Abstract
Innate lymphoid cells (ILCs) are important mediators of the immune response and homeostasis in barrier tissues of mammals. However, the existence and function of ILCs in other vertebrates is poorly understood. Here, we use single-cell RNA sequencing to generate a comprehensive atlas of zebrafish lymphocytes during tissue homeostasis and following immune challenge. We profiled 14,080 individual cells from the gut of wild-type zebrafish, as well as of rag1-deficient zebrafish which lack T and B cells, and discovered populations of ILC-like cells. We uncovered a rorc-positive subset of innate lymphoid cells that could express cytokines associated with type 1, 2 and 3 responses upon immune challenge. Specifically, these ILC-like cells expressed il22 and tnfa following exposure to inactivated bacteria, or il13 following exposure to helminth extract. Cytokine-producing ILC-like cells express a specific repertoire of novel immune-type receptors (NITRs), likely involved in recognition of environmental cues. We identified additional novel markers of zebrafish ILCs and generated a cloud repository for their in-depth exploration.
Introduction
Vertebrate immune systems consist of the innate arm, which responds immediately to challenge, and the adaptive arm, which responds via acquired antigen receptors. In mammals, myeloid cells (granulocytes, mast cells, monocytes/macrophages, dendritic cells) form the innate immune system, whereas B and T lymphocytes contribute to the adaptive immune response (1, 2). Recently discovered innate lymphoid cells (ILCs) represent a rare population of lymphocytes (3–5). Unlike T and B cells, ILCs do not express antigen receptors or undergo clonal expansion when stimulated. Instead, in the absence of adaptive antigen receptors, ILCs sense environmental cues mostly through cytokine receptors, and promptly respond to signals by producing distinct cytokines. More recently, it has been demonstrated that both murine and human ILCs express a receptor for the neuropeptide neuromedin, secreted by cholinergic neurons which directly sense worm products and control the expression of innate type 2 cytokines (6). During homeostasis, humans and mice contain four populations of ILCs: natural killer (NK) cells, and three subsets of helper ILCs (ILC1, ILC2 and ILC3). NK cells bear similarity to cytotoxic T cells (CD8+ cells), which directly kill cells infected with intracellular pathogens. Helper ILCs in human and mouse are classified as ILC1, ILC2 and ILC3 based on their transcription factor (TF) and cytokine secretion profiles, as well as phenotypic cell-surface markers (3–5, 7). Both Th1 and ILC1 express T-bet (encoded by tbx21) as well as so-called “Th1 cytokines” such as interferon gamma (IFNg) and tumour necrosis factor alpha (TNFα), and act against intracellular pathogens. Th2 and ILC2 express GATA-3 and secrete IL-4, IL-13 and amphiregulin, and contribute to defence against helminths and venoms. Th17 and ILC3s express RORγt (encoded by rorc) as well as IL-17a, IL-17f and IL-22 and promote immunity against extracellular bacteria and fungi (3, 4, 8, 9). To date, the bulk of our knowledge of ILCs comes from studies in humans and in mice (3, 4, 10, 11).
The different immune cell types are usually distinguished based on expression of specific CD (cluster of differentiation) markers. However, a homogenous population of blood cells, as defined by surface markers, may include many distinct transcriptional states with different functional properties (12–15). In addition, the surface markers used to define distinct human and murine leukocyte subsets are not the same, making it difficult to compare cell types across different species. Therefore, there is a need for unbiased methodologies that define immune cell types based on cellular state rather than cell surface markers. This is particularly relevant for species other than mouse and human, where specific antibodies for distinct blood and immune cell types are not readily available.
In zebrafish, the heterogeneity of haematopoietic cells has mostly been investigated with fluorescent transgenic reporter lines, as very few antibodies for surface markers are available (16). These approaches have confirmed the presence of erythrocytes, thrombocytes, neutrophils, macrophages, eosinophils, T cells, B cells and NK cells in zebrafish. Comprehensive transcriptome atlases exist for many of these cell types (17–19). Because these studies focused on steady state conditions, they were limited in their ability to characterise the response mechanisms following immune challenge. Compared to mice and humans, little is known about the diversity of cytokine-producing innate lymphoid cells in zebrafish, and a detailed characterization of their transcriptional profiles is still lacking.
Here, we characterized the repertoire of innate and adaptive lymphocytes in zebrafish. Using single-cell RNA sequencing, we generated a comprehensive atlas of cellular states of lymphocytes collected from various organs in steady state and following immune challenge. By studying cytokine-expression of lymphocytes in rag1-/- zebrafish, we have identified cells that resemble ILC2 and ILC3 cells described in mice and in humans.
Results
rag1-/- mutants lack T and B cells but have cytokine-producing cells in the gut
Rag1- and Rag2-deficient mouse strains, which lack adaptive but retain innate lymphoid cells (20–22), have provided substantial insight into ILCs. These mice showed expression of many cytokines previously considered to be T cell-specific and therefore provided the first evidence of the existence of helper ILCs (20–22). Thus, to focus on innate lymphoid cells in zebrafish, we turned to rag1-/- mutants. As in mice, rag1-/- zebrafish lack T and B lymphocytes (23) but retain NK cells (24).
In line with previous reports (23–25), rag1-/- zebrafish displayed a reduced population of lymphoid cells in the gut as defined by FSC/SSC gating on FACS (Figure 1A-B). Further, bulk qPCR on FACS-sorted cells from the lymphoid population of rag1-/- zebrafish showed two- and four-fold decreases in the expression of T cell markers such as cd3z and trac, respectively, compared to the wild-type zebrafish, whereas the expression level of il7r and lck remained the same (Figure 1C). To verify that rag1-/- zebrafish indeed lack adaptive lymphocytes, we sequenced 171 lck:EGFP+ single cells collected from gut and kidney of the rag1-/- zebrafish and applied TraCeR (26), a novel method for reconstruction of TCR sequences from single-cell RNA-seq data to search for V(D)J recombination events in individual cells. No TCR rearrangements were detected in cells isolated from rag1-/- zebrafish (Table S1). These data confirm that the rag1-/- provides an excellent tool to examine the innate lymphocytes in zebrafish.
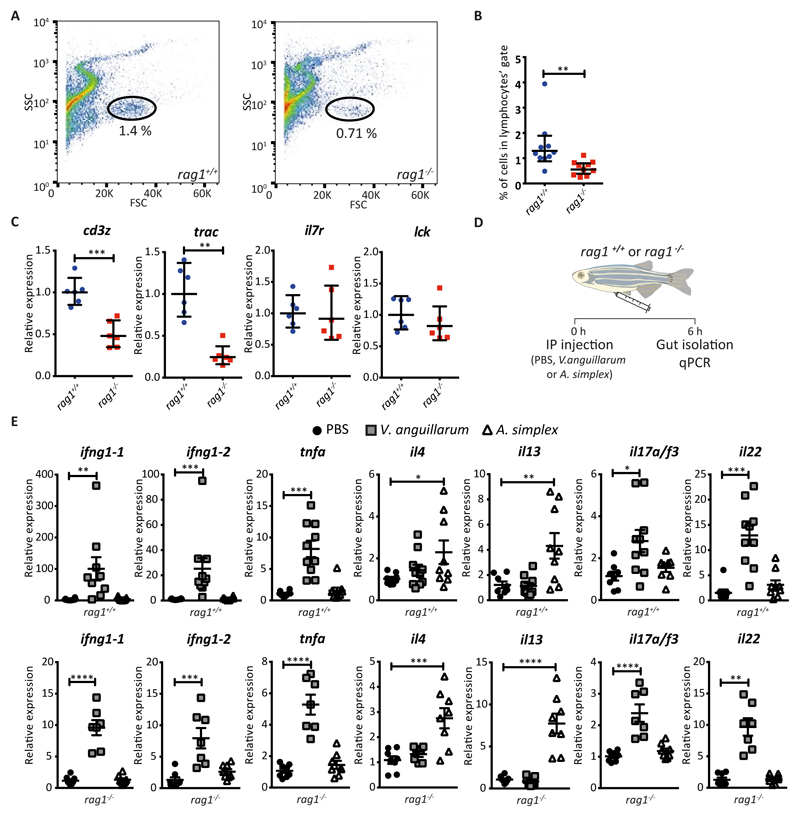
Figure 1. Rag1-/- zebrafish have cytokine producing cells in the gut.
A. Representative FACS plots showing the percentage of cells in the lymphocytes’ gate (as defined by FSC/SSC gating) in the gut of wild-type zebrafish (left) and rag1-/-mutant (right). B. Percentage of cells in the lymphocytes’ gate within 50 000 recorded events in the gut of wild-type and rag1-/-mutant zebrafish. Bars represent the geometric mean ± 95% confidence interval to estimate total number of lymphocytes. Mann-Whitney test. C. qPCR expression of T cells associated markers (cd3z, trac) and lymphocytes’ markers (il7r, lck) in mutant and wild-type zebrafish. Bars represent the geometric mean ± 95% confidence interval to estimate fold changes. Mann-Whitney test. D. Scheme of short-term inflammation experiment. E. qPCR expression of immune type 1 (ifng1-1, ifng1-2), immune type 2 (il4, il13) and immune type 3 (il17a/f3, il22) signature cytokines in the gut of the wild-type (rag1+/+) and mutant (rag1-/-) zebrafish following six hours challenge with V. anguillarumor A. simplex. Bars represent the geometric mean ± 95% confidence interval to estimate fold changes. One-way ANOVA test.
