We utilized pulsed-field gel electrophoresis (PFGE) to purify high-molecular-weight DNA from HIV-infected cells. This purification, in combination with our previously described droplet digital PCR (ddPCR) assay, was used to develop a method to quantify proviral integrated HIV DNA free of lower-molecular-weight species of HIV DNA.
KEYWORDS: HIV, latent reservoir, pulsed-field gel electrophoresis, integrated DNA
ABSTRACT
We utilized pulsed-field gel electrophoresis (PFGE) to purify high-molecular-weight DNA from HIV-infected cells. This purification, in combination with our previously described droplet digital PCR (ddPCR) assay, was used to develop a method to quantify proviral integrated HIV DNA free of lower-molecular-weight species of HIV DNA. Episomal 2-long-terminal-repeat (2-LTR) circles were completely cleared from HIV DNA samples. Technical replicates of the complete assay, starting with the same specimens, resulted in no statistical differences in quantification of integrated HIV gag sequences in cellular DNA from cells from HIV-infected subjects after prolonged treatment with antiretroviral therapy (ART). The PFGE ddPCR assay was compared to the Alu-gag quantitative PCR (qPCR) assay, the most widely used assay to measure proviral integrated HIV DNA. Spearman's rho nonparametric correlation determined PFGE ddPCR results to be positively correlated with Alu-gag qPCR results (r = 0.7052; P = 0.0273). In summary, PFGE ddPCR is a sensitive, reproducible, and robust method to measure proviral integrated HIV DNA and is theoretically more accurate than previously described assays, because it is a direct measure of integrated HIV DNA.
INTRODUCTION
Antiretroviral therapy (ART) dramatically reduces the morbidity and deaths resulting from infection with HIV by suppressing viral replication; however, it does not cure the infection. The establishment of latently infected cells provides a source for resumption of active virus replication upon the discontinuation of ART (1–3).
Assays to measure this latent reservoir are critical, both for characterizing its composition and for quantifying the efficacy of candidate interventions to reduce it (4). Proviral HIV DNA represents the largest component of the latent reservoir, permitting the greatest sensitivity and precision for measuring the reservoir (5, 6). Most of this integrated HIV DNA, however, is defective for replication and shows little correlation with measures of the replication-competent latent reservoir (7, 8).
Quantifying truly integrated HIV DNA in the latent reservoir has proved challenging. Failed HIV integration events accumulate in the nuclei of infected cells as lower-molecular-weight HIV species with the same sequences as proviral HIV DNA (9, 10). Attempts to measure integrated HIV DNA have involved utilizing primer/probe sets measuring host cell Alu sequences, in addition to HIV sequences. Alu-gag quantitative PCR (qPCR) is a technically complex assay requiring DNA from a large number of host cells. In addition, the variable distances of Alu sequences from each proviral sequence, ranging up to thousands of base pairs, has required assumptions about amplification efficiencies, resulting in the use of a multiplication factor to calculate integrated HIV DNA copies (11, 12).
To address the challenge of distinguishing proviral DNA from lower-molecular-weight species of HIV DNA, we investigated the use of two methods. First, we applied pulsed-field gel electrophoresis (PFGE) to physically remove lower-molecular-weight HIV species from total cellular DNA, to purify integrated HIV DNA. PFGE has been reported to be an improvement over traditional gel electrophoresis for resolving genomic DNA sizes (13, 14). Second, we utilized droplet digital PCR (ddPCR), which is a sensitive and precise platform for quantifying DNA and RNA sequences of multiple viral pathogens, including HIV and herpesviruses (15, 16). Historically, gel electrophoresis and gel purification have been used to isolate proviral DNA (17); however, they are slow, inefficient for recovery, and prone to cross-contamination. We have optimized and validated a protocol using an automated PFGE platform to purify integrated HIV DNA from HIV-infected peripheral blood mononuclear cells (PBMCs) and CD4+ T cells, followed by quantification using ddPCR.
MATERIALS AND METHODS
Study subjects.
Peripheral blood was obtained from HIV-infected donors and processed for PBMCs or CD4+ T cells before being frozen and eventually subjected to cellular DNA extraction. Pre-ART samples were obtained from chronically HIV-infected donors with unknown dates of infection. Samples from chronically infected donors receiving ART were obtained from donors with viral loads of less than 50 copies/ml for at least 6 months.
U1 cell line.
The U1 HIV cell line, derived from U937, was obtained from the NIH AIDS Reagent Program. U1 cells contain integrated proviral HIV DNA as well as unintegrated episomal HIV species, such as 2-long-terminal-repeat (2-LTR) circles. The integrated HIV genome is defective in Tat function but highly inducible with various cytokines. The U1 cell line is used as a model for HIV latency (18). U1 cells were cultured and expanded using NIH AIDS Reagent Program guidelines.
Purification of high-molecular-weight DNA using PFGE.
Cellular DNA was extracted using a QIAamp DNA Blood kit (Qiagen), following the manufacturer's protocol. DNA was precipitated with ethanol following elution, to increase the concentration. DNA concentrations were estimated from the A260/A280 absorptivity ratio using a NanoDrop One spectrophotometer (Thermo Scientific). Pulsed-field high-molecular-weight extraction utilized the BluePippin platform (Sage Science). The platform was cleaned before every run, using the provided wash cassette and deionized water. Up to 5 μg of total DNA was diluted in a total volume of 30 μl (166 ng/μl); 10 μl of loading solution (Sage Science) was added to the DNA, for a final volume of 40 μl. A BluePippin 0.75% agarose dye-free cassette was prepared and loaded according to the manufacturer's protocol. A 5-lane cassette runs 4 samples, with 1 lane reserved for S1 molecular weight markers (Sage Science). The 0.75% DF Marker S1 high-pass 15- to 20-kb automated protocol was used, with 15 kb being set as the DNA molecular weight cutoff value. The protocol run time was approximately 4.5 h. Once the protocol was completed, 40 μl of DNA eluate was collected and the elution chambers were washed with 0.1% Tween 20 buffer (Sage Science), which was added to the initial eluate for 80 μl of total eluate.
