Abstract
Extracellular vesicles (EVs) released by virus-infected cells typically incorporate host and viral components inside the vesicles (cargo molecules). Here, we investigated if human cytomegalovirus (HCMV) proteins are incorporated in EV outer membrane released by HCMV-infected cells. We separated EVs from HCMV using an iodixanol step-gradient and found that the separated vesicles carried EV markers such as the tetraspanin CD63 and Rab27A. Flow analysis of individual EVs demonstrated that on average, 15 ± 3.7% of EVs were positive for gB, 5.3 ± 2.3% were positive for gH and 3.74 ± 1.5% were positive for both gB and gH. In light of previous findings demonstrating HIV envelope proteins in EV membranes, the presence of viral protein at the surface of EVs released by HCMV-infected cells indicated that viral membrane proteins incorporated in EVs released by virus-infected cells may be a general phenomenon.
Keywords: Extracellular vesicles, HCMV, gB, gH
1. Introduction
Extracellular vesicles (EVs) are released by many, if not by all, cells in humans. Initially identified as platelet products and thought to be “platelet dust” (Wolf, 1967), EVs are now considered to be an important part of cell–cell communication (Hoshino et al., 2015; Meckes, 2015; Tkach and Thery, 2016; van Dongen et al., 2016; van Niel et al., 2018). EVs incorporate from the cell of origin various cellular molecules such as proteins as well as messenger RNA, small non-coding microRNAs, and mitochondrial DNA (Bang and Thum, 2012; van Niel et al., 2018). It has been shown that EVs generated from virus-infected cells, in particular infected with HIV-1, may incorporate viral proteins which, upon interaction with cells, may trigger various physiological responses (Barclay et al., 2017; Vojtech et al., 2014). Some of these responses may be similar to those triggered by an infectious virus (Anderson et al., 2016). Although, the presence of viral proteins inside EVs has been widely reported for many viruses (van Dongen et al., 2016), the presence of these proteins at the surface of EVs and their function have not been studied extensively. We recently described the incorporation of gp120 in EVs released by HIV-infected cells (Arakelyan et al., 2017) and have now extended this work to a member of the herpesvirus (HHV) family, human cytomegalovirus (HCMV). Infections with HCMV are often associated with significant immune activation that have in turn, been proposed to contribute to the clinical phenotypes of disease that are associated with HCMV infections (Deeks, 2011; Margolis, 2015).
EVs released from HHV-infected cells contain various cellular and viral-encoded molecules (reviewed in Liu et al. (2017), Sadeghipour and Mathias (2017)) in particular viral mRNAs and miRNAs (Canitano et al., 2013; Han et al., 2016; Kalamvoki et al., 2014; Meckes et al., 2010; Pegtel et al., 2010; Yogev et al., 2017) as well as viral and host proteins (Kalamvoki et al., 2014; Meckes et al., 2013; Miettinen et al., 2012; Ota et al., 2014; Temme et al., 2010). Here, we report on the presence of two HCMV envelope proteins, gB and gH that are essential for HCMV infectivity, on the surface of EVs secreted by HCMV-infected cells.
2. Results
We studied EVs isolated from the cell-free supernatant of UL32-EGFP-HCMV-infected human lung fibroblast (MRC-5 cells), or AD169 HCMV-infected primary dermal fibroblast cells using an iodixanol step-gradient. These EVs were concentrated in the upper fraction of the io-dixanol gradient (between 10% and 15%). We determined the purity of this fraction by measuring HCMV DNA by qPCR and the size and distribution using Nanosight and transmission electron microscopy (TEM). Also, to identify contaminating HCMV virions, we stained all lipid containing particles in a UL32-EGFP-HCMV viral preparations with a fluorescent dye, DiI. Therefore, by thresholding on DiI, any event positive for DiI and GFP represented a virion and any DiI-positive / GFP-negative event represented a vesicle. Thus, the analysis of individual particles allowed us to distinguish HCMV virions from EVs. Finally, using specific fluorescent antibodies, we analyzed the expression of gB and gH, two abundant envelope glycoproteins of HCMV, on EVs.
2.1. Evaluation of HCMV in isolated fractions
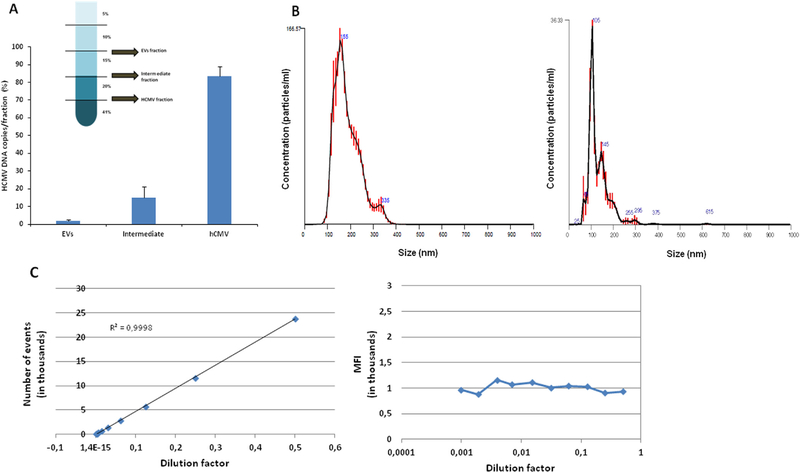
To evaluate the efficiency of separation of the viral stocks in EV and HCMV fractions, we measured UL32-EGFP-HCMV and AD169 HCMV DNA by qPCR. One ml of viral preparation was separated into fractions by iodixanol gradient centrifugation. The EV fraction, the intermediate fraction, and the HCMV viral preparation fraction were collected (Fig. 1A) and the genome copies of HCMV were quantified. In case of UL32-EGFP-HCMV, 1.7 ± 0.5%, 15 ± 3.5% and 83.3 ± 3.2% of the initial viral preparation DNA copy numbers were found respectively in the EV, intermediate and HCMV virion fraction (Fig. 1A). A similar distribution was found for AD169 HCMV, as 2.6 ± 0.2%, 6.6 ± 0.2% and 90.8 ± 0.3% of the initial viral preparation DNA copy numbers were found in the EV, intermediate and HCMV virion fraction, respectively. EVs isolated from the supernatant of UL32-EGFP-HCMV-infected cells were then characterized with NanoSight. The average particle size of the initial HCMV preparation was 182 ± 5.3 nm, while the size of isolated EVs (EV fraction) was on average 127 ± 1.7 nm. The size distribution data from the nanoparticle tracking analysis of the HCMV preparation (left panel) and EV fraction (right panel) are presented in Fig. 1B.
