [DOI] [PMC free article] [PubMed]
- 25.Alió JL, Piñero DP, Alesón A, Teus MA, Barraquer RI, Murta J, et al. Keratoconus-integrated characterization considering anterior corneal aberrations, internal astigmatism, and corneal biomechanics. J Cataract Refract Surg. 2011;37(3):552–568. doi: 10.1016/j.jcrs.2010.10.046. [DOI] [PubMed] [Google Scholar]
- 26.Ambrósio R, Alonso RS, Luz A, Velarde LGC. Corneal-thickness spatial profile and corneal-volume distribution: tomographic indices to detect keratoconus. J Cataract Refract Surg. 2006;32(11):1851–1859. doi: 10.1016/j.jcrs.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 27.Barsam A, Petrushkin H, Brennan N, Bunce C, Xing W, Foot B, et al. Acute corneal hydrops in keratoconus: a national prospective study of incidence and management. Eye. 2015;29(4):469–474. doi: 10.1038/eye.2014.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bourges JL, Savoldelli M, Dighiero P, et al. Recurrence of keratoconus characteristics: a clinical and histological follow-up analysis of donor grafts. Ophthalmology. 2003;110(10):1920–1925. doi: 10.1016/S0161-6420(03)00617-1. [DOI] [PubMed] [Google Scholar]
- 29.Droitcourt C, Touboul D, Ged C, et al. A prospective study of filaggrin null mutations in keratoconus patients with or without atopic disorders. Dermatology. 2011;4:336–341. doi: 10.1159/000328408. [DOI] [PubMed] [Google Scholar]
- 30.Fontes BM, Ambrosio R, Jr, Jardim D, Velarde GC, Nosé W. Corneal biomechanical metrics and anterior segment parameters in mild keratoconus. Ophthalmology. 2010;117(4):673–679. doi: 10.1016/j.ophtha.2009.09.023. [DOI] [PubMed] [Google Scholar]
- 31.Karolak JA, Kulinska K, Nowak DM, et al. Sequence variants in COL4A1 and COL4A2 genes in Ecuadorian families with keratoconus. Mol Vis. 2011;17:827–843. [PMC free article] [PubMed] [Google Scholar]
- 32.Kenney MC, Chwa M, Atilano SR, Tran A, Carballo M, Saghizadeh M, et al. Increased levels of catalase and cathepsin V/L2 but decreased TIMP-1 in keratoconus corneas: evidence that oxidative stress plays a role in this disorder. Investig Opthalmol Vis Sci. 2005;46(3):823–832. doi: 10.1167/iovs.04-0549. [DOI] [PubMed] [Google Scholar]
- 33.Lema I, Durán JA, Ruiz C, Díez-Feijoo E, Acera A, Merayo J. Inflammatory response to contact lenses in patients with keratoconus compared with myopic subjects. Cornea. 2008;27(7):758–763. doi: 10.1097/ICO.0b013e31816a3591. [DOI] [PubMed] [Google Scholar]
- 34.Li X, Bykhovskaya Y, Canedo ALC, Haritunians T, Siscovick D, Aldave AJ, et al. Genetic association of COL5A1 variants in keratoconus patients suggests a complex connection between corneal thinning and keratoconus. Investig Opthalmol Vis Sci. 2013;54(4):2696–2704. doi: 10.1167/iovs.13-11601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li X, Bykhovskaya Y, Tang YG, Picornell Y, Haritunians T, Aldave AJ, et al. An association between the calpastatin (CAST) gene and keratoconus. Cornea. 2013;32(5):696–701. doi: 10.1097/ICO.0b013e3182821c1c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li X, Yang H, Rabinowitz YS. Keratoconus: classification scheme based on videokeratography and clinical signs. J Cataract Refract Surg. 2009;35(9):1597–1603. doi: 10.1016/j.jcrs.2009.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maeda N, Klyce SD, Smolek MK, et al. Automated keratoconus screening with corneal topography analysis. Invest Ophthalmol Vis Sci. 1994;35(6):2749–2757. [PubMed] [Google Scholar]
- 38.Matthews FJ, Cook SD, Majid MA, Dick AD, Smith VA. Changes in the balance of the tissue inhibitor of matrix metalloproteinases (TIMPs)-1 and -3 may promote keratocyte apoptosis in keratoconus. Exp Eye Res. 2007;84(6):1125–1134. doi: 10.1016/j.exer.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 39.Pathak D, Nayak B, Singh M, et al. Mitochondrial complex 1 gene analysis in keratoconus. Mol Vis. 2011;17:1514–1525. [PMC free article] [PubMed] [Google Scholar]
- 40.Piñero DP, Alió JL, Alesón A, Vergara ME, Miranda M. Corneal volume, pachymetry, and correlation of anterior and posterior corneal shape in subclinical and different stages of clinical keratoconus. J Cataract Refract Surg. 2010;36(5):814–825. doi: 10.1016/j.jcrs.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 41.Scarcelli G, Pineda R, Yun SH. Brillouin optical microscopy for corneal biomechanics. Invest Ophthalmol Vis Sci. 2012;53(1):185–190. doi: 10.1167/iovs.11-8281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shah S, Laiquzzaman M. Comparison of corneal biomechanics in pre and post-refractive surgery and keratoconic eyes by ocular response analyser. Cont Lens Anterior Eye. 2009;32(3):129–132. doi: 10.1016/j.clae.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 43.Sherwin T, Brookes NH, Loh IP, et al. Cellular incursion into Bowman’s membrane in the peripheral cone of the keratoconic cornea. Exp Eye Res. 2002;74(4):473–482. doi: 10.1006/exer.2001.1157. [DOI] [PubMed] [Google Scholar]
- 44.Sutton G, Madigan M, Roufas A, Mcavoy J. Secreted frizzled-related protein 1 (SFRP1) is highly upregulated in keratoconus epithelium: a novel finding highlighting a new potential focus for keratoconus research and treatment. Clin Exp Ophthalmol. 2010;38(1):43–48. doi: 10.1111/j.1442-9071.2009.02216.x. [DOI] [PubMed] [Google Scholar]
- 45.Suzuki M, Amano S, Honda N, et al. Longitudinal changes in corneal irregular astigmatism and visual acuity in eyes with KCN. Jpn J Ophthalmol. 2007;51(4):265–269. doi: 10.1007/s10384-007-0453-2. [DOI] [PubMed] [Google Scholar]
- 46.Wilson SE, Lin DT, Klyce SD. Corneal topography of keratoconus. Cornea. 1991;10(1):2–8. [PubMed] [Google Scholar]
- 47.Lass JH, Lembach RG, Park SB, et al. Clinical management of keratoconus. A multicenter analysis. Ophthalmology. 1990;97:433–445. doi: 10.1016/s0161-6420(90)32569-1. [DOI] [PubMed] [Google Scholar]
- 48.Crews MJ, Driebe WT, Stern GA. The clinical management of keratoconus: a 6 year retrospective study. CLAO J. 1994;20(3):194–197. doi: 10.1097/00140068-199407000-00013. [DOI] [PubMed] [Google Scholar]
- 49.Karseras AG, Ruben M. Aetiology of keratoconus. Br J Ophthalmol. 1976;60(7):522–525. doi: 10.1136/bjo.60.7.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bikbov MM, Bikbova GM, Khabibullin AF. Corneal collagen cross-linking in keratoconus management. Vestnik Oftalmologii. 2011;127:21–25. [PubMed] [Google Scholar]
- 51.Nielsen K, Hjortdal J, Nohr EA, Ehlers N. Incidence and prevalence of keratoconus in Denmark. Acta Ophthalmol Scand. 2007;85(8):890–892. doi: 10.1111/j.1600-0420.2007.00981.x. [DOI] [PubMed] [Google Scholar]
- 52.Karamichos D, Hjortdal J. Keratoconus: tissue engineering and biomaterials. J Funct Biomater. 2014;5(3):111–134. doi: 10.3390/jfb5030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jonas Jost B., Nangia Vinay, Matin Arshia, Kulkarni Maithili, Bhojwani Krishna. Prevalence and Associations of Keratoconus in Rural Maharashtra in Central India: The Central India Eye and Medical Study. American Journal of Ophthalmology. 2009;148(5):760–765. doi: 10.1016/j.ajo.2009.06.024. [DOI] [PubMed] [Google Scholar]
- 54.Warren C. Understanding keratoconus. Insight. 2008;33(4):28–29. [PubMed] [Google Scholar]
- 55.Li X, Yang H, Rabinowitz YS. Longitudinal study of keratoconus progression. Exp Eye Res. 2007;85(4):502–507. doi: 10.1016/j.exer.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Andreassen TT, Simonsen AH, Oxlund H. Biomechanical properties of keratoconus and normal corneas. Exp Eye Res. 1980;31(4):435–441. doi: 10.1016/s0014-4835(80)80027-3. [DOI] [PubMed] [Google Scholar]
- 57.Chaerkady R, Shao H, Scott S-G, Pandey A, Jun AS, Chakravarti S. The keratoconus corneal proteome: loss of epithelial integrity and stromal degeneration. J Proteom. 2013;87:122–131. doi: 10.1016/j.jprot.2013.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meek KM, Tuft SJ, Huang Y, et al. Changes in collagen orientation and distribution in keratoconus corneas. Investig Ophthalmol Vis Sci. 2005;46:1948–1956. doi: 10.1167/iovs.04-1253. [DOI] [PubMed] [Google Scholar]
- 59.Zadnik K, Barr JT, Edrington TB, Everett DF, Jameson M, McMahon TT, et al. Baseline findings in the Collaborative Longitudinal Evaluation of Keratoconus (CLEK) study. Investig Ophthalmol Vis Sci. 1998;39(13):2537–46. [PubMed]
- 60.Sherwin T, Brookes NH. Morphological changes in keratoconus: pathology or pathogenesis. Clin Exp Ophthalmol. 2004;32(2):211–217. doi: 10.1111/j.1442-9071.2004.00805.x. [DOI] [PubMed] [Google Scholar]
- 61.Cohen EJ. Keratoconus and normal-tension glaucoma: a study of the possible association with abnormal biomechanical properties as measured by corneal hysteresis. Trans Am Ophthalmol Soc. 2009;107:282–299. [PMC free article] [PubMed] [Google Scholar]
- 62.Abu-Amero KK, Helwa I, Al-Muammar A, Strickland S, Hauser MA, Allingham RR, et al. Case-control association between CCT-associated variants and keratoconus in a Saudi Arabian population. J Negat Results BioMed. 2015;14(1):10. doi: 10.1186/s12952-015-0029-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee KE. Association of age, stature, and education with ocular dimensions in an older white population. Arch Ophthalmol. 2009;127(1):88–93. doi: 10.1001/archophthalmol.2008.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dirani Mohamed, Islam Amirul, Shekar Sri N., Baird Paul N. Dominant Genetic Effects on Corneal Astigmatism: The Genes in Myopia (GEM) Twin Study. Investigative Opthalmology & Visual Science. 2008;49(4):1339. doi: 10.1167/iovs.07-1011. [DOI] [PubMed] [Google Scholar]
- 65.Maumenee IH. The eye in the Marfan syndrome. Trans Am Ophthalmol Soc. 1981;79:684–733. [PMC free article] [PubMed] [Google Scholar]
- 66.Zadnik K, Barr JT, Edrington TB, Nichols JJ, Wilson BS, Siegmund K, Gordon MO. Corneal scarring and vision in keratoconus: a baseline report from the Collaborative Longitudinal Evaluation of Keratoconus (CLEK) Study. Cornea. 2000;19(6):804–812. doi: 10.1097/00003226-200011000-00009. [DOI] [PubMed] [Google Scholar]
- 67.Davidson AE, Hayes S, Hardcastle AJ, Tuft SJ. The pathogenesis of keratoconus. Eye (Lond) 2014;28:189–195. doi: 10.1038/eye.2013.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Panikkar K, Manayath G, Rajaraman R, Saravanan V. Progressive keratoconus, retinal detachment, and intracorneal silicone oil with obsessive-compulsive eye rubbing. Oman J Ophthalmol. 2016;9(3):170. doi: 10.4103/0974-620X.192285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou L, Sawaguchi S, Twining SS, Sugar J, Feder RS, Yue BY. Expression of degradative enzymes and protease inhibitors in corneas with keratoconus. Investig Ophthalmol Vis Sci. 1998;7:1117–1124. [PubMed] [Google Scholar]
- 70.Rong SS, Ma STU, Yu XT, Ma L, Chu WK, Chan TCY, et al. Genetic associations for keratoconus: a systematic review and meta-analysis. Sci Rep. 2017;7(1):4620. doi: 10.1038/s41598-017-04393-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gomes JA, Tan D, Rapuano CJ, et al. Global consensus on keratoconus and ectatic disease. Cornea. 2015;34:359–369. doi: 10.1097/ICO.0000000000000408. [DOI] [PubMed] [Google Scholar]
- 72.Georgiou T, Funnell CL, Cassels-Brown A, O’Conor R. Influence of ethnic origin on the incidence of keratoconus and associated atopic disease in Asians and white patients. Eye Lond. 2004;18:379–383. doi: 10.1038/sj.eye.6700652. [DOI] [PubMed] [Google Scholar]
- 73.Burnside RD, Lose EJ, Domínguez MG, Sánchez-Corona J, Rivera H, Carroll AJ, et al. Molecular cytogenetic characterization of two cases with constitutional distal 11q duplication/triplication. Am J Med Genet A. 2009;149A(7):1516–1522. doi: 10.1002/ajmg.a.32906. [DOI] [PubMed] [Google Scholar]
- 74.Kok Yee Onn, Ling Tan Grace Feng, Loon Seng Chee. Review. Cornea. 2012;31(5):581–593. doi: 10.1097/ICO.0b013e31820cd61d. [DOI] [PubMed] [Google Scholar]
- 75.Pearson AR, Soneji B, Sarvananthan N, Sandford-Smith JH. Does ethnic origin influence the incidence or severity of keratoconus? Eye. 2000;14(4):625–628. doi: 10.1038/eye.2000.154. [DOI] [PubMed] [Google Scholar]
- 76.Cornes BK, Khor CC, Nongpiur ME, Xu L, Tay W-T, Zheng Y, et al. Identification of four novel variants that influence central corneal thickness in multi-ethnic Asian populations. Hum Mol Genet. 2012;21(19):437–445. doi: 10.1093/hmg/ddr463. [DOI] [PubMed] [Google Scholar]
- 77.Tay KH, Chan WK. Penetrating keratoplasty for keratoconus. Ann Acad Med Singap. 1997;26:132–137. [PubMed] [Google Scholar]
- 78.Gokhale NS. Epidemiology of keratoconus. Indian J Ophthalmol. 2013;61(8):382. doi: 10.4103/0301-4738.116054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ziaei H, Jafarinasab MR, Javadi MA, Karimian F, Poorsalman H, Mahdavi M, et al. Epidemiology of keratoconus in an Iranian population. Cornea. 2012;31(9):1044–1047. doi: 10.1097/ICO.0b013e31823f8d3c. [DOI] [PubMed] [Google Scholar]
- 80.Li SW, Li ZX, Shi WY, et al. Clinical features of 233 cases of keratoconus. Zhonghua Yan Ke Za Zhi. 2005;41(7):610–613. [PubMed] [Google Scholar]
- 81.Hao Xiao-Dan, Chen Peng, Chen Zhao-Li, Li Su-Xia, Wang Ye. Evaluating the Association between Keratoconus and Reported Genetic Loci in a Han Chinese Population. Ophthalmic Genetics. 2015;36(2):132–136. doi: 10.3109/13816810.2015.1005317. [DOI] [PubMed] [Google Scholar]
- 82.Gorskova EN, Sevost’ianov EN. Epidemiology of keratoconus in the Urals. Vestn Oftalmol. 1998;114:38–40. [PubMed] [Google Scholar]
- 83.Millodot Michel, Shneor Einat, Albou Sophie, Atlani Esther, Gordon-Shaag Ariela. Prevalence and Associated Factors of Keratoconus in Jerusalem: A Cross-sectional Study. Ophthalmic Epidemiology. 2011;18(2):91–97. doi: 10.3109/09286586.2011.560747. [DOI] [PubMed] [Google Scholar]
- 84.Assiri A A. Incidence and severity of keratoconus in Asir province, Saudi Arabia. British Journal of Ophthalmology. 2005;89(11):1403–1406. doi: 10.1136/bjo.2005.074955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ihalainen A. Clinical and epidemiological features of keratoconus genetic and external factors in the pathogenesis of the disease. Acta Ophthalmol Scand. 1986;178(Suppl):55–64. [PubMed] [Google Scholar]
- 86.Tanabe U, Fujiki K, Ogawa A, Ueda S, Kanai A. Prevalence of keratoconus patients in Japan. J Jpn Opthalmol Soc. 1985;89:407–411. [PubMed] [Google Scholar]
- 87.Lee JE, Oum BS, et al. Evaluation of differentially expressed genes identified in keratoconus. Mol Vis. 2009;15:2480–2487. [PMC free article] [PubMed] [Google Scholar]
- 88.Takahashi A, Nakayasu K, Okisaka S, et al. Quantitative analysis of collagen fiberin keratoconus. J Jpn Opthalmol Soc. 1990;94:1068–1073. [PubMed] [Google Scholar]
- 89.Lema I, Duran JA. Inflammatory molecules in the tears of patients with keratoconus. Ophthalmology. 2005;4:654–659. doi: 10.1016/j.ophtha.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 90.Wojcik KA, Kaminska A, Blasiak J, Szaflik J, Szaflik JP. Oxidative stress in the pathogenesis of keratoconus and Fuchs endothelial corneal dystrophy. Int J Mol Sci. 2013;14:19294–19308. doi: 10.3390/ijms140919294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wilson SE, He YG, Weng J, Li Q, McDowall AW, Vital M, Chwang EL. Epithelial injury induces keratocyte apoptosis: hypothesized role for the interleukin-1 system in the modulation of corneal tissue organization. Exp Eye Res. 1996;62:325–327. doi: 10.1006/exer.1996.0038. [DOI] [PubMed] [Google Scholar]
- 92.Kim W-J, Rabinowitz YS, Meisler DM, Wilson SE. Keratocyte apoptosis associated with keratoconus. Exp Eye Res. 1999;69:475–481. doi: 10.1006/exer.1999.0719. [DOI] [PubMed] [Google Scholar]
- 93.Brown DJ, Lin B, Chwa M, Atilano SR, Kim DW, Kenney MC. Elements of the nitric oxide pathway can degrade TIMP-1 and increase gelatinase activity. Mol Vis. 2004;10:281–288. [PubMed] [Google Scholar]
- 94.Gaskin JCF, Patel DV, McGhee CNJ. Acute corneal hydrops in keratoconus—new perspectives. Am J Ophthalmol. 2014;157(5):921–8. [DOI] [PubMed]
- 95.Abu-Amero KK, Al-Muammar AM, Kondkar AA. Genetics of keratoconus: where do we stand? J Ophthalmol. 2014;2014:1–11. doi: 10.1155/2014/641708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.El Dib RP, de Freitas D. A systematic review of Ferrara’s ring in the treatment of keratoconus. J Refract Surg. 2008;24(9):865–866. doi: 10.3928/1081597X-20081101-25. [DOI] [PubMed] [Google Scholar]
- 97.Ertan A, Colin J. Intracorneal rings for keratoconus and keratectasia. J Cataract Refract Surg. 2007;33(7):1303–1314. doi: 10.1016/j.jcrs.2007.02.048. [DOI] [PubMed] [Google Scholar]
- 98.Wilson SE, Verity SM, Conger DL. Accuracy and reproducibility of the corneal analysis system and topographic modeling system. Cornea. 1992;11:28–35. doi: 10.1097/00003226-199201000-00004. [DOI] [PubMed] [Google Scholar]
- 99.Bawazeer AM. Atopy and keratoconus: a multivariate analysis. Br J Ophthalmol. 2000;84(8):834–836. doi: 10.1136/bjo.84.8.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Williams KA, Hornsby NB, Bartlett CM, Holland HK, Esterman AJ, Coster DJ. The Australian Corneal Graft Registry: 2004 report. Adelaide: Snap Printing; 2004.
- 101.Ashwin PT, Mcdonnell PJ. Collagen cross-linkage: a comprehensive review and directions for future research. Br J Ophthalmol. 2009;94(8):965–970. doi: 10.1136/bjo.2009.164228. [DOI] [PubMed] [Google Scholar]
- 102.Espandar L, Meyer J. Keratoconus: overview and update on treatment. Middle East Afr J Ophthalmol. 2010;17(1):15–20. doi: 10.4103/0974-9233.61212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jones-Jordan LA, Walline JJ, Sinnott LT, Kymes SM, Zadnik K. Asymmetry in keratoconus and vision-related quality of life. Cornea. 2013;32(3):267–272. doi: 10.1097/ICO.0b013e31825697c4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rabinowitz Y. The genetics of keratoconus. Ophthalmol Clin N Am. 2003;16(4):607–620. doi: 10.1016/s0896-1549(03)00099-3. [DOI] [PubMed] [Google Scholar]
- 105.Stabuc-Silih M, Strazisar M, Ravnik-Glavac M, Hawlina M, Glavac D. Genetics and clinical characteristics of keratoconus. Acta Dermatovenerol Alp Pannonica Adriat. 2010;19:3–10. [PubMed]
- 106.Fink Barbara A, Sinnott Loraine T, Wagner Heidi, Friedman Chad, Zadnik Karla. The Influence of Gender and Hormone Status on the Severity and Progression of Keratoconus. Cornea. 2010;29(1):65–72. doi: 10.1097/ICO.0b013e3181ac0518. [DOI] [PubMed] [Google Scholar]
- 107.Wong TY, Foster PJ, Johnson GJ, Klein BE, Seah SK. The relationship between ocular dimensions and refraction with adult stature: the Tanjong Pagar Survey. Invest Ophthalmol Vis Sci. 2001;42(6):1237–42. [PubMed]
- 108.Fotedar R, Wang JJ, Burlutsky G, Morgan IG, Rose K, Wong TY, et al. Distribution of axial length and ocular biometry measured using partial coherence laser interferometry (IOL Master) in an older white population. Ophthalmology. 2010;117(3):417–423. doi: 10.1016/j.ophtha.2009.07.028. [DOI] [PubMed] [Google Scholar]
- 109.Burdon KP, Vincent AL. Insights into keratoconus from a genetic perspective. Clin Exp Optom. 2013;96(2):146–154. doi: 10.1111/cxo.12024. [DOI] [PubMed] [Google Scholar]
- 110.Chwa M, Atilano SR, Hertzog D, Zheng H, Langberg J, Kim DW, et al. Hypersensitive response to oxidative stress in keratoconus corneal fibroblasts. Investig Opthalmol Vis Sci. 2008;49(10):4361–4369. doi: 10.1167/iovs.08-1969. [DOI] [PubMed] [Google Scholar]
- 111.Buddi Rajeev, Lin Brian, Atilano Shari R., Zorapapel Nadia C., Kenney M. Cristina, Brown Donald J. Evidence of Oxidative Stress in Human Corneal Diseases. Journal of Histochemistry & Cytochemistry. 2002;50(3):341–351. doi: 10.1177/002215540205000306. [DOI] [PubMed] [Google Scholar]
- 112.Kenney MC, Brown DJ. The cascade hypothesis of keratoconus. Cont Lens Anterior Eye. 2003;26(3):139–146. doi: 10.1016/S1367-0484(03)00022-5. [DOI] [PubMed] [Google Scholar]
- 113.Shoham A, Hadziahmetovic M, Dunaief JL, Mydlarski MB, Schipper HM. Oxidative stress in diseases of the human cornea. Free Radic Biol Med. 2008;45:1047–1055. doi: 10.1016/j.freeradbiomed.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 114.Hao XD, Chen P, et al. Evaluating the association between keratoconus and reported genetic loci in a Han Chinese population. Ophthalmic Genet. 2015;36(2):132–136. doi: 10.3109/13816810.2015.1005317. [DOI] [PubMed] [Google Scholar]
- 115.Tuft SJ, Hassan H, George S, Frazer DG, Willoughby CE, Liskova P. Keratoconus in 18 pairs of twins. Acta Ophthalmol. 2012;90:482–486. doi: 10.1111/j.1755-3768.2012.02448.x. [DOI] [PubMed] [Google Scholar]
- 116.Davidson AE, Borasio E, Liskova P, Khan AO, Hassan H, Cheetham ME, et al. Brittle cornea syndrome ZNF469 mutation carrier phenotype and segregation analysis of rare ZNF469 variants in familial keratoconus. Invest Ophthalmol Vis Sci. 2015;56:578–586. doi: 10.1167/iovs.14-15792. [DOI] [PubMed] [Google Scholar]
- 117.Burdon KP, Coster DJ, Charlesworth JC, Mills RA, Laurie KJ, Giunta C, et al. Apparent autosomal dominant keratoconus in a large Australian pedigree accounted for by digenic inheritance of two novel loci. Hum Genet. 2008;124(4):379–386. doi: 10.1007/s00439-008-0555-z. [DOI] [PubMed] [Google Scholar]
- 118.Tyynismaa H, Sistonen P, Tuupanen S, Tervo T, Dammert A, Latvala T, et al. A locus for autosomal dominant keratoconus: linkage to 16q22.3-q23.1 in Finnish families. Investig Ophthalmol Vis Sci. 2002;43(10):3160–3164. [PubMed] [Google Scholar]
- 119.Hughes AE, Dash DP, Jackson AJ, Frazer DG, Silvestri G. Familial keratoconus with cataract: linkage to the long arm of chromosome 15 and exclusion of candidate genes. Investig Opthalmol Vis Sci. 2003;44(12):5063–5066. doi: 10.1167/iovs.03-0399. [DOI] [PubMed] [Google Scholar]
- 120.Synowiec E, Wójcik KA, et al. Lack of association between polymorphisms of the DNA base excision repair genes MUTYH and hOGG1 and keratoconus in a Polish subpopulation. Arch Med Sci. 2015;11(5):1101–10. [DOI] [PMC free article] [PubMed]
- 121.Bisceglia L, Bonis PD, Pizzicoli C, Fischetti L, Laborante A, Perna MD, et al. Linkage analysis in keratoconus: replication of locus 5q21.2 and identification of other suggestive loci. Investig Opthalmol Vis Sci. 2009;50(3):1081–1086. doi: 10.1167/iovs.08-2382. [DOI] [PubMed] [Google Scholar]
- 122.Owens H, Gamble G. A profile of keratoconus in New Zealand. Cornea. 2003;22(2):122–125. doi: 10.1097/00003226-200303000-00008. [DOI] [PubMed] [Google Scholar]
- 123.Falls HF. Dominantly inherited keratoconus: report of a family. J Hum Genet. 1969;17:317–324. [PubMed] [Google Scholar]
- 124.Edwards M, Mcghee CN, Dean S. The genetics of keratoconus. Clin Exp Ophthalmol. 2001;29(6):345–351. doi: 10.1046/j.1442-9071.2001.d01-16.x. [DOI] [PubMed] [Google Scholar]
- 125.Wang Y, Rabinowitz Y, Rotter JHY. Genetic epidemiological study of keratoconus: evidence for major gene determination. Am J Med Genet. 2000;93(5):403–409. [PubMed] [Google Scholar]
- 126.Forstot SL, Goldstein JH, Damiano RE, Dukes DK. Familial keratoconus. Am J Ophthalmol. 1988;105(1):92–93. doi: 10.1016/0002-9394(88)90130-4. [DOI] [PubMed] [Google Scholar]
- 127.Elder M. Leber congenital amaurosis and its association with keratoconus and keratoglobus. J Pediatr Ophthalmol Strabismus. 1994;31(1):38–40. doi: 10.3928/0191-3913-19940101-08. [DOI] [PubMed] [Google Scholar]
- 128.Gajecka M, Nowak D. The genetics of keratoconus. Middle East Afr J Ophthalmol. 2011;18(1):2–6. doi: 10.4103/0974-9233.75876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Arbab M, Tahir S, et al. TNF-α genetic predisposition and higher expression of inflammatory pathway components in keratoconus. Investig Ophthalmol Vis Sci. 2017;58(9):3481–3487. doi: 10.1167/iovs.16-21400. [DOI] [PubMed] [Google Scholar]
- 130.Karimian F, Aramesh S, Rabei HM, Javadi MA, Rafati N. Topographic evaluation of relatives of patients with keratoconus. Cornea. 2008;27(8):874–878. doi: 10.1097/ICO.0b013e31816f5edc. [DOI] [PubMed] [Google Scholar]
- 131.Buehren J, Kook D, Yoon G, Kohnen T. Detection of subclinical keratoconus by using corneal anterior and posterior surface aberrations and thickness spatial profiles. Investig Ophthalmol Vis Sci. 2010;51(7):3424–3432. doi: 10.1167/iovs.09-4960. [DOI] [PubMed] [Google Scholar]
- 132.Cullen J. F., Butler H. G. MONGOLISM (DOWN'S SYNDROME) AND KERATOCONUS. British Journal of Ophthalmology. 1963;47(6):321–330. doi: 10.1136/bjo.47.6.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bechara SJ, Waring GO, Insler MS. Keratoconus in two pairs of identical twins. Cornea. 1996;15(1):90–93. [PubMed] [Google Scholar]
- 134.Mcmahon TT, Shin JA, Newlin A, Edrington TB, Sugar J, Zadnik K. Discordance for keratoconus in two pairs of monozygotic twins. Cornea. 1999;18(4):444–451. doi: 10.1097/00003226-199907000-00010. [DOI] [PubMed] [Google Scholar]
- 135.Parker J, Ko WW, Pavlopoulos G, Wolfe PJ, Rabinowitz YS, Feldman ST. Videokeratography of keratoconus in monozygotic twins. J Refract Surg. 1996;12(1):180–183. doi: 10.3928/1081-597X-19960101-31. [DOI] [PubMed] [Google Scholar]
- 136.Han S, Chen P, Fan Q, Khor C-C, Sim X, Tay W-T, et al. Association of variants in FRAP1 and PDGFRA with corneal curvature in Asian populations from Singapore. Hum Mol Genet. 2011;20(18):3693–3698. doi: 10.1093/hmg/ddr269. [DOI] [PubMed] [Google Scholar]
- 137.Klein Alison P. Heritability Analysis of Spherical Equivalent, Axial Length, Corneal Curvature, and Anterior Chamber Depth in the Beaver Dam Eye Study. Archives of Ophthalmology. 2009;127(5):649. doi: 10.1001/archophthalmol.2009.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Biino G, Palmas MA, Corona C, Prodi D, Fanciulli M, Sulis R, et al. Ocular refraction: heritability and genome-wide search for eye morphometry traits in an isolated Sardinian population. Hum Genet. 2004;116(3):152–159. doi: 10.1007/s00439-004-1231-6. [DOI] [PubMed] [Google Scholar]
- 139.Hammond CJ, Duncan DD, Snieder H, de Lange M, West SK, Spector TD, et al. The heritability of age-related cortical cataract: the twin eye study. Investig Ophthalmol Vis Sci. 2001;42(3):601–5. [PubMed]
- 140.Lyhne N. The importance of genes and environment for ocular refraction and its determiners: a population based study among 20–45 year old twins. Br J Ophthalmol. 2001;85(12):1470–1476. doi: 10.1136/bjo.85.12.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lim Laurence Shen, Saw Seang-Mei, Jeganathan V. Swetha E., Tay Wan Ting, Aung Tin, Tong Louis, Mitchell Paul, Wong Tien Yin. Distribution and Determinants of Ocular Biometric Parameters in an Asian Population: The Singapore Malay Eye Study. Investigative Opthalmology & Visual Science. 2010;51(1):103. doi: 10.1167/iovs.09-3553. [DOI] [PubMed] [Google Scholar]
- 142.Ip JM, Huynh SC, Robaei D, Kifley A, Rose KA, Morgan IG, et al. Ethnic differences in refraction and ocular biometry in a population-based sample of 11–15-year-old Australian children. Eye. 2007;22(5):649–656. doi: 10.1038/sj.eye.6702701. [DOI] [PubMed] [Google Scholar]
- 143.Kleinstein RN. Refractive error and ethnicity in children. Arch Ophthalmol. 2003;121(8):1141–1147. doi: 10.1001/archopht.121.8.1141. [DOI] [PubMed] [Google Scholar]
- 144.Yuen Leonard H., He Mingguang, Aung Tin, Htoon Hla M., Tan Donald T., Mehta Jodhbir S. Biometry of the Cornea and Anterior Chamber in Chinese Eyes: An Anterior Segment Optical Coherence Tomography Study. Investigative Opthalmology & Visual Science. 2010;51(7):3433. doi: 10.1167/iovs.09-4307. [DOI] [PubMed] [Google Scholar]
- 145.Saw S-M, Carkeet A, Chia K-S, Stone RA, Tan DT. Component dependent risk factors for ocular parameters in Singapore Chinese children. Ophthalmology. 2002;109(11):2065–2071. doi: 10.1016/s0161-6420(02)01220-4. [DOI] [PubMed] [Google Scholar]
- 146.Wong TY, Klein R, Klein BE, Tomany SC. Refractive errors and 10-year incidence of age-related maculopathy. Investig Ophthalmol Vis Sci. 2002;43(9):2869–2873. [PubMed] [Google Scholar]
- 147.Udar N, Atilano SR, Small K, Nesburn AB, Kenney MC. SOD1 haplotypes in familial keratoconus. Cornea. 2009;28(8):902–907. doi: 10.1097/ICO.0b013e3181983a0c. [DOI] [PubMed] [Google Scholar]
- 148.Gajecka Marzena, Radhakrishna Uppala, Winters Daniel, Nath Swapan K., Rydzanicz Malgorzata, Ratnamala Uppala, Ewing Kimberly, Molinari Andrea, Pitarque Jose A., Lee Kwanghyuk, Leal Suzanne M., Bejjani Bassem A. Localization of a Gene for Keratoconus to a 5.6-Mb Interval on 13q32. Investigative Opthalmology & Visual Science. 2009;50(4):1531. doi: 10.1167/iovs.08-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Tanwar M, Kumar M, Nayak B, et al. VSX1 gene analysis in keratoconus. Mol Vis. 2010;16:2395–2401. [PMC free article] [PubMed] [Google Scholar]
- 150.De Bonis P, Laborante A, Pizzicoli C, et al. Mutational screening of VSX1, SPARC, SOD1, LOX, and TIMP3 in keratoconus. Mol Vis. 2011;17:2482–2494. [PMC free article] [PubMed] [Google Scholar]
- 151.Ambekar R, Toussaint KC, Jr, Johnson AW. The effect of keratoconus on the structural, mechanical, and optical properties of the cornea. J Mech Behav Biomed Mater. 2011;4(3):223–236. doi: 10.1016/j.jmbbm.2010.09.014. [DOI] [PubMed] [Google Scholar]
- 152.Saee-Rad S, Raoofian R, Mahbod M, et al. Analysis of superoxide dismutase 1, dual-specificity phosphatase 1, and transforming growth factor, beta 1 genes expression in keratoconic and non-keratoconic corneas. Mol Vis. 2013;19:2501–2507. [PMC free article] [PubMed] [Google Scholar]
- 153.Paliwal P, Singh A, Tandon R, Titiyal JS, Sharma A. A novel VSX1 mutation identified in an individual with keratoconus in India. Mol Vis. 2009;15:2475–2479. [PMC free article] [PubMed] [Google Scholar]
- 154.Arnal E, Peris-Martínez C, Menezo JL, Johnsen-Soriano S, Romero FJ. Oxidative stress in keratoconus? Investig Opthalmol Vis Sci. 2011;52(12):8592–8597. doi: 10.1167/iovs.11-7732. [DOI] [PubMed] [Google Scholar]
- 155.Cremona FA, Ghosheh FR, Rapuano CJ, Eagle RC, Hammersmith KM, Laibson PR, et al. Keratoconus associated with other corneal dystrophies. Cornea. 2009;28(2):127–135. doi: 10.1097/ICO.0b013e3181859935. [DOI] [PubMed] [Google Scholar]
- 156.Mcmonnies CW. Mechanisms of rubbing-related corneal trauma in keratoconus. Cornea. 2009;28(6):607–615. doi: 10.1097/ICO.0b013e318198384f. [DOI] [PubMed] [Google Scholar]
- 157.Lindsay RG, Bruce AS, Gutteridge IF. Keratoconus associated with continual eye rubbing due to punctal agenesis. Cornea. 2000;19(4):567–569. doi: 10.1097/00003226-200007000-00034. [DOI] [PubMed] [Google Scholar]
- 158.Goldich Y, Marcovich AL, Barkana Y, Mandel Y, Hirsh A, Morad Y, et al. Clinical and corneal biomechanical changes after collagen cross-linking with riboflavin and UV irradiation in patients with progressive keratoconus. Cornea. 2012;31(6):609–614. doi: 10.1097/ICO.0b013e318226bf4a. [DOI] [PubMed] [Google Scholar]
- 159.Jafri B, Lichter H, Stulting RD. Asymmetric keratoconus attributed to eye rubbing. Cornea. 2004;23(6):560–564. doi: 10.1097/01.ico.0000121711.58571.8d. [DOI] [PubMed] [Google Scholar]
- 160.Barr Joseph T, Wilson Brad S, Gordon Mae O, Rah Marjorie J, Riley Colleen, Kollbaum Pete S, Zadnik Karla. Estimation of the Incidence and Factors Predictive of Corneal Scarring in the Collaborative Longitudinal Evaluation of Keratoconus (CLEK) Study. Cornea. 2006;25(1):16–25. doi: 10.1097/01.ico.0000164831.87593.08. [DOI] [PubMed] [Google Scholar]
- 161.Grünauer-Kloevekorn C, Duncker GI. Keratoconus: epidemiology, risk factors and diagnosis. Klin Monabl Augenheilkd. 2006;223(6):493–502. doi: 10.1055/s-2005-859021. [DOI] [PubMed] [Google Scholar]
- 162.Francois M, Chassaing N, Calvas P. Chapter 2: Genetics of keratoconus. In: Barbara A, editor. Textbook on keratoconus: new insights. New Delhi: Jaypee Brothers Medical Publishers Pvt.; 2012. p. 12–17.
- 163.Cosar CB, Sridhar MS, Cohen EJ, Held EL, Alvim Pde T, Rapuano CJ, et al. Indications for penetrating keratoplasty and associated procedures, 1996–2000. Cornea. 2002;21(2):148–151. doi: 10.1097/00003226-200203000-00003. [DOI] [PubMed] [Google Scholar]
- 164.Bareja U, Vajpayee RB. Posterior keratoconus due to iron nail injury—a case report. Indian J Ophthalmol. 1991;39(1):30. [PubMed]
- 165.Sharma Reena, Titiyal Jeewan S, Prakash Gaurav, Sharma Namrata, Tandon Radhika, Vajpayee Rasik B. Clinical Profile and Risk Factors for Keratoplasty and Development of Hydrops in North Indian Patients With Keratoconus. Cornea. 2009;28(4):367–370. doi: 10.1097/ICO.0b013e31818cd077. [DOI] [PubMed] [Google Scholar]
- 166.Heon E. VSX1: a gene for posterior polymorphous dystrophy and keratoconus. Hum Mol Genet. 2002;11(9):1029–1036. doi: 10.1093/hmg/11.9.1029. [DOI] [PubMed] [Google Scholar]
- 167.McGhee CN. 2008 Sir Norman McAlister Gregg Lecture: 150 years of practical observations on the conical cornea—what have we learned? Clin Exp Ophthalmol. 2009;37(2):160–76. [DOI] [PubMed]
- 168.Shapiro Michael B., France Thomas D. The Ocular Features of Down's Syndrome. American Journal of Ophthalmology. 1985;99(6):659–663. doi: 10.1016/s0002-9394(14)76031-3. [DOI] [PubMed] [Google Scholar]
- 169.Austin MG, Schaefer RF. Marfan’s syndrome, with unusual blood vessel manifestations. AMA Arch Pathol. 1957;64:205–209. [PubMed] [Google Scholar]
- 170.Beckh U, Schonherr U, Naumann GO. Autosomal dominant keratoconus as the chief ocular symptom in Lobstein osteogenesis imperfecta tarda (in German) Klin Monbl Augenheilkd. 1995;206(04):268–272. doi: 10.1055/s-2008-1035438. [DOI] [PubMed] [Google Scholar]
- 171.Wajntal A, Koiffmann CP, Mendonça BB, Epps-Quaglia D, Sotto MN, Rati PBM, et al. GAPO syndrome (McKusick 23074)—a connective tissue disorder: report on two affected sibs and on the pathologic findings in the older. Am J Med Genet. 1990;37(2):213–223. doi: 10.1002/ajmg.1320370210. [DOI] [PubMed] [Google Scholar]
- 172.Kuming BS, Joffe L. Ehlers–Danlos syndrome associated with keratoconus: a case report. S Afr Med J. 1977;52(10):403–405. [PubMed] [Google Scholar]
- 173.Robertson I. Keratoconus and the Ehlers–Danlos syndrome: a new aspect of keratoconus. Med J Aust. 1975;1:571–573. doi: 10.5694/j.1326-5377.1975.tb111590.x. [DOI] [PubMed] [Google Scholar]
- 174.Freedman J, Gombos GM. Bilateral macular coloboma, keratoconus, and retinitis pigmentosa. Ann Ophthalmol. 1971;3:664–665. [PubMed] [Google Scholar]
- 175.McMahon Timothy T., Kim Linda S., Fishman Gerald A., Stone Edwin M., Zhao Xinping C., Yee Richard W., Malicki Jarema. CRB1Gene Mutations Are Associated with Keratoconus in Patients with Leber Congenital Amaurosis. Investigative Opthalmology & Visual Science. 2009;50(7):3185. doi: 10.1167/iovs.08-2886. [DOI] [PubMed] [Google Scholar]
- 176.Totan Y, Hep B, Cekic O, Gündüz A, Aydin E. Incidence of keratoconus in subjects with vernal keratoconjunctivitis: a videokeratographic study. Ophthalmology. 2001;108:824–827. doi: 10.1016/s0161-6420(00)00664-3. [DOI] [PubMed] [Google Scholar]
- 177.Stoiber Josef, Muss Wolfgang H., Ruckhofer Josef, Thaller-Antlanger Helga, Alzner Egon, Grabner Günther. Recurrent Keratoconus in a Patient with Leber Congenital Amaurosis. Cornea. 2000;19(3):395–398. doi: 10.1097/00003226-200005000-00028. [DOI] [PubMed] [Google Scholar]
- 178.Dharmaraj Sharola. The Phenotype of Leber Congenital Amaurosis in Patients With AIPL1 Mutations. Archives of Ophthalmology. 2004;122(7):1029. doi: 10.1001/archopht.122.7.1029. [DOI] [PubMed] [Google Scholar]
- 179.Wollensak G, Green WR, Temprano J. Keratoconus associated with corneal granular dystrophy in a patient of Italian origin. Cornea. 2002;21(1):121–122. doi: 10.1097/00003226-200201000-00026. [DOI] [PubMed] [Google Scholar]
- 180.Igarashi S. Association of keratoconus and Avellino corneal dystrophy. Br J Ophthalmol. 2003;87(3):367–368. doi: 10.1136/bjo.87.3.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Nucci P, Brancato R. Keratoconus and congenital hip dysplasia. Am J Ophthalmol. 1991;111(6):775–776. doi: 10.1016/s0002-9394(14)76792-3. [DOI] [PubMed] [Google Scholar]
- 182.Greenfield G, Stein R, Romano A, Goodman RM. Blue sclerae and keratoconus: key features of a distinct heritable disorder of connective tissue. Clin Genet. 1973;4:8–16. doi: 10.1111/j.1399-0004.1973.tb01115.x. [DOI] [PubMed] [Google Scholar]
- 183.Zala L, Ettlin C, Krebs A. Focal dermal hypoplasia with keratoconus, papillomatosis of esophagus and hidrocystomas (author’s transl) Dermatologica. 1975;150(3):176–185. [PubMed] [Google Scholar]
- 184.Perlman Jeffrey M., Zaidman Gerald W. Bilateral Keratoconus in Crouzon??s Syndrome. Cornea. 1994;13(1):80–81. doi: 10.1097/00003226-199401000-00014. [DOI] [PubMed] [Google Scholar]
- 185.Lee WJ, Kim JC, Shyn KH. Clinical evaluation of corneal diseases associated with floppy eyelid syndrome. Korean J Ophthalmol. 1996;10(2):116–121. doi: 10.3341/kjo.1996.10.2.116. [DOI] [PubMed] [Google Scholar]
- 186.Stabuc-Silih M, Strazisar M, Hawlina M, et al. Absence of pathogenic mutations in VSX1 and SOD1 genes in patients with keratoconus. Cornea. 2010;29(2):172–176. doi: 10.1097/ICO.0b013e3181aebf7a. [DOI] [PubMed] [Google Scholar]
- 187.Abu-Amero KK, Hellani AM, Al Mansouri SM, Kalantan H, Al Muammar A-M. High-resolution analysis of DNA copy number alterations in patients with isolated sporadic keratoconus. Mol Vis. 2011;17:822–826. [PMC free article] [PubMed] [Google Scholar]
- 188.Bisceglia Luigi, Ciaschetti Marilena, De Bonis Patrizia, Campo Pablo Alberto Perafan, Pizzicoli Costantina, Scala Costanza, Grifa Michele, Ciavarella Pio, Noci Nicola Delle, Vaira Filippo, Macaluso Claudio, Zelante Leopoldo. VSX1Mutational Analysis in a Series of Italian Patients Affected by Keratoconus: Detection of a Novel Mutation. Investigative Opthalmology & Visual Science. 2005;46(1):39. doi: 10.1167/iovs.04-0533. [DOI] [PubMed] [Google Scholar]
- 189.Burmeister M, Novak J, Liang M-Y, Basu S, Ploder L, Hawes NL, et al. Ocular retardation mouse caused by Chx10 homeobox null allele: impaired retinal progenitor proliferation and bipolar cell differentiation. Nat Genet. 1996;12(4):376–384. doi: 10.1038/ng0496-376. [DOI] [PubMed] [Google Scholar]
- 190.Chow RL, Snow B, Novak J, Looser J, Freund C, Vidgen D, et al. Vsx1, a rapidly evolving paired -like homeobox gene expressed in cone bipolar cells. Mech Dev. 2001;109(2):315–322. doi: 10.1016/s0925-4773(01)00585-8. [DOI] [PubMed] [Google Scholar]
- 191.Sherwin JC, Hewitt AW, Ruddle JB, Mackey DA. Genetic isolates in ophthalmic diseases. Ophthalmic Genet. 2008;29(4):149–161. doi: 10.1080/13816810802334341. [DOI] [PubMed] [Google Scholar]
- 192.Hutchings H. Identification of a new locus for isolated familial keratoconus at 2p24. J Med Genet. 2005;42(1):88–94. doi: 10.1136/jmg.2004.022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Brancati F. A locus for autosomal dominant keratoconus maps to human chromosome 3p14-q13. J Med Genet. 2004;41(3):188–192. doi: 10.1136/jmg.2003.012872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Fullerton J, Paprocki P, Foote S, Mackey DA, Williamson R, Forrest S. Identity-by-descent approach to gene localisation in eight individuals affected by keratoconus from north-west Tasmania, Australia. Hum Genet. 2002;110(5):462–470. doi: 10.1007/s00439-002-0705-7. [DOI] [PubMed] [Google Scholar]
- 195.Tang YG, Rabinowitz YS, Taylor KD, Li X, Hu M, Picornell Y, et al. Genomewide linkage scan in a multigeneration Caucasian pedigree identifies a novel locus for keratoconus on chromosome 5q14.3-q21.1. Genet Med. 2005;7(6):397–405. doi: 10.1097/01.gim.0000170772.41860.54. [DOI] [PubMed] [Google Scholar]
- 196.Li X, Rabinowitz YS, Tang YG, Picornell Y, Taylor KD, Hu M, et al. Two-stage genome-wide linkage scan in keratoconus sib pair families. Investig Opthalmol Vis Sci. 2006;47(9):3791–3795. doi: 10.1167/iovs.06-0214. [DOI] [PubMed] [Google Scholar]
- 197.Nielsen K, Heegaard S, Vorum H, Birkenkamp-Demtröder K, Ehlers N, Orntoft TF. Altered expression of CLC, DSG3, EMP3, S100A2, and SLPI in corneal epithelium from keratoconus patients. Cornea. 2005;24(6):661–668. doi: 10.1097/01.ico.0000153556.59407.69. [DOI] [PubMed] [Google Scholar]
- 198.Nielsen K, Vorum H, Fagerholm P, Birkenkamp-Demtröder K, Honoré B, Ehlers N, et al. Proteome profiling of corneal epithelium and identification of marker proteins for keratoconus, a pilot study. Exp Eye Res. 2006;82(2):201–209. doi: 10.1016/j.exer.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 199.Nielsen K, Birkenkamp-Demtröder K, Ehlers N, Orntoft TF. Identification of differentially expressed genes in keratoconus epithelium analyzed on microarrays. Investig Opthalmol Vis Sci. 2003;44(6):2466–2476. doi: 10.1167/iovs.02-0671. [DOI] [PubMed] [Google Scholar]
- 200.Ha NT, Nakayasu K, Murakami A, Ishidoh K, Kanai A. Microarray analysis identified differentially expressed genes in keratocytes from keratoconus patients. Curr Eye Res. 2004;28(6):373–379. doi: 10.1080/02713680490502201. [DOI] [PubMed] [Google Scholar]
- 201.Lu Y, Vitart V, Burdon KP, Khor CC, Bykhovskaya Y, Mirshahi A, et al. Genome-wide association analyses identify multiple loci associated with central corneal thickness and keratoconus. Nat Genet. 2013;45(2):155–163. doi: 10.1038/ng.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Hamilton J. B. THE SIGNIFICANCE OF HEREDITY IN OPHTHALMOLOGY. PRELIMINARY SURVEY OF HEREDITARY EYE DISEASES IN TASMANIA . British Journal of Ophthalmology. 1938;22(2):83–108. doi: 10.1136/bjo.22.2.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Vincent AL, Jordan CA, Cadzow MJ, Merriman TR, Mcghee CN. Mutations in the zinc finger protein gene, ZNF469, contribute to the pathogenesis of keratoconus. Investig Opthalmol Vis Sci. 2014;55(9):5629–5635. doi: 10.1167/iovs.14-14532. [DOI] [PubMed] [Google Scholar]
- 204.Gordon MO. The ocular hypertension treatment study. Arch Ophthalmol. 2002;120(6):714–720. doi: 10.1001/archopht.120.6.714. [DOI] [PubMed] [Google Scholar]
- 205.Vitart V, Benčić G, Hayward C, Herman JŠ, Huffman J, Campbell S, et al. New loci associated with central cornea thickness include COL5A1, AKAP13 and AVGR8. Hum Mol Genet. 2010;19(21):4304–4311. doi: 10.1093/hmg/ddq349. [DOI] [PubMed] [Google Scholar]
- 206.Vithana EN, Aung T, Khor CC, Cornes BK, Tay WT, Sim X, et al. Collagen-related genes influence the glaucoma risk factor, central corneal thickness. Hum Mol Genet. 2011;20(4):649–658. doi: 10.1093/hmg/ddq511. [DOI] [PubMed] [Google Scholar]
- 207.Lu Yi, Dimasi David P., Hysi Pirro G., Hewitt Alex W., Burdon Kathryn P., Toh Tze'Yo, Ruddle Jonathan B., Li Yi Ju, Mitchell Paul, Healey Paul R., Montgomery Grant W., Hansell Narelle, Spector Timothy D., Martin Nicholas G., Young Terri L., Hammond Christopher J., Macgregor Stuart, Craig Jamie E., Mackey David A. Common Genetic Variants near the Brittle Cornea Syndrome Locus ZNF469 Influence the Blinding Disease Risk Factor Central Corneal Thickness. PLoS Genetics. 2010;6(5):e1000947. doi: 10.1371/journal.pgen.1000947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Hoehn R, Zeller T, Verhoeven VJM, Grus F, Adler M, Wolfs RC, et al. Population-based meta-analysis in Caucasians confirms association with COL5A1 and ZNF469 but not COL8A2 with central corneal thickness. Hum Genet. 2012;131(11):1783–1793. doi: 10.1007/s00439-012-1201-3. [DOI] [PubMed] [Google Scholar]
- 209.Gao X, Gauderman WJ, Liu Y, Marjoram P, Torres M, Haritunians T, et al. A genome-wide association study of central corneal thickness in latinos. Investig Opthalmol Vis Sci. 2013;54(4):2435–2443. doi: 10.1167/iovs.13-11692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Abu A, Frydman M, Marek D, Pras E, Nir U, Reznik-Wolf H, et al. Deleterious mutations in the zinc-finger 469 gene cause brittle cornea syndrome. Am J Hum Genet. 2008;82(5):1217–1222. doi: 10.1016/j.ajhg.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Karolak JA, Gambin T, Rydzanicz M, Szaflik JP, Polakowski P, Frajdenberg A, et al. Evidence against ZNF469 being causative for keratoconus in Polish patients. Acta Ophthalmol. 2016;94(3):289–294. doi: 10.1111/aos.12968. [DOI] [PubMed] [Google Scholar]
- 212.Li X, Bykhovskaya Y, Haritunians T, Siscovick D, Aldave A, Szczotka-Flynn L, et al. A genome-wide association study identifies a potential novel gene locus for keratoconus, one of the commonest causes for corneal transplantation in developed countries. Hum Mol Genet. 2011;21(2):421–429. doi: 10.1093/hmg/ddr460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Burdon KP, Macgregor S, Bykhovskaya Y, Javadiyan S, Li X, Laurie KJ, et al. Association of polymorphisms in the hepatocyte growth factor gene promoter with keratoconus. Investig Ophthalmol Vis Sci. 2011;52(11):8514–8519. doi: 10.1167/iovs.11-8261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Bykhovskaya Y, Li X, Epifantseva I, Haritunians T, Siscovick D, Aldave A, et al. Variation in the lysyl oxidase (LOX) gene is associated with keratoconus in family-based and case-control studies. Investig Opthalmol Vis Sci. 2012;53(7):4152–4157. doi: 10.1167/iovs.11-9268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Sahebjada S, Schache M, Richardson AJ, Snibson G, Daniell M, Baird PN. Association of the hepatocyte growth factor gene with keratoconus in an Australian population. PLoS One. 2014;9(1):e84067. doi: 10.1371/journal.pone.0084067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Yang X, Liu X, Peng J, Zheng H, Lu F, Gong B, et al. Evaluation ofMYOC, ACAN, HGF, andMETas Candidate genes for high myopia in a Han Chinese population. Genet Test Mol Biomark. 2014;18(6):446–452. doi: 10.1089/gtmb.2013.0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Dudakova L, Palos M, Jirsova K, Stranecky V, Krepelova A, Hysi PG, et al. Validation of rs2956540:G > C and rs3735520:G > A association with keratoconus in a population of European descent. Eur J Hum Genet. 2015;23(11):1581–1583. doi: 10.1038/ejhg.2015.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Teri M. Genomewide association studies and assessment of the risk of disease. N Engl J Med. 2010;363(2):166–176. doi: 10.1056/NEJMra0905980. [DOI] [PubMed] [Google Scholar]
- 219.Liskova P, Dudakova L, Krepelova A, Klema J, Hysi PG. Replication of SNP associations with keratoconus in a Czech cohort. Plos One. 2017;12(2):e0172365. doi: 10.1371/journal.pone.0172365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Lechner Judith, Dash Durga P., Muszynska Dorota, Hosseini Mohsen, Segev Fani, George Sonia, Frazer David G., Moore Jonathan E., Kaye Stephen B., Young Terri, Simpson David A., Churchill Amanda J., Héon Elise, Willoughby Colin E. Mutational Spectrum of theZEB1Gene in Corneal Dystrophies Supports a Genotype–Phenotype Correlation. Investigative Opthalmology & Visual Science. 2013;54(5):3215. doi: 10.1167/iovs.13-11781. [DOI] [PubMed] [Google Scholar]
- 221.Bem Danai, Yoshimura Shin-Ichiro, Nunes-Bastos Ricardo, Bond Frances F., Kurian Manju A., Rahman Fatima, Handley Mark T.W., Hadzhiev Yavor, Masood Imran, Straatman-Iwanowska Ania A., Cullinane Andrew R., McNeill Alisdair, Pasha Shanaz S., Kirby Gail A., Foster Katharine, Ahmed Zubair, Morton Jenny E., Williams Denise, Graham John M., Dobyns William B., Burglen Lydie, Ainsworth John R., Gissen Paul, Müller Ferenc, Maher Eamonn R., Barr Francis A., Aligianis Irene A. Loss-of-Function Mutations in RAB18 Cause Warburg Micro Syndrome. The American Journal of Human Genetics. 2011;88(4):499–507. doi: 10.1016/j.ajhg.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Udar N, Atilano SR, Brown DJ, Holguin B, Small K, Nesburn AB, et al. SOD1: a candidate gene for keratoconus. Investig Opthalmol Vis Sci. 2006;47(8):3345–3351. doi: 10.1167/iovs.05-1500. [DOI] [PubMed] [Google Scholar]
- 223.Czugala M, Karolak JA, Nowak DM, Polakowski P, Pitarque J, Molinari A, et al. Novel mutation and three other sequence variants segregating with phenotype at keratoconus 13q32 susceptibility locus. Eur J Hum Genet. 2011;20(4):389–397. doi: 10.1038/ejhg.2011.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Guan T, Liu C, Ma Z, Ding S. The point mutation and polymorphism in keratoconus candidate gene TGFBI in Chinese population. Gene. 2012;503(1):137–139. doi: 10.1016/j.gene.2012.04.061. [DOI] [PubMed] [Google Scholar]
- 225.Stabuc-Silih M, Ravnik-Glavac M, Glavac D, Hawlina M, Strazisar M. Polymorphisms in COL4A3 and COL4A4 genes associated with keratoconus. Mol Vis. 2009;15:2848–2860. [PMC free article] [PubMed] [Google Scholar]
- 226.Abu-Amero KK, Kondkar AA, Azad TA, Sultan T, Kalantan H, Al-Muammar AM. Keratoconus is associated with increased copy number of mitochondrial DNA. Mol Vis. 2014;20:1203–1208. [PMC free article] [PubMed] [Google Scholar]
- 227.Mikami T, Meguro A, Teshigawara T, Takeuchi M, Uemoto R, Kawagoe T, et al. Interleukin 1 beta promoter polymorphism is associated with keratoconus in a Japanese population. Mol Vis. 2013;19:845–851. [PMC free article] [PubMed] [Google Scholar]
- 228.Saee-Rad S, Hashemi H, Miraftab M, Noori-Daloii MR, Chaleshtori MH, Raoofian R, et al. Mutation analysis of VSX1 and SOD1 in Iranian patients with keratoconus. Mol Vis. 2011;17(336–37):3128–3136. [PMC free article] [PubMed] [Google Scholar]
- 229.Hughes AE, Bradley DT, Campbell M, Lechner J, Dash DP, Simpson DA, et al. Mutation altering the miR-184 seed region causes familial keratoconus with cataract. Am J Hum Genet. 2011;89(5):628–633. doi: 10.1016/j.ajhg.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Lechner J, Porter LF, Rice A, Vitart V, Armstrong DJ, Schorderet DF, et al. Enrichment of pathogenic alleles in the brittle cornea gene, ZNF469, in keratoconus. Hum Mol Genet. 2014;23(20):5527–5535. doi: 10.1093/hmg/ddu253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Fukui K, Sasaki T, Imazumi K, Matsuura Y, Nakanishi H, Takai Y. Isolation and characterization of a GTPase activating protein specific for the Rab3 subfamily of small G proteins. J Biol Chem. 1997;272(8):4655–4658. doi: 10.1074/jbc.272.8.4655. [DOI] [PubMed] [Google Scholar]
- 232.Borck G, Wunram H, Steiert A, Volk AE, Körber F, Roters S, et al. A homozygous RAB3GAP2 mutation causes Warburg Micro syndrome. Hum Genet. 2010;129(1):45–50. doi: 10.1007/s00439-010-0896-2. [DOI] [PubMed] [Google Scholar]
- 233.Sahebjada S, Schache M, Richardson AJ, Snibson G, Macgregor S, Daniell M, et al. Evaluating the association between keratoconus and the corneal thickness genes in an independent Australian population. Investig Opthalmol Vis Sci. 2013;54(13):8224–8228. doi: 10.1167/iovs.13-12982. [DOI] [PubMed] [Google Scholar]
- 234.Rabinowitz YS, Maumenee IH, Lundergan MK, Puffenberger E, Zhu D, Antonarakis S, et al. Molecular genetic analysis in autosomal dominant keratoconus. Cornea. 1992;11(4):302–308. doi: 10.1097/00003226-199207000-00005. [DOI] [PubMed] [Google Scholar]
- 235.Rosenfeld JA, Drautz JM, Clericuzio CL, Cushing T, Raskin S, Martin J, et al. Deletions and duplications of developmental pathway genes in 5q31 contribute to abnormal phenotypes. Am J Med Genet. 2011;155A(8):1906–1916. doi: 10.1002/ajmg.a.34100. [DOI] [PubMed] [Google Scholar]
- 236.Aldave AJ, Bourla N, Yellore VS, Rayner SA, Khan MA, Salem AK, et al. Keratoconus is not associated with mutations in COL8A1 and COL8A2. Cornea. 2007;26(8):963–965. doi: 10.1097/ICO.0b013e31811dfaf7. [DOI] [PubMed] [Google Scholar]
- 237.Bradshaw AD, Puolakkainen P, Wight TN, Sage EH, Dasgupta J, Davidson JM. SPARC-null mice display abnormalities in the dermis characterized by decreased collagen fibril diameter and reduced tensile strength. J Investig Dermatol. 2003;120(6):949–955. doi: 10.1046/j.1523-1747.2003.12241.x. [DOI] [PubMed] [Google Scholar]
- 238.Wang Y, Jin T, Zhang X, Wei W, Cui Y, Geng T, et al. Common single nucleotide polymorphisms and keratoconus in the Han Chinese population. Ophthalmic Genet. 2013;34(3):160–166. doi: 10.3109/13816810.2012.743569. [DOI] [PubMed] [Google Scholar]
- 239.Mok J-W, Baek S-J, Joo C-K. VSX1 gene variants are associated with keratoconus in unrelated Korean patients. J Hum Genet. 2008;53(9):842–849. doi: 10.1007/s10038-008-0319-6. [DOI] [PubMed] [Google Scholar]
- 240.Tang YG, Picornell Y, Su X, Li X, Yang H, Rabinowitz YS. Three VSX1 gene mutations, L159M, R166W, and H244R, are not associated with keratoconus. Cornea. 2008;27(2):189–192. doi: 10.1097/ICO.0b013e31815a50e7. [DOI] [PubMed] [Google Scholar]
- 241.Dash DP, George S, Oprey D, Burns D, Nabili S, Donnelly U, et al. Mutational screening of VSX1 in keratoconus patients from the European population. Eye. 2009;24(6):1085–1092. doi: 10.1038/eye.2009.217. [DOI] [PubMed] [Google Scholar]
- 242.Macé M, Galiacy SD, Erraud A, Mejía JE, Etchevers H, Allouche M, et al. Comparative transcriptome and network biology analyses demonstrate antiproliferative and hyperapoptotic phenotypes in human keratoconus corneas. Investig Opthalmol Vis Sci. 2011;52(9):6181–6191. doi: 10.1167/iovs.10-70981. [DOI] [PubMed] [Google Scholar]
- 243.Maruyama Y, Li Y, Zhang Y, Wang X, Sugar J, Yue BY. Mapping of Sp1 regulation sites in the promoter of the human alpha1-proteinase inhibitor gene. J Cell Biochem. 2002;85(3):482–489. doi: 10.1002/jcb.10149. [DOI] [PubMed] [Google Scholar]
- 244.Paliwal P, Tandon R, Dube D, Kaur P, Sharma A. Familial segregation of a VSX1 mutation adds a new dimension to its role in the causation of keratoconus. Mol Vis. 2011;17:481–485. [PMC free article] [PubMed] [Google Scholar]
- 245.Abu-Amero KK, Kalantan H, Al-Muammar AM. Analysis of the VSX1 gene in keratoconus patients from Saudi Arabia. Mol Vis. 2011;17:667–672. [PMC free article] [PubMed] [Google Scholar]
- 246.Aldave AJ, Yellore VS, Principe AH. Candidate gene screening for posterior polymorphous dystrophy. Cornea. 2005;24:151–155. doi: 10.1097/01.ico.0000141235.26096.1d. [DOI] [PubMed] [Google Scholar]
- 247.Vincent AL, Jordan C, Sheck L, Niederer R, Patel DV, McGhee CN. Screening the visual system homeobox 1 gene in keratoconus and posterior polymorphous dystrophy cohorts identifies a novel variant. Mol Vis. 2013;19:852–860. [PMC free article] [PubMed] [Google Scholar]
- 248.Eran P, Almogit A, David Z, Wolf HR, Hana G, Yaniv B, et al. The D144E substitution in the VSX1 gene: a non-pathogenic variant or a disease causing mutation? Ophthalmic Genet. 2008;29:53–59. doi: 10.1080/13816810802008242. [DOI] [PubMed] [Google Scholar]
- 249.Semina EV, Mintz-Hittner HA, Murray JC. Isolation and characterization of a novel human paired-like homeodomain-containing transcription factor gene, VSX1, expressed in ocular tissues. Genomics. 2000;63:289–293. doi: 10.1006/geno.1999.6093. [DOI] [PubMed] [Google Scholar]
- 250.Shi Z, Jervis D, Nickerson PE, Chow RL. Requirement for the paired-like homeodomain transcription factor VSX1 in type 3a mouse retinal bipolar cell terminal differentiation. J Comp Neurol. 2011;520(1):117–129. doi: 10.1002/cne.22697. [DOI] [PubMed] [Google Scholar]
- 251.Aldave AJ, Yellore VS, Salem AK, Yoo GL, Rayner SA, Yang H, et al. No VSX1 gene mutations associated with keratoconus. Investig Ophthalmol Vis Sci. 2006;47:2820–2822. doi: 10.1167/iovs.05-1530. [DOI] [PubMed] [Google Scholar]
- 252.Liskova P, Ebenezer ND, Hysi PG, Gwilliam R, El-Ashry MF, Moodaley LC, et al. Molecular analysis of the VSX1 gene in familial keratoconus. Mol Vis. 2007;13:1887–1891. [PMC free article] [PubMed] [Google Scholar]
- 253.Mintz-Hittner HA, Semina EV, Frishman LJ, Prager TC, Murray JC. VSX1 (RINX) mutation with craniofacial anomalies, empty sella, corneal endothelial changes, and abnormal retinal and auditory bipolar cells. Ophthalmology. 2004;111:828–836. doi: 10.1016/j.ophtha.2003.07.006. [DOI] [PubMed] [Google Scholar]
- 254.Watson TC, Chow RL. Absence of Vsx1 expression in the normal and damaged mouse cornea. Mol Vis. 2011;17:737–744. [PMC free article] [PubMed] [Google Scholar]
- 255.Al-Muammar Abdulrahman M., Kalantan Hatem, Azad Taif Anwar, Sultan Tahira, Abu-Amero Khaled K. Analysis of theSOD1Gene in Keratoconus Patients from Saudi Arabia. Ophthalmic Genetics. 2014;36(4):373–375. doi: 10.3109/13816810.2014.889173. [DOI] [PubMed] [Google Scholar]
- 256.Jeoung JW, Kim MK, Park SS, Kim SY, Ko HS, Wee WR, et al. VSX1 gene and keratoconus. Cornea. 2012;31(7):746–750. doi: 10.1097/ICO.0b013e3181e16dd0. [DOI] [PubMed] [Google Scholar]
- 257.Barbaro Vanessa, Di Iorio Enzo, Ferrari Stefano, Bisceglia Luigi, Ruzza Alessandro, De Luca Michele, Pellegrini Graziella. Expression ofVSX1in Human Corneal Keratocytes during Differentiation into Myofibroblasts in Response to Wound Healing. Investigative Opthalmology & Visual Science. 2006;47(12):5243. doi: 10.1167/iovs.06-0185. [DOI] [PubMed] [Google Scholar]
- 258.Hayashi T, Huang J, Deeb SS. RINX(VSX1), a novel homeobox gene expressed in the inner nuclear layer of the adult retina. Genomics. 2000;67(2):128–139. doi: 10.1006/geno.2000.6248. [DOI] [PubMed] [Google Scholar]
- 259.Chow RL, Volgyi B, Szilard RK, Ng D, Mckerlie C, Bloomfield SA, et al. Control of late off-center cone bipolar cell differentiation and visual signaling by the homeobox gene Vsx1. Proc Natl Acad Sci. 2004;101(6):1754–1759. doi: 10.1073/pnas.0306520101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Ohtoshi A, Wang SW, Maeda H, Saszik SM, Frishman LJ, Klein WH, et al. Regulation of retinal cone bipolar cell differentiation and photopic vision by the CVC homeobox gene Vsx1. Curr Biol. 2004;14(6):530–536. doi: 10.1016/j.cub.2004.02.027. [DOI] [PubMed] [Google Scholar]
- 261.Hosseini SM, Herd S, Vincent AL, Heon E. Genetic analysis of chromosome 20-related posterior polymorphous corneal dystrophy: genetic heterogeneity and exclusion of three candidate genes. Mol Vis. 2008;14:71–80. [PMC free article] [PubMed] [Google Scholar]
- 262.Guan T, Wang X, Zheng L-B, Wu H-J, Yao Y-F. Analysis of the VSX1 gene in sporadic keratoconus patients from China. BMC Ophthalmol. 2017;17(1):173. doi: 10.1186/s12886-017-0567-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Heon E, Mathers WD, Alward WL, Weisenthal RW, Sunden SL, Fishbaugh JA, et al. Linkage of posterior polymorphous corneal dystrophy to 20q11. Hum Mol Genet. 1995;4(3):485–488. doi: 10.1093/hmg/4.3.485. [DOI] [PubMed] [Google Scholar]
- 264.Valleix S, Nedelec B, Rigaudiere F, Dighiero P, Pouliquen Y, Renard G, et al. H244R VSX1 is associated with selective cone on bipolar cell dysfunction and macular degeneration in a PPCD family. Investig Opthalmol Vis Sci. 2006;47(1):48–54. doi: 10.1167/iovs.05-0479. [DOI] [PubMed] [Google Scholar]
- 265.Gwilliam Rhian, Liskova Petra, Filipec Martin, Kmoch Stanislav, Jirsova Katerina, Huckle Elizabeth J., Stables Catherine L., Bhattacharya Shomi S., Hardcastle Alison J., Deloukas Panos, Ebenezer Neil D. Posterior Polymorphous Corneal Dystrophy in Czech Families Maps to Chromosome 20 and Excludes theVSX1Gene. Investigative Opthalmology & Visual Science. 2005;46(12):4480. doi: 10.1167/iovs.05-0269. [DOI] [PubMed] [Google Scholar]
- 266.Baratz KH, Tosakulwong N, Ryu E, Brown WL, Branham K, Chen W, et al. E2-2 protein and Fuchs’s corneal dystrophy. N Engl J Med. 2010;11:1016–1024. doi: 10.1056/NEJMoa1007064. [DOI] [PubMed] [Google Scholar]
- 267.Waring GO, Rodrigues MM, Laibson PR. Corneal dystrophies. II. Endothelial dystrophies. Surv Ophthalmol. 1978;23(3):147–168. doi: 10.1016/0039-6257(78)90151-0. [DOI] [PubMed] [Google Scholar]
- 268.Bechara SJ, Grossniklaus HE, Waring GO, Wells JA. Keratoconus associated with posterior polymorphous dystrophy. Am J Ophthalmol. 1991;112(6):729–731. doi: 10.1016/s0002-9394(14)77284-8. [DOI] [PubMed] [Google Scholar]
- 269.Lam HY, Wiggs JL, Jurkunas UV. Unusual presentation of presumed posterior polymorphous dystrophy associated with iris heterochromia, band keratopathy, and keratoconus. Cornea. 2010;29(10):1180–1185. doi: 10.1097/ICO.0b013e3181d007e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Krafchak CM, Pawar H, Moroi SE, Sugar A, Lichter PR, Mackey DA, et al. Mutations in TCF8 cause posterior polymorphous corneal dystrophy and ectopic expression of COL4A3 by corneal endothelial cells. Am J Hum Genet. 2005;77(5):694–708. doi: 10.1086/497348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Biswas S. Missense mutations in COL8A2, the gene encoding the alpha2 chain of type VIII collagen, cause two forms of corneal endothelial dystrophy. Hum Mol Genet. 2001;10(21):2415–2423. doi: 10.1093/hmg/10.21.2415. [DOI] [PubMed] [Google Scholar]
- 272.Kawata A, Kato S, Shimizu T, Hayashi H, Hirai S, Misawa H, et al. Aberrant splicing of human Cu/Zn superoxide dismutase (SODI) RNA transcripts. NeuroReport. 2000;11(12):2649–2653. doi: 10.1097/00001756-200008210-00009. [DOI] [PubMed] [Google Scholar]
- 273.Hirano M, Hung W-Y, Cole N, Azim A, Deng H-X, Siddique T. Multiple transcripts of the human Cu, Zn superoxide dismutase gene. Biochem Biophys Res Commun. 2000;276(1):52–56. doi: 10.1006/bbrc.2000.3427. [DOI] [PubMed] [Google Scholar]
- 274.Noor R, Mittal S, Iqbal J. Superoxide dismutase—applications and relevance to human diseases. Med Sci Monit. 2002;8(9):RA210–5. [PubMed]
- 275.Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362(64 15):59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 276.Behndig A, Svensson B, Marklund SL, Karlsson K. Superoxide dismutase isoenzymes in the human eye. Investig Ophthalmol Vis Sci. 1998;39:471–475. [PubMed] [Google Scholar]
- 277.Zelko IN, Mariani TJ, Folz RJ. Superoxide dismutase multigene family: a comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression. Free Radic Biol Med. 2002;33(3):337–349. doi: 10.1016/s0891-5849(02)00905-x. [DOI] [PubMed] [Google Scholar]
- 278.Behding A, Karlsson K, Johansson BO, Brannstrom T, Marklund SL. Superoxide dismutase isoenzymes in the normal and diseased human cornea. Investig Ophthalmol Vis Sci. 2001;42:2293–2296. [PubMed] [Google Scholar]
- 279.Gondhowiardjo TD, van Haeringen NJ, Volker-Dieben HJ, Beekhuis HW, Kok JH, Van Rij G, et al. Analysis of corneal aldehyde dehydrogenase patterns in pathologic corneas. Cornea. 1993;12:146–154. doi: 10.1097/00003226-199303000-00010. [DOI] [PubMed] [Google Scholar]
- 280.Kenny MC, Brown DJ, Rajeev B. Abstract: The elusive causes of keratoconus: a working hypothesis (CLAO J. 2000;26:10–13). Am J Ophthalmol. 2000;130(2):263. [PubMed]
- 281.Chwa M, Atilano SR, Reddy V, Jordan N, Kim DW, Kenney MC. Increased stress-induced generation of reactive oxygen species and apoptosis in human keratoconus fibroblasts. Investig Opthalmol Vis Sci. 2006;47(5):1902–1910. doi: 10.1167/iovs.05-0828. [DOI] [PubMed] [Google Scholar]
- 282.Ballinger SW, Houten BV, Conklin CA, Jin G-F, Godley BF. Hydrogen peroxide causes significant mitochondrial DNA damage in human RPE cells. Exp Eye Res. 1999;68(6):765–772. doi: 10.1006/exer.1998.0661. [DOI] [PubMed] [Google Scholar]
- 283.Ballinger SW, Patterson C, Yan C-N, Doan R, Burow DL, Young CG, et al. Hydrogen peroxide- and peroxynitrite-induced mitochondrial DNA damage and dysfunction in vascular endothelial and smooth muscle cells. Circ Res. 2000;86(9):960–966. doi: 10.1161/01.res.86.9.960. [DOI] [PubMed] [Google Scholar]
- 284.Wang X, Simpkins JW, Dykens JA, Cammarata PR. Oxidative damage to human lens epithelial cells in culture: estrogen protection of mitochondrial potential, ATP, and cell viability. Investig Opthalmol Vis Sci. 2003;44(5):2067–2075. doi: 10.1167/iovs.02-0841. [DOI] [PubMed] [Google Scholar]
- 285.Rohrbach M, Spencer HL, Porter LF, Burkitt-Wright EM, Bürer C, Janecke A, et al. ZNF469 frequently mutated in the brittle cornea syndrome (BCS) is a single exon gene possibly regulating the expression of several extracellular matrix components. Mol Genet Metab. 2013;109(3):289–295. doi: 10.1016/j.ymgme.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Burkitt Wright EM, Spencer HL, Daly SB, Manson FD, Zeef LA, Urquhart J, et al. Mutations in PRDM5 in brittle cornea syndrome identify a pathway regulating extracellular matrix development and maintenance. Am J Hum Genet. 2011;88:767–777. doi: 10.1016/j.ajhg.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Al-Hussain H, Zeisberger SM, Huber PR, Giunta C, Steinmann B. Brittle cornea syndrome and its delineation from the kyphoscoliotic type of Ehlers–Danlos syndrome (EDS VI): report on 23 patients and review of the literature. Am J Med Genet A. 2004;124A(1):28–34. doi: 10.1002/ajmg.a.20326. [DOI] [PubMed] [Google Scholar]
- 288.Christensen AE, Knappskog PM, Midtbø M, Gjesdal CG, Mengel-From J, Morling N, et al. Brittle cornea syndrome associated with a missense mutation in the zinc-finger 469 gene. Investig Opthalmol Vis Sci. 2010;51(1):47–52. doi: 10.1167/iovs.09-4251. [DOI] [PubMed] [Google Scholar]
- 289.Maier P, Broszinski A, Heizmann U, Bohringer D, Reinhardau T. Active transforming growth factor-beta2 is increased in the aqueous humor of keratoconus patients. Mol Vis. 2007;13:1198–1202. [PubMed] [Google Scholar]
- 290.Saghizadeh M, Chwa M, Aoki A, Lin B, Pirouzmanesh A, Brown D, et al. Altered expression of growth factors and cytokines in keratoconus, bullous keratopathy and diabetic human corneas. Exp Eye Res. 2001;73(2):179–189. doi: 10.1006/exer.2001.1028. [DOI] [PubMed] [Google Scholar]
- 291.Bykhovskaya Yelena, Caiado Canedo Ana L., Wright Kenneth W., Rabinowitz Yaron S. C.57 C > T Mutation in MIR 184 is Responsible for Congenital Cataracts and Corneal Abnormalities in a Five-generation Family from Galicia, Spain. Ophthalmic Genetics. 2013;36(3):244–247. doi: 10.3109/13816810.2013.848908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Chen J, Chen Y, Han SN. Comparison of TGF-β1 in tears and corneal haze following Epi-LASIK with and without mitomycin C. Int J Ophthalmol. 2013;6:312–315. doi: 10.3980/j.issn.2222-3959.2013.03.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Takács L, Csutak A, Balázs E, Módis L, Berta A. Expression of ßig-h3 is lower than normal in keratoconus corneas but increases with scarring. Cornea. 1999;18(5):599–605. [PubMed] [Google Scholar]
- 294.Pahuja N, Kumar NR, Shroff R, Shetty R, Nuijts RMMA, Ghosh A, et al. Differential molecular expression of extracellular matrix and inflammatory genes at the corneal cone apex drives focal weakening in keratoconus. Investig Opthalmol Vis Sci. 2016;57(13):5372–5382. doi: 10.1167/iovs.16-19677. [DOI] [PubMed] [Google Scholar]
- 295.Bhola NE, Balko JM, Dugger TC, Kuba MG, Sánchez V, Sanders M, et al. TGF-β inhibition enhances chemotherapy action against triple-negative breast cancer. J Clin Investig. 2013;123(3):1348–1358. doi: 10.1172/JCI65416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Gonzalo-Gil E, Galindo-Izquierdo M. Role of transforming growth factor-beta (TGF) beta in the physiopathology of rheumatoid arthritis. Reumatol Clín (Engl Ed). 2014;10(3):174–179. doi: 10.1016/j.reuma.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 297.Engler C, Chakravarti S, Doyle J, Eberhart CG, Meng H, Stark WJ, et al. Transforming growth factor-β signaling pathway activation in keratoconus. Am J Ophthalmol. 2011;151(5):752–759. doi: 10.1016/j.ajo.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Yan X, Liu Z, Chen Y. Regulation of TGF-beta signaling by Smad7. Acta Biochim Biophys Sin (Shanghai) 2009;41(4):263–272. doi: 10.1093/abbs/gmp018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Bai Shuting, Cao Xu. A Nuclear Antagonistic Mechanism of Inhibitory Smads in Transforming Growth Factor-β Signaling. Journal of Biological Chemistry. 2001;277(6):4176–4182. doi: 10.1074/jbc.M105105200. [DOI] [PubMed] [Google Scholar]
- 300.Schiller M, Javelaud D, Mauviel A. TGF-β-induced SMAD signaling and gene regulation: consequences for extracellular matrix remodeling and wound healing. J Dermatol Sci. 2004;35:83–92. doi: 10.1016/j.jdermsci.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 301.Gomes LR, Terra LF, Wailemann RA, Labriola L, Sogayar MC. TGF-β1 modulates the homeostasis between MMPs and MMP inhibitors through p38 MAPK and ERK1/2 in highly invasive breast cancer cells. BMC Cancer. 2012;12(1):26. doi: 10.1186/1471-2407-12-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Evans RA, Tian YC, Steadman R, Phillips AO. TGF-β1-mediated fibroblast–myofibroblast terminal differentiation—the role of smad proteins. Exp Cell Res. 2003;282(2):90–100. doi: 10.1016/s0014-4827(02)00015-0. [DOI] [PubMed] [Google Scholar]
- 303.Samarakoon R, Overstreet JM, Higgins PJ. TGF-β signaling in tissue fibrosis: redox controls, target genes and therapeutic opportunities. Cell Signal. 2013;25(1):264–268. doi: 10.1016/j.cellsig.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Karamichos D, Hutcheon AEK, Zieske JD. Transforming growth factor-β3 regulates assembly of a non-fibrotic matrix in a 3D corneal model. J Tissue Eng Regen Med. 2011;5(8):228–238. doi: 10.1002/term.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Waddington SN, Crossley R, Sheard V, Howe SJ, Buckley SMK, Coughlan L, et al. Gene delivery of a mutant TGF beta 3 reduces markers of scar tissue formation after cutaneous wounding. Mol Ther. 2010;18:2104–2111. doi: 10.1038/mt.2010.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Szcześniak MW, Kabza M, Karolak JA, Rydzanicz M, Nowak DM, Ginter-Matuszewska B, et al. KTCNlncDB—a first platform to investigate lncRNAs expressed in human keratoconus and non-keratoconus corneas. Database. 2017. [DOI] [PMC free article] [PubMed]
- 307.Khaled Mariam Lofty, Bykhovskaya Yelena, Yablonski Sarah E. R., Li Hanzhou, Drewry Michelle D., Aboobakar Inas F., Estes Amy, Gao X. Raymond, Stamer W. Daniel, Xu Hongyan, Allingham R. Rand, Hauser Michael A., Rabinowitz Yaron S., Liu Yutao. Differential Expression of Coding and Long Noncoding RNAs in Keratoconus-Affected Corneas. Investigative Opthalmology & Visual Science. 2018;59(7):2717. doi: 10.1167/iovs.18-24267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Xu J, Bai J, Zhang X, Lv Y, Gong Y, Liu L, et al. A comprehensive overview of lncRNA annotation resources. Brief Bioinform. 2017;18(2):236–249. doi: 10.1093/bib/bbw015. [DOI] [PubMed] [Google Scholar]
- 309.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11):1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 310.DOSTIE J. Numerous microRNPs in neuronal cells containing novel microRNAs. RNA. 2003;9(2):180–186. doi: 10.1261/rna.2141503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Meola N, Gennarino V, Banfi S. microRNAs and genetic diseases. PathoGenetics. 2009;2(1):7. doi: 10.1186/1755-8417-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Mencía A, Modamio-Høybjør S, Redshaw N, Morin M, Mayo-Merino F, Olavarrieta L, et al. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat Genet. 2009;41(5):609–613. doi: 10.1038/ng.355. [DOI] [PubMed] [Google Scholar]
- 313.Sun Dandan, Lee Yong Sun, Malhotra Ankit, Kim Hak Kyun, Matecic Mirela, Evans Clive, Jensen Roderick V., Moskaluk Christopher A., Dutta Anindya. miR-99 Family of MicroRNAs Suppresses the Expression of Prostate-Specific Antigen and Prostate Cancer Cell Proliferation. Cancer Research. 2011;71(4):1313–1324. doi: 10.1158/0008-5472.CAN-10-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Karali M, Peluso I, Gennarino VA, Bilio M, Verde R, Lago G, et al. miRNeye: a microRNA expression atlas of the mouse eye. BMC Genom. 2010;11(1):715. doi: 10.1186/1471-2164-11-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129(7):1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Ryan DG, Oliveira-Fernandes M, Lavker RM. MicroRNAs of the mammalian eye display distinct and overlapping tissue specificity. Mol Vis. 2006;12(134–135):1175–1184. [PubMed] [Google Scholar]
- 317.Nowak DM, Gajecka M. Nonrandom distribution of miRNAs genes and single nucleotide variants in keratoconus loci. PLos One. 2015;10(7):e0132143. doi: 10.1371/journal.pone.0132143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Lechner Judith, Bae Ha Ae, Guduric-Fuchs Jasenka, Rice Aine, Govindarajan Gowthaman, Siddiqui Salina, Abi Farraj Layal, Yip Shea Ping, Yap Maurice, Das Manoranjan, Souzeau Emmanuelle, Coster Doug, Mills Richard A., Lindsay Richard, Phillips Tony, Mitchell Paul, Ali Manir, Inglehearn Chris F., Sundaresan Periasamy, Craig Jamie E., Simpson David A., Burdon Kathryn P., Willoughby Colin E. Mutational Analysis ofMIR184in Sporadic Keratoconus and Myopia. Investigative Opthalmology & Visual Science. 2013;54(8):5266. doi: 10.1167/iovs.13-12035. [DOI] [PubMed] [Google Scholar]
- 319.Dash DP, Silvestri G, Hughes AE. Fine mapping of the keratoconus with cataract locus on chromosome 15q and candidate gene analysis. Mol Vis. 2006;12:499–505. [PubMed] [Google Scholar]
- 320.McBride HM, Neuspiel M, Wasiak S. Mitochondria: more than just a powerhouse. Curr Biol. 2006;16(14):R551–R560. doi: 10.1016/j.cub.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 321.Nohl H. Generation of superoxide radicals as byproduct of cellular respiration. Ann Biol Clin. 1994;52(3):199–204. [PubMed] [Google Scholar]
- 322.Torroni A, Achilli A, Macaulay V, Richards M, Bandelt H. Harvesting the fruit of the human mtDNA tree. Trends Genet. 2006;22(6):339–345. doi: 10.1016/j.tig.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 323.Carelli Valerio, Achilli Alessandro, Valentino Maria Lucia, Rengo Chiara, Semino Ornella, Pala Maria, Olivieri Anna, Mattiazzi Marina, Pallotti Francesco, Carrara Franco, Zeviani Massimo, Leuzzi Vincenzo, Carducci Carla, Valle Giorgio, Simionati Barbara, Mendieta Luana, Salomao Solange, Belfort Rubens, Sadun Alfredo A., Torroni Antonio. Haplogroup Effects and Recombination of Mitochondrial DNA: Novel Clues from the Analysis of Leber Hereditary Optic Neuropathy Pedigrees. The American Journal of Human Genetics. 2006;78(4):564–574. doi: 10.1086/501236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Fuku Noriyuki, Park Kyong Soo, Yamada Yoshiji, Nishigaki Yutaka, Cho Young Min, Matsuo Hitoshi, Segawa Tomonori, Watanabe Sachiro, Kato Kimihiko, Yokoi Kiyoshi, Nozawa Yoshinori, Lee Hong Kyu, Tanaka Masashi. Mitochondrial Haplogroup N9a Confers Resistance against Type 2 Diabetes in Asians. The American Journal of Human Genetics. 2007;80(3):407–415. doi: 10.1086/512202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Hofmann S. Wolfram (DIDMOAD) Syndrome and Leber Hereditary Optic Neuropathy (LHON) Are Associated with Distinct Mitochondrial DNA Haplotypes. Genomics. 1997;39(1):8–18. doi: 10.1006/geno.1996.4474. [DOI] [PubMed] [Google Scholar]
- 326.Abu-Amero KK, Bosley TM. Increased relative mitochondrial DNA content in leucocytes of patients with NAION. Br J Ophthalmol. 2006;90(7):823–825. doi: 10.1136/bjo.2006.090332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Schrier SA, Falk MJ. Mitochondrial disorders and the eye. Curr Opin Ophthalmol. 2011;22(5):325–331. doi: 10.1097/ICU.0b013e328349419d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Abu-Amero KK, Azad TA, Sultan T, Kalantan H, Kondkar AA, Al-Muammar AM. Association of mitochondrial haplogroups H and R with keratoconus in Saudi Arabian patients. Investig Opthalmol Vis Sci. 2014;55(5):2827–2831. doi: 10.1167/iovs.14-14300. [DOI] [PubMed] [Google Scholar]
- 329.Aktekin M, Sargon MF, Cakar P, Celik HH, Firat E. Ultrastructure of the cornea epithelium in keratoconus. Okajimas Folia Anat Jpn. 1998;75(1):45–53. doi: 10.2535/ofaj1936.75.1_45. [DOI] [PubMed] [Google Scholar]
- 330.Atilano SR, Coskun P, Chwa M, Jordan N, Reddy V, Le K, et al. Accumulation of mitochondrial DNA damage in keratoconus corneas. Investig Opthalmol Vis Sci. 2005;46(4):1256–1263. doi: 10.1167/iovs.04-1395. [DOI] [PubMed] [Google Scholar]
- 331.Hao X-D, Chen P, Wang Y, Li S-X, Xie L-X. Mitochondrial DNA copy number, but not haplogroup is associated with keratoconus in Han Chinese population. Exp Eye Res. 2015;132:59–63. doi: 10.1016/j.exer.2015.01.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.