Agilent TapeStation quality control.
For the initial optimization of the assay, but not for the patient data, DNA concentrations were measured by the Qubit PicoGreen fluorometer assay (Thermo Fisher Scientific) and normalized before loading onto the Agilent 2200 TapeStation platform. The Agilent Genomic DNA ScreenTape assay analyzed the quality of high-molecular-weight DNA purification.
ddPCR for HIV DNA.
After PFGE, the high-molecular-weight samples were treated with the BanII restriction enzyme (New England BioLabs), according to the manufacturer's protocol, in preparation for ddPCR. The Bio-Rad QX200 ddPCR system was run according to the manufacturer's protocol, using an annealing temperature of 60°C, with the following primer-probe assays: 2-LTR-6-carboxyfluorescein (FAM) (MH535 2-LTR forward, 5′-AACTAGGGAACCCACTGCTTAAG-3′; MH535 2LTR reverse, 5′-TCCACAGATCAAGGATATCTTGTC-3′; MH603 2-LTR probe, 5′-ACACTACTTGAAGCACTCAAGGCAAGCTTT-3′), skGag-HEX (SK462 gag forward, 5′-AGTTGGAGGACATCAAGCAGCCATGCAAAT-3′; SK431 gag reverse, 5′-TGCTATGTCAGTTCCCCTTGGTTCTCT-3′; SK102 gag probe, 5′-AGACCATCAATGAGGAAGCTGCAGAATGGGAT-3′), and RPP30-HEX (RPP30 forward, 5′-GATTTGGACCTGCGAGCG-3′; RPP30 reverse, 5′-GCGGCTGTCTCCACAAGT-3′; RPP30 probe, 5′-CTGACCTGAAGGCTCT-3′). Digested DNA was diluted 1:10 to measure the RPP30 gene within the dynamic range of the ddPCR platform. The RPP30 gene was the host cell gene used to normalize HIV copies to host cell numbers. These gag primers are useful for all HIV-1 subtypes.
Alu-gag qPCR for HIV DNA.
DNA was isolated from PBMCs using the Gentra Puregene Blood kit and was treated with RNase according to the manufacturer's instructions (Qiagen). The resulting DNA solutions were assayed by qPCR for the albumin gene in order to quantify the number of cell equivalents of DNA per volume (albumin forward, 5′-GCTGTCATCTCTTGTGGGCTGT-3′; albumin reverse, 5′-AAACTCATGGGAGCTGCTGGTT-3′; albumin probe, 5′-CCTGTCATGCCCACACAAATCTCTCC-3′) (19). The total amount of HIV DNA present in the samples was determined by qPCR for the R-U5 region of the 5′ long terminal repeat (LTR) (RU5 forward, 5′-TTAAGCCTCAATAAAGCTTGCC-3′; RU5 reverse, 5′-GTTCGGGCGCCACTGCTAGA-3′; Yun's probe, 5′-CCAGAGTCACACAACAGACGGGCACA-3′) (20).
Alu-gag PCR was performed at a limiting dilution, such that at least 20% of the wells had no detectable HIV DNA (Alu forward, 5′-GCCTCCCAAAGTGCTGGGATTACAG-3′; SK431 reverse, 5′-TGCTATGTCAGTTCCCCTTGGTTCTCT-3′). The amount of integrated HIV DNA was determined using a binomial Poisson distribution (21). Specifically, using the calculated number of HIV molecules per cell, samples were appropriately diluted at two concentrations so that 20 to 80% of the wells of a 96-well PCR plate were positive for HIV DNA by Alu-gag PCR. Eight reaction mixtures containing only the gag primer, resulting in linear amplification, were used as controls for the identification of true-positive results.
Reaction components were as described previously (11, 22), with the following modifications. We utilized a new gag primer (SK462) for a lower linear amplification signal (23). Also, slightly different thermocycler conditions provided enhanced amplification. Specifically, samples were preamplified on an endpoint PCR instrument for 40 cycles of 95°C for 15 s, 56.2°C for 45 s, and 72°C for 3.5 min; initial denaturation at 95°C for 2 min and final extension at 72°C for 10 min were also included. Reaction mixtures were held at 10°C until they were analyzed by qPCR, using the R-U5 total HIV assay, in which 15 μl from each well of the Alu-gag PCR preamplification reaction was transferred for analysis. A minimum of 80 reactions were performed in Alu-gag assays, and samples were assayed at two DNA concentrations to compensate for PCR inhibitors.
These changes to the previously established Alu-gag qPCR assay resulted in increased sensitivity. The new version detected 20.3% of proviruses, corresponding to a correction factor of 4.93, which represents approximately 2-fold enhancement over our previous assay. Alu-gag PCR preamplification reactions were performed in 50-μl volumes, and R-U5 qPCRs were performed in 30-μl volumes.
Statistical analysis.
GraphPad Prism 7.0 software was used for statistical analysis and was verified by the San Diego Center for AIDS Research Biostatistics and Modeling Core. For all nonparametric analyses, medians and 95% confidence intervals (CIs) are reported. Agilent TapeStation integration percentages before and after PFGE were compared with the Mann-Whitney unpaired nonparametric t test. Normalized HIV gag copy numbers per 1 million cells before and after PFGE ddPCR were compared with Wilcoxon's paired nonparametric t test. Statistical correlations between values obtained before and after PFGE ddPCR and Alu-gag qPCR were Spearman's rho nonparametric correlations.