Fig. 1. Characterization of EVs in the EV fraction.
(A) HCMV DNA quantification in iodixanol fractions. After separation on a discontinuous iodixanol gradient using 1 ml of UL32-EGFP-HCMV as input (insert), the amount of virus present in each fraction was evaluated with qPCR and presented as a percentage of the viral input. (B) NanoSight analysis of HCMV preparation (left panel) and of isolated EV fraction (right panel). (C) Flow analysis of single EVs. DiI-stained EVs were serially diluted two-fold from 1:2–1:1024. The events were acquired in an LSRII flow cytometer, set to be triggered by DiI fluorescence. Presented are the numbers of events as a function of the dilution factor (left panel) and the median fluorescence intensity (MFI) of each dilution (right panel).
2.2. Detection and characterization of single particles by flow virometry
We used the GFP label in UL32-EGFP-HCMV combined with flow cytometry to distinguish EVs from viral particles on an individual basis. First, we verified that the acquired events represented single particles rather than their aggregates. Similar to the approach reported earlier (Arakelyan et al., 2013), we performed serial two-fold dilutions of the DiI-labeled EVs isolated from the UL32-EGFP-HCMV viral preparation. We acquired events using HTS on a LSRII flow cytometer. We used a gating strategy previously described in (Zicari et al., 2016) and plotted the number of events as a function of the dilution factor. Fig. 1C (left panel) shows a linear relation between the number of events and the dilution factor while the mean fluorescence intensity (MFI) of the events remained constant (Fig. 1C, right panel) confirming that the visualized events represent single particles according to the earlier published criterion (van der Pol et al., 2012).
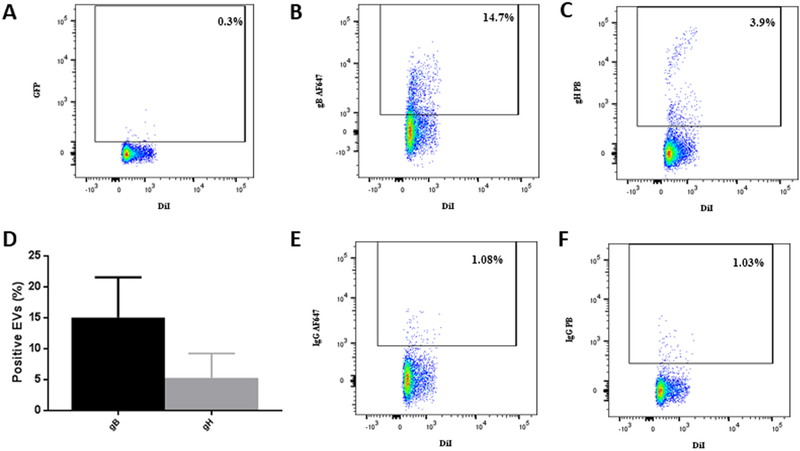
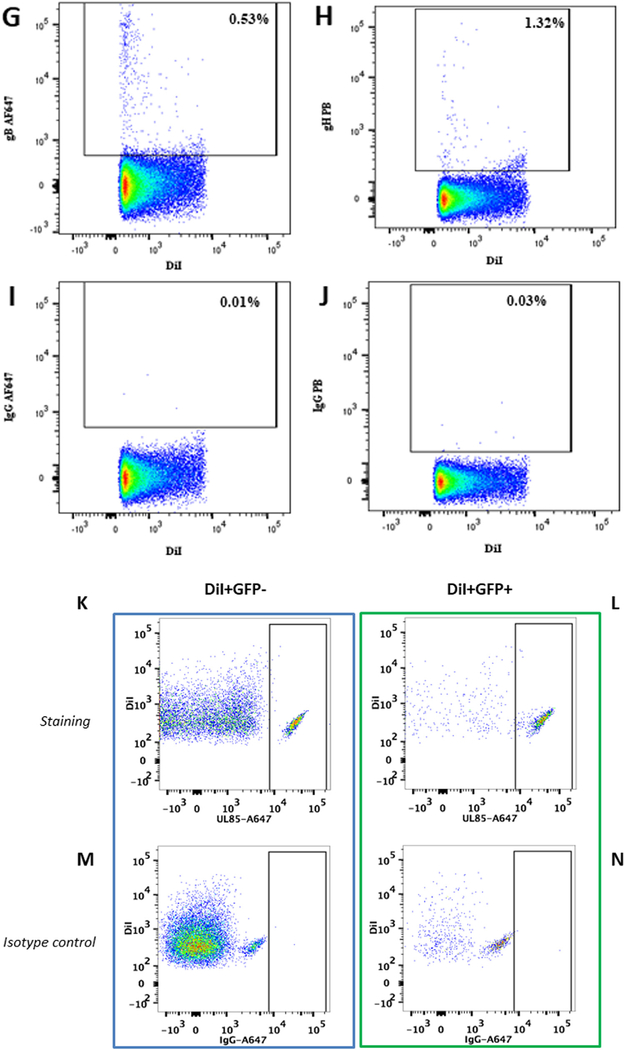
Next, we analyzed EVs for the presence of viral proteins. EVs, de-fined as DiI-positive/GFP-negative events, constituted 99.7 ± 0.1% (n = 3) of the total events, while 0.3 ± 0.1% (n = 3) were double labeled, thus constituting UL32-EGFP-HCMV (Fig. 2A). We assayed EVs (DiI-positive/GFP-negative particles) released by MRC-5 cells infected with UL32-EGFP-HCMV for the presence of HCMV envelope proteins. Specifically, EVs isolated by iodixanol step centrifugation and labeled with DiI were incubated with anti-gB AF647 and anti-gH PB antibodies or with their isotype controls, IgG AF647 and IgG PB, respectively. Labeled EVs were analyzed with flow cytometry setting the threshold on DiI fluorescence. In a typical experiment presented in Fig. 2B-C, 14.7% of EVs were positive for gB and 3.9% were positive for gH. On average, 15 ± 3.7% (n = 3) of EVs were positive for gB, 5.3 ± 2.3% (n = 3) were positive for gH (Fig. 2D) and 3.74 ± 1.5% (n = 3) were positive for both gB and gH. The specificity of this staining was confirmed in two types of control experiments: (i) by staining with isotype control antibodies (Fig. 2E–F) and (ii) by analysis of EVs released by uninfected cells using the same procedure as described above (Fig. 2G–J). Using a similar strategy and the anti-UL-85-AF647 anti-body that specifically recognized the HCMV minor capsid protein (UL85), we found that the HCMV capsid protein was present not only in the HCMV fraction (DiI+/GFP+) but also in the EV fraction (DiI +/GFP-) (Fig. 2K–N).