Mammals contain three populations of helper ILCs (ILC-1, ILC-2 and ILC-3) that rapidly respond to different tissue signals by producing effector cytokines (27–29). To study this process in zebrafish, we established short-term inflammation models that trigger cytokine expression of potential ILCs in zebrafish gut (Figure 1D). Formalin-inactivated Vibrio anguillarum has been used as a fish vaccine and is known to induce type 3 immunity (30); whereas the nematode Anisakis simplex, a common fish parasite, is expected to induce type 2 immunity (31). We injected wild-type and rag1-/- zebrafish intraperitoneally with PBS (control) or extracts of inactivated V. anguillarum or of lyophilised A. simplex. Six hours post-injection, we dissected the guts and evaluated the expression of signature cytokines by qRT-PCR (Figure 1D, E). We found that, in both wild-type and rag1-/- zebrafish, injection of V. anguillarum extract induced the expression of Th1/ILC1 cytokines such as ifng1-1, ifng1-2 as well as Th17/ILC3 cytokines, il17a/f3 and il22 (Figure 1E). The expression levels of the Th2/ILC2 cytokines il4 and il13 remained unchanged in V. anguillarum-compared to PBS-injected zebrafish. Conversely, injection of A. simplex extract induced the expression of Th2/ILC2 cytokines il4 and il13 but not of ifng1-1, ifng1-2, tnfa, il17a/f3 and il22 (Figure 1E).
These findings have two important implications. First, they confirm that intraperitoneal injection of V. anguillarum extract induces a type 1/type 3 immune response in zebrafish gut, and of A. simplex extract induce a type 2 immune response. Second, they reveal the presence of cytokine-producing cells in the gut of immune-challenged rag1-/- zebrafish, in the context of T cell deficiency. Given that mammalian ILCs have phenotypes that mirror polarized Th subsets in their expression of effector cytokines, our data suggest that the gut in zebrafish contains bona fide ILC subtypes.
Single-cell RNA sequencing reveals ILC2- and ILC3-like cells in zebrafish
ILCs comprise around 0.5-5% of lymphocytes in barrier tissues in mammals and as such represent a rare population of cells (9, 32). As the LCK gene is expressed in all three ILC subtypes in humans (33) (Figure S1), we reasoned that its expression pattern could be conserved in zebrafish. To capture ILCs subtypes in zebrafish, we utilised our short-term inflammation protocol on Tg(lck:EGFP) rag1-/- zebrafish. Single-cell RNA sequencing of thousands of lck:EGFP+ cells isolated from a gut of immune-challenged rag1-/- mutants provided a powerful approach to study cytokine-producing ILCs in zebrafish.
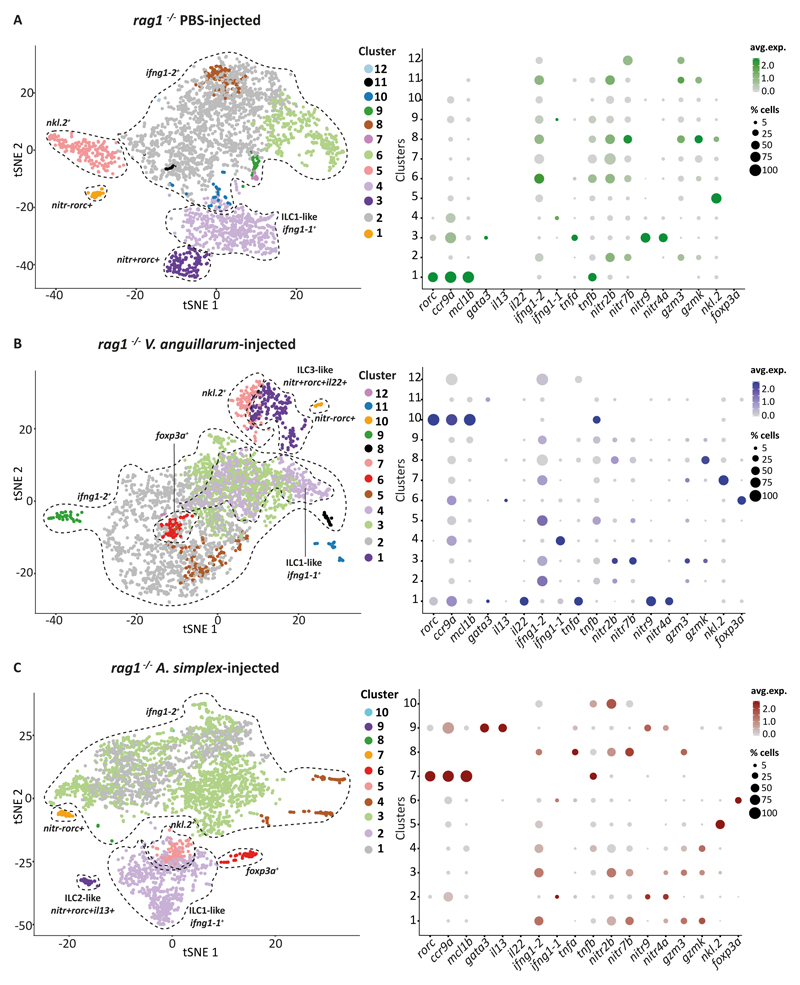
10x Genomics captures single cells in droplets, such that 5000 cells can be captured and subsequently sequenced within a single run (34). As above, we injected Tg(lck:EGFP) rag1-/- mutant zebrafish intraperitoneally with PBS, inactivated V. anguillarum or lyophilised A. simplex extracts and sorted lck:EGFP+ cells from the gut six hours post-injection. To ensure that a sufficient number of cells were loaded on 10x, we combined an equal number of lck:EGFP+ cells for each condition (PBS, A. simplex and V. anguillarum) from three different zebrafish (nine zebrafish in total). By using this approach, we generated a comprehensive data set that included 3211 single lck:EGFP+ cells from the guts of PBS-injected, 3626 cells from A. simplex-injected and 3487 cells from V. anguillarum-injected rag1-/- zebrafish (Figure 2). On average, we detected 600 genes per cell (Figure S2). This relatively modest number of detected genes was observed in all our data sets and could be linked with the small size of lymphocytes and their low RNA content (35). Clustering, followed by unbiased identification of marker genes for each cluster (see Methods section), revealed ILC-like cells in all three datasets (Figure 2, Figure S3-4, Figure S5A, Table S2).
Figure 2. Analysis of the lck+cells, collected from the gut of rag1-/- zebrafish.
2D projection of tSNE analysis of 10x RNAseq data showing heterogeneity of innate lymphoid cells. Dotplots show the level of expression of marker genes and percentage of cells per cluster that express the gene of interest.
We first analysed lck:EGFP+ cells from PBS-injected zebrafish and identified several clusters of lymphocytes that expressed ifng1-2 and granzyme genes (gzm3, gzmk) (Figure 2A). This transcriptional signature resembles mammalian NK cells. In addition, we identified cells that exclusively expressed ifng1-1 but not granzymes or nk lysins (cluster 4 on Figure 2A, Table S2). Cells in cluster 4 could be considered ILC1-like cells in zebrafish.
Importantly, our analysis revealed two rare (0.8% and 3.8%) populations of rorc+ cells (clusters 1 and 3 on Figure 2A and Figure S3-4, Figure S5A, Table S2). The TF RORγt (encoded by rorc) is expressed at low levels in circulating and tissue resident ILC precursors in human, as well as in mature ILC3 in human and mouse (36, 37). Rorc is required for the development and function of ILC3 (38, 39). We found that the rorc+ clusters did not express cytotoxicity-associated genes such as granzymes (gzm3, gzmk) or nk lysins (nkl.2). To investigate the potential function of these two rorc+ clusters, we performed differential expression (DE) analysis followed by gene ontology (GO) enrichment analysis (see Methods). We found that rorc+ cluster 1 was associated with GO-terms like “response to stress” and “protein folding” (Figure S5B) and showed high expression of the pro-survival gene mcl1b, transcription factor sox13, and tumour necrosis factor beta (tnfb), an orthologue of human lymphotoxin alpha (LTA), but was negative/low for novel immune-type receptor genes (e.g. nitr2b, nitr7b, nitr9, nitr4a, etc.), as well as ifng1-2/ifng1-1 (from here on nitr-rorc+ cluster) (Figure 2, Figure S3-4). In contrast, rorc+ cluster 3 was associated with terms like “immune system process”, “response to interferon gamma” and “response to other organisms” (Figure S5B) and was positive for tumour necrosis factor alpha (tnfa) as well as novel immune-type receptors nitr9 and nitr4a, but also negative for ifng1-2/ifng1-1 (from here on nitr+rorc+ cluster) (40, 41), (Figure 2A, Figure S3-4). Cells within this cluster also expressed gata3, which has been shown to be indispensable for development of all helper ILCs and expressed by subsets of ILCs at different levels (42).
In V. anguillarum-injected zebrafish, nitr+rorc+ cells (cluster 1), but not nitr-rorc+ cells (cluster 10), expressed il22 and tnfa (Figure 2B, Figure S3-4, Table S2). In humans and mice, ILC3 cells produce the cytokines IL-22 and TNF-α cells upon stimulation with bacteria, triggering antimicrobial response and repair programs in epithelial cells during infection (43–46). These data strongly suggest the existence of ILC3-like cells in the zebrafish gut that respond to immune challenge by producing relevant cytokines.
Interestingly, in zebrafish injected with lyophilised A. simplex, nitr+rorc+ cells (cluster 9) expressed il13 and gata3 (Figure 2C, Figure S3-4, Table S2). The gata3 TF is highly expressed in ILC2 cells and is required for their development (46); it plays a critical role in activating IL-13 production in ILC2 upon stimulation, thus promoting anti-helminth immunity. Therefore, these cells (Figure 2C) resemble mammalian ILC2 cells. Again, il13-producing cells were nitr+ but lacked expression of granzymes, nkl.2 and ifng1-2.