RESULTS
Removal of lower-molecular-weight DNA using PFGE purification.
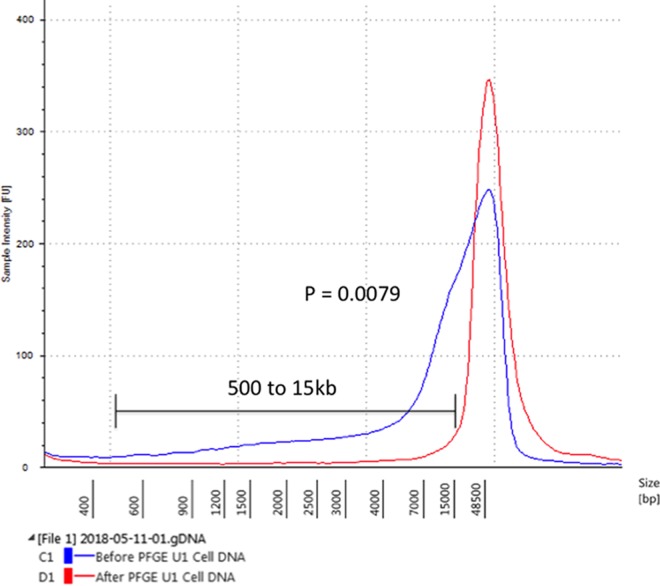
DNA from the U1 HIV cell line was assayed before and after PFGE removal of lower-molecular-weight DNA. U1 cells contain low-molecular-weight HIV species such as 2-LTR circles, as well as at least 1 provirus per cell. The area under the curve for the electropherogram showed a substantial decrease in lower-molecular-weight DNA (less than 15 kb in size) after PFGE across 5 replicates (P = 0.0079) (Fig. 1). It is possible to collect lower-molecular-weight DNA after PFGE enrichment. Lower-molecular-weight DNA appears in the flowthrough electrophoresis buffer of the BluePippin platform. The flowthrough DNA is highly diluted but can be concentrated by ethanol precipitation or speed vacuum protocols. The lower-molecular-weight flowthrough DNA contained 2-LTR circles and HIV gag, as detected by ddPCR (data not shown).
FIG 1.
Agilent TapeStation electropherogram, showing a representative plot of DNA from the U1 cell line before and after PFGE. From 500 bp to 15 kb, statistical analysis by the Mann-Whitney unpaired nonparametric t test determined a significant difference for results before versus after PFGE (P = 0.0079).
Removal of HIV 2-LTR circular DNA by PFGE purification from DNA from cells from untreated HIV-infected subjects.
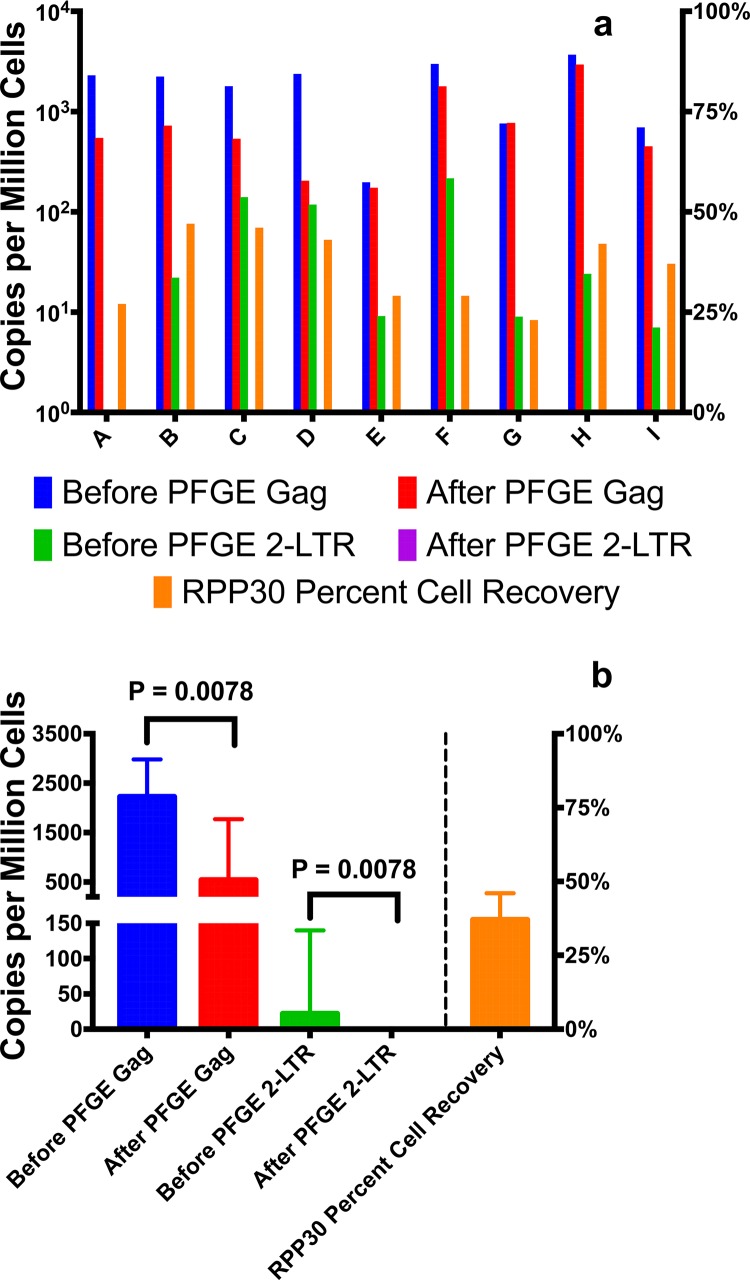
DNA was extracted from 9 PBMC samples collected from HIV-infected subjects who were not receiving ART. 2-LTR circles and HIV gag were measured by ddPCR before and after PFGE purification (Fig. 2a). In all 9 samples, no 2-LTR circles were detected after PFGE. HIV gag was readily detected and quantified after PFGE. The recovery of HIV gag copies per million cells decreased after PFGE (P = 0.0078) (Fig. 2b). This is expected and consistent with the removal of low-molecular-weight unintegrated HIV DNA species such as 2-LTR circles, which also contain HIV gag sequences, as does chromosomal DNA that is sheared to sizes of less than 15 kb. With the removal of 2-LTR circles and other lower-molecular-weight forms of HIV DNA, PFGE purification permits the measurement of HIV DNA present only in higher-molecular-weight chromosomal DNA, which contains the latent HIV reservoir. The median recovery of host genomic DNA was 37%, as measured with the RPP30 gene. This loss is likely attributable to shearing of chromosomal DNA and removal by PFGE, as well as loss of higher-molecular-weight DNA during the purification process.