Fig. 2. EVs carry HCMV surface proteins.
DiI-labeled EVs isolated from UL32-EGFP-HCMV were stained with anti-gB AF647 and anti-gH PB antibodies or with their isotype controls. (A) Visualization of EVs as DiI-positive/GFP-negative events. (B) EVs stained with anti-gB AF647 antibodies or (C) with anti-gH PB antibodies. (D) Distribution of viral antigens gB and gH on EVs isolated from UL32-EGFP-HCMV viral preparation. Presented are means ( ± SEM) of EVs evaluated in three experiments. (E) Isotype control to B. (F) Isotype control to C. (G–J) DiI-labeled EVs, isolated from supernatant of control uninfected cells, were stained with anti-gB AF647 antibodies and anti-gH PB antibodies or with their isotype controls. (G) EV fraction stained with anti-gB AF647 antibodies. (H) EV fraction stained with anti-gH PB antibodies. (I) Isotype control to G. (J) Isotype control to H. (K-N) Detection of HCMV minor capsid protein UL-85 in EVs and HCMV by flow cytometry. EV fraction (left panel) or HCMV fraction (right panel) were stained with (upper row) or with its corresponding isotype control (lower row). (K) DiI+GVP- particles (EVs) stained with anti UL-85-AF647 antibody (L) DiI+GFP+ particles (HCMV) stained with anti UL-85-AF647 antibody. (M) Isotype control to K. (N) Isotype control to L.
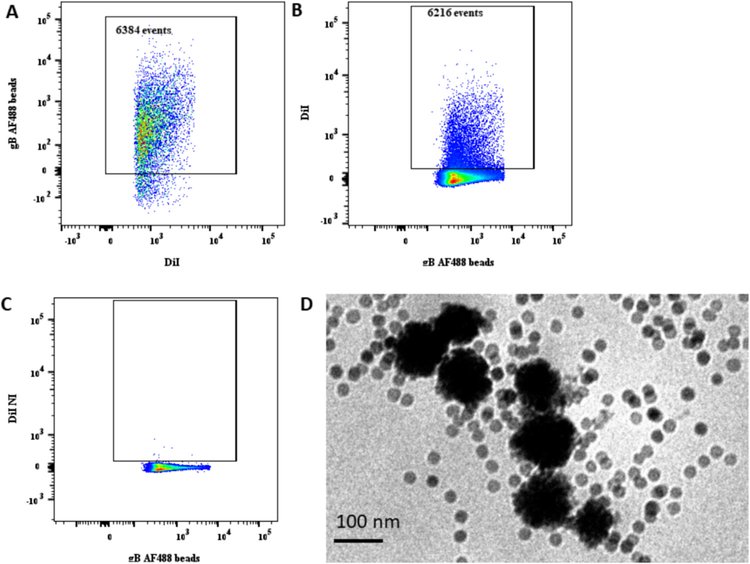
To confirm the presence of HCMV proteins on EVs released by HCMV-infected cells, we used our flow virometry nanotechnology (Arakelyan et al., 2013). DiI-stained EVs, collected from infected or control uninfected MRC-5 cells and purified on iodixanol gradient, were captured with 15-nm magnetic nanoparticles (MNPs) coupled to specific anti-gB antibodies and labeled with Zenon AF488. As described in the original protocol (Arakelyan et al., 2013), MNPs were used in large excess compared with the number of virions or EVs to avoid ag-gregation. The EV-MNP complexes were isolated on magnetic columns, eluted, and visualized with a flow cytometer. In a typical experiment, most of the GFP-negative events (6384 events) were double-positive for DiI and anti-gB AF488 antibodies (Fig. 3A), representing captured gB+ EVs. We detected a similar number of EVs using another method of quantification of the same samples: by thresholding on AF488 fluorescence, we found that 6216 events were double-positive (Fig. 3B), representing gB+ EVs. We repeated this experiment with three different samples of EVs and confirmed the accuracy of our quantification: 6178.6 ± 105.4 EVs were gB+ when we thresholded on DiI and 6160 ± 30.7 EVs were gB+ when we used AF488 threshold (n = 3). In our control experiments, we applied the capture protocol on EVs isolated from uninfected MRC-5 cells and found no EVs carrying gB (Fig. 3C).
Fig. 3. EVs captured by anti-gB-magnetic nano-particles.
DiI labeled EVs were captured by AF488-labeled anti-gB-magnetic nanoparticles (MNPs). A typical experiment out of three is shown. (A) DiI-labeled EVs released by HCMV-infected cells and captured by AF488labeled anti-gB-MNPs; thresholding on DiI fluorescence. (B) DiI-labeled EVs released by HCMV-infected cells captured by AF488-la-beled anti-gB-MNPs; thresholding on AF488 fluorescence. (C) DiI-labeled EVs released by control uninfected cells and captured by AF488-labeled anti-gB-MNPs; thresholding on AF488 fluorescence. (D) Visualization in transmission electron microscopy of EVs from HCMV-infected cells captured by anti-gB-MNPs. Note the excess of MNPs that according to the protocol (Arakelyan et al., 2013) should be applied to capture virions and exclude their aggregation.
2.3. Visualization of gB-carrying EVs with electron microscopy
To confirm that MNPs coupled to specific antibodies actually captured EVs, we visualized them using TEM. We analyzed EVs that were captured with MNPs coupled to anti-gB-specific antibody. TEM revealed EVs with a diameter ranging from 80 to 100 nm, of dark appearance due to the DiI and AsO4 contrasting (Belazi et al., 2009). These EVs were attached to anti-gB-MNPs (Fig. 3D).
2.4. Detection of HCMV and EV markers in HCMV and EV fractions by western blot
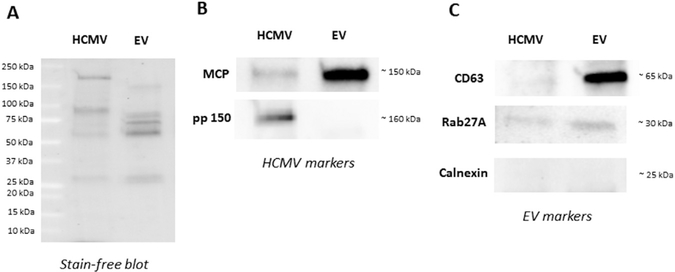
Following the iodixanol gradient step purification, both HCMV and EV fractions obtained from AD169 HCMV viral preparation were analyzed for HCMV and EV markers by western blot. For comparison, we loaded similar amounts of proteins for both EV and HCMV fractions (Fig. 4A). First, we probed HCMV and EV fractions for the presence of the tetraspanin CD63, Rab27A, which controls different steps of the exosome secretion pathway, and for the presence of calnexin. Fig. 4B demonstrates the presence of CD63 and Rab27A in the EV fraction while calnexin was absent. In the HCMV fraction, the expression of CD63 and Rab27A was either absent of very weak.