Immune challenged zebrafish (V. anguillarum- or A. simplex-injected) also had a population of foxp3a+ cells (Figure 2B-C, Figure S3-4, Table S2). Cells in this cluster were negative for interferon gamma genes, nitr genes and granzymes as well as cd4-1. It is tempting to speculate that these cells might represent the zebrafish equivalent of recently reported mammalian regulatory innate lymphoid cells (47). However, it should be noted that we did not detect expression of il10 in this cluster.
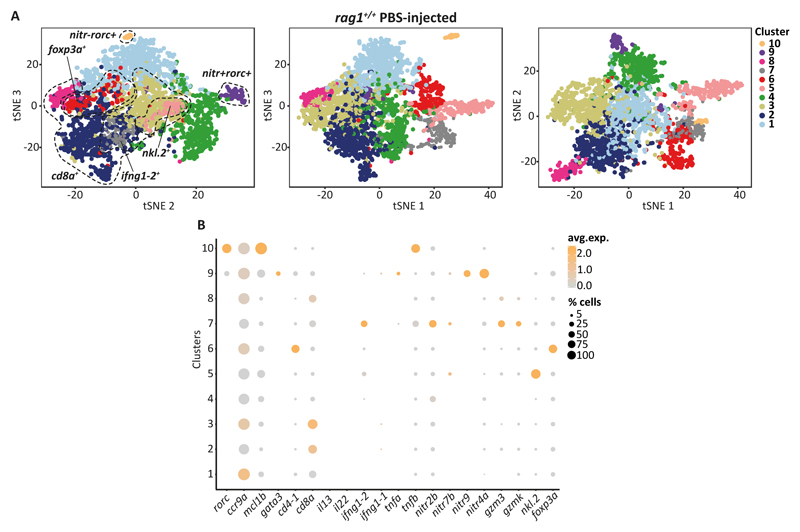
To validate that the rorc+ ILCs are a genuine constituent of the zebrafish gut, and not only present in rag1-/- zebrafish, we sequenced and analysed additional 3756 GFP+ cells from the gut of PBS-injected wild-type lck:EGFP zebrafish. As expected, most of these cells were T cells that expressed cd8a or cd4-1 (specifically Tregs). However, we also identified two clusters of rorc+ cells that were negative for T cell marker genes: namely, nitr-rorc+ cells (cluster 10, Figure 3) that expressed mcl1b and tnfb, and nitr+rorc+ cells (cluster 9, Figure 3) that expressed tnfa (Figure 3). These cells represented 0.8% and 1.9% of the lck:EGFP+ cell population of wild-type zebrafish (Table S2). Taken together, our transcriptional profiling of innate lymphocytes in rag1-deficient and wild type zebrafish identified ILC-like populations.
Figure 3. Analysis of the lck+cells collected from the gut of wild-type zebrafish.
A. 2D projection of tSNE analysis of 10x RNAseq data showing heterogeneity of innate and adaptive lymphocytes’ pool. B. Dotplot shows the level of expression of signature genes and percentage of cells per cluster that express the gene of interest.
ILC-like subtypes have distinct response to immune challenge
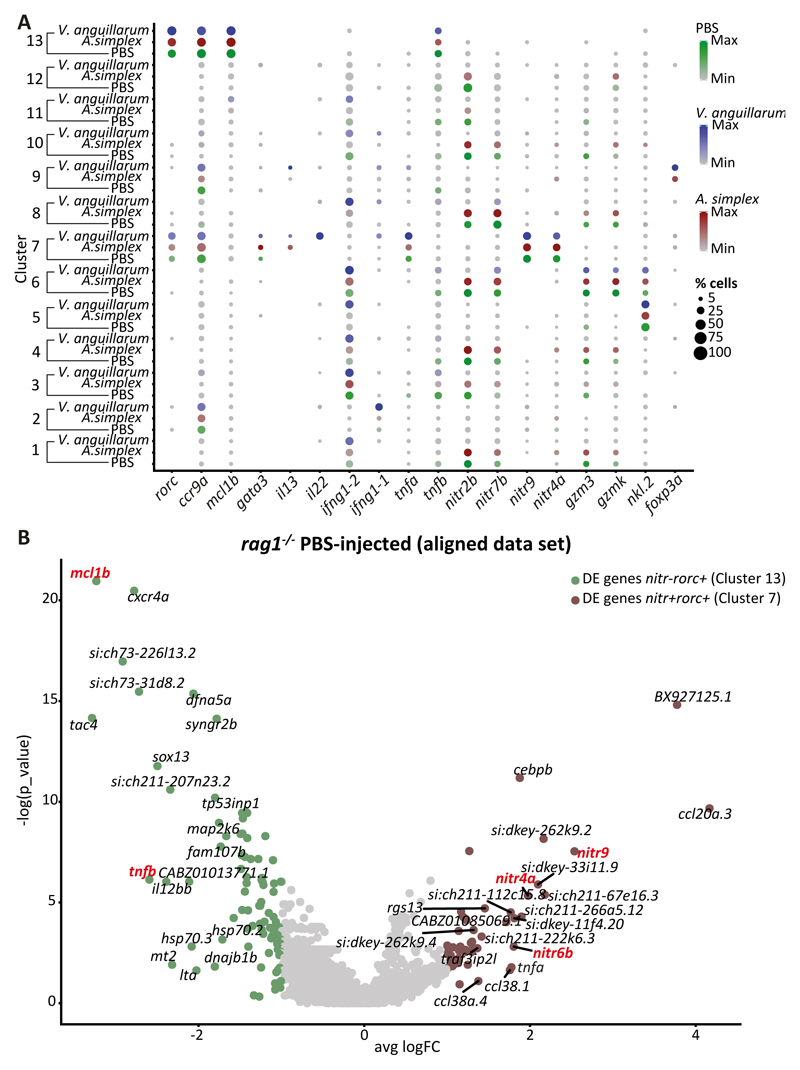
Our dataset revealed distinct, heterogeneous populations of innate lymphoid cells within the guts of PBS-, V. anguillarum-, and A. simplex-injected zebrafish. We hypothesized that these distinct populations are biologically relevant, reflecting the response of immune cells to disparate stimuli. To test this hypothesis, we evaluated whether the distinct cell types identified in control zebrafish matched those present in V. anguillarum- and A. simplex-injected zebrafish, and whether the specific treatments led to transcriptional changes in these cell types. We performed integrated analysis of PBS-, V. anguillarum-, and A. simplex-injected zebrafish using a recently developed computational strategy for scRNA-seq alignment (48). This methodology was specifically designed to allow comparison of RNA-seq datasets across different conditions (Figure 4A). By following the Seurat alignment workflow, we uncovered “shared” cell types across all three datasets and compared their gene expression profiles.
Figure 4. Integrated analysis of PBS - V. anguillarum- A. simplex-injected rag1-/- zebrafish.
A. Dotplot with the expression level of selected marker genes in each of the clusters. The size of the dots indicates the percentage of cells within the cluster that express the gene of interest; each cluster contains cells from three different conditions. B. Volcano plot showing the top 20 differentially expressed genes between nitr+rorc+(Cluster 7) and nitr-rorc+(Cluster 13) cells originated from rag1-/- PBS-injected zebrafish using aligned dataset.
Our analysis showed that the nitr+rorc+ cells found separately in the three conditions (clusters 3, 1 and 9 on Figures 2A-C, respectively) corresponded to a unique cell subtype (cluster 7 on Figure 4A; Figure S6A). Cells in this cluster upregulated il22, rorc and tnfa following in vivo stimulation with inactivated V. anguillarum (Figure 4A, Figure S6B). In contrast, injection of A. simplex resulted in upregulation of il13 and gata3 (Figure 4A), but ifng1-1, ifng1-2, tnfb and nitr genes remained unaltered relative to the control (Figure 4A).
Similarly, the nitr-rorc+ population of cells identified separately in PBS-, V. anguillarum- and A. simplex-injected zebrafish (clusters 1, 10 and 7 on Figures 2A-C, respectively) grouped as distinct cluster (cluster 13 in Figure 4A). These cells expressed mcl1b, sox13 and tnfb (orthologue of human lymphotoxin alpha, LTA) (Figure 4A, B). Mcl1 is pro-survival gene relevant for maintenance of viability but not of proliferation and is often expressed in long-lived cells (49–51); whereas the human tnfb ortholog LTA is expressed in lymphoid tissue inducer (LTi) cells and is involved in the regulation of cell proliferation, differentiation and survival (52, 53). Unlike nitr+rorc+ ILCs, nitr-rorc+ cells did not respond to immune stimuli by expressing cytokines (Figure 4B, Figure S6B).
We next asked whether unstimulated nitr+rorc+ cells express unique surface receptors that enable them to respond to the immune challenge. In addition to nitr9 and nitr4a, nitr+rorc+ ILCs specifically expressed novel immune-type receptors nitr6b and nitr5 as shown by DE analysis (Figure 4B). In contrast with human unstimulated ILC subsets, less than 10% of unstimulated nitr+rorc+ cells expressed cytokine receptors, toll-like and other pattern recognition receptors. The developmental origins and hierarchical relationship between nitr-rorc+ and nitr+rorc+ populations, however, remains unclear.
Finally, cells identified in cluster 4 in PBS-injected zebrafish, cluster 4 in V. anguillarum-injected zebrafish and cluster 2 in A. simplex-injected zebrafish (Figures 2A-C, respectively) grouped as cluster 2 (Figure 4A). These cells showed clear upregulation of ifng1-1 following immune challenge with V. anguillarum and no expression of granzymes or nk lysins. These data further support that these cells potentially represent ILC1-like cells in zebrafish.
Altogether, our analyses of over 10,000 single cells collected from the gut of rag1-/- zebrafish identified previously unappreciated diversity of innate lymphoid cells in zebrafish and revealed how this heterogeneity translates to cell-specific immune responses.