FIG 2.
HIV sequence copies per million cells using ddPCR with DNA from PBMCs from 9 subjects not receiving ART, before or after PFGE. HIV gag was measured to capture both proviral DNA and HIV episomes. 2-LTR primers were used to measure 2-LTR circular episomes. (a) HIV gag and 2-LTR circle copy numbers for the individual subjects. (b) Median values for the 9 subjects. The median gag copy number before PFGE was 2,227 copies per million cells (95% CI, 1,006 to 2,769 copies per million cells). The median gag copy number after PFGE was 542 copies per million cells (95% CI, 210 to 1,584 copies per million cells) (P = 0.0078). The median 2-LTR circle copy number before PFGE was 22 copies per million cells (95% CI, 0.76 to 120 copies per million cells); after PFGE, no 2-LTR circles were detected in any of the samples (P = 0.0078). The median recovery of genomic DNA of 37% (95% CI, 28.9 to 42.8%) was determined by measuring the host cell RPP30 gene.
Reproducibility between replicates in virally suppressed primary samples.
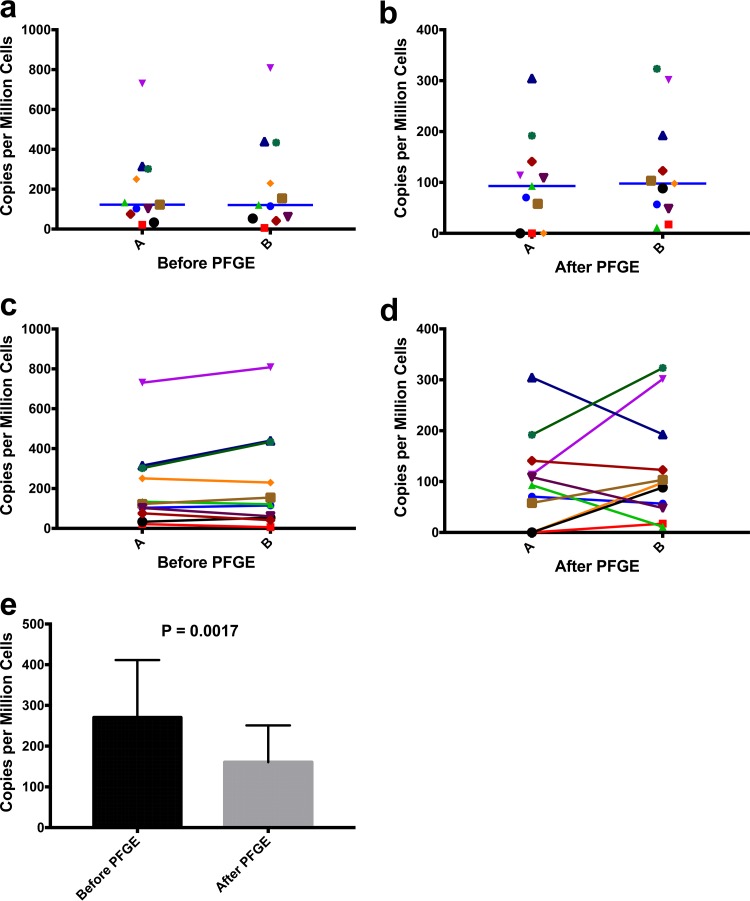
HIV gag was measured by ddPCR before and after PFGE purification of DNA from PBMC samples collected from 11 subjects who were receiving ART and had suppressed viral loads. Two complete technical replicates were performed for each PBMC sample, to measure reproducibility and significant variance of the entire workflow, from DNA extraction from PBMCs to ddPCR before and after PFGE purification. Before PFGE, ddPCR measurements were not statistically different between technical replicates for HIV gag, using Wilcoxon's paired nonparametric t test (Fig. 3a). Similarly, technical replicates for HIV gag after PFGE were also not statistically different using Wilcoxon's paired nonparametric t test (Fig. 3b), demonstrating reproducibility. As with pre-ART samples (Fig. 2b), the HIV gag copies per million cells decreased with PFGE purification, consistent with the presence of unintegrated HIV DNA (P = 0.0017) (Fig. 3e). Even with virological suppression with ART for at least 6 months, 2-LTR circles and other lower-molecular-weight HIV DNA could be detected in the cells, consistent with the prolonged half-life of episomal DNA during the life of cells or in one of their proliferated progeny (24, 25). PFGE purification eliminates these HIV DNA species, ensuring direct measurement of integrated HIV DNA, even in DNA from subjects with viral suppression with ART. Physical isolation of chromosomal HIV DNA from episomal HIV DNA suggests the frequent presence of unintegrated DNA in this small cohort of HIV-infected individuals receiving prolonged ART.