Fig. 4. Detection of HCMV and EV markers in HCMV and EV fractions by western blot.
(A) stain-free blot representing the spectrum of proteins obtained for HCMV and EV fractions after separation of AD169 HCMV pre-paration using a iodixanol step-gradient centrifugation. (B) Detection of EV-associated proteins CD63 (~60 kDa), Rab27A (~30 kDa), and calnexin (~25 kDa) proteins in both fractions. (C) Detection of HCMV capsid protein MCP (~150 kDa) and HCMV tegument protein, pp150 (~160 kDa) in HCMV and EV fractions after separation of AD169 viral preparation using a iodixanol step-gradient centrifugation.
Next, we probed both fractions for HCMV markers, the major capsid protein (UL86; MCP) and the tegument protein pp150 (UL32) (Fig. 4C). In confirmation of the flow cytometry findings that show the presence of capsid protein on EVs, we found MCP in the EV fraction by western blot. As expected both MCP and pp150 were found in the HCMV fraction (Fig. 4C).
3. Discussion
Human cytomegalovirus, a member of the Herpesvirus family (Dolan et al., 2004) encodes a large number of viral envelope proteins (Varnum et al., 2004), including two major glycoproteins that are essential for viral entry into all cell types, gB and gH, which together with a third glycoprotein, gL, represent the fusion complex of this virus (Vanarsdall and Johnson, 2012). Both gB and gH are important targets of antiviral antibody responses and have been viewed as potential components of prophylactic vaccines to limit HCMV infection and disease (Wussow et al., 2014). Moreover, HCMV gB and gH are abundant virion envelope proteins (Varnum et al., 2004).
Recently, it became clear that viral suspensions, in particular HIV-1 and HTLV-1, are in fact complex mixtures of virions and small EVs released by both infected and uninfected cells (Nolte-’t Hoen et al., 2016; van Dongen et al., 2016). Here, we studied EVs released by HCMV-infected cells.
Many EVs released by virus-infected cells are similar in size and other physical properties to viruses such as HIV, and therefore in many cases, it is very difficult to separate these two types of particles into pure fractions. HCMV is considerably larger than HIV, and as a result we were able to separate EVs from virions using step-gradient cen-trifugation. While the lower fraction (“HCMV” fraction) may contain mature and immature virions, defective viruses, EVs that carry enough viral components so that they acquired densities similar to that of the virions, as well as some other unidentified particles of cellular origin, the upper fraction contained predominantly EVs and almost no viral DNA. Although qPCR measurements report about DNA rather than in-tegral virions, the purity of the EV fraction was also confirmed by flow analysis using a GFP+ HCMV virus and therefore, we focused on particles of this upper fraction.
To confirm that the particles in this fraction were actually EVs, we probed with western blot the EV fraction for EV markers as defined by the International Society for Extracellular Vesicles (Lotvall et al., 2014). In accordance with this definition, we demonstrated the presence of CD63 and Rab27A and the absence of calnexin in the EV fraction. Also, we found MCP in the EV fraction confirming the findings from flow cytometry demonstrating the presence of the minor capsid protein, UL85, in EVs.
As far as the HCMV fraction is concerned, as expected, we detected both capsid (MCP) and tegument (pp150) viral proteins. We also detected Rab27A but no CD63. The presence of Rab27A in the HCMV fraction was not unexpected as Rab27A, which is recruited to the assembly site, has been shown to be associated to virus-wrapping membranes and incorporated into the viral envelope (Fraile-Ramos et al., 2010). Also, the absence of CD63 on HCMV was reported earlier (Walker et al., 2009), although other authors did find CD63 on HCMV (Cepeda et al., 2010; Fraile-Ramos et al., 2007). The controversy in these data may be explained by the difference in the sensitivity of the assays used in different works or by differential expression of CD63 in various cells infected by HCMV.
Although the same amount of proteins was loaded for each fraction, it is difficult to compare the amount of MCP or pp150 in HCMV with that in EV fraction due to the different protein composition of each fraction as well as the difference in the size and quantities of EVs and virions.
The above-described western blot data represent a bulk analysis that may reflect the presence of small amounts of concomitant viral particles in the EV fraction. For individual analysis of EVs, we used flow cytometry. First, we showed using NanoSight technology that the average size of the initial HCMV preparation was ~ 180 nm reflecting the presence of virions that are approximately of 200 nm in diameter (Arvin et al., 2007) and some smaller particles. The average size of the purified EV fraction was ~ 130 nm.
Although the above-described data evidence that EVs were isolated from the initial HCMV preparation, the purity of this fraction was further accessed by flow analysis which allows to focus on EVs excluding even small contamination with HCMV virions. We stained all particles with lipidic dye DiI and gated on GFP-negative/DiI-positive events to exclude virions from our flow analysis, and therefore focused only on events representing EVs. However, to analyze their antigens, we first confirmed that the acquired events represented individual particles rather than aggregates. By demonstrating that the MFI of the events remained constant in serially diluted samples while their concentration was decreased according to the dilution factor, we fulfilled a single-particle criterion (van der Pol et al., 2012).
Next, we investigated whether EVs released by HCMV-infected cells carried viral membrane proteins. EVs were stained with specific fluorescent antibodies against two HCMV glycoproteins: gB and gH. gB and gH as noted previously are two of the most abundant HCMV envelope glycoproteins (Varnum et al., 2004). We found that ∼15% of EVs were positive for gB and∼5% were positive for gH, a proportion consistent with the relative representation of these glycoproteins in the envelope of the mature cell-free virus (Varnum et al., 2004). A smaller fraction (∼4%) was positive for both viral proteins.
Thus, HCMV-infected cells release EVs that contain viral antigens (Liu et al., 2017; Sadeghipour and Mathias, 2017). To confirm this conclusion, we investigated the antigenic composition of EVs using a flow technique originally developed in our laboratory, which allows to analyze antigenic composition of individual EVs (Arakelyan et al., 2013, 2017, 2015; Zicari et al., 2016). Other techniques able to analyze individual EVs have been described as well (Morales-Kastresana et al., 2017; van der Vlist et al., 2012).
We coupled antibodies against gB with 15-nm magnetic nano-particles (MNPs), incubated them with DiI-labeled EVs, and then isolated them in a high magnetic field (Arakelyan et al., 2013). Flow analysis revealed EVs released by HCMV-infected cells captured through their gB antigen. No such EVs were present in control pre-parations of EVs isolated from uninfected cells.
These EVs captured by anti-gB MNPs were visualized with TEM. We found that EVs visualized by TEM were in the 50–100 nm range, which is the characteristic of exosomes (40–150 nm) (Colombo et al., 2014; van Dongen et al., 2016; van Niel et al., 2018). Since these captured EVs containing HCMV gB and gH constitute a minority of the total EV population, the imaged vesicles may constitute a specific subset of EVs, in particular slightly smaller than the average EV as evaluated by Nano-Sight.
Thus, similar to EVs released by HIV-infected cells, EVs released by HCMV-infected cells also carry viral surface proteins. Such EVs may contribute to various physiological effects in which viruses have been implicated since these EVs and HCMV should target the same cells., i.e. cells expressing HCMV receptors. In particular, they may contribute to the mechanisms by which exosomes secreted by HCMV-infected HUVEC cells activate allogeneic T cells via antigen presenting cells reported by Walker et al. (2009).
Here, we found that EVs released by HCMV-infected cells in-corporate not only membrane HCMV proteins but also capsid proteins. This result is in agreement with the notion that exosomes and HHVs share biogenesis pathways (Sadeghipour and Mathias, 2017) and that in the course of this biogenesis, EVs acquire viral proteins. As a result, viral stock is composed of EVs, non-infectious particles and infectious virions (Nolte-’t Hoen et al., 2016; van Dongen et al., 2016).
4. Materials and methods
4.1. Viruses, cells, and antibodies
The recombinant UL32-EGFP-HCMV virus (with GFP gene inserted into the C terminus of the inner tegument protein pUL32 (pp150GFP-HCMV)) was kindly provided by Drs. David Johnson (Oregon Health Sciences University, Portland, Oregon) and Jeffrey Cohen (NIAID, NIH, Bethesda, MD). It was generated as described in (Sampaio et al., 2005) from the HCMV strain TB40 (Sinzger et al., 1999), then propagated in human diploid fibroblasts (MRC-5) in minimum essential medium (MEM) containing 10% fetal bovine serum (FBS). Wild type AD169 CMV was produced in primary dermal fibroblasts that were obtained from discarded newborn dermal tissue. HCMV antibodies used were monoclonal anti-gB IgG1 (Virusys Corporation, Taneytown, MD), monoclonal anti-gH IgG1 (Thermo Fisher Scientific, Asheville, NC), monoclonal anti-gH IgG2b (14–4b) (Bogner et al., 1992), and their purified mouse IgG1 and IgG2b isotype control antibodies (Biolegend, San Diego, CA).
4.2. Labeling of HCMV and EVs with DiI and separation of EVs
HCMV virions and EVs were labeled with the lipophilic tracer DiI (Thermo Fisher Scientific): 1 ml of UL32-EGFP-HCMV viral preparation and 1 ml of AD169 HCMV were incubated with 1 μM DiI in the dark for 30 min at room temperature (RT). Iodixanol (Sigma, St. Louis, MO) discontinuous gradient was obtained by addition of four iodixanol solutions prepared in phosphate buffered saline (PBS) (10%, 15%, 20%, and 41%) in one 4-ml Seton open top tube (Thermo Fisher Scientific). 1 ml of DiI labeled UL32-EGFP-HCMV or WT AD169 in 5% iodixanol were overlaid in the tube. The tube was ultracentrifuged at 130,000g for 18 h at 4 °C with no brake. After centrifugation, the EV (between 10% and 15%), the intermediate (between 15% and 20%), and the HCMV (between 20% and 41%) fractions were collected (Fig. 1). The densities of the four separate iodixanol fractions (10%, 15%, 20% and 41%) used in the gradient step purification were respectively 1.05 ± 0.005, 1.10 ± 0.007, 1.12 ± 0.008 and 1.26 ± 0.04 (n = 4). These are in the same range of densities previously reported for similar iodixanol gradient (Walker et al., 2009).
4.3. Extraction and quantification of HCMV DNA
200 μl aliquots of EV fraction, intermediate fraction, and HCMV fraction were subjected to nucleic acid extraction using a NucliSENS easyMAG 2.0 instrument (BioMérieux, Durham, NC). Quantitative real-time PCR (qPCR) was performed with a PerfeCTa FastMix II Low ROX kit (Quanta BioSciences, Gaithersburg, MD) using the following primer set: HHV-5 FWD: AACCAAGATGCAGGTGATAGG, HHV-5 REV: AGCG TGACGTGCATAAAGA, and the probe: /56-FAM/TACCTGGAG/ZEN/ TCCTTCTGCGAGGA/3IABkFQ/. Amplifications were carried out on a BioRad CFX96 Touch Thermocycler according to the following cycling parameters: 2 min at 95 °C followed by 44 cycles of 10 s at 95 °C and 30 s at 60 °C.
4.4. Quantification of EV fractions on NanoSight
EVs isolated from the cell-free supernatant of UL32-EGFP-HCMV-infected cells and from supernatant of uninfected MRC-5 cells were evaluated for concentration and size distribution on a NanoSight NS 300 (Salisbury, United Kingdom) equipped with a 405-nm laser and analyzed with NTA 3.0 software (NanoSight NS300, Malvern Instruments, UK). The measurements were performed with constant sample flow using a syringe pump for 180 s at camera level 13.
4.5. Serial dilutions of DiI-labeled EV fraction
Serial two-fold dilutions from 1:2–1:1024 of the EV fraction, isolated from the UL32-EGFP-HCMV viral preparation, were performed to show that the number of events detected with the flow cytometer was inversely linearly dependent on the dilutions analyzed. We ran samples using a high throughput sampler (HTS) at a flow rate of 1 μl/sec on a LSRII flow cytometer (BD Bioscience, San Jose, CA) that was set to acquire 80 μl of each sample in duplicate with a threshold set at 300 fluorescence units on the DiI fluorescence channel.
4.6. Protein extraction, SDS-PAGE, and western blotting
Total proteins were extracted from both EV and HCMV fractions, according to RIPA Lysis and Extraction Buffer (Thermo Fisher Scientific, Waltham, MA). 10 μg of proteins were loaded on a 4–20% precast polyacrylamide gel (Bio-Rad Laboratories, Hercules, CA) and separated by SDS-PAGE, then transferred to PVDF membranes and probed with anti-CD63 (1 μg/ml, Thermo Fisher Scientific, Waltham, MA), anti-Calnexin (1 μg/ml, Thermo Fisher Scientific, Waltham, MA), anti-Rab27A (1 μg/ml, Thermo Fisher Scientific, Waltham, MA), anti-HCMV capsid MCP (2 μg/ml), and anti-HCMV tegument pp150 (2 μg/ml) monoclonal primary anti-mouse monoclonal antibodies (clones 28–4 and 36–14) and then goat peroxidase-conjugated anti-mouse IgG secondary anti-body (Bio-Rad Laboratories, Hercules, CA). Peroxidase activity and digital images were detected by using V3 Western Workflow™ (Bio-Rad Laboratories, Hercules, CA).
4.7. Labeling of anti-gB and anti-gH detection antibodies and their isotype controls with fluorophores
Labeling of monoclonal antibodies was performed according to the Thermo Fischer Scientific protocol. Briefly, 10 μl of 1 M solution of sodium bicarbonate were added in each vial containing 100 μg of antibodies (anti-gB, anti-gH, or isotype controls). Then, anti-gB and its isotype control were labeled with Alexa Fluor 647 (AF647) while anti-gH and its isotype control were labeled with Pacific Blue (PB) in rotation for 1 h at RT in the dark. During the incubation, Zeba spin desalting columns (Thermo Fisher Scientific) were twice washed with PBS to remove the storage buffer solution. After the incubation, the antibodies were added to Zeba columns and centrifuged at 2000g for 3 min, recovered in the flow-through, and stored at 4 °C. The concentrations of fluorescent-labeled antibodies were measured with a Nanodrop 1000 (Thermo Scientific).
4.8. Detection of EV fractions with flow cytometry
To stain EVs for gB and gH, an aliquot of DiI-labeled EVs (60 μl) was incubated with 1 μg of anti-gB AF647 antibody and with 1 μg of anti-gH PB antibody for 20 min at RT. Another aliquot (60 μl) was similarly stained with the isotype control antibodies. Also, 60 μl of DiI-labeled EVs from uninfected MRC-5 were stained with specific antibodies or isotype controls as an additional negative control. After 20 min, the mixture of EVs and antibodies was diluted with 240 μl of 4% paraformaldehyde (PFA). Labeled EVs were analyzed with a LSRII flow cytometer equipped with 355-, 407-, 488-, 532-, and 638-nm laser lines. Compensation beads (BD) were used to perform compensation controls.
4.9. Intravesicular staining by flow cytometry
100 μl of DiI-labeled EV fraction were incubated with 50 μl Fix and Perm Buffer A (ThermoFisher Scientific) for 30 min at RT, then 50 μl of component B of Fix and Perm buffer were added followed by immediate addition of either 1 μg of UL85-AF647 (in house labeled antibody (Alexa Fluor™ 647 Antibody Labeling Kit, ThermoFisher Scientific) or isotype control antibody in 50 μl of staining buffer. The mixture was incubated for 30 min at RT, fixed with PBS-PFA 1% and analyzed with flow cytometer by thresholding on DiI.
4.10. Preparation of anti-gB 15-nm MNPs
1 mg of 15-nm carboxyl-terminated magnetic iron oxide nano-particles (MNPs) (OceanNanoTech, Springdale, AZ) was coupled to IgG1 mouse monoclonal antibodies against HCMV glycoprotein gB (Virusys Corporation, Taneytown, MD) following the Ocean Nanotech protocol for conjugation. After the procedure, the anti-gB-coupled MNPs were resuspended in 2 ml of wash/storage buffer and stored at 4 °C.
4.11. Detection of EVs captured by anti-gB-MNPs with flow cytometry
To visualize the fraction of EVs captured by anti-gB-MNPs, we la-beled 60 μl of the latter with 5 μl of Zenon Alexa Fluor 488 (AF488) mouse IgG (Thermo Fisher Scientific). Then, the fluorescent anti-gB-MNPs were incubated with 60 μl of DiI-labeled EVs from supernatant of infected or uninfected MRC-5 cells. After 40 min of incubation at 37 °C with continuous mixing, the complexes were washed on magnetic columns to remove unbound EVs, eluted in 600 μl of PBS-PFA 1%, and analyzed using a BD LSRII flow cytometer thresholding on DiI fluorescence or on AF488 fluorescence.
4.12. Preparation of EV samples captured by anti-gB-MNPs for transmission electron microscopy
DiI-labeled EVs (240 μl) were incubated with 240 μl of anti-gBMNPs (~2.6 ×1012 MNPs/ml) for 40 min at 37 °C with continuous mixing. After the incubation, the complexes were washed three times on MS MACS columns on an MS MACS magnet (Miltenyi Biotech, Auburn, CA) to remove the unbound EVs and were then eluted in 240 μl of PBS. The complexes were divided between two wells of a 24-well plate containing 14-mm round coverslips. The plate was placed on a flat magnet for the rest of the procedure in order to keep the EV-MNPs in the wells during all the washes. The complexes were then fixed for 1 h with 2.5% glutaraldehyde (Electron Microscopy Sciences, Hatfield, PA) in 0.1 M phosphate buffer (PB) at pH 7.2. After 1 h, the wells were washed three times with 0.1 M PB and post-fixed with 4% osmium tetroxide (Electron Microscopy Sciences, Hatfield, PA) diluted V/V in deionized water and again V/V in 0.2 M PB to obtain a solution of 1% osmium tetroxide in 0.1 M PB. After 1 h, wells were washed three times with 0.1 M PB, and then the samples were dehydrated sequentially with two 10-min incubations in 70% ethanol, two 10-min incubations in 90% ethanol, and two 10-min incubations in 100% ethanol. We performed the embedding steps using the EMbed Resin previously prepared with the EMbed-812 kit (Electron Microscopy Sciences, Hatfield, PA). The first 10-min incubation was done with EMbed Resin diluted V/ V in 100% ethanol. The second 10-min incubation was done with pure EMbed Resin. After this step, two Beem Embedding Capsules per well were filled with pure EMbed Resin and then flipped vertically over the beads in the wells. The polymerization step was done at 60 °C for 24 h, avoiding humidity. Capsules were detached mechanically from the plastic and 80-nm sections were then cut with an ultramicrotome (Reichert Ultracut S) as described in Ganor et al. (Ganor et al., 2010) and observed with a transmission electron microscope (JEOL 1011) equipped with a GATAL numerical camera. Pictures were taken and digitalized with Digital Micrograph software at the Electron Microscopy Facility of the Cochin Institute. At least 50 randomly chosen fields were observed per duplicate.
4.13. Statistical analysis
The results are presented as means ± standard errors of the mean (SEM), and n, the number of replicates, is indicated.
Acknowledgements
We thank Drs. David Johnson and Jeffrey Cohen for providing UL32-EGFP-HCMV and MRC-5 cells and for helpful advice.
Fundings
The work of SZ, AA, RANP, WF, CV, AL and LM was supported by the NICHD/NIH Intramural Program (USA). The work of AL was also supported by the Russian Federation Government #14.B25.31.0016 agreement #18–15-00420 and RFBR grant #16–04-017/16. The work of AS and MB was supported by ANRS, France (AO2015–2-17046). The work of WB was supported by NIH (1RO1AI089956–01A1).
Footnotes
Conflict of interests
The authors declare that they have no competing financial interests.
References
- Anderson MR, Kashanchi F, Jacobson S, 2016. Exosomes in viral disease. Neurotherapeutics 13, 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakelyan A, Fitzgerald W, Margolis L, Grivel JC, 2013. Nanoparticle-based flow virometry for the analysis of individual virions. J. Clin. Invest 123, 3716–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakelyan A, Fitzgerald W, Zicari S, Vanpouille C, Margolis L, 2017. Extracellular vesicles carry HIV Env and facilitate hiv infection of human lymphoid tissue. Sci. Rep 7, 1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakelyan A, Ivanova O, Vasilieva E, Grivel JC, Margolis L, 2015. Antigenic composition of single nano-sized extracellular blood vesicles. Nanomedicine 11, 489–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvin A, Campadelli-Fiume G, Mocarski E, Moore P, Roizman B, Whitley R, Yamanishi K, 2007. Human Herpesviruses. Cambridge University Press. [PubMed] [Google Scholar]
- Bang C, Thum T, 2012. Exosomes: new players in cell-cell communication. Int. J. Biochem. Cell Biol 44, 2060–2064. [DOI] [PubMed] [Google Scholar]
- Barclay RA, Schwab A, DeMarino C, Akpamagbo Y, Lepene B, Kassaye S, Iordanskiy S, Kashanchi F, 2017. Exosomes from uninfected cells activate transcription of latent HIV-1. J. Biol. Chem 292, 11682–11701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belazi D, Sole-Domenech S, Johansson B, Schalling M, Sjovall P, 2009. Chemical analysis of osmium tetroxide staining in adipose tissue using imaging ToF-SIMS. Histochem Cell Biol. 132, 105–115. [DOI] [PubMed] [Google Scholar]
- Bogner E, Reschke M, Reis B, Reis E, Britt W, Radsak K, 1992. Recognition of compartmentalized intracellular analogs of glycoprotein H of human cytomegalovirus. Arch. Virol 126, 67–80. [DOI] [PubMed] [Google Scholar]
- Canitano A, Venturi G, Borghi M, Ammendolia MG, Fais S, 2013. Exosomes released in vitro from Epstein-Barr virus (EBV)-infected cells contain EBV-encoded latent phase mRNAs. Cancer Lett. 337, 193–199. [DOI] [PubMed] [Google Scholar]
- Cepeda V, Esteban M, Fraile-Ramos A, 2010. Human cytomegalovirus final envelopment on membranes containing both trans-Golgi network and endosomal markers. Cell Microbiol 12, 386–404. [DOI] [PubMed] [Google Scholar]
- Colombo M, Raposo G, Thery C, 2014. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev. Cell Dev. Biol 30, 255–289. [DOI] [PubMed] [Google Scholar]
- Deeks SG, 2011. HIV infection, inflammation, immunosenescence, and aging. Annu Rev. Med 62, 141–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan A, Cunningham C, Hector RD, Hassan-Walker AF, Lee L, Addison C, Dargan DJ, McGeoch DJ, Gatherer D, Emery VC, Griffiths PD, Sinzger C, McSharry BP, Wilkinson GW, Davison AJ, 2004. Genetic content of wild-type human cytomegalovirus. J. Gen. Virol 85, 1301–1312. [DOI] [PubMed] [Google Scholar]
- Fraile-Ramos A, Cepeda V, Elstak E, van der Sluijs P, 2010. Rab27a is required for human cytomegalovirus assembly. PLoS One 5, e15318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraile-Ramos A, Pelchen-Matthews A, Risco C, Rejas MT, Emery VC, HassanWalker AF, Esteban M, Marsh M, 2007. The ESCRT machinery is not required for human cytomegalovirus envelopment. Cell Microbiol 9, 2955–2967. [DOI] [PubMed] [Google Scholar]
- Ganor Y, Zhou Z, Tudor D, Schmitt A, Vacher-Lavenu MC, Gibault L, Thiounn N, Tomasini J, Wolf JP, Bomsel M, 2010. Within 1h, HIV-1 uses viral synapses to enter efficiently the inner, but not outer, foreskin mucosa and engages Langerhans-T cell conjugates. Mucosal Immunol. 3, 506–522. [DOI] [PubMed] [Google Scholar]
- Han Z, Liu X, Chen X, Zhou X, Du T, Roizman B, Zhou G, 2016. miR-H28 and miR-H29 expressed late in productive infection are exported and restrict HSV-1 replication and spread in recipient cells. Proc. Natl. Acad. Sci. USA 113, E894–E901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di Giannatale A, Ceder S, Singh S, Williams C, Soplop N, Uryu K, Pharmer L, King T, Bojmar L, Davies AE, Ararso Y, Zhang T, Zhang H, Hernandez J, Weiss JM, Dumont-Cole VD, Kramer K, Wexler LH, Narendran A, Schwartz GK, Healey JH, Sandstrom P, Labori KJ, Kure EH, Grandgenett PM, Hollingsworth MA, de Sousa M, Kaur S, Jain M, Mallya K, Batra SK, Jarnagin WR, Brady MS, Fodstad O, Muller V, Pantel K, Minn AJ, Bissell MJ, Garcia BA, Kang Y, Rajasekhar VK, Ghajar CM, Matei I, Peinado H, Bromberg J, Lyden D, 2015. Tumour exosome integrins determine organotropic metastasis. Nature 527, 329–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalamvoki M, Du T, Roizman B, 2014. Cells infected with herpes simplex virus 1 export to uninfected cells exosomes containing STING, viral mRNAs, and microRNAs. Proc. Natl. Acad. Sci. USA 111, E4991–E4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Zhou Q, Xie Y, Zuo L, Zhu F, Lu J, 2017. Extracellular vesicles: novel vehicles in herpesvirus infection. Virol. Sin 32, 349–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotvall J, Hill AF, Hochberg F, Buzas EI, Di Vizio D, Gardiner C, Gho YS, Kurochkin IV, Mathivanan S, Quesenberry P, Sahoo S, Tahara H, Wauben MH, Witwer KW, Thery C, 2014. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J. Extracell. Vesicles 3, 26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis L, 2015. Immunoactivation at the crossroads of human disease. Am. J. Med 128, 562–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meckes DG Jr., 2015. Exosomal communication goes viral. J. Virol 89, 5200–5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meckes DG Jr., Gunawardena HP, Dekroon RM, Heaton PR, Edwards RH, Ozgur S, Griffith JD, Damania B, Raab-Traub N, 2013. Modulation of B-cell exosome proteins by gamma herpesvirus infection. Proc. Natl. Acad. Sci. USA 110, E2925–E2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meckes DG Jr., Shair KH, Marquitz AR, Kung CP, Edwards RH, Raab-Traub N, 2010. Human tumor virus utilizes exosomes for intercellular communication. Proc.Natl. Acad. Sci. USA 107, 20370–20375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen JJ, Matikainen S, Nyman TA, 2012. Global secretome characterization of herpes simplex virus 1-infected human primary macrophages. J. Virol 86, 12770–12778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Kastresana A, Telford B, Musich TA, McKinnon K, Clayborne C, Braig Z, Rosner A, Demberg T, Watson DC, Karpova TS, Freeman GJ, DeKruyff RH, Pavlakis GN, Terabe M, Robert-Guroff M, Berzofsky JA, Jones JC, 2017.Labeling extracellular vesicles for nanoscale flow cytometry. Sci. Rep 7, 1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte-’t Hoen E, Cremer T, Gallo RC, Margolis LB, 2016. Extracellular vesicles and viruses: are they close relatives? Proc. Natl. Acad. Sci. USA 113, 9155–9161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota M, Serada S, Naka T, Mori Y, 2014. MHC class I molecules are incorporated into human herpesvirus-6 viral particles and released into the extracellular environment.Microbiol Immunol. 58, 119–125. [DOI] [PubMed] [Google Scholar]
- Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, de Gruijl TD, Wurdinger T, Middeldorp JM, 2010. Functional delivery of viral miRNAs via exosomes. Proc. Natl. Acad. Sci. USA 107, 6328–6333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeghipour S, Mathias RA, 2017. Herpesviruses hijack host exosomes for viral pathogenesis. Semin Cell Dev. Biol. 67, 91–100. [DOI] [PubMed] [Google Scholar]
- Sampaio KL, Cavignac Y, Stierhof YD, Sinzger C, 2005. Human cytomegalovirus labeled with green fluorescent protein for live analysis of intracellular particle movements. J. Virol 79, 2754–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinzger C, Schmidt K, Knapp J, Kahl M, Beck R, Waldman J, Hebart H, Einsele H, Jahn G, 1999. Modification of human cytomegalovirus tropism through propagation in vitro is associated with changes in the viral genome. J. Gen. Virol 80 (Pt 11), 2867–2877. [DOI] [PubMed] [Google Scholar]
- Temme S, Eis-Hubinger AM, McLellan AD, Koch N, 2010. The herpes simplex virus1 encoded glycoprotein B diverts HLA-DR into the exosome pathway. J. Immunol 184, 236–243. [DOI] [PubMed] [Google Scholar]
- Tkach M, Thery C, 2016. Communication by extracellular vesicles: where we are and where we need to go. Cell 164, 1226–1232. [DOI] [PubMed] [Google Scholar]
- van der Pol E, van Gemert MJ, Sturk A, Nieuwland R, van Leeuwen TG, 2012.Single vs. swarm detection of microparticles and exosomes by flow cytometry. J.Thromb. Haemost 10, 919–930. [DOI] [PubMed] [Google Scholar]
- van der Vlist EJ, Nolte-’t Hoen EN, Stoorvogel W, Arkesteijn GJ, Wauben MH, 2012. Fluorescent labeling of nano-sized vesicles released by cells and subsequent quantitative and qualitative analysis by high-resolution flow cytometry. Nat. Protoc 7, 1311–1326. [DOI] [PubMed] [Google Scholar]
- van Dongen HM, Masoumi N, Witwer KW, Pegtel DM, 2016. Extracellular vesicles exploit viral entry routes for cargo delivery. Microbiol Mol. Biol. Rev 80, 369–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Niel G, D’Angelo G, Raposo G, 2018. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol 19, 213–228. [DOI] [PubMed] [Google Scholar]
- Vanarsdall AL, Johnson DC, 2012. Human cytomegalovirus entry into cells. Curr. Opin. Virol 2, 37–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnum SM, Streblow DN, Monroe ME, Smith P, Auberry KJ, Pasa-Tolic L, Wang D, Camp 2nd DG, Rodland K, Wiley S, Britt W, Shenk T, Smith RD, Nelson JA, 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol 78, 10960–10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojtech L, Woo S, Hughes S, Levy C, Ballweber L, Sauteraud RP, Strobl J, Westerberg K, Gottardo R, Tewari M, Hladik F, 2014. Exosomes in human semen carry a distinctive repertoire of small non-coding RNAs with potential regulatory functions. Nucleic Acids Res 42, 7290–7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JD, Maier CL, Pober JS, 2009. Cytomegalovirus-infected human endothelial cells can stimulate allogeneic CD4+ memory T cells by releasing antigenic exosomes. J. Immunol 182, 1548–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf P, 1967. The nature and significance of platelet products in human plasma. Br. J. Haematol 13, 269–288. [DOI] [PubMed] [Google Scholar]
- Wussow F, Chiuppesi F, Martinez J, Campo J, Johnson E, Flechsig C, Newell M,Tran E, Ortiz J, La Rosa C, Herrmann A, Longmate J, Chakraborty R, Barry PA, Diamond DJ, 2014. Human cytomegalovirus vaccine based on the envelope gH/gL pentamer complex. PLoS Pathog. 10, e1004524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogev O, Henderson S, Hayes MJ, Marelli SS, Ofir-Birin Y, Regev-Rudzki N, Herrero J, Enver T, 2017. Herpesviruses shape tumour microenvironment through exosomal transfer of viral microRNAs. PLoS Pathog. 13, e1006524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zicari S, Arakelyan A, Fitzgerald W, Zaitseva E, Chernomordik LV, Margolis L, Grivel JC, 2016. Evaluation of the maturation of individual Dengue virions with flow virometry. Virology 488, 20–27. [DOI] [PMC free article] [PubMed] [Google Scholar]