Innate immune response shows high degree of heterogeneity between individuals
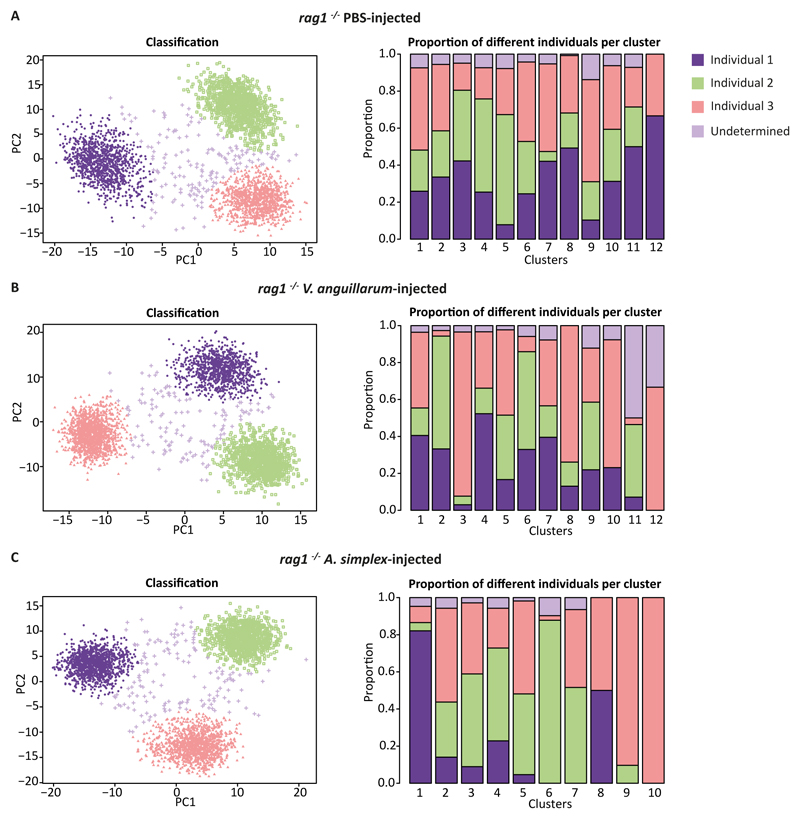
Since immune response can vary between individuals upon challenge (54) we tested the robustness of our findings by investigating whether individual zebrafish are particularly over- or under-represented within each of the identified clusters. The identification of ILC-like populations required the analysis of thousands of cells from multiple zebrafish injected with either PBS, V. anguillarum, or A. simplex. In order to assign a likely donor ID to each cell we utilised somatic mutations present within transcripts to locate genomic sites which vary between cells (for details please see Materials and Methods). Using these sites, we assigned a genotype to each cell at each genomic site and subsequently clustered cells based on shared mutational profiles. This allowed us to assign each cell to a likely donor (Figure 5).
Figure 5. Genotype clustering of cells from the rag1-/- PBS-,V. anguillarum- and A. simplex-injected zebrafish.
The first three principal components were used and the clusters were generated using the probabilistic PCA algorithm. Individual 1 - violet colour; Individual 2 - green colour; Individual 3 - red colour; Undetermined - light violet colour. Bar plots showing frequency (log scaled) of different donor within the cell type clusters under different challenge conditions.
Our analysis revealed that within the ILC1-like clusters from each experiment there was a significant contribution from all donors (Figure 5). Given the size of the ILC1-like clusters (558 cells, 652 cells and 677 cells in PBS-, V. anguillarum- and A. simplex-injected zebrafish, respectively), the distribution of donor cells suggests that ILC1-like cells are a stable population of cells in zebrafish gut. Similar conclusions could be made about the nitr+rorc+ ILC population in PBS (123 cells) and the ILC3-like population of cells in V. anguillarum-injected zebrafish (195 cells). However, the ILC2-like cells within the A. simplex-injected zebrafish were only detectable in individual 2 and 3 suggesting that the stimulus was not strong enough to trigger response in individual 1.
We also observed an unequal donor contribution across other clusters in PBS, V. anguillarum, or A. simplex-injected zebrafish. Interestingly, the genotype composition of clusters showed a more significant skew towards individual donors within the immune challenge experiments (Figure 5B, C) than for the PBS control (Figure 5A). In particular, cluster 3 (923 cells) within the V. anguillarum experiment seemed largely dominated by individual 3 (820 cells, 88.8%) whilst individual 1 (28 cells, 3%) and individual 2 (43 cells, 4.6%) were less present. Conversely, in cluster 2 (1263 cells), individual 3 contributed 38 cells (3%) whilst individuals 1 and 2 contributed 420 (33.2%) and 771 (61%) cells respectively. Examining the transcriptional properties of these clusters showed that the two clusters are actually very similar (Figure 2). In addition, an analysis of A. simplex-injected zebrafish (clusters 1 and 3) yielded very similar results whereby the apparent disparity of individual 1 in cluster 3 is accounted for by the relative abundance of individual 1 cells within the transcriptionally similar cluster 1. It is therefore possible that the two cell type clusters, 2 and 3 in V. anguillarum- and 1 and 3 in A. simplex-injected zebrafish actually represent the functionally similar biological cell type and the observed differences correspond to individual immune response.
Single cell atlas of innate and adaptive lymphocytes in zebrafish
To allow easy retrieval of sequencing data from zebrafish innate and adaptive lymphocytes we generated a cloud repository (https://www.sanger.ac.uk/science/tools/lymphocytes/lymphocytes/) with transcriptional profiles of over 14,000 single cells collected from healthy and immune challenged zebrafish using 10x genomics and Smart-seq2 methodology (please see Explanatory Note in Supplementary Material).
To capture the diversity of lymphoid cell types, we purified and sequenced the RNA from single cells collected from primary lymphoid organs (kidney and thymus), secondary lymphoid organs (spleen) as well as barrier tissues (gut and gills) of healthy, unstimulated adult zebrafish. We used three different transgenic lines: Tg(lck:EGFP) (55), which labels T cells as well as NK cells (18); Tg(cd4-1:mCherry) (56), which labels CD4 T cells and macrophages, and Tg(mhc2dab:GFP, cd45:dsRed) (57) (Figure S7A) that is expected to label B cells (when sorted as GFP+/DsRed-) (Table S3).
We performed single-cell RNA sequencing (Smart-seq2) of reporter-positive cells; 542 cells out of 796 passed quality control (QC) and were subjected to further analysis (Figure S8). Based on 3,374 highly variable genes inferred from biological cell-to-cell variation (Figure S9A), we generated Diffusion Maps and clustered cells within the 3D diffusion space. Our hierarchical clustering approach revealed three main populations.
The cells in the first cluster (C1) appeared to be T cells with high expression of cd4-1, cd8a and lck (Figure S7B). As expected, cells in this cluster originated from cd4-1:mCherry and lck:EGFP transgenic cells collected from kidney, gills, gut, thymus and spleen (Figure S9B). To further confirm our computational prediction that cells in C1 are indeed T cells, we applied TraCeR (26). We were able to unambiguously detect V(D)J recombination events in 224 cells out of 362 (Figure S7C, Table S1) and all observed TCR rearrangements were different. The second cluster (C2) had a signature of B lymphocytes and cells in this cluster originated from kidneys of the Tg(mhc2dab:GFP, cd45:dsRed) line (Figure S9B,C). They showed expression of immunoglobulin-heavy variable 1-4 (ighv1-4), an orthologue of human immunoglobulin heavy constant mu gene (IGHM). Importantly, we detected BCR rearrangements in 36 cells from this cluster using BraCer, therefore confirming their B cell identity (58) (Figure S7C, Table S1). The cluster three (C3) was exclusively comprised of cells that originated from cd4-1:mCherry transgenic cells collected from gills, gut and spleen (Figure S9B, C). These cells had a high expression of macrophage receptor with collagenous structure (marco) and macrophage expressed gene 1 (mpeg1.1), strongly indicative of their macrophage identity. This was not surprising, as cd4-1:mCherry has been found to label both CD4 T cells and macrophages (56).
Thus, single-cell RNA-Seq of lck:EGFP+, mhc2dab:GFP+/cd45:dsRed- and cd4-1:mCherry+ cells identified the adaptive lymphocytes in zebrafish, namely T and B cells, and their transcriptional signatures. Although these reporter lines might not label the entire spectrum of indicated cell types this is the most comprehensive transcriptional atlas of blood cell types in zebrafish to date.
Discussion
Our work provides a comprehensive atlas of both adaptive and innate lymphocytes across different organs in healthy and immune challenged zebrafish. Importantly, we identified populations of innate lymphocytes in rag1-deficient and wild-type zebrafish that resemble helper ILC subtypes in mammals. By analysing 14,080 lck:EGFP+ single cells collected from gut of unstimulated and stimulated zebrafish, we discovered two previously unknown populations of rorc+ ILC-like cells in zebrafish, nitr+rorc+ and nitr-rorc+, which appear in some ways to recapitulate NCR+ ILC3 and NCR- ILC3 subsets, respectively, in humans and mice (5). We obtained functional insight into these two distinct populations of rorc+ cells by exposing adult zebrafish to specific stimuli that rapidly induce corresponding cytokines in the gut.
The survey of cell surface receptors also suggested significant differences in the expression of cytokine receptors in ILC-like cells in zebrafish compared to their mammalian counterparts. Whereas both human and mouse ILCs constitutively express receptors for cytokines and are mainly activated by cytokines released by the epithelium or antigen presenting cells, in our dataset zebrafish nitr+rorc+ ILC-like cells express cytokine receptors in less than 10% of cells. More recently, other receptors such as aryl hydrocarbon receptor, Toll-like receptors, and other pattern recognition receptors, as well as NCRs have been reported to enable mouse and human ILC3 cells to directly sense environmental cues and induce cytokine expression (59, 60). Again, zebrafish ILC-like cells did not express the orthologues of these receptors but instead expressed NITRs. Teleost genomes contain multiple nitr genes which are considered to be the functional homologues of mammalian NCRs and killer cell immunoglobulin like receptors (KIRs) (40). However, our data should be interpreted in the light of methodology we used. Deeper sequencing or analysis of protein expression might provide additional information on the level of expression of various receptors in ILCs in zebrafish. Given the prominent expression of nitr genes in zebrafish ILCs, it remains to be seen whether and how these receptors modulate ILC functions.
In addition to ILC3 cells, another population of Rorc-expressing ILCs has been identified in mammals - lymphoid tissue inducer cells (LTis) (61). LTis contribute to the formation of lymph nodes and gut-associated lymphoid tissue, including Peyer’s patches, isolated lymphoid follicles and cryptopatches. Zebrafish gut does not have such organized lymphoid structures (62) and consequently it is presumed that they don’t have LTis (52). In our datasets, we identified a population of nitr-rorc+ cells that exclusively expressed tnfb, a zebrafish orthologue of human LTA, a marker gene for LTi cells. Although nitr-rorc+ share some transcriptional features with LTis it is hard to speculate on the function of nitr-rorc+ cells in zebrafish and whether they are evolutionarily linked with mammalian LTi cells. Interestingly, zebrafish nitr-rorc+ cells expressed mcl1b which is pro-survival gene relevant for maintenance of viability but not of proliferation and is often expressed in long-lived cells (49–51).
We also identified ifng producing cells. Zebrafish have two ifng genes (ifng1-1 and ifng1-2) which show mutually exclusive expression in our data set. The products of two ifng genes bind to different receptors in zebrafish and are thus functionally specialized (63). The ifng1-2 was co-expressed with granzymes or granulysins in the population transcriptionally resembling NK-like cells. In contrast, ifng1-1 expressing cells had no expression of granzymes nor granulysins and thus represent a distinct subtype of innate lymphocytes, possibly ILC1-like cells. Similar to human ILCs from tonsils, ifng producing cells showed no expression of the Th1 master regulator T-bet (33).
Like humans and mice, zebrafish appear to contain established ILC1-like population that responds to immune challenge. In addition, zebrafish have NK-like cells which are more numerous than helper ILCs in the gut of rag1 deficient zebrafish and show basal level of expression of ifng1-2. However, in contrast to mammals, zebrafish appear to display population of ILCs which do not produce detectable level cytokines in homeostasis. Circulating and tissue resident ILC precursors (ILCPs) in human and mouse also express Rorc at low level, and differentiate into multiple ILC subtypes in vitro (36, 37). ILCPs cannot be stimulated to produce effector cytokines; only already differentiated ILC1, ILC2 and ILC3 respond to immune challenge by producing relevant cytokines (36, 37). Indeed, scRNA-Seq of ILCs isolated from the gut of unstimulated mice (64) revealed that every subset of ILCs contained cells expressing cytokine transcripts, a feature we did not observe in PBS-injected zebrafish. It remains possible, however, that due to the modest number of detected genes we were not able to detect low basal level of cytokine production in these cells. While we document several distinct populations of ILCs in this study, further studies are needed to understand if there are precursor-progenitor relationships and/or plasticity between these lineages. Furthermore, our studies have focused on rag1-/- zebrafish. Developing tools would be essential for studying how ILCs cooperate with other lymphocytes to drive immune responses.
Materials and Methods
Study design
The aim of this study was to characterised innate and adaptive lymphocytes in zebrafish in steady state and following the immune challenge, using scRNA-seq. Multiple zebrafish, either in steady state or exposed to immune challenge, were used to collect cells for sequencing. To verify that all cells were intermixed based on their transcriptional similarities, Principal Component Analysis (PCA) and diffusion maps were used. Following the sequencing zebrafish were genotyped using probabilistic PCA on single nucleotide polymorphisms (SNPs) identified based on de novo variant calling.
Zebrafish strains and maintenance
The maintenance of zebrafish wild-type line (AB), transgenic lines Tg(lck:EGFP) (55), Tg(cd4-1:mcherry) (56), Tg(mhc2dab:GFP, cd45:dsRed) (57) and rag1hu1999mutants (also known as rag1t26683/26683 (23)) was performed in accordance with EU regulations on laboratory animals. Hu1999 mutation results in a premature stop codon in the middle of the catalytic domain of the Rag1 protein and is considered a null allele (23).
FACS sorting
Kidneys from heterozygote transgenic zebrafish either wild-type or rag1-/- mutant, were dissected and processed as previously described (17). The guts, spleens, gills and thymuses were dissected and placed in ice cold PBS/5% foetal bovine serum. Single cell suspensions were generated by first passing through a 40 µm strainer using the plunger of a 1 ml syringe as a pestle. These were then passed through a 20 µm strainer before adding 4',6-diamidino-2-phenylindole (DAPI, Beckman Coulter, cat no B30437) to the samples. For Smart-seq2 experiment individual cells were index sorted into 96 well plates using a BD Influx Index Sorter. Cells from kidney, gut, gills, spleen and thymus from non-transgenic zebrafish line were used for gating.
For the 10x experiment, guts from either Tg(lck:EGFP) rag1-/- mutant or wild-type zebrafish were isolated and single cell suspensions were prepared as described above. Three zebrafish, per each condition (i.e. zebrafish intraperitoneally injected with PBS, lyophilised Anisakis simplex or inactivated Vibrio anguillarum), were used to collect the total of 12,000 lck+cells (4000 per zebrafish) for 10x experiment.
Plate-based single-cell RNA processing
The Smart-seq2 protocol (65) was used for whole transcriptome amplification and library preparation as previously described. Generated libraries were sequenced in pair-end mode on Hi-Seq4000 platform.
Droplet-based single-cell RNA processing
Following the sorting, cells were spun down and resuspended in ice cold PBS with 0.04% bovine serum albumin at the concentration of 500 cells/μl. Libraries were constructed using Chromium™ Controller and Chromium™ Single Cell 3’ Library & Gel Bead Kit v2 (10x Genomics) according to the manufacturer’s protocol for 5000 cells recovery. Briefly, cellular suspension was added to the master mix containing nuclease-free water, RT Reagent Mix, RT Primer, Additive A and RT Enzyme Mix. Master mix with cells was transferred to the wells in the row labelled 1 on the Chromium™ Single Cell A Chip (10x Genomics). Single Cell 3’ Gel Beads were transferred into the row labelled 2 and Partitioning Oil was transferred into the row labelled 3. The chip was loaded on Chromium™ Controller to generate single-cell GEMs. GEM-RT was performed in a C1000 Touch Thermal cycler (Bio-Rad) at the following conditions: 53°C for 45 min, 85°C for 5 min, held at 4°C. Post GEM-RT cleanup was performed with DynaBeads MyOne Silane Beads (Thermo Fisher Scientific). cDNA was amplified using C1000 Touch Thermal cycler at the following conditions: 98°C for 3 min, 12 cycles of (90°C for 15 s, 67°C for 20 s and 72°C for 1 min), 72°C for 1 min, held 4°C. Amplified cDNA was cleaned with the SPRIselect Reagent Kit (Beckman Coulter) and quality was assessed using 2100 Bioanalyser (Agilent). Libraries were constructed following the manufacturer’s protocol and sequenced in pair-end mode on Hi-Seq4000 platform.
Short-term inflammation experiments
Vibrio anguillarum strain 1669 was grown in TSA broth medium to OD600 1.5. Bacterial pellet (9 mL of full grown culture) was resuspended in NaCl 9 g/L, 0.35% formaldehyde, and incubated overnight at 20°C. The suspension was washed four times in NaCl 9 g/l and resuspended in 800 μl of the same isotonic solution.
Anisakis simplex larvae, extracted from wild herring (Clupea harengus), were lyophilized using a freeze dryer/lyophiliser Alpha 1-2 LD plus (Martin Christ) following manufacturer's instructions: samples were placed in glass vials with vented rubber caps, placed in a freeze dryer holding tray and placed at -80°C until ready to lyophilise. The freeze dryer machine was cooled down before use and worms were exposed to lyophilisation for 18 hours at -44 °C to -45°C, and pressure at 0.071 to 0.076 mbar. Lyophilized larvae were homogenized in 1 mL PBS using a FastPrep-24 instrument (MP Biomedicals) with 1/4" ceramic sphere in 2 mL tubes for 20 seconds at 6 g.
Five microliters of each extract were mixed with 15 μl of sterile PBS and transferred to a 1.5 mL Eppendorf tube. Micro-Fine U-100 insulin syringes were loaded with the suspension mix and injected intraperitoneally into the midline between the pelvic fins.
RNA isolation and qPCR experiment
RNA was isolated with Trizol reagent (Invitrogen) according to manufacturer’s instructions. One μg of RNA was reverse-transcribed using M-MLV Reverse Transcriptase Kit (Invitrogen). Real-time PCR was performed using Rox SYBR Green MasterMix dTTP Blue Kit (Takyon) and run on a QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems).
The following primers were used:
ifng-1-1: forward, 5’- ACCAGCTGAATTCTAAGCCAA -3’
reverse, 5’-TTTTCGCCTTGACTGAGTGAA -3’
ifng-1-2: forward, 5’- CATCGAAGAGCTCAAAGCTTACTA -3’
reverse, 5’-TGCTCACTTTCCTCAAGATTCA -3’
tnfa: forward, 5’- TTCACGCTCCATAAGACCCA -3’
reverse, 5’-CAGAGTTGTATCCACCTGTTA -3’
il13: forward, 5’- GAAGTGTGAGCATGATTATTTC -3’
reverse, 5’-CTCGTCTTGGTGGTTGTAAG -3’
il4: forward, 5’- CCTGACATATATGAGACAGGACACTAC -3’
reverse, 5’-TTACCCTTCAAAGCCATTCC -3’
il17a/f3: forward, 5’- AAGATGTTCTGGTGTGAAGAAGTG -3’
reverse, 5’-ACCCAAGCTGTCTTTCTTTGAC -3’
il22: forward, 5’- TGCAGAATCACTGTAAACACGA -3’
reverse, 5’-CTCCCCGATTGCTTTGTTAC -3’
cd3z: forward, 5’- CCGGTGGAGGAGTCTCATTA -3’
reverse, 5’-CTCCAGATCTGCCCTCCTC -3’
ef1a: forward, 5’ - ACCTACCCTCCTCTTGGTCG - 3’
reverse, 5’ - GGAACGGTGTGATTGAGGGAA - 3’
Samples were analysed using ΔΔCt method. The mean Ct value of housekeeping gene (ef1a) was used for normalization. 2-ΔΔCt values were graphed with the geometric mean ±95% confidence intervals to estimate the fold change. The raw qPCR data for the short-term inflammation experiment can be found at the online repository, https://zenodo.org/record/1437804.
Alignment and quantification of single-cell RNA-sequencing data
For the samples that were processed using the Smart-seq2 protocol, the reads were aligned to the zebrafish reference genome (Ensemble BioMart version 89) combined with the sequences for EGFP, mCherry, mhc2dab and ERCC spike-ins. Salmon v0.8.2 (66) was used for both alignment and quantification of reads with the default paired-end parameters, while library type was set to inward (I) relative orientation (reads face each other) with unstranded (U) protocol (parameter –l IU).
For the samples that were processed using the Chromium Single Cell 3’ protocol, Cell Ranger v2.1 was used in order to de-multiplex raw base call (BCL) files generated by Illumina sequencers into FASTQ files, perform the alignment, barcode counting, and UMI counting. Ensembl BioMart version 91 was used to generate the reference genome.
Quality control of single-cell data
For the Smart-seq2 protocol transcript per million (TPM) values reported by Salmon were used for the quality control (QC). Wells with fewer than 900 expressed genes (TPM > 1) or having more than either 60% of ERCC or 45% of mitochondrial content were annotated as poor quality cells. As a result, 322 cells failed QC and 542 single cells were selected for the further study.
Chromium Single Cell 3’ samples were filtered based on the Median Absolute Deviation (MAD) of the distribution of the number of detected genes. In addition, the percentage of mitochondrial content was set to less than 10%. Following QC, 3,211 single cells from the rag1-/-PBS-injected sample, 3,626 single cells from the rag1-/- A. simplex-injected samples, 3,487 single cells from the rag1-/- V. anguillarum-injected samples, and 3,756 from the rag1+/+ PBS-injected samples were used in downstream analysis.
Downstream analysis of Smart-seq2 data
For each of the 542 single cells, counts reported by Salmon were transformed into normalised counts per million (CPM) and used for the further analysis. This was performed by dividing the number of counts for each gene with the total number of counts for each cell and by multiplying the resulting number by a factor of 1,000,000. Genes that were expressed in less than 1% of cells (e.g. 5 single cells with CPM > 1) were filtered out. In the final step we ended up using 16,059 genes across the 542 single cells. The scran R package (version 1.6.7) (67) was then used to normalise the data and remove differences due to the library size or capture efficiency and sequencing depth.
In order to identify the highly variable genes (HVGs) we utilised the Brennecke Method (68). We inferred the noise model from the ERCCs and selected genes that vary higher than 20% percentage of variation. This was performed by using the “BrenneckeGetVariableGenes” command of M3Drop v1.4.0 R package setting fdr equal to 0.01 and minimum percentage of variance due to biological factors (minBiolDisp) equal to 0.2. In total, 3,374 were annotated as HVGs.
To verify that all cells were intermixed (in the reconstructed 3D component space) based on their transcriptional similarities and not based on the zebrafish of origin, we used Principal Component Analysis (PCA) and diffusion maps (destiny R package (version 2.6.1)).
The first 3 diffusion components were clustered using shared nearest neighbour (SNN) modularity optimization-based clustering algorithm implemented by Seurat Package. We used the "FindClusters" command. Three clusters were selected for the further analysis.
BraCeR and TraCeR analysis
We have used TraCeR (26) and BraCeR (58) tools in order to reconstruct the sequences of rearranged T and B cell receptor genes (TCR and BCR, respectively), from our Smart-seq2 single-cell RNA-seq data. In order to build combinatorial recombinomes (tracer/bracer build command) for the Danio rerio species, fasta files describing all V, J, C, D sequences were collected from the international ImMunoGeneTics information system (http://www.imgt.org) (69). For TCR, complete information of the alpha and beta chain location was available, while for the BCR, H and L location sequences were available. Using a threshold of 50 TPMs for gene expression, we identified a total of 244 single cells as TCR positive and 36 as BCR positive.
Downstream analysis of 10x Genomics data
The downstream analysis of the 10x data was performed using the Seurat (version 2.2.0) and the cellranger (version 1.1.0) R packages. Briefly, raw counts that passed the QC were centered by a factor of 1000 and log transformed. HVGs were detected based on their average expression against their dispersion, by means of the “FindVariableGenes” Seurat command with the following parameters: mean.function equal to ExpMean, dispersion.function equal to LogVMR, x.low.cutoff equal to 0.0125, x.high.cutoff equal to 3, and y.cutoff equal to 0.5. The number of HVGs across samples varied between 1500 and 2500 genes, accordingly.
HVGs were used for the calculation of the Principal Components (PCs) using Seurat’s “RunPCA” command. For the 3D tSNE transformation (“RunTSNE” command) we used PCs with JackStraw statistics lower than 0.01. The later statistics were estimated using the Seurat’s “JackStraw” command with 200 replicate samplings. The proportion of the data that was randomly permuted for each replicate was set to 1%.
Clustering in the 3D tSNE space was performed (pheatmap version 1.0.8) using euclidean distance and centroid linkage. The silhouette scores were used to estimate the optimal number of clusters. However, the final decision on the number of clusters was made on case by case basis. Positive marker genes that expressed in at least half of genes within the cluster were calculated with “FindAllMarkers” Seurat command, using Wilcoxon rank sum test with threshold set to 0.25. DotPlots in Figures 2, 3 and 4 were generated using the Seurat’s “DotPlot” and “SplitDotPlotGG” command, respectively.
Seurat Alignment Strategy
In order to perform direct comparison of clusters that belong to the same cell type across different conditions, we adopted the Seurat Alignment workflow (48). We calculated Highly Variable Genes (HVGs), for each of the three different conditions, and selected 961 HVGs that were expressed in at least 2 datasets. Canonical Correlation Analysis (CCA) was then performed in order to identify shared correlation structures across the different conditions using the “RunMultiCCA” command. Twenty significant CCA components were selected by means of the shared correlation strength, using the “MetageneBicorPlot” command. Aligned CCA space was then generated with the “AlignSubspace” Seurat command. Thirteen Clusters were identified using the shared nearest neighbor (SNN) modularity optimization based clustering algorithm (“FindClusters” command) on the 20 significant CCA aligned components at 0.5 resolution. Dotplot of genes at different clusters on the aligned data was generated by using the SplitDotPlotGG command.
Barcode Extraction and Initial Variant Calling
For each cell barcode, transcript extraction and indexing of the generated files was done using SAMtools package (https://github.com/samtools/samtools). The cellular data sets were merged for subsequent analysis. Variant genomic sites were identified de novo within each experiment condition through the use of the mpileup and call functions from the BCF tools package (https://github.com/samtools/bcftools). The resulting variant call format (VCF) file contained all genomic sites which showed variation amongst the transcripts from single cells. Poor quality variants were filtered out.
Variant Calling per Cell
For each experiment condition, the VCF file produced by the initial variant calling was used as a reference to genotype each cell at each quality-controlled variant site. The cellular VCFs were merged to create a single VCF file containing the genotype of each cell at each variant site identified in the experiment. The variants were filtered to exclude those variant sites present in less than 5% of cells. This resulted in 2592 variants identified within the rag1-/- PBS-injected zebrafish, 2778 variants within the rag1-/- V. anguillarum-injected zebrafish and 2879 variants within the rag1-/-A. simplex-injected zebrafish.
Genotype Clustering
To process the VCF file, the VariantAnnotation package (70) was used to filter out variants which did not originate from single-nucleotide polymorphisms (SNPs). Using the snpStats package (71), for each processed experiment (Ncells × NSNPs) matrix was created containing the genotype of each cell at each variant site. Genotypes were numerically encoded as: 0 = Homozygous reference allele, 1 = Heterozygous, 2 = Homozygous alternative allele, NA = Missing genotype. For dimensionality reduction of the matrix probabilistic PCA (72) from the pcaMethods package (73) was used and for clustering of the cells by principle component Mclust package (74) was used.
All relevant scripts have been uploaded to an online repository at https://github.com/dhall1995/genotyping_scRNAseq.
Statistics
Statistical analyses were conducted using R, or Python, or GraphPad prism. The types of statistical tests and significance levels are described in respective figure legends. The results were considered statistically significant when p value was lower than .05 and were marked in the figures as: ****p < .0001; ***p < .001; **p < .01; *p < .05
Supplementary Material
Acknowledgements
The authors would like to thank WTSI Cytometry Core Facility for their help with index cell sorting and Nick Boughton and the Core Sanger Web Team for hosting the cloud web application. The authors would also like to thank the CRUK Cambridge Institute Genomics Core Facility for their contribution in sequencing the data and WTSI Single Cell Genomics Core Facility for their assistance with 10x experiments. We would also like to thank Ida Lindeman and Michael Stubbington for their help with using BraCeR and TraCeR tools and Gérard Eberl for reading the manuscript and providing valuable suggestions. The authors gratefully acknowledge the support of Philippe Herbomel (Institut Pasteur), in whose laboratory zebrafish rearing and inflammation assays were carried out. Authors would like to thank Davis McCarthy from the European Bioinformatics Institute (EBI) for his support in the implementation of the zebrafish genotype analysis.