FIG 3.
(a and c) HIV gag DNA copies per million cells before PFGE for complete technical replicates of PBMCs from 11 ART-suppressed subjects. The median for the first replicate (A) was 122.3 copies per million cells (95% CI, 62.6 to 335.5 copies per million cells), and that for the second replicate (B) was 120.9 copies per million cells (95% CI, 59.95 to 388 copies per million cells). By Wilcoxon's paired nonparametric t test, no significant difference was observed between replicates. (b and d) HIV gag DNA copies per million cells after PFGE for complete technical replicates of cells from 11 ART-suppressed subjects. The median for the first replicate (A) was 93 copies per million cells (95% CI, 36.4 to 160.2 copies per million cells), and that for the second replicate (B) was 97.8 copies per million cells (95% CI, 52.7 to 195.3 copies per million cells). By Wilcoxon's paired nonparametric t test, no significant difference was observed between replicates. (e) HIV gag DNA copies per million cells before or after PFGE for PBMCs from 22 ART-suppressed subjects. The median gag copy number before PFGE was 270.4 copies per million cells (95% CI, 193.7 to 486.6 copies per million cells), and that after PFGE was 160.6 copies per million cells (95% CI, 89.6 to 399.5 copies per million cells). The median genomic DNA recovery percentage was 22.5% (95% CI, 20.4 to 29.6%), as determined with the RPP30 gene. Wilcoxon's paired nonparametric t test determined a significant difference for results before versus after PFGE (P = 0.0017).
Comparison of measurements of integrated HIV DNA using PFGE and Alu-gag qPCR.
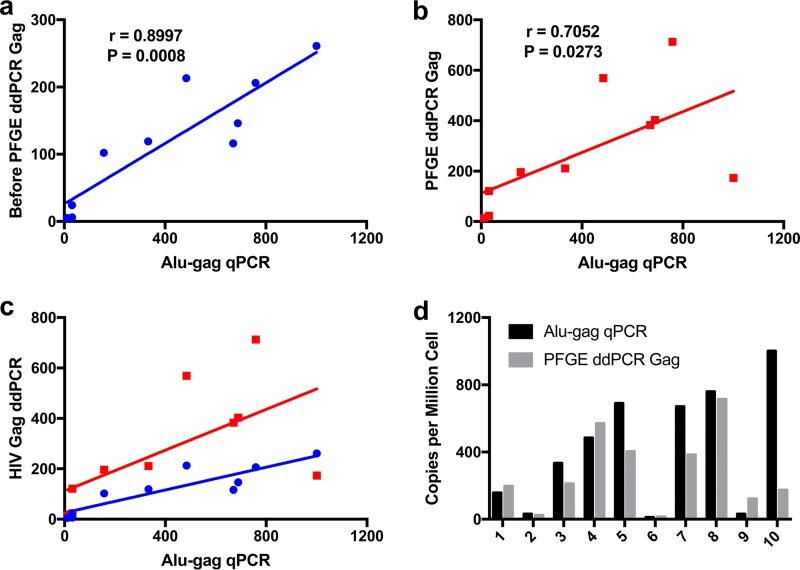
DNA was extracted from PBMCs collected from 10 participants who were receiving ART and had suppressed viral loads. HIV 2-LTR circles were detected by ddPCR for 6 of the 10 ART-suppressed persons (data not shown). Integrated HIV gag was quantified by the Alu-gag qPCR assay and by ddPCR before and after PFGE purification. Levels of integrated HIV gag measured by Alu-gag qPCR and ddPCR before (Fig. 4a) and after (Fig. 4b) PFGE were highly correlated by nonparametric Spearman's rho correlation (r = 0.8997 and r = 0.7052, respectively). Integrated HIV DNA levels were greater in 6 of the 10 samples by Alu-gag qPCR, compared with PFGE ddPCR, but the values were not considered statistically different, as determined with Wilcoxon's paired nonparametric t test (Fig. 4d). Our findings suggest that physical isolation of integrated DNA before PCR provides more robust direct measurements of integrated DNA than does Alu-gag qPCR.
FIG 4.
(a and b) Correlations between values obtained using Alu-gag qPCR and HIV gag ddPCR with samples from 10 ART-suppressed subjects, normalized to copies per million cells, before (a) and after (b) PFGE. The HIV gag PFGE and ddPCR were performed on the DNA with blinding to the results of the Alu-gag qPCR. The r and P values were determined using Spearman's rho correlations. (c) Values in panels a and b plotted together, to provide visual comparison. (d) Comparison of integrated HIV DNA copies per million cells determined using Alu-gag qPCR and gag ddPCR after PFGE, with DNA from PBMCs collected from 10 ART-suppressed subjects. Wilcoxon's paired nonparametric t test determined no significant difference between the methods.
DISCUSSION
We optimized an automated PFGE platform to purify high-molecular-weight genomic DNA by removing DNA species smaller than 15,000 bp. We utilized this PFGE platform to analyze DNA from primary PBMCs from HIV-infected persons, to quantitate integrated HIV DNA in the latent HIV reservoir by excluding any signal from episomal HIV DNA. This PFGE ddPCR assay was comparable to Alu-gag qPCR, which is currently the most used method for quantifying integrated HIV DNA.
The automated PFGE platform eliminated DNA species below 15,000 bp, thus improving the analysis of cellular chromosomal DNA. Unlike the more common direct-current agarose gel electrophoresis, PFGE uses an alternating-current mechanism that constantly switches electrical direction at specifically timed intervals, to allow better separation between large and small DNA fragments through the gel. In Fig. 1, lower-molecular-weight DNA is swept entirely through the gel cassette, while genomic DNA is diverted for collection. Because individual gel lanes are separated and enclosed in plastic, cross-contamination between samples does not occur. Also, the size-specific DNA fragment is eluted from the gel and into a collection reservoir with no need for gel purification. This automated platform allows users to start a PFGE enrichment protocol overnight and have eluted size-selected DNA in the morning.
In the PFGE purification protocol, up to 5 μg of total DNA (the DNA in about 0.7 million diploid cells) can be loaded. The purified higher-molecular-weight (chromosomal) DNA is recovered with efficiency of 25 to 35% or more, as measured with the host cell RPP30 gene. This method works with primary PBMCs collected from HIV-infected subjects. In untreated HIV-infected PBMCs, it is difficult to measure integrated HIV DNA because of the abundance of preintegration complexes and 1-LTR or 2-LTR circles. The removal of these lower-molecular-weight species of HIV DNA by PFGE, however, provides only high-molecular-weight chromosomal DNA for the measurement of integrated HIV DNA. Figure 2a shows that 2-LTR circles were no longer detectable after PFGE. Also, HIV detection was substantially decreased after PFGE of DNA from HIV-infected PBMCs from subjects not treated with ART. Even with ART, however, episomal HIV DNA is usually detectable in PBMC DNA (7, 15).
PFGE ddPCR is highly reproducible. Starting at the step of DNA extraction from HIV-infected PBMCs obtained from persons who were receiving ART and had experienced viral suppression for at least 6 months, we performed complete technical replicates with 11 samples through the entire assay workflow. There were no statistical differences between PFGE technical replicates of the workflow (Fig. 3a and b). As with PBMCs from untreated HIV-infected persons shown in Fig. 2a, PBMCs and CD4+ T cells from virally suppressed HIV-infected persons decreased in HIV DNA after PFGE purification, as shown in Fig. 3e. The decrease in detection in samples suppressed by ART was smaller than the decrease in untreated samples, which was expected because fewer HIV DNA species were present in samples obtained from subjects after ART.
Currently, the most widely used method for measuring integrated HIV DNA in latent HIV samples is the Alu-gag qPCR assay. We were interested in comparing the PFGE ddPCR assay with the Alu-gag qPCR assay using blinded samples. The PFGE ddPCR assay results were positively correlated with Alu-gag qPCR assay results both before (Fig. 4a) and after (Fig. 4b) PFGE. The greater correlation of ddPCR results with Alu-gag qPCR results before versus after PFGE (Fig. 4a to c) may relate to the use of a correction factor to calculate integrated DNA. This correction factor assumes that only a fraction of integration events are detected, because of diminished efficiency of Alu-gag qPCR amplification as the distance between the 2 primers increases. In 6 of the 10 samples tested, HIV gag levels were greater with the Alu-gag qPCR assay than the PFGE ddPCR assay.
A major limitation of the Alu-gag qPCR assay is its inherent indirect measurement. The Alu-gag qPCR assay described herein is twice as sensitive as prior assays (22); nonetheless, it detects only 20% of integration events within a standard integration cell line. For this reason, a correction factor about 5-fold is applied to each measurement. However, this correction can lead to inaccurate results, because the correction factor was determined using a standard integration cell line (12), which likely experienced less selection pressure than the HIV-infected cells of an individual living with HIV for years. The standard integration cell line was generated by infecting T cells with a vesicular stomatitis virus G protein (VSV-G) pseudotyped replication-incompetent (∆env) virus engineered to express hygromycin resistance and express green fluorescent protein (GFP). These infected cells were then cultured for a few weeks under antibiotic selection. During this culture period, the unintegrated HIV DNA diminished. Thus, the standard integration cell line was generated by exposing the cells to minimal selection pressure for a short time. In contrast, proviruses in vivo in HIV-infected individuals likely undergo much stronger selection pressures over many years. Given that Alu sites are enriched in introns, significant selection may occur, with clonal expansion of cells containing an HIV provirus in an intron. Thus, over time clonal proviruses that are near an Alu site might be positively selected. This, in turn, could lead to overestimation of the integration level, as discussed in more detail elsewhere (26).
An unexpected advantage of the PFGE ddPCR assay may be its ability to detect unintegrated HIV DNA sensitively. Surprisingly, there was often less HIV DNA per cell after PFGE, consistent with the presence of unintegrated HIV DNA. Future studies could be designed to exploit this aspect of PFGE. Prior studies that attempted to capture the presence of integrated HIV DNA were either laborious (17) or insensitive, due to the errors in measurement of integrated and total DNA (27, 28). The direct robust measurement of integrated and unintegrated HIV DNA by PFGE may prove useful in HIV eradication trials, especially for monitoring therapies that have potential to induce ongoing replication (28).
Although the PFGE ddPCR assay is easier to perform and provides more direct and robust measurements than the Alu-gag qPCR assay, there are some limitations to the PFGE ddPCR approach. The cost of an instrument and commercial kits is a consideration. Also, ddPCR provides more precise quantitative measurements than qPCR but at a higher price than qPCR (nearly 5 times the cost per well). With PFGE and separation of DNA fragments, the assay throughput is slower than with other PCR methods. The BluePippin PFGE cassettes run only 4 samples simultaneously, with a 4.5-hour run time. Each cassette is limited to 5 μg of total DNA (about 1 million cells) per lane. It is possible to measure more than 5 μg of total DNA from a single sample by running replicate lanes and combining the eluted enrichments, but this contributes to an even slower assay throughput. Most subjects treated with ART have 101 to 104 copies of integrated HIV DNA per million cells. One example of the sensitivity and ability to assay proviral DNA in small cell numbers was observed with the myeloid mouse model of HIV infection. An average of 10 proviral copies per million cells could be demonstrated with only 100,000 macrophages available from mice fully suppressed with ART after HIV infection (29).
In summary, the PFGE ddPCR assay is a sensitive and more direct measure of proviral integrated HIV DNA. Unlike the Alu-gag qPCR assay, the PFGE ddPCR assay physically separates contaminating lower-molecular-weight DNA such as 2-LTR circles, which provide sites for PCR amplification and confound integrated HIV DNA measurements. The method uses fewer cells and less DNA than the Alu-gag qPCR assay and is less technically difficult to perform; nevertheless, the method can provide sensitive and accurate integrated HIV DNA quantitation comparable to that of Alu-gag qPCR. This assay can provide insights into the existence of integrated HIV DNA in subsets and cell types in ART-suppressed people and animal models not previously well characterized, such as macrophages in the study by Honeycutt et al. (29). Its applications may prove useful for cure strategies, especially those designed to target HIV sequences, such as those using clustered regularly interspaced short palindromic repeat (CRISPR) methods.
ACKNOWLEDGMENTS
The valuable contributions of Matt Strain in initiating this project are greatly appreciated. We acknowledge the Biostatistics and Modeling Core and the Genomics and Sequencing Core of the San Diego Center for AIDS Research, for advice and support. HIV-1 infected U937 cells (U1 cells) from Thomas Folks were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH.
This work was supported by the Department of Veterans Affairs and grants from the National Institutes of Health (grants AI126619, AI126620, P30 AI036214, R33 AI104283, R01 AI120011, R33 AI104280, and P30 AI045008, R01 AI12001, and UM1 AI126617). This work was performed with the support of the James B. Pendleton Charitable Trust and the Philadelphia Foundation.
We report no conflicts of interest.
S.M.L. and D.D.R. conceived and designed the experiments, L.J.M. and U.O. provided clinical specimens, S.M.L., K.H., and D.J.V. performed the experiments, S.M.L., U.O., and D.D.R. analyzed the data, and S.M.L. and D.D.R. wrote the paper. All authors read and approved the final manuscript.
REFERENCES
- 1.Richman DD, Margolis DM, Delaney M, Greene WC, Hazuda D, Pomerantz RJ. 2009. The challenge of finding a cure for HIV infection. Science 323:1304–1307. doi: 10.1126/science.1165706. [DOI] [PubMed] [Google Scholar]
- 2.Archin NM, Sung JM, Garrido C, Soriano-Sarabia N, Margolis DM. 2014. Eradicating HIV-1 infection: seeking to clear a persistent pathogen. Nat Rev Microbiol 12:750–764. doi: 10.1038/nrmicro3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Margolis DM, Garcia JV, Hazuda DJ, Haynes BF. 2016. Latency reversal and viral clearance to cure HIV-1. Science 353:aaf6517. doi: 10.1126/science.aaf6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Massanella M, Richman DD. 2016. Measuring the latent reservoir in vivo. J Clin Invest 126:464–472. doi: 10.1172/JCI80567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pierson T, McArthur J, Siliciano RF. 2000. Reservoirs for HIV-1: mechanisms for viral persistence in the presence of antiviral immune responses and antiretroviral therapy. Annu Rev Immunol 18:665–708. doi: 10.1146/annurev.immunol.18.1.665. [DOI] [PubMed] [Google Scholar]
- 6.Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, Boucher G, Boulassel MR, Ghattas G, Brenchley JM, Schacker TW, Hill BJ, Douek DC, Routy JP, Haddad EK, Sekaly RP. 2009. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med 15:893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eriksson S, Graf EH, Dahl V, Strain MC, Yukl SA, Lysenko ES, Bosch RJ, Lai J, Chioma S, Emad F, Abdel-Mohsen M, Hoh R, Hecht F, Hunt P, Somsouk M, Wong J, Johnston R, Siliciano RF, Richman DD, O'Doherty U, Palmer S, Deeks SG, Siliciano JD. 2013. Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog 9:e1003174. doi: 10.1371/journal.ppat.1003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ho Y-C, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DIS, Lai J, Blankson JN, Siliciano RF. 2013. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 155:540–551. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Avettand-Fènoël V, Hocqueloux L, Ghosn J, Cheret A, Frange P, Melard A, Viard JP, Rouzioux C. 2016. Total HIV-1 DNA, a marker of viral reservoir dynamics with clinical implications. Clin Microbiol Rev 29:859–880. doi: 10.1128/CMR.00015-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamid FB, Kim J, Shin C-G. 2017. Distribution and fate of HIV-1 unintegrated DNA species: a comprehensive update. AIDS Res Ther 14:9. doi: 10.1186/s12981-016-0127-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Doherty U, Swiggard WJ, Jeyakumar D, McGain D, Malim MH. 2002. A sensitive, quantitative assay for human immunodeficiency virus type 1 integration. J Virol 76:10942–10950. doi: 10.1128/JVI.76.21.10942-10950.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liszewski MK, Yu JJ, O'Doherty U. 2009. Detecting HIV-1 integration by repetitive-sampling Alu-gag PCR. Methods 47:254–260. doi: 10.1016/j.ymeth.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herschleb J, Ananiev G, Schwartz DC. 2007. Pulsed-field gel electrophoresis. Nat Protoc 2:677–684. doi: 10.1038/nprot.2007.94. [DOI] [PubMed] [Google Scholar]
- 14.Parizad EG, Parizad EG, Valizadeh A. 2016. The application of pulsed field gel electrophoresis in clinical studies. J Clin Diagn Res 10:DE1–DE4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strain MC, Lada SM, Luong T, Rought SE, Gianella S, Terry VH, Spina CA, Woelk CH, Richman DD. 2013. Highly precise measurement of HIV DNA by droplet digital PCR. PLoS One 8:e55943. doi: 10.1371/journal.pone.0055943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gianella S, Massanella M, Richman DD, Little SJ, Spina CA, Vargas MV, Lada SM, Daar ES, Dube MP, Haubrich RH, Morris SR, Smith DM. 2014. Cytomegalovirus replication in semen is associated with higher levels of proviral HIV DNA and CD4+ T cell activation during antiretroviral treatment. J Virol 88:7818–7827. doi: 10.1128/JVI.00831-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koelsch KK, Liu L, Haubrich R, May S, Havlir D, Günthard HF, Ignacio CC, Campos-Soto P, Little SJ, Shafer R, Robbins GK, D'Aquila RT, Kawano Y, Young K, Dao P, Spina CA, Richman DD, Wong JK. 2008. Dynamics of total, linear nonintegrated, and integrated HIV-1 DNA in vivo and in vitro. J Infect Dis 197:411–419. doi: 10.1086/525283. [DOI] [PubMed] [Google Scholar]
- 18.Folks TM, Justement J, Kinter A, Dinarello CA, Fauci AS. 1987. Cytokine-induced expression of HIV-1 in a chronically infected promonocyte cell line. Science 238:800–802. doi: 10.1126/science.3313729. [DOI] [PubMed] [Google Scholar]
- 19.Graf EH, Mexas AM, Yu JJ, Shaheen F, Liszewski MK, Di Mascio M, Migueles SA, Connors M, O'Doherty U. 2011. Elite suppressors harbor low levels of integrated HIV DNA and high levels of 2-LTR circular HIV DNA compared to HIV+ patients on and off HAART. PLoS Pathog 7:e1001300. doi: 10.1371/journal.ppat.1001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu JJ, Wu TL, Liszewski MK, Dai J, Swiggard WJ, Baytop C, Frank I, Levine BL, Yang W, Theodosopoulos T, O'Doherty U. 2008. A more precise HIV integration assay designed to detect small differences finds lower levels of integrated DNA in HAART treated patients. Virology 379:78–86. doi: 10.1016/j.virol.2008.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Spiegelaere W, Malatinkova E, Lynch L, Van Nieuwerburgh F, Messiaen P, O'Doherty U, Vandekerckhove L. 2014. Quantification of integrated HIV DNA by repetitive-sampling Alu-HIV PCR on the basis of Poisson statistics. Clin Chem 60:886–895. doi: 10.1373/clinchem.2013.219378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agosto LM, Yu JJ, Dai J, Kaletsky R, Monie D, O'Doherty U. 2007. HIV-1 integrates into resting CD4+ T cells even at low inoculums as demonstrated with an improved assay for HIV-1 integration. Virology 368:60–72. doi: 10.1016/j.virol.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michael NL, Herman SA, Kwok S, Dreyer K, Wang J, Christopherson C, Spadoro JP, Young KK, Polonis V, McCutchan FE, Carr J, Mascola JR, Jagodzinski LL, Robb ML. 1999. Development of calibrated viral load standards for group M subtypes of human immunodeficiency virus type 1 and performance of an improved AMPLICOR HIV-1 MONITOR test with isolates of diverse subtypes. J Clin Microbiol 37:2557–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pierson TC, Kieffer TL, Ruff CT, Buck C, Gange SJ, Siliciano RF. 2002. Intrinsic stability of episomal circles formed during human immunodeficiency virus type 1 replication. J Virol 76:4138–4144. doi: 10.1128/JVI.76.8.4138-4144.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Butler SL, Johnson EP, Bushman FD. 2002. Human immunodeficiency virus cDNA metabolism: notable stability of two-long terminal repeat circles. J Virol 76:3739–3747. doi: 10.1128/JVI.76.8.3739-3747.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pinzone MR, O'Doherty U. 2018. Measuring integrated HIV DNA ex vivo and in vitro provides insights about how reservoirs are formed and maintained. Retrovirology 15:22. doi: 10.1186/s12977-018-0396-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agosto LM, Liszewski MK, Mexas A, Graf E, Pace M, Yu JJ, Bhandoola A, O'Doherty U. 2011. Patients on HAART often have an excess of unintegrated HIV DNA: implications for monitoring reservoirs. Virology 409:46–53. doi: 10.1016/j.virol.2010.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mexas AM, Graf EH, Pace MJ, Yu JJ, Papasavvas E, Azzoni L, Busch MP, Foulkes AS, Migueles SA, Montaner LJ, O'Doherty U. 2012. Concurrent measures of total and integrated HIV DNA monitor reservoirs and ongoing replication in eradication trials. AIDS 26:2295–2306. doi: 10.1097/QAD.0b013e32835a5c2f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Honeycutt JB, Thayer WO, Baker CE, Ribeiro RM, Lada SM, Cao Y, Cleary RA, Hudgens MG, Richman DD, Garcia JV. 2017. HIV persistence in tissue macrophages of humanized myeloid-only mice during antiretroviral therapy. Nat Med 23:638–643. doi: 10.1038/nm.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]