Abstract
With their essential regulatory roles in gene expression and high abundance in the brain, circular RNAs (circRNAs) have recently attracted considerable attention. Many studies have shown that circRNAs play important roles in the pathology of CNS diseases, but whether circRNAs participate in E. coli-induced bacterial meningitis is unclear. We used high-throughput sequencing to analyze the transcriptional profiles of host circRNAs in primary brain microvascular endothelial cells in response to meningitic E. coli. A total of 308 circRNAs were significantly altered, including 140 upregulated and 168 downregulated ones (p < 0.05). Kyoto Encyclopedia of Genes and Genomes (KEGG) and gene ontology enrichment of the parental genes of these altered circRNAs indicated that they are likely to be involved in diverse biological processes via influencing the expression of their parental genes. Coupled with our previous mRNA and microRNA sequencing data, a competing endogenous RNA analysis was performed, and the potential regulatory network was preliminarily constructed and validated. By revealing the transcriptional profiles of the host circRNAs involved in E. coli meningitis, it is envisaged that the novel insight gained into the regulatory mechanisms of circRNAs in the development of bacterial meningitis will lead to better understanding of how to prevent and treat bacterial CNS infections.
Keywords: circular RNAs, CNS, brain microvascular endothelial cells, meningitic Escherichia coli, competing endogenous RNAs
Introduction
Bacterial meningitis, an important cause of CNS dysfunction, has high morbidity and mortality, and most survivors have lifelong sequelae.1 As the most important physiological barrier in mammals, the blood-brain barrier (BBB), which consists of brain microvascular endothelial cells (BMECs), pericytes, and astrocytes, maintains homeostasis in the CNS by regulating the transport of nutrients, molecules, and cells, while also blocking the entry of circulating pathogens from the blood to the brain.2, 3 Unfortunately, an increasing variety of bacterial pathogens are capable of breaking and damaging the BBB.4, 5 Among these, the common bacterial pathogen Escherichia coli possesses the ability to invade the CNS and cause meningitis, particularly in neonates.6, 7 We have previously shown that meningitic E. coli infection could disrupt and enhance BBB permeability, and several cellular factors were highly induced and involved in this process.8 However, the underlying mechanism for meningitic E. coli penetration of the BBB remains largely unknown.
Circular RNA (circRNA) transcripts were first identified in the early 1990s, and they have a special covalent loop structure without a 5′ cap or 3′ polyA tail.9 circRNAs have several distinct features, and they are usually transcribed from one or multiple exons in their parental gene, but they are more stable than their linear parental gene. They exhibit tissue-specific patterns, with high expressional abundance in brain tissues, and they are present mainly in the cytoplasm and can be transported and sorted into exosomes.10, 11, 12 Thus far, it has been reported that circRNAs function across multiple biological processes, such as acting as scaffolds during protein complex assembly,13 modulating parental gene expression,14, 15 or acting as microRNA (miRNA) sponges.16, 17 Numerous cancer researchers have reported that circRNAs can participate in the proliferation, migration, and invasion of tumor cells in different cancers.18, 19, 20, 21 Notably, many studies have evidenced the essential roles played by circRNAs, because of the their high abundances in the brain, in the pathological processes of CNS diseases, such as multiple sclerosis, Alzheimer’s disease, stroke, and depression.22, 23, 24, 25 However, it is not known whether cellular circRNAs are involved in regulating the CNS disorder induced by infection with meningitic E. coli. Therefore, the transcriptional profiles of circRNAs in BMECs and their potential regulatory mechanisms during E. coli meningitis are worthy of investigation.
Here we used circRNA transcriptomic sequencing as well as bioinformatic approaches to identify the significantly altered circRNAs and their potential regulatory effects in primary human BMECs (hBMECs) in response to meningitic E. coli infection. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment were also performed to predict the potential functions of the parental genes of the significantly changed circRNAs. Combined with our previous mRNA and miRNA sequencing data for hBMECs after challenge with meningitic E. coli, we preliminarily evaluated and validated the potential of these circRNAs as competitive endogenous RNAs (ceRNAs) in E. coli-induced meningitis. By identifying and characterizing the circRNAs that were altered during the interaction between meningitic E. coli and primary hBMECs, our current understanding about how meningitic E. coli regulates its penetration of the BBB will be enhanced, making possible new molecular targets and concepts for better prevention, diagnosis, and therapy for E. coli-induced meningitis.
Results
Meningitic E. coli Infection Altered the Profiles of circRNAs in Primary hBMECs
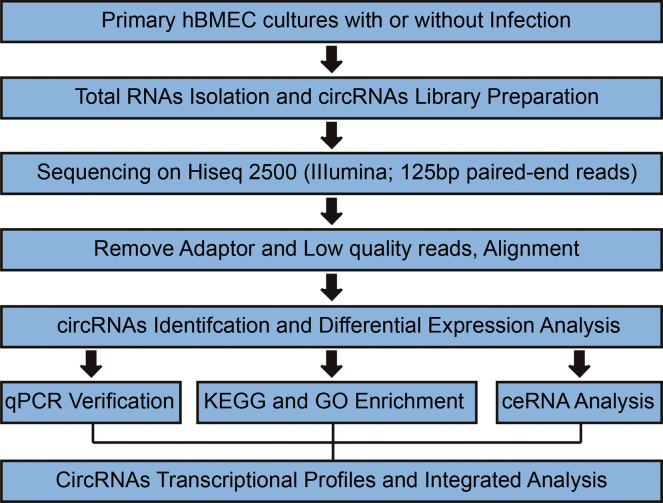
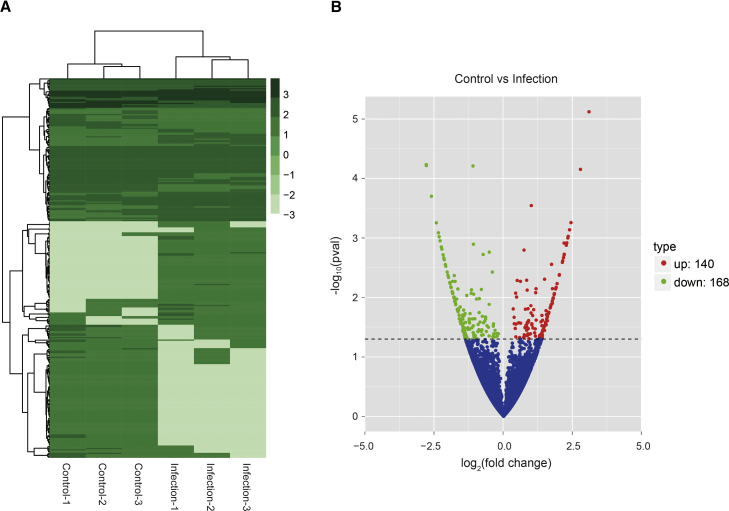
To investigate the profiles of the whole circRNA population in primary hBMECs in response to meningitic E. coli, the cells were challenged with or without meningitic E. coli strain PCN033 for 3 hr. After rRNA depletion and RNase R treatment (see the Materials and Methods), the purified cellular RNA products were subjected to circRNA sequencing, as shown in the Figure 1 flow chart. The sequencing reads were generated from three control groups and three infection groups, in which 8.61–11.9 million raw reads per sample were obtained (Table S1). The error rate distribution (Figure S1), base content (Figure S2), and the raw read classification (Figure S3) were analyzed for each sample. The heatmap and volcano plot revealed the alteration trends of these circRNAs in the cells upon meningitic E. coli infection (Figures 2A and 2B). A total of 41,504 circRNAs were obtained, as detailed in Table S2. Among these, 140 circRNAs were upregulated and 168 circRNAs were downregulated (p < 0.05) (Figure 2B; Table S3). The most significantly upregulated and downregulated circRNAs are listed in Table 1, respectively.
Figure 1.
Overview of Sample Preparation, Circular RNA Sequencing, and Bioinformatic Analysis
Figure 2.
Expression Profiling of the Changes in circRNAs in Meningitic E. coli-Infected Primary hBMECs
(A) Heatmap showing unsupervised clustering of significantly expressed circRNAs from primary hBMECs in the meningitic E. coli infection groups compared with the control groups. The expression profiles are displayed with three samples in each group. Dark green represents high and pale green represents low relative expression. (B) Volcano plot of the upregulated and downregulated circRNAs from primary hBMECs in the meningitic E. coli infection group compared with the control group. p < 0.05 was considered statistically significant.
Table 1.
The Most Significantly Changed circRNAs in Primary hBMECs in Response to Meningitic E. coli (>4-fold)
| circRNA ID | Fold Change | Chromosome | Start Site | End Site | Strand | Genomic Length (bp) | Spliced Length (bp) | Gene ID |
|---|---|---|---|---|---|---|---|---|
| circ_0014254 | 8.48 | chr18 | 37,084,124 | 37,173,081 | + | 88,957 | 76,366 | ENSG00000150477 |
| circ_0030423 | 6.86 | chr4 | 39,737,419 | 39,774,933 | + | 37,514 | 336 | ENSG00000078140 |
| circ_0001409 | 5.41 | chr10 | 76,036,007 | 76,058,783 | + | 22,776 | 22,776 | – |
| circ_0003433 | 5.21 | chr11 | 68,429,574 | 68,433,838 | + | 4,264 | 4,138 | ENSG00000162337 |
| circ_0035876 | 5.04 | chr7 | 152,273,704 | 152,315,338 | − | 41,634 | 5,776 | ENSG00000055609 |
| circ_0030790 | 5.00 | chr4 | 83,272,086 | 83,279,114 | − | 7,028 | 7,028 | – |
| circ_0029917 | 4.87 | chr4 | 143,415,476 | 143,415,771 | + | 295 | 295 | ENSG00000109458 |
| circ_0038415 | 4.80 | chr8 | 55,941,854 | 55,947,723 | + | 5,869 | 5,869 | – |
| circ_0003467 | 4.57 | chr11 | 68,583,042 | 68,596,218 | + | 13,176 | 13,176 | ENSG00000110075 |
| circ_0019064 | 4.55 | chr1 | 233,198,938 | 233,227,371 | − | 28,433 | 8,711 | ENSG00000135749 |
| circ_0024628 | 4.55 | chr2 | 174,223,032 | 174,229,451 | − | 6,419 | 6,419 | – |
| circ_0001582 | 4.48 | chr10 | 92,648,211 | 92,650,517 | + | 2,306 | 223 | ENSG00000138160 |
| circ_0001726 | 4.40 | chr10 | 97,656,284 | 97,662,968 | + | 6,684 | 6,684 | – |
| circ_0006937 | 4.36 | chr14 | 20,996,208 | 20,996,711 | + | 503 | 503 | ENSG00000165792 |
| circ_0028050 | 4.04 | chr3 | 179,378,340 | 179,386,629 | + | 8,289 | 8,289 | ENSG00000171109 |
| circ_0000206 | 4.03 | chr10 | 110,593,072 | 110,598,290 | + | 5,218 | 1,740 | ENSG00000108055 |
| circ_0036105 | −6.95 | chr7 | 22,219,865 | 22,318,037 | − | 98,172 | 69,625 | ENSG00000136237 |
| circ_0001639 | −6.93 | chr10 | 94,441,895 | 94,500,327 | + | 58,432 | 58,432 | – |
| circ_0024411 | −6.10 | chr2 | 15,488,893 | 15,539,356 | − | 50,463 | 17,793 | ENSG00000151779 |
| circ_0033002 | −5.39 | chr6 | 107,503,656 | 107,533,610 | + | 29,954 | 29,663 | ENSG00000112320 |
| circ_0020478 | −5.13 | chr1 | 63,478,763 | 63,490,218 | − | 11,455 | 11,455 | ENSG00000142856 |
| circ_0037938 | −5.03 | chr8 | 15,623,079 | 15,673,836 | + | 50,757 | 11,622 | ENSG00000104723 |
| circ_0008815 | −4.92 | chr15 | 63,529,013 | 63,563,008 | + | 33,995 | 33,995 | ENSG00000140455 |
| circ_0000395 | −4.76 | chr10 | 124,038,514 | 124,046,724 | − | 8,210 | 7,159 | ENSG00000182022 |
| circ_0020149 | −4.70 | chr1 | 47,260,288 | 47,260,539 | − | 251 | 251 | ENSG00000123473 |
| circ_0018492 | −4.56 | chr1 | 21,001,198 | 21,050,994 | − | 49,796 | 49,796 | ENSG00000075151 |
| circ_0017073 | −4.47 | chr1 | 1,495,484 | 1,529,331 | + | 33,847 | 17,157 | ENSG00000160072 |
| circ_0005524 | −4.43 | chr12 | 56,643,402 | 56,645,353 | − | 1,951 | 1,951 | ENSG00000110955 |
| circ_0021024 | −4.31 | chr20 | 18,441,592 | 18,443,464 | − | 1,872 | 1,872 | ENSG00000089091 |
| circ_0007911 | −4.26 | chr14 | 90,578,105 | 90,618,045 | − | 39,940 | 39,940 | ENSG00000165914 |
| circ_0014803 | −4.12 | chr19 | 11,397,243 | 11,397,597 | − | 354 | 354 | – |
| circ_0040941 | −4.04 | chrX | 155,506,897 | 155,545,277 | − | 38,380 | 31,714 | ENSG00000185973 |
Features of the circRNAs
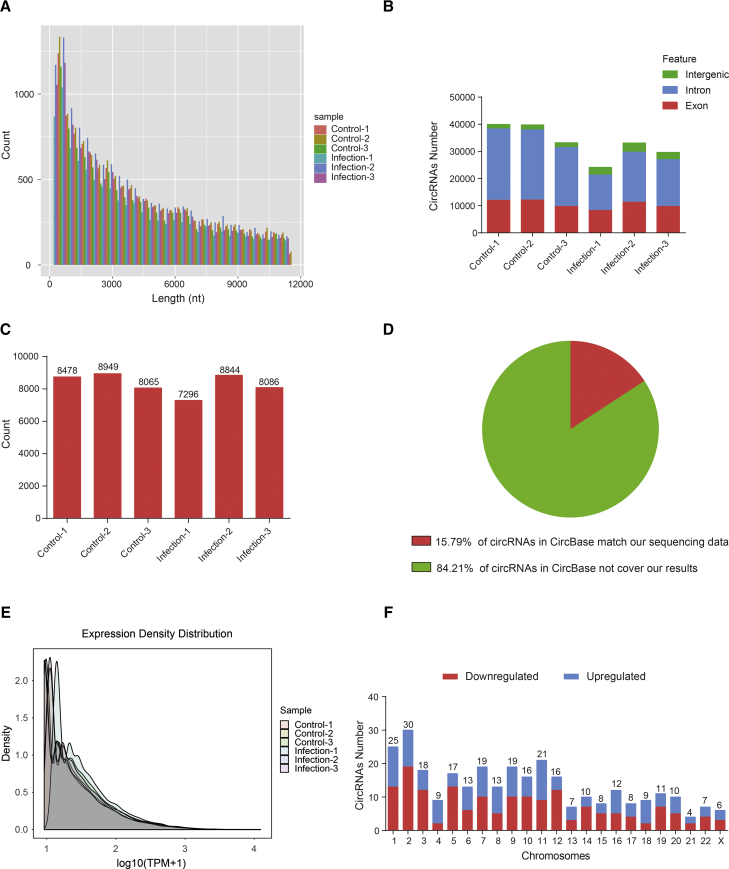
The circRNA lengths that were calculated for the control and infection samples were further analyzed. As shown in Figure 3A, the number of circRNAs decreased with an accompanying increase in the transcript length (Figure 3A). As it is known that circRNAs result from the diverse splicing of exons or introns, we subsequently analyzed the sources of the circRNAs in the different samples. We found most circRNAs identified herein were formed from exon or intron splicing, with the number of intron-derived circRNAs higher than that derived from exons. Only a small number of circRNAs were derived from intergenic regions (Figure 3B).
Figure 3.
Characterization of circRNAs in Primary hBMECs
(A) Transcript lengths for the 41,504 circRNAs from the primary hBMECs in six samples. (B) Genomic origins of the circRNAs from the primary hBMECs in six samples. (C) For each sample, the identities of the circRNAs were confirmed by the CircBase database. (D) The proportion of circRNAs in CircBase matching our sequencing data. (E) The expression density distribution of the circRNAs in each sample. (F) The distributions of the differentially expressed circRNAs on all 23 chromosomes. Red is the number of downregulated circRNAs and blue is the number of upregulated circRNAs.
We also compared all of the identified circRNAs with CircBase (http://www.circbase.org/), a database of known human circRNAs. At least 7,296 circRNAs in each sample could be matched in CircBase (Figure 3C). However, only approximately 15.79% of the human circRNAs recorded in CircBase matched our circRNA data, but the majority of the circRNAs in this database (84.21%) did not cover our results (Figure 3D), suggesting that novel circRNAs had been identified in abundance during the meningitic E. coli infection of primary hBMECs. When we compared the density distribution of circRNA expression, no significant differences among the samples were observed (Figure 3E). Finally, we found that all the chromosomes contained significantly upregulated and downregulated circRNAs, with chromosome 2 having the most circRNA transcripts (Figure 3F).
Real-Time qPCR Verification of the Significantly Changed circRNAs
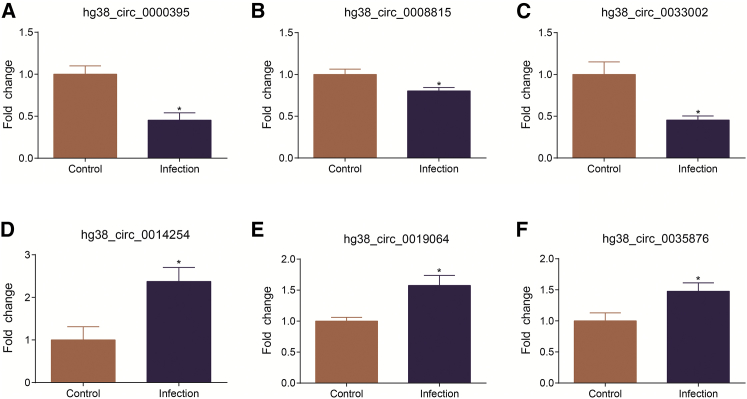
We next employed real-time qPCR to verify the results of the differentially altered circRNAs from the sequencing data. To do this, we selected 3 significantly upregulated and 3 significantly downregulated circRNAs from the sequencing data (hg38_circ_0000395, hg38_circ_0008815, hg38_circ_0033002, hg38_circ_0014254, hg38_circ_0019064, and hg38_circ_0035876). Divergent primers were designed to amplify the specific joint area of each circRNA, thus distinguishing the circRNAs from their linear parental mRNAs transcripts. The primers used for circRNA quantitation are listed in Table S4. The qPCR results showed that hg38_circ_0000395, hg38_circ_0008815, and hg38_circ_0033002 were significantly downregulated and hg38_circ_0014254, hg38_circ_0019064, and hg38_circ_0035876 were significantly upregulated upon meningitic E. coli infection (Figure 4), the trend of which was the same as that of the circRNA sequencing (circRNA-seq) data.
Figure 4.
qPCR Verification of the circRNA Transcription in Primary hBMECs in Response to Meningitic E. coli Infection
(A–F) The relative expression levels of 6 circRNAs, including 3 downregulated ones (A, hg38_circ_0000395; B, hg38_circ_0008815; C, hg38_circ_0033002) and 3 upregulated ones (D, hg38_circ_0014254; E, hg38_circ_0019064; F, hg38_circ_0035876), were examined by qPCR in primary hBMECs treated with or without meningitic E. coli for 3 hr. The data from circRNA-seq were highly consistent with the qPCR results. Data are expressed as the mean ± SEM from three independent experiments. *p < 0.05.
Altered circRNAs Participate in Multiple Biological Functions and Reflect the Regulation Patterns of Their Parental Genes
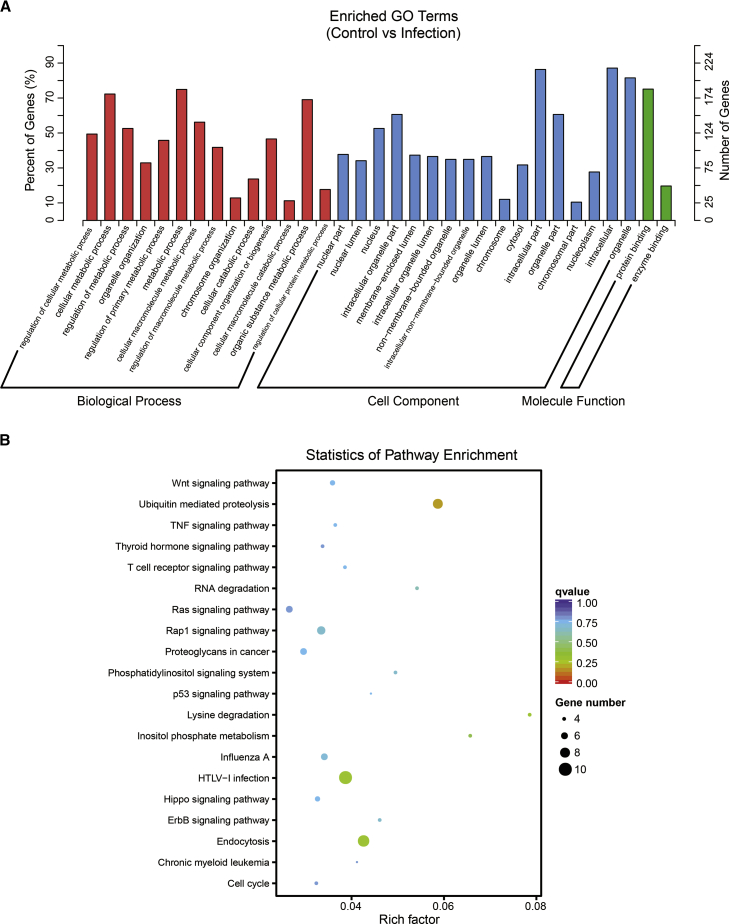
Most circRNAs are splice products of their parental genes, and they usually have a negative correlation with their linear parental mRNAs.26, 27 To better understand the potential roles as well as the regulatory effects of these circRNAs in the development of E. coli meningitis, bioinformatic approaches were used to analyze the potential function of the parental genes for the altered circRNAs in primary hBMECs upon infection. Via GO annotation and enrichment analyses, we observed that the biological processes of the circRNAs (or their parental mRNAs) mainly involved metabolism, including metabolic process and catabolic process. In the cell component category, the parental genes of the circRNAs were widely distributed in the cells. Interestingly, for molecule function, these altered circRNAs (or parental genes) were only enriched in two categories: protein binding and enzyme binding (Figure 5A).
Figure 5.
Summary of the Gene Ontology and KEGG Pathway Analysis for the Parental Genes of the Differentially Expressed circRNAs
(A) Gene ontology of the parental genes for the differentially expressed circRNAs. The y axis is the targeted gene numbers corresponding to the GO terms. (B) KEGG analysis of the 20 most enriched pathways. The coloring of the q-values indicates the significance of the rich factor. Circles indicate the target genes that are involved, and their sizes are proportional to the number of genes. The x axis represents the enrichment pathway name. The y axis represents the rich factor.
These results collectively indicate that the circRNAs that were altered upon meningitic E. coli infection were largely transcribed from metabolism-associated parental genes and are mainly involved in regulating protein interactions and enzyme activity. Moreover, the enriched signaling pathways by the parental genes of these altered circRNAs were determined with KEGG analysis, and the results revealed that several canonical signaling pathways were significantly enriched (e.g., endocytosis; ubiquitin-mediated proteolysis; and Rap1-, Ras-, Wnt-, tumor necrosis factor (TNF)-, p53-, Hippo-, and ErbB-signaling pathways), among which some signaling cascades have already been evidenced to be involved in meningitic E. coli infection (Figure 5B). More importantly, we elucidated the expression regulation patterns of the significantly altered circRNAs along with the significantly altered parental genes (Table 2). Among these, 32 pairs of circRNAs and parental mRNAs were positively correlated, while the other 42 pairs were negatively correlated. Notably, the parental early growth response 1 gene (egr-1, ENSG00000120738), which has been reported to show a variety of biological functions,28, 29 was the most significantly upregulated one (fold change, 36.61), implying that the circ_0031410-related positive regulation of egr-1 plays an important role during meningitic E. coli interaction with the BBB.
Table 2.
Both Significantly Changed circRNAs and Parental Genes (p values < 0.05)
| circRNA ID | circRNA Fold Change | mRNA ID | mRNA Fold Change | circRNA ID | circRNA Fold Change | mRNA ID | mRNA Fold Change |
|---|---|---|---|---|---|---|---|
| circ_0003433 | 5.21 | ENSG00000162337 | 1.21 | circ_0002276 | −1.91 | ENSG00000148925 | −1.23 |
| circ_0001582 | 4.48 | ENSG00000138160 | −1.22 | circ_0032477 | −1.98 | ENSG00000130449 | 1.28 |
| circ_0029541 | 3.71 | ENSG00000114480 | −1.20 | circ_0020568 | −2.06 | ENSG00000118454 | −1.24 |
| circ_0011368 | 3.55 | ENSG00000187741 | 1.38 | circ_0014954 | −2.11 | ENSG00000071564 | 1.21 |
| circ_0025677 | 3.29 | ENSG00000084676 | 1.16 | circ_0000613 | −2.13 | ENSG00000148468 | −1.43 |
| circ_0002057 | 3.24 | ENSG00000172273 | 1.21 | circ_0002902 | −2.14 | ENSG00000149187 | 1.21 |
| circ_0025824 | 2.95 | ENSG00000021574 | −1.20 | circ_0028659 | −2.24 | ENSG00000172939 | 1.18 |
| circ_0038073 | 2.94 | ENSG00000184661 | −1.28 | circ_0005816 | −2.32 | ENSG00000067798 | 1.45 |
| circ_0031410 | 2.90 | ENSG00000120738 | 36.61 | circ_0031368 | −2.35 | ENSG00000031003 | −1.64 |
| circ_0011744 | 2.87 | ENSG00000007168 | −1.33 | circ_0036597 | −2.38 | ENSG00000011275 | 1.14 |
| circ_0017427 | 2.85 | ENSG00000116580 | 1.23 | circ_0029371 | −2.50 | ENSG00000136068 | 1.22 |
| circ_0021902 | 2.78 | ENSG00000244274 | 1.15 | circ_0017198 | −2.51 | ENSG00000143398 | 1.28 |
| circ_0009167 | 2.76 | ENSG00000169375 | 1.28 | circ_0029484 | −2.55 | ENSG00000163378 | −1.24 |
| circ_0012892 | 2.73 | ENSG00000136450 | −1.53 | circ_0013583 | −2.55 | ENSG00000055483 | 1.46 |
| circ_0019739 | 2.65 | ENSG00000162591 | 1.47 | circ_0040511 | −2.60 | ENSG00000130956 | 1.15 |
| circ_0015873 | 2.60 | ENSG00000125753 | 1.38 | circ_0007338 | −2.62 | ENSG00000020577 | 1.34 |
| circ_0014050 | 2.55 | ENSG00000101752 | −1.23 | circ_0024599 | −2.63 | ENSG00000128708 | −1.30 |
| circ_0014152 | 2.52 | ENSG00000101596 | −1.19 | circ_0015638 | −2.68 | ENSG00000160410 | 1.18 |
| circ_0033186 | 2.51 | ENSG00000079819 | 1.25 | circ_0007369 | −2.72 | ENSG00000178974 | 1.18 |
| circ_0018931 | 2.48 | ENSG00000154370 | 1.58 | circ_0022363 | −2.77 | ENSG00000101187 | 1.37 |
| circ_0006595 | 2.18 | ENSG00000125257 | −1.14 | circ_0014713 | −2.78 | ENSG00000130816 | 1.16 |
| circ_0009677 | 2.13 | ENSG00000103222 | 1.24 | circ_0006251 | −2.83 | ENSG00000102781 | −1.21 |
| circ_0039245 | 1.99 | ENSG00000119487 | −1.26 | circ_0023875 | −3.25 | ENSG00000196576 | 1.20 |
| circ_0002067 | 1.96 | ENSG00000110395 | 1.26 | circ_0014054 | −3.34 | ENSG00000101752 | −1.23 |
| circ_0038782 | 1.86 | ENSG00000136891 | 1.35 | circ_0002026 | −3.38 | ENSG00000110367 | −1.30 |
| circ_0037202 | 1.84 | ENSG00000004864 | −1.28 | circ_0017793 | −3.41 | ENSG00000117115 | 1.17 |
| circ_0026699 | 1.82 | ENSG00000114978 | −1.29 | circ_0019085 | −3.45 | ENSG00000143674 | −1.28 |
| circ_0014023 | 1.78 | ENSG00000101557 | −1.18 | circ_0022441 | −3.62 | ENSG00000101161 | 1.17 |
| circ_0041472 | 1.67 | ENSG00000165288 | −1.11 | circ_0037683 | −3.62 | ENSG00000167632 | 1.26 |
| circ_0038029 | 1.53 | ENSG00000120889 | 1.61 | circ_0022523 | −3.75 | ENSG00000142192 | 1.18 |
| circ_0022192 | 1.31 | ENSG00000124164 | −1.30 | circ_0035593 | −3.86 | ENSG00000105939 | −2.32 |
| circ_0003309 | 1.30 | ENSG00000175592 | 3.69 | circ_0017073 | −4.47 | ENSG00000197785 | 1.18 |
| circ_0012974 | −1.29 | ENSG00000062716 | 1.45 | circ_0017073 | −4.47 | ENSG00000160072 | 1.29 |
| circ_0027134 | −1.31 | ENSG00000082701 | −1.27 | circ_0000395 | −4.76 | ENSG00000182022 | 1.43 |
| circ_0001286 | −1.33 | ENSG00000198879 | −1.30 | circ_0037938 | −5.03 | ENSG00000104723 | −1.15 |
| circ_0014655 | −1.55 | ENSG00000168502 | 1.18 | circ_0020478 | −5.13 | ENSG00000142856 | −1.15 |
| circ_0007114 | −1.85 | ENSG00000198604 | 1.22 | circ_0036105 | −6.95 | ENSG00000136237 | 1.23 |
ceRNA Analysis
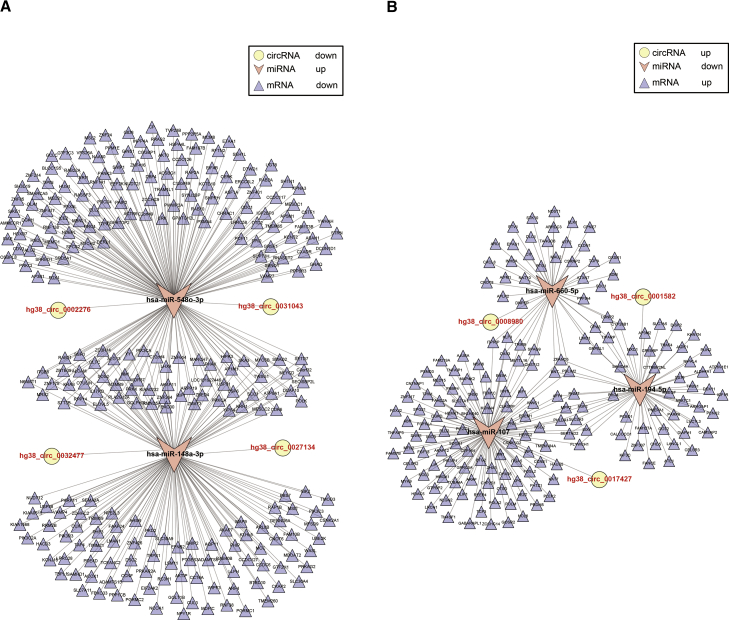
A classic function of circRNA regulation is the ability to act as ceRNAs, a process whereby cytosolic miRNAs are absorbed and prevented from binding to mRNAs, thereby influencing the expression or stability of the target mRNA.17 We therefore evaluated the ceRNA potential of the significantly altered circRNAs in hBMECs in response to meningitic E. coli. Here, the miRNA-binding sites on the circRNAs were predicted using miRanda.30 In combination with our previous mRNA transcriptional profiles,31 as well as our miRNA sequencing data (unpublished data), we constructed a whole circRNA-miRNA-mRNA transcriptional regulatory network (Tables S5 and S6), in which the circRNAs act as sponges, the miRNAs as mediators, and the mRNAs as the targets. Because there is support that circRNAs with the ability to sponge miRNAs are largely transcribed from exons,9 we paid particular attention to the exon-derived circRNAs by performing a circRNA-miRNA-mRNA integrated analysis of the circRNAs, as shown in Figure 6.
Figure 6.
circRNA-miRNA-mRNA Regulatory Network Analysis
(A) ceRNA analysis of the downregulated circRNAs derived from exons. (B) ceRNA analysis of the upregulated circRNAs derived from exons. In the circRNA-miRNA-mRNA network, circles represent circRNAs, recessed triangles represent miRNAs, and standard triangles represent mRNAs.
The results shown in Figure 6A reveal a down-up-down regulation pattern, in which downregulated hg38_circ_0027134 and hg38_circ_0032477 could sponge hsa-miR-148a-3p and downregulated hg38_circ_0031043 and hg38_circ_0002276 could sponge hsa-miR-548o-3p, thereby downregulating all of their target mRNAs. Conversely, Figure 6B reveals the up-down-up pattern, in which hg38_circ_0017427 could sponge has-miR-107, hg38_circ_0008980 could sponge hsa-miR-660-5p, and hg38_circ_0001582 could sponge hsa-miR-194-5p to exert positive regulation on these target mRNAs. Together, all the integrated data support the notion that, when acting as regulatory ceRNAs, the significantly altered circRNAs possess a strong ability to regulate gene expression.
Experimental Evidence of the Regulatory Effect of the circRNA on Its Target mRNA by Sponging Specific miRNA
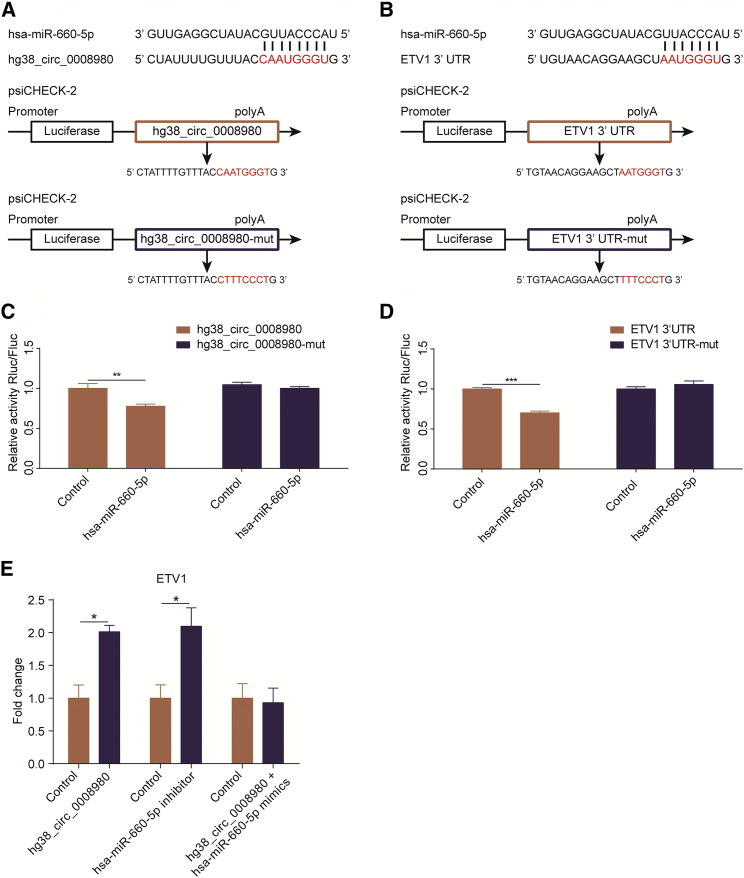
Based on the bioinformatic network shown in Figure 6, we sought to prove this regulatory pattern among circRNA, miRNA, and mRNA by experimental approaches. Since hg38_circ_0008980, hg38_circ_0001582, and hg38_circ_0017427 were significantly increased upon infection and included in the network (Figure 6B), we first tried to overexpress these three circRNAs by transfecting into 293T cells. The hg38_circ_0008980 showed an extremely significant overexpression in 293T cells, while the other two were not successfully circled and overexpressed (Figure S4). By sequence analysis, we did find that the hg38_circ_0008980, as well as the 3′ UTR of one target mRNA (such as ETV1, used as an example), contained the possible miRNA response elements (MREs) of hsa-miR-660-5p. Meanwhile, these hsa-miR-660-5p-binding regions on both hg38_circ_0008980 and the 3′ UTR of ETV1 were mutated (Figures 7A and 7B).
Figure 7.
The circRNA hg38_circ_0008980 Positively Regulates the Expression of ETV1 via Targeting the miRNA hsa-miR-660-5p
(A and B) The miRNA response elements (MREs) of hsa-miR-660-5p were shown on both the sequence of hg38_circ_0008980 (A) and ETV1 3′ UTR (B), and mutations were introduced on these MREs, respectively. Both wild-type and the mutated sequences were cloned into psiCHECK-2 plasmid. (C and D) Dual-luciferase reporter assays showed that hsa-miR-660-5p transfection significantly decreased the luciferase activity of the wild-type hg38_circ_0008980 (C) or ETV1 3′ UTR (D), while luciferase activities of the mutated constructs were not affected. The relative activities were expressed as the renilla luciferase activity normalized to the firefly luciferase activity. (E) The hg38_circ_0008980 overexpression as well as hsa-miR-660-5p inhibition significantly increased the transcription of ETV1 mRNA, and, in contrast, the hsa-miR-660-5p mimics completely blocked hg38_circ_0008980 overexpression-mediated upregulation of ETV1. Data were expressed as the mean ± SEM from three independent assays. *p < 0.05, **p < 0.01, and ***p < 0.001.
Through dual-luciferase reporter assays, we observed that transfection of the hsa-miR-660-5p significantly decreased the transcriptional activity of the wild-type hg38_circ_0008980, but not that of the mutated circRNA. Likewise, hsa-miR-660-5p significantly decreased the luciferase activity of ETV1 3′ UTR, but the mutated 3′ UTR was not affected at all (Figures 7C and 7D). These results indicated a direct association and regulation between hg38_circ_0008980 and hsa-miR-660-5p, as well as between hsa-miR-660-5p and the ETV1 3′ UTR. By real-time PCR, we moreover observed that both hg38_circ_0008980 overexpression and the hsa-miR-660-5p inhibition could significantly increase the ETV1 expression, while hg38_circ_0008980 overexpression-mediated upregulation of ETV1 was totally prevented by hsa-miR-660-5p mimics (Figure 7E), indicating that the positive regulation of hg38_circ_0008980 on its target gene ETV1 is likely dependent on hsa-miR-660-5p. These results further supported that the abovementioned circRNA-miRNA-mRNA network is likely to be a new molecular regulation mechanism and should exert certain important regulatory effects on meningitic E. coli penetration of the BBB.
Discussion
With their high abundance in the brain, circRNAs have recently been discovered to be important cellular regulators of diverse biological processes, especially in multiple CNS-associated diseases.12 We have previously reported on the transcriptional profiling of host mRNAs, miRNAs, and long noncoding RNAs (lncRNAs) in primary hBMECs in response to meningitis E. coli, and we have identified several of the key molecules involved in the interaction of meningitic E. coli with the BBB.6 To further supplement our whole transcriptional profiling and investigate the potential regulatory network among the noncoding and coding RNAs, the circRNA libraries from primary hBMECs with or without meningitic E. coli challenge were constructed and sequenced. A total of 41,504 circRNAs were identified in six samples from two groups, among which 140 were found to be significantly upregulated while 168 were significantly downregulated in the infected samples compared with the controls. Also, the expression features of these circRNAs, the GO and KEGG enrichment for the parental genes of the differentially expressed circRNAs, and the ceRNA analysis integrated with our previous mRNA and miRNA data as well as the experimental approaches were performed to further illustrate the potential roles of the altered circRNAs in the process of E. coli meningitis. As far as we know, this is the first report on circRNA transcription in primary hBMECs upon meningitic E. coli infection.
By expression feature analysis of the circRNAs in hBMECs, we found that most of the circRNAs were derived from intron and exon splicing. When comparing all the circRNAs we identified with the known human circRNAs in CircBase, a total of 17,275 identified circRNAs in this study were pairable, and 15.79% of the human circRNAs recorded in this database were identified by our circRNA sequencing. The result reflects the specificity of circRNA expression in different cell lines, and also it extends the existing number of circRNAs in the current database. In addition, by comparing the density distribution of the circRNAs, we found that infection with meningitic E. coli did not influence the density distribution of the circRNAs in the primary hBMECs.
It was recently reported that blood contains a higher abundance of circRNAs than mRNAs, indicating the potential of circRNAs to act as novel biomarkers in standard clinical blood samples, especially in cancer research.32 One report showed that circRNA hsa_circ_0001895 was significantly downregulated in five gastric cancer cell lines (AGS, BGC-823, HGC-27, MGC-803, and SGC-7901) compared with normal gastric epithelial cells (GES-1), and the in vivo experiments further indicated that hsa_circ_0001895 was significantly downregulated in cancer tissues and gastric precancerous lesions alike, which supports hsa_circ_0001895 as a potential biomarker for clinical prognosis prediction.33 Likewise, hsa_circ_0001649 was found to be significantly downregulated in hepatocellular carcinoma tissues compared with the paired adjacent liver tissues, indicating the possibility that hsa_circ_0001649 might serve as a novel potential biomarker during hepatocellular carcinoma development.34 In multiple sclerosis, circ_0005402 and circ_0035560 exhibit low expressional abundances in patients, making them potential biomarkers of this disease.22 Furthermore, studies have reported that circRNA_103636 is a potential novel diagnostic and therapeutic biomarker for major depressive disorder.35 Our study identified hundreds of circRNAs as being differentially expressed in primary hBMECs upon meningitic E. coli infection. Whether these differentially expressed circRNAs can be persistently detected from the bloodstream, and thus used as potential diagnostic biomarkers for E. coli meningitis, remains to be seen.
circRNAs are known to regulate the expression of their parental genes,36, 37 and a previous study reported that circRNA production and pre-mRNA splicing compete with each other.26 For example, a circRNA molecule called Circ-Mbl modulates the linear mRNA ratio by competing with the linear muscleblind gene from which it is synthesized.38 Similarly, two studies have revealed that circ-ITCH can act as a sponge for oncogenic miR-7 and miR-214 to enhance ITCH expression, thus suppressing the activation of Wnt/β-catenin signaling.15, 39 A recent study on stroke showed that the host genes that form stroke-responsive circRNAs participate in the process of metabolism, response to stimulus, protein binding, ion binding, and nucleic acid binding, and all these processes are necessary for the survival of healthy neurons.24 Here, our bioinformatic analysis indicated that the parental genes of the altered circRNAs in the primary hBMECs upon meningitic E. coli infection are involved mainly in the Wnt/β-catenin-signaling pathway and in ubiquitin-mediated proteolysis in the TNF-signaling pathway, for example. Both of these are essential molecular events in the inflammatory response during host and pathogen interactions.
The function of circRNAs as miRNA sponges in the regulation of gene expression has been widely reported.40 The most well-known circRNA, ciRS-7 (also called CDR1), is highly expressed in the brain and contains more than 70 conventional binding sites as the sponge for miR-7.16 The biological functions of ciRS-7 in cancer (e.g., hepatocellular carcinoma and colorectal cancer) have been widely reported. For example, overexpression of ciRS-7 can promote hepatocellular carcinoma proliferation and invasion by targeting miR-7,41 and ciRS-7 regulates the epidermal growth factor receptor (EGFR)-RAF1-mitogen-activated protein kinase (MAPK) pathway by inhibiting miR-7 activity in colorectal cancer.42 circHIPK3, another well-known circRNA, is derived from the second exon of the HIPK3 gene and is particularly highly abundant with a high back-spliced ratio.43, 44 It has been shown that circHIPK3 can bind directly to miR-124 and inhibit miR-124 activity to regulate cell growth.45 Also, circHIPK3 was able to sponge miR-558 to suppress heparanase expression in bladder cancer cells.19
Moreover, there is support that circRar1 can induce upregulation of the apoptosis-associated factors caspase 8 and p38 by modulating its target miRNA, miR-671,46 and that circ-HRCR acts as an endogenous miR-223 sponge to inhibit cardiac hypertrophy and heart failure.47 In a recent study, circRNAs were also reported to regulate the BBB permeability by acting as miRNA sponge. CircHECW2 can competitively sponge mir-30d to upregulate the expression of ATG5, promote the transformation of BMECs into mesenchymal cells, and destroy the BBB function.48 In a stroke model, circDLGAP4 was significantly downregulated, and overexpression of circDLGAP4 significantly attenuated neurological deficits and decreased infarct areas and BBB damage in the transient middle cerebral artery occlusion mouse stroke model by competitively binding miR-143 to regulate the tight junction protein expression.49 In our study, the ceRNA potential of the altered circRNAs was evaluated, and partial networks for the significantly altered miRNAs, mRNAs, and circRNAs in primary hBMECs in response to meningitic E. coli were preliminarily deduced as well as evidenced by experimental approaches. These results provide potential novel nucleic acid targets for further elucidating the molecular mechanisms and the underlying regulation of bacterial meningitis development.
circRNAs are prominently involved in numerous diseases, and their transcriptional profiles have been increasingly reported in different disease models, such as bladder carcinoma,50 hypopharyngeal squamous cell carcinoma,51 and multiple system atrophy,52 etc. Here, our work showed that bacterial infection of hBMECs caused transcriptional alteration of the circRNA profiles in these cells. Identification of thousands of novel circRNAs in this study hugely supplements those already known in the circRNA database. More importantly, KEGG and GO enrichment of the parental genes of these altered circRNAs and their ceRNA analysis indicate that circRNAs could be the important nucleic acid targets involved in meningitic E. coli infection of primary hBMECs. Collectively, our data augment the number of known circRNAs and deepen the current understanding of circRNAs, thus providing a theoretical basis for the future study of circRNAs as diagnostic and therapeutic targets in bacterial meningitis.
Materials and Methods
Cell Culture
The primary hBMECs we used in this study were kindly provided by Professor Kwang Sik Kim (Johns Hopkins University School of Medicine, USA). They were routinely cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), 10% Nu-Serum, 2 mM L-glutamine, 1 mM sodium pyruvate, nonessential amino acids, vitamins, penicillin, and streptomycin (100 U/mL).53 Cells were incubated in a 37°C incubator under 5% CO2 until monolayer confluence. Confluent cells were washed three times with Hank’s balanced salt solution (Corning Cellgro, Manassas, VA, USA) and starved in serum-free medium (1:1 mixture of Ham’s F-12 and M-199) for 16–18 hr before further treatment. The 293T cells were routinely cultured in DMEM (Invitrogen, Carlsbad, CA, USA) supplemented with 10% FBS in a 37°C incubator under 5% CO2 until 70% confluence prior to the transfection.
Preparation of Meningitic E. coli-Infected hBMECs
The meningitic E. coli strain used in this study, PCN033, was isolated from swine cerebrospinal fluid in Hunan Province, China, in 2006.54 The strain was routinely grown aerobically at 37°C in Luria-Bertani (LB) medium overnight. PCN033-induced infections of primary hBMECs were performed in accordance with the methods described previously.31 Briefly, an overnight E. coli culture was resuspended and diluted in serum-free medium, and then it was added to a starved confluent primary hBMEC monolayer grown in 10-cm dishes at an MOI of 10 (approximately 108 colony-forming units per dish) to allow invasion at 37°C for 3 hr. This infection model was validated by real-time PCR analysis of multiple host molecules, such as vascular endothelial growth factor A (VEGFA), interleukin (IL)-6, MCP-1, and those tight junction proteins, all of which have been evidenced to be involved in response to the infection.8 The cells were then washed three times with chilled PBS and subjected to RNA extraction with TRIzol reagent (Aidlab Biotech, Beijing, China), in accordance with the manufacturer’s instructions.
circRNA Library Preparation and Sequencing
Six libraries were constructed and sequenced by Novogene (Beijing, China). RNA sample preparation was based on 5 μg RNA for each sample. First, the Epicenter Ribo-zero rRNA Removal Kit (Epicenter, Madison, WI, USA) was used to deplete the rRNAs. The rRNA-depleted RNAs were then subjected to TRIzol extraction after RNase R (Epicenter, Madison, WI, USA) treatment. Next, the sequencing libraries were constructed using the RNase R-digested RNAs and the rRNA-depleted RNAs by NEBNext Ultra Directional RNA Library Prep Kit for Illumina (New England Biolabs, Beverly, MA, USA), following the manufacturer’s guidelines. In short, divalent cations were used to accomplish fragmentation in the NEBNext First Strand Synthesis Reaction Buffer, under an elevated temperature. First-strand cDNA was prepared by using a random hexamer primer and M-MuLV Reverse Transcriptase (RNaseH−), and the second-strand cDNA synthesis was performed using DNA polymerase I and RNase H. dUTP was replaced with dTTP in the dNTPs in the reaction buffer, while other overhangs were transformed into blunt ends via exonuclease and polymerase activities. For hybridization, a NEBNext adaptor containing a hairpin loop structure was ligated, after adenylating the 3′ ends of the DNA fragments. Library fragments were purified using the AMPure XP system (Beckman Coulter, Beverly, MA, USA) to select the best cDNA fragments of 150–200 bp. Before proceeding with PCR, 3 μL USER Enzyme (New England Biolabs) with size-selected, adaptor-ligated cDNA was added and the reaction was incubated 37°C for 15 min followed by 95°C for 5 min. PCR was equipped with Index (X) Primer, Phusion High-Fidelity DNA polymerase, and Universal PCR primer. After library purification (AMPure XP system), DNA fragments were qualified by the Agilent Bioanalyzer 2100 system.
The cBot Cluster Generation System using HiSeq PE Cluster Kit version (v.)4 cBot (Illumina) was used for clustering the index-coded samples, according to the manufacturer’s recommendations. After cluster generation, the library preparations were sequenced on an Illumina Hiseq 2500 platform, and 125-bp paired-end reads were generated.
circRNA Identification and Differential Expression Analysis
Raw data in fastq format were first treated with a custom perl script. Clean reads were acquired after eliminating the adaptor-containing reads, the ploy-N-containing reads, and the low-quality reads from the raw data. The Q20, Q30, and GC contents of the selected reads were calculated. All downstream analysis was based on the high-quality, clean data generated in this step. The reference genome and gene annotations were downloaded from the genome website (ftp://ftp.ensembl.org/pub/release-84/fasta/homo_sapiens/dna/). The reference genome index was built using Bowtie v.2.0.6,55 and the paired-end clean reads were aligned to the reference genome with TopHat v.2.0.9.
20-mers from the 5′ and 3′ ends of the unmapped reads were removed and placed independently of the reference sequences by Bowtie v.2.0.6. Anchor sequences were expanded by find_circ16 such that the complete read was aligned and the breakpoints were flanked by GU/AG splice sites. The back-spliced reads with at least two supporting reads were then annotated as circRNAs. The circRNA expression levels were normalized by TPM (transcript per million) using the following criteria:56 normalized expression = (mapped reads)/(total reads) * 1,000,000.
Differential expression analysis for the control and infection groups was performed using DESeq2 (v. 1.6.3).57 p values were adjusted using the Benjamini-Hochberg method.58 By default, the threshold of the corrected p value for each differential expression result was set to 0.05.
GO and KEGG Enrichment Analysis
GO enrichment analysis for the source genes of the differentially expressed circRNAs was performed by GOseq (v. 1.18.0).59 KEGG60 is a database for understanding the high-level functions and utilities of a biological system, such as the cell, the organism, and the ecosystem, from the molecular-level information, especially the large-scale molecular datasets generated by genome sequencing and other high-throughput experimental technologies (https://www.genome.jp/kegg/). KOBAS61 was used for KEGG pathway enrichment analysis.
Transfection
The circRNA overexpression was performed by cloning the whole circRNA sequence into a commercial vector purchased from BersinBio (Guangzhou, China). The hsa-miR-660-5p mimics and inhibitor were obtained from GenePharma (Shanghai, China). Transfections were performed using jetPRIME reagent (Polyplus Transfection, Illkirch, France), following the manufacturer’s protocol. The medium was replaced into the complete medium after 5 hr of transfection, and the total RNAs were collected after another 48 hr of incubation.
Dual-Luciferase Assay
The hg38_circ_0008980, hg38_circ_0008980-mut, ETV1 3′ UTR, and ETV1 3′ UTR-mut were synthesized by Genscript (Nanjing, China) and cloned into psiCHECK-2 plasmid for the dual-luciferase assay. The 293T cells were seeded in 24-well plates and cultured until 70% confluence before transfection. For each experimental group, 200 ng plasmids and 50 μmol miRNA mimics or negative control were used. After 24 hr of incubation, the Dual-Luciferase system was used to detect both firefly and renilla luciferase activities, following the manufacturer’s instructions (Promega, Madison, WI, USA). The results were calculated as the ratio of renilla luciferase activity and the firefly luciferase activity, and they were recorded as the mean ± SEM from three replicated wells.
RNA Isolation and Real-Time qPCR Analysis
Total RNA was extracted from primary hBMECs using TRIzol reagent. The linear-stranded RNA was removed by RNase R (Epicenter, Madison, WI, USA). RNA (1 μg) was reverse transcribed into cDNA with HiScript II Q RT SuperMix for qPCR (Vazyme, Nanjing, China).
Changes in the circRNAs in the primary hBMECs in response to meningitic E. coli were validated by real-time qPCR for 3 downregulated (hg38_circ_0000395, hg38_circ_0008815, and hg38_circ_0033002) and 3 upregulated (hg38_circ_0014254, hg38_circ_0019064, and hg38_circ_0035876) circRNAs. The circRNAs were amplified with circRNA-specific outward-facing divergent primers (listed in Table S4). Real-time PCR was performed with the CFX96 Real-Time System (Bio-Rad, Hercules, CA, USA) using FastStart Universal SYBR Green Master (Roche, Basel, Switzerland), according to the manufacturer’s recommendations. The amplification conditions were 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Melting curve analysis of the products was conducted with denaturation at 95°C for 15 s and annealing at 60°C for 1 min, with slow dissociation by ramping from 60°C to 95°C at 0.05°C/s to ensure the specificities of the PCR products. GAPDH was used as the internal control to normalize the data.
Statistical Analysis
Data are expressed as the mean ± SEM, and the statistical significance of the differences between the groups was evaluated by the Student’s t test. p values < 0.05 (*) were considered significant, and p values < 0.01 (**) as well as p values < 0.001 (***) were all considered extremely significant. Graphs were plotted and analyzed using GraphPad Prism v. 6.0 (GraphPad, La Jolla, CA, USA).
The transcriptomic data have been deposited at the BioProject database of the National Center for Biotechnology Information (NCBI): SRP116604.
Author Contributions
R.Y. performed the experiments, analyzed the data, and drafted the whole manuscript. C.T., H.C., B.X., and A.C. provided the technical support and helped with the ceRNA analysis. B.Y., J.F., L.L., and N.A. participated in the experiments. X.W. conceived and designed the experiments and revised the manuscript.
Conflicts of Interest
The authors declare no competing financial interests.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (NSFC) (31772736 and 31502062), the National Key R&D Program of China (2016YFD0500406), the Outstanding Youth Project of Natural Science Foundation in Hubei Province (2018CFA070), and the Fundamental Research Funds for the Central Universities (2662018PY032).
Footnotes
Supplemental Information includes four figures and six tables and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.10.013.
Supplemental Information
References
- 1.Kim K.S. Mechanisms of microbial traversal of the blood-brain barrier. Nat. Rev. Microbiol. 2008;6:625–634. doi: 10.1038/nrmicro1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burkhart A., Thomsen L.B., Thomsen M.S., Lichota J., Fazakas C., Krizbai I., Moos T. Transfection of brain capillary endothelial cells in primary culture with defined blood-brain barrier properties. Fluids Barriers CNS. 2015;12:19. doi: 10.1186/s12987-015-0015-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abbott N.J., Patabendige A.A., Dolman D.E., Yusof S.R., Begley D.J. Structure and function of the blood-brain barrier. Neurobiol. Dis. 2010;37:13–25. doi: 10.1016/j.nbd.2009.07.030. [DOI] [PubMed] [Google Scholar]
- 4.Schubert-Unkmeir A., Konrad C., Slanina H., Czapek F., Hebling S., Frosch M. Neisseria meningitidis induces brain microvascular endothelial cell detachment from the matrix and cleavage of occludin: a role for MMP-8. PLoS Pathog. 2010;6:e1000874. doi: 10.1371/journal.ppat.1000874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nau R., Djukic M., Spreer A., Ribes S., Eiffert H. Bacterial meningitis: an update of new treatment options. Expert Rev. Anti Infect. Ther. 2015;13:1401–1423. doi: 10.1586/14787210.2015.1077700. [DOI] [PubMed] [Google Scholar]
- 6.Wang X., Maruvada R., Morris A.J., Liu J.O., Wolfgang M.J., Baek D.J., Bittman R., Kim K.S. Sphingosine 1-phosphate activation of EGFR as a novel target for meningitic Escherichia coli penetration of the blood-brain barrier. PLoS Pathog. 2016;12:e1005926. doi: 10.1371/journal.ppat.1005926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim K.S. Pathogenesis of bacterial meningitis: from bacteraemia to neuronal injury. Nat. Rev. Neurosci. 2003;4:376–385. doi: 10.1038/nrn1103. [DOI] [PubMed] [Google Scholar]
- 8.Yang R., Liu W., Miao L., Yang X., Fu J., Dou B., Cai A., Zong X., Tan C., Chen H., Wang X. Induction of VEGFA and Snail-1 by meningitic Escherichia coli mediates disruption of the blood-brain barrier. Oncotarget. 2016;7:63839–63855. doi: 10.18632/oncotarget.11696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeck W.R., Sharpless N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y., Zheng Q., Bao C., Li S., Guo W., Zhao J., Chen D., Gu J., He X., Huang S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Danan M., Schwartz S., Edelheit S., Sorek R. Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res. 2012;40:3131–3142. doi: 10.1093/nar/gkr1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen W., Schuman E. Circular RNAs in brain and other tissues: a functional enigma. Trends Neurosci. 2016;39:597–604. doi: 10.1016/j.tins.2016.06.006. [DOI] [PubMed] [Google Scholar]
- 13.Du W.W., Fang L., Yang W., Wu N., Awan F.M., Yang Z., Yang B.B. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24:357–370. doi: 10.1038/cdd.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y., Zhang X.O., Chen T., Xiang J.F., Yin Q.F., Xing Y.H., Zhu S., Yang L., Chen L.L. Circular intronic long noncoding RNAs. Mol. Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 15.Li F., Zhang L., Li W., Deng J., Zheng J., An M., Lu J., Zhou Y. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget. 2015;6:6001–6013. doi: 10.18632/oncotarget.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 17.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 18.Chen J., Li Y., Zheng Q., Bao C., He J., Chen B., Lyu D., Zheng B., Xu Y., Long Z. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208–219. doi: 10.1016/j.canlet.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 19.Li Y., Zheng F., Xiao X., Xie F., Tao D., Huang C., Liu D., Wang M., Wang L., Zeng F., Jiang G. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017;18:1646–1659. doi: 10.15252/embr.201643581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang X.Y., Huang Z.L., Xu Y.H., Zheng Q., Chen Z., Song W., Zhou J., Tang Z.Y., Huang X.Y. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-100338/miR-141-3p pathway in hepatitis B-related hepatocellular carcinoma. Sci. Rep. 2017;7:5428. doi: 10.1038/s41598-017-05432-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nair A.A., Niu N., Tang X., Thompson K.J., Wang L., Kocher J.P., Subramanian S., Kalari K.R. Circular RNAs and their associations with breast cancer subtypes. Oncotarget. 2016;7:80967–80979. doi: 10.18632/oncotarget.13134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iparraguirre L., Muñoz-Culla M., Prada-Luengo I., Castillo-Triviño T., Olascoaga J., Otaegui D. Circular RNA profiling reveals that circular RNAs from ANXA2 can be used as new biomarkers for multiple sclerosis. Hum. Mol. Genet. 2017;26:3564–3572. doi: 10.1093/hmg/ddx243. [DOI] [PubMed] [Google Scholar]
- 23.Lukiw W.J. Circular RNA (circRNA) in Alzheimer’s disease (AD) Front. Genet. 2013;4:307. doi: 10.3389/fgene.2013.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mehta S.L., Pandi G., Vemuganti R. Circular RNA expression profiles alter significantly in mouse brain after transient focal ischemia. Stroke. 2017;48:2541–2548. doi: 10.1161/STROKEAHA.117.017469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang G., Ma Y., An T., Pan Y., Mo F., Zhao D., Liu Y., Miao J.N., Gu Y.J., Wang Y., Gao S.H. Relationships of circular RNA with diabetes and depression. Sci. Rep. 2017;7:7285. doi: 10.1038/s41598-017-07931-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 27.Kelly S., Greenman C., Cook P.R., Papantonis A. Exon skipping is correlated with exon circularization. J. Mol. Biol. 2015;427:2414–2417. doi: 10.1016/j.jmb.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 28.Koldamova R., Schug J., Lefterova M., Cronican A.A., Fitz N.F., Davenport F.A., Carter A., Castranio E.L., Lefterov I. Genome-wide approaches reveal EGR1-controlled regulatory networks associated with neurodegeneration. Neurobiol. Dis. 2014;63:107–114. doi: 10.1016/j.nbd.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang L., Jiang Y., Wen Z., Xu X., Xu X., Zhu J., Xie X., Xu L., Xie Y., Liu X., Xu G. Over-expressed EGR1 may exaggerate ischemic injury after experimental stroke by decreasing BDNF expression. Neuroscience. 2015;290:509–517. doi: 10.1016/j.neuroscience.2015.01.020. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y.C., Liao J.Y., Li Z.Y., Yu Y., Zhang J.P., Li Q.F., Qu L.H., Shu W.S., Chen Y.Q. Genome-wide screening and functional analysis identify a large number of long noncoding RNAs involved in the sexual reproduction of rice. Genome Biol. 2014;15:512. doi: 10.1186/s13059-014-0512-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang R., Huang F., Fu J., Dou B., Xu B., Miao L., Liu W., Yang X., Tan C., Chen H., Wang X. Differential transcription profiles of long non-coding RNAs in primary human brain microvascular endothelial cells in response to meningitic Escherichia coli. Sci. Rep. 2016;6:38903. doi: 10.1038/srep38903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dong Y., He D., Peng Z., Peng W., Shi W., Wang J., Li B., Zhang C., Duan C. Circular RNAs in cancer: an emerging key player. J. Hematol. Oncol. 2017;10:2. doi: 10.1186/s13045-016-0370-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shao Y., Chen L., Lu R., Zhang X., Xiao B., Ye G., Guo J. Decreased expression of hsa_circ_0001895 in human gastric cancer and its clinical significances. Tumour Biol. 2017;39 doi: 10.1177/1010428317699125. 1010428317699125. [DOI] [PubMed] [Google Scholar]
- 34.Qin M., Liu G., Huo X., Tao X., Sun X., Ge Z., Yang J., Fan J., Liu L., Qin W. Hsa_circ_0001649: A circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016;16:161–169. doi: 10.3233/CBM-150552. [DOI] [PubMed] [Google Scholar]
- 35.Cui X., Niu W., Kong L., He M., Jiang K., Chen S., Zhong A., Li W., Lu J., Zhang L. hsa_circRNA_103636: potential novel diagnostic and therapeutic biomarker in Major depressive disorder. Biomarkers Med. 2016;10:943–952. doi: 10.2217/bmm-2016-0130. [DOI] [PubMed] [Google Scholar]
- 36.Chen L.L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 2016;17:205–211. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 37.Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., Zhong G., Yu B., Hu W., Dai L. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 38.Kumar L., Shamsuzzama, Haque R., Baghel T., Nazir A. Circular RNAs: the emerging class of non-coding RNAs and their potential role in human neurodegenerative diseases. Mol. Neurobiol. 2017;54:7224–7234. doi: 10.1007/s12035-016-0213-8. [DOI] [PubMed] [Google Scholar]
- 39.Wan L., Zhang L., Fan K., Cheng Z.X., Sun Q.C., Wang J.J. Circular RNA-ITCH suppresses lung cancer proliferation via inhibiting the Wnt/β-catenin pathway. Biomed Res. Int. 2016;2016:1579490. doi: 10.1155/2016/1579490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meng X., Li X., Zhang P., Wang J., Zhou Y., Chen M. Circular RNA: an emerging key player in RNA world. Brief. Bioinform. 2017;18:547–557. doi: 10.1093/bib/bbw045. [DOI] [PubMed] [Google Scholar]
- 41.Yu L., Gong X., Sun L., Zhou Q., Lu B., Zhu L. The circular RNA Cdr1as act as an oncogene in hepatocellular carcinoma through targeting miR-7 expression. PLoS ONE. 2016;11:e0158347. doi: 10.1371/journal.pone.0158347. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Weng W., Wei Q., Toden S., Yoshida K., Nagasaka T., Fujiwara T., Cai S., Qin H., Ma Y., Goel A. Circular RNA ciRS-7-A promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin. Cancer Res. 2017;23:3918–3928. doi: 10.1158/1078-0432.CCR-16-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liang D., Wilusz J.E. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28:2233–2247. doi: 10.1101/gad.251926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng Q., Bao C., Guo W., Li S., Chen J., Chen B., Luo Y., Lyu D., Li Y., Shi G. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nan A., Chen L., Zhang N., Liu Z., Yang T., Wang Z., Yang C., Jiang Y. A novel regulatory network among LncRpa, CircRar1, MiR-671 and apoptotic genes promotes lead-induced neuronal cell apoptosis. Arch. Toxicol. 2017;91:1671–1684. doi: 10.1007/s00204-016-1837-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang K., Long B., Liu F., Wang J.X., Liu C.Y., Zhao B., Zhou L.Y., Sun T., Wang M., Yu T. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur. Heart J. 2016;37:2602–2611. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- 48.Yang L., Han B., Zhang Y., Bai Y., Chao J., Hu G., Yao H. Engagement of circular RNA HECW2 in the nonautophagic role of ATG5 implicated in the endothelial-mesenchymal transition. Autophagy. 2018;14:404–418. doi: 10.1080/15548627.2017.1414755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bai Y., Zhang Y., Han B., Yang L., Chen X., Huang R., Wu F., Chao J., Liu P., Hu G. Circular RNA DLGAP4 ameliorates ischemic stroke outcomes by targeting miR-143 to regulate endothelial-mesenchymal transition associated with blood-brain barrier integrity. J. Neurosci. 2018;38:32–50. doi: 10.1523/JNEUROSCI.1348-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhong Z., Lv M., Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci. Rep. 2016;6:30919. doi: 10.1038/srep30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cao S., Wei D., Li X., Zhou J., Li W., Qian Y., Wang Z., Li G., Pan X., Lei D. Novel circular RNA expression profiles reflect progression of patients with hypopharyngeal squamous cell carcinoma. Oncotarget. 2017;8:45367–45379. doi: 10.18632/oncotarget.17488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen B.J., Mills J.D., Takenaka K., Bliim N., Halliday G.M., Janitz M. Characterization of circular RNAs landscape in multiple system atrophy brain. J. Neurochem. 2016;139:485–496. doi: 10.1111/jnc.13752. [DOI] [PubMed] [Google Scholar]
- 53.Zhu L., Pearce D., Kim K.S. Prevention of Escherichia coli K1 penetration of the blood-brain barrier by counteracting the host cell receptor and signaling molecule involved in E. coli invasion of human brain microvascular endothelial cells. Infect. Immun. 2010;78:3554–3559. doi: 10.1128/IAI.00336-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu C., Zheng H., Yang M., Xu Z., Wang X., Wei L., Tang B., Liu F., Zhang Y., Ding Y. Genome analysis and in vivo virulence of porcine extraintestinal pathogenic Escherichia coli strain PCN033. BMC Genomics. 2015;16:717. doi: 10.1186/s12864-015-1890-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou L., Chen J., Li Z., Li X., Hu X., Huang Y., Zhao X., Liang C., Wang Y., Sun L. Integrated profiling of microRNAs and mRNAs: microRNAs located on Xq27.3 associate with clear cell renal cell carcinoma. PLoS ONE. 2010;5:e15224. doi: 10.1371/journal.pone.0015224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Benjamini Y., Drai D., Elmer G., Kafkafi N., Golani I. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 59.Young M.D., Wakefield M.J., Smyth G.K., Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M., Itoh M., Katayama T., Kawashima S., Okuda S., Tokimatsu T., Yamanishi Y. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mao X., Cai T., Olyarchuk J.G., Wei L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics. 2005;21:3787–3793. doi: 10.1093/bioinformatics/bti430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.