Funding
The study was supported by Cancer Research UK grant number C45041/A14953 (to A.C. and E.I.A.), European Research Council project 677501 – ZF_Blood (to A.C. and P.M.S.), EMBO Long-Term Fellowship ALTF-807-2015 (to P.P.H), ANR grant 17-CE15-0017-01 – ZF-ILC (to P.P.H) and ANR-16-CE20-0002-03 (to J.-P.L), H2020-MSCA-IF-2015 grant 708128 – ZF-ILC (to P.P.H), ANR-10-LABX-73 (‘revive’ to P. Herbomel) and a core support grant from the Wellcome Trust and MRC to the Wellcome Trust – Medical Research Council Cambridge Stem Cell Institute.
Footnotes
Author Contributions
P.P.H conceived and proposed the original idea of identifying ILCs in zebrafish, under supervision of J.-P. L; C.M.C provided lyophilized A. simplex; P.B generated V. anguillarum extract. P.P.H and J.-P. L designed, and P.P.H and A.F.R performed inflammation experiments; E.I.A carried out the complete single-cell analysis and created the cloud repository under supervision of A.C; P.M.S performed single-cells experiments, designed and generated all figures with input from A.C.; DH performed genotyping analysis; E.I.A, P.M.S, P.P.H, P.B, J.-P.L. and A.C contributed to the discussion of the results. A.C wrote the manuscript with input from P.M.S, E.I.A, J.-P. L and P.P.H.
Competing interests
The authors declare that they have no competing interests.
Data and materials availability
Raw data can be found under the accession number E-MTAB-7117 and E-MTAB-7159 on ArrayExpress.
References
- 1.Boehm T. Evolution of Vertebrate Immunity. Curr Biol. 2012;22:R722–R732. doi: 10.1016/j.cub.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 2.Riera Romo M, Pérez-Martínez D, Castillo Ferrer C. Innate immunity in vertebrates: an overview. Immunology. 2016;148:125–39. doi: 10.1111/imm.12597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Artis D, Spits H. The biology of innate lymphoid cells. Nature. 2015;517:293–301. doi: 10.1038/nature14189. [DOI] [PubMed] [Google Scholar]
- 4.Eberl G, Colonna M, Di Santo JP, McKenzie ANJ. Innate lymphoid cells. Innate lymphoid cells: a new paradigm in immunology. Science. 2015;348 doi: 10.1126/science.aaa6566. aaa6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walker JA, Barlow JL, McKenzie ANJ. Innate lymphoid cells — how did we miss them? Nat Rev Immunol. 2013;13:75–87. doi: 10.1038/nri3349. [DOI] [PubMed] [Google Scholar]
- 6.Klose CSN, Mahlakõiv T, Moeller JB, Rankin LC, Flamar AL, Kabata H, Monticelli LA, Moriyama S, Putzel GG, Rakhilin N, Shen X, et al. The neuropeptide neuromedin U stimulates innate lymphoid cells and type 2 inflammation. Nature. 2017;549:282–286. doi: 10.1038/nature23676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, Koyasu S, Locksley RM, McKenzie AN, Mebius RE, Powrie F, et al. Innate lymphoid cells — a proposal for uniform nomenclature. Nat Rev Immunol. 2013;13:145–9. doi: 10.1038/nri3365. [DOI] [PubMed] [Google Scholar]
- 8.Annunziato F, Romagnani C, Romagnani S. The 3 major types of innate and adaptive cell-mediated effector immunity. J Allergy Clin Immunol. 2015;135:626–35. doi: 10.1016/j.jaci.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 9.Simoni Y, Fehlings M, Kløverpris HN, McGovern N, Koo SL, Loh CY, Lim S, Kurioka A, Fergusson JR, Tang CL, Kam MH, et al. Human Innate Lymphoid Cell Subsets Possess Tissue-Type Based Heterogeneity in Phenotype and Frequency. Immunity. 2017;46:148–161. doi: 10.1016/j.immuni.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hardman CS, Chen YL, Salimi M, Jarrett R, Johnson D, Jarvinen VJ, Owens RJ, Repapi E, Cousins DJ, Barlow JL, McKenzie ANJ, et al. Sci Immunol. 2017;18 doi: 10.1126/sciimmunol.aan5918. aan5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lo BC, Gold MJ, Hughes MR, Antignano F, Valdez Y, Zaph C, Harder KW, McNagny KM. Sci Immunol. 2016;3 doi: 10.1126/sciimmunol.aaf8864. aaf8864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo G, Luc S, Marco E, Lin TW, Peng C, Kerenyi MA, Beyaz S, Kim W, Xu J, Das PP, Neff T, et al. Mapping Cellular Hierarchy by Single-Cell Analysis of the Cell Surface Repertoire. Cell Stem Cell. 2013;13:492–505. doi: 10.1016/j.stem.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaitin DA, Kenigsberg E, Keren-Shaul H, Elefant N, Paul F, Zaretsky I, Mildner A, Cohen N, Jung S, Tanay A, Amit I. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343:776–9. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Villani AC, Satija R, Reynolds G, Sarkizova S, Shekhar K, Fletcher J, Griesbeck M, Butler A, Zheng S, Lazo S, Jardine L, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017;356:eaah4573. doi: 10.1126/science.aah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson NK, Kent DG, Buettner F, Shehata M, Macaulay IC, Calero-Nieto FJ, Sánchez Castillo M, Oedekoven CA, Diamanti E, Schulte R, Ponting CP, et al. Combined Single-Cell Functional and Gene Expression Analysis Resolves Heterogeneity within Stem Cell Populations. Cell Stem Cell. 2015;16:712–724. doi: 10.1016/j.stem.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carradice D, Lieschke GJ. Zebrafish in hematology: sushi or science? Blood. 2008;111:3331–3342. doi: 10.1182/blood-2007-10-052761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Athanasiadis EI, Botthof JG, Andres H, Ferreira L, Lio P, Cvejic A. Single-cell RNA-sequencing uncovers transcriptional states and fate decisions in haematopoiesis. Nat Commun. 2017;8 doi: 10.1038/s41467-017-02305-6. 2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carmona SJ, Teichmann SA, Ferreira L, Macaulay IC, Stubbington MJ, Cvejic A, Gfeller D. Single-cell transcriptome analysis of fish immune cells provides insight into the evolution of vertebrate immune cell types. Genome Res. 2017;27:451–461. doi: 10.1101/gr.207704.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang Q, Iyer S, Lobbardi R, Moore JC, Chen H, Lareau C, Hebert C, Shaw ML, Neftel C, Suva ML, Ceol CJ, et al. Dissecting hematopoietic and renal cell heterogeneity in adult zebrafish at single-cell resolution using RNA sequencing. J Exp Med. 2017;214:2875–2887. doi: 10.1084/jem.20170976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fort MM, Cheung J, Yen D, Li J, Zurawski SM, Lo S, Menon S, Clifford T, Hunte B, Lesley R, Muchamuel T, et al. IL-25 induces IL-4, IL-5, and IL-13 and Th2-associated pathologies in vivo. Immunity. 2001;15:985–95. doi: 10.1016/s1074-7613(01)00243-6. [DOI] [PubMed] [Google Scholar]
- 21.Hurst SD, Muchamuel T, Gorman DM, Gilbert JM, Clifford T, Kwan S, Menon S, Seymour B, Jackson C, Kung TT, Brieland JK, et al. New IL-17 family members promote Th1 or Th2 responses in the lung: in vivo function of the novel cytokine IL-25. J Immunol. 2002;169:443–53. doi: 10.4049/jimmunol.169.1.443. [DOI] [PubMed] [Google Scholar]
- 22.Sawa S, Lochner M, Satoh-Takayama N, Dulauroy S, Bérard M, Kleinschek M, Cua D, Di Santo JP, Eberl G. RORγt+ innate lymphoid cells regulate intestinal homeostasis by integrating negative signals from the symbiotic microbiota. Nat Immunol. 2011;12:320–326. doi: 10.1038/ni.2002. [DOI] [PubMed] [Google Scholar]
- 23.Wienholds E, Schulte-Merker S, Walderich B, Plasterk RHA. Target-Selected Inactivation of the Zebrafish rag1 Gene. Science. 2002;297:99–102. doi: 10.1126/science.1071762. [DOI] [PubMed] [Google Scholar]
- 24.Petrie-Hanson L, Hohn C, Hanson L. Characterization of rag1 mutant zebrafish leukocytes. BMC Immunol. 2009;10:8. doi: 10.1186/1471-2172-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tokunaga Y, Shirouzu M, Sugahara R, Yoshiura Y, Kiryu I, Ototak M, Nagasaw T, Somamoto T, Nakao M. Comprehensive validation of T- and B-cell deficiency in rag1-null zebrafish: Implication for the robust innate defense mechanisms of teleosts. Sci Rep. 2017;7 doi: 10.1038/s41598-017-08000-2. 7536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stubbington MJT, Lönnberg T, Proserpio V, Clare S, Speak AO, Dougan G, Teichmann SA. T cell fate and clonality inference from single-cell transcriptomes. Nat Methods. 2016;13:329–332. doi: 10.1038/nmeth.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hernández PP, Mahlakoiv T, Yang I, Schwierzeck V, Nguyen N, Guendel F, Gronke K, Ryffel B, Hoelscher C, Dumoutier L, Renauld JC, et al. Interferon-λ and interleukin 22 act synergistically for the induction of interferon-stimulated genes and control of rotavirus infection. Nat Immunol. 2015;16:698–707. doi: 10.1038/ni.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang YJ, Kim HY, Albacker LA, Baumgarth N, McKenzie AN, Smith DE, Dekruyff RH, Umetsu DT. Innate lymphoid cells mediate influenza-induced airway hyper-reactivity independently of adaptive immunity. Nat Immunol. 2011;12:631–8. doi: 10.1038/ni.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takatori H, Kanno Y, Watford WT, Tato CM, Weiss G, Ivanov II, Littman DR, O'Shea JJ. Lymphoid tissue inducer-like cells are an innate source of IL-17 and IL-22. J Exp Med. 2009;206:35–41. doi: 10.1084/jem.20072713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corripio-Miyar Y, Zou J, Richmond H, Secombes CJ. Identification of interleukin-22 in gadoids and examination of its expression level in vaccinated fish. Mol Immunol. 2009;46:2098–2106. doi: 10.1016/j.molimm.2009.01.024. [DOI] [PubMed] [Google Scholar]
- 31.Nieuwenhuizen N, Lopata AL, Jeebhay MF, Herbert DR, Robins TG, Brombacher F. Exposure to the fish parasite Anisakis causes allergic airway hyperreactivity and dermatitis. J Allergy Clin Immunol. 2006;117:1098–1105. doi: 10.1016/j.jaci.2005.12.1357. [DOI] [PubMed] [Google Scholar]
- 32.Halim YF, Takei F. Isolation and characterization of mouse innate lymphoid cells. Curr Protoc Immunol. 2014;25:1–13. doi: 10.1002/0471142735.im0325s106. [DOI] [PubMed] [Google Scholar]
- 33.Björklund ÅK, Forkel M, Picelli S, Konya V, Theorell J, Friberg D, Sandberg R, Mjösberg J. The heterogeneity of human CD127+ innate lymphoid cells revealed by single-cell RNA sequencing. Nat Immunol. 2016;17:451–460. doi: 10.1038/ni.3368. [DOI] [PubMed] [Google Scholar]
- 34.Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, Gregory MT, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8 doi: 10.1038/ncomms14049. 14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baran-Gale J, Chandra T, Kirschner K. Experimental design for single-cell RNA sequencing. Brief Funct Genomics. 2018;17:233–239. doi: 10.1093/bfgp/elx035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim AI, Li Y, Lopez-Lastra S, Stadhouders R, Paul F, Casrouge A, Serafini N, Puel A, Bustamante J, Surace L, Masse-Ranson G, et al. Systemic Human ILC Precursors Provide a Substrate for Tissue ILC Differentiation. Cell. 2017;168:1086–1100.e10. doi: 10.1016/j.cell.2017.02.021. [DOI] [PubMed] [Google Scholar]
- 37.Scoville SD, Mundy-Bosse BL, Zhang MH, Chen L, Zhang X, Keller KA, Hughes T, Chen L, Cheng S, Bergin SM, Mao HC, et al. A Progenitor Cell Expressing Transcription Factor RORγt Generates All Human Innate Lymphoid Cell Subsets. Immunity. 2016;44:1140–50. doi: 10.1016/j.immuni.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Satoh-Takayama N, Vosshenrich CA, Lesjean-Pottier S, Sawa S, Lochner M, Rattis F, Mention JJ, Thiam K, Cerf-Bensussan N, Mandelboim O, Eberl G, et al. Microbial Flora Drives Interleukin 22 Production in Intestinal NKp46+ Cells that Provide Innate Mucosal Immune Defense. Immunity. 2008;29:958–970. doi: 10.1016/j.immuni.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 39.Sanos SL, Bui VL, Mortha A, Oberle K, Heners C, Johner C, Diefenbach A. RORγt and commensal microflora are required for the differentiation of mucosal interleukin 22–producing NKp46+ cells. Nat Immunol. 2009;10:83–91. doi: 10.1038/ni.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoder JA, Turner PM, Wright PD, Wittamer V, Bertrand JY, Traver D, Litman GW. Developmental and tissue-specific expression of NITRs. Immunogenetics. 2010;62:117–22. doi: 10.1007/s00251-009-0416-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei S, Zhou JM, Chen X, Shah RN, Liu J, Orcutt TM, Traver D, Djeu JY, Litman GW, Yoder JA. The zebrafish activating immune receptor Nitr9 signals via Dap12. Immunogenetics. 2007;59:813–821. doi: 10.1007/s00251-007-0250-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yagi R, Zhong C, Northrup DL, Yu F, Bouladoux N, Spencer S, Hu G, Barron L, Sharma S, Nakayama T, Belkaid Y, et al. The Transcription Factor GATA3 Is Critical for the Development of All IL-7Rα-Expressing Innate Lymphoid Cells. Immunity. 2014;40:378–388. doi: 10.1016/j.immuni.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, Abbas AR, Modrusan Z, Ghilardi N, de Sauvage FJ, Ouyang W. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 44.Wolk K, Kunz S, Witte E, Friedrich M, Asadullah K, Sabat R. IL-22 Increases the Innate Immunity of Tissues. Immunity. 2004;21:241–254. doi: 10.1016/j.immuni.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 45.Lindemans CA, Calafiore M, Mertelsmann AM, O'Connor MH, Dudakov JA, Jenq RR, Velardi E, Young LF, Smith OM, Lawrence G, Ivanov JA, et al. Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature. 2015;528:560–564. doi: 10.1038/nature16460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoyler T, Klose CS, Souabni A, Turqueti-Neves A, Pfeifer D, Rawlins EL, Voehringer D, Busslinger M, Diefenbach A. The Transcription Factor GATA-3 Controls Cell Fate and Maintenance of Type 2 Innate Lymphoid Cells. Immunity. 2012;37:634–648. doi: 10.1016/j.immuni.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang S, Xia P, Chen Y, Qu Y, Xiong Z, Ye B, Du Y, Tian Y, Yin Z, Xu Z, Fan Z. Regulatory Innate Lymphoid Cells Control Innate Intestinal Inflammation In Brief Regulatory Innate Lymphoid Cells Control Innate Intestinal Inflammation. Cell. 2017;171:201–216.e18. doi: 10.1016/j.cell.2017.07.027. [DOI] [PubMed] [Google Scholar]
- 48.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vikström IB, Slomp A, Carrington EM, Moesbergen LM, Chang C, Kelly GL, Glaser SP, Jansen JH, Leusen JH, Strasser A, Huang DC, et al. MCL-1 is required throughout B-cell development and its loss sensitizes specific B-cell subsets to inhibition of BCL-2 or BCL-XL. Cell Death Dis. 2016;7:e2345–e2345. doi: 10.1038/cddis.2016.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang-Yen H-F. Mcl-1: a highly regulated cell death and survival controller. J Biomed Sci. 2006;13:201–204. doi: 10.1007/s11373-005-9064-4. [DOI] [PubMed] [Google Scholar]
- 51.Sathe P, Delconte RB, Souza-Fonseca-Guimaraes F, Seillet C, Chopin M, Vandenberg CJ, Rankin LC, Mielke LA, Vikstrom I, Kolesnik TB, Nicholson SE, et al. Innate immunodeficiency following genetic ablation of Mcl1 in natural killer cells. Nat Commun. 2014;5:4539. doi: 10.1038/ncomms5539. [DOI] [PubMed] [Google Scholar]
- 52.Lane PJL, Gaspal FM, McConnell FM, Withers DR, Anderson G. Lymphoid Tissue Inducer Cells: Pivotal Cells in the Evolution of CD4 Immunity and Tolerance? Front Immunol. 2012;3:24. doi: 10.3389/fimmu.2012.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lane PJL, McConnell FM, Withers D, Gaspal F, Saini M, Anderson G. Lymphoid tissue inducer cells: bridges between the ancient innate and the modern adaptive immune systems. Mucosal Immunol. 2009;2:472–477. doi: 10.1038/mi.2009.111. [DOI] [PubMed] [Google Scholar]
- 54.Shen-Orr SS, Furman D. Variability in the immune system: of vaccine responses and immune states. Curr Opin Immunol. 2013;25:542–7. doi: 10.1016/j.coi.2013.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Langenau DM, Ferrando AA, Traver D, Kutok JL, Hezel JP, Kanki JP, Zon LI, Look AT, Trede NS. In vivo tracking of T cell development, ablation, and engraftment in transgenic zebrafish. Proc Natl Acad Sci U S A. 2004;101:7369–74. doi: 10.1073/pnas.0402248101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dee CT, Nagaraju RT, Athanasiadis EI, Gray C, Fernandez Del Ama L, Johnston SA, Secombes CJ, Cvejic A, Hurlstone AF. CD4-Transgenic Zebrafish Reveal Tissue-Resident Th2- and Regulatory T Cell-like Populations and Diverse Mononuclear Phagocytes. J. Immunol. 2016;197:3520–3530. doi: 10.4049/jimmunol.1600959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wittamer V, Bertrand JY, Gutschow PW, Traver D. Characterization of the mononuclear phagocyte system in zebrafish. Blood. 2011;117:7126–7135. doi: 10.1182/blood-2010-11-321448. [DOI] [PubMed] [Google Scholar]
- 58.Lindeman I, Emerton G, Mamanova L, Snir O, Polanski K, Qiao SW, Sollid LM, Teichmann SA, Stubbington MJT. BraCeR: B-cell-receptor reconstruction and clonality inference from single-cell RNA-seq. Nat Methods. 2018;15:563–565. doi: 10.1038/s41592-018-0082-3. [DOI] [PubMed] [Google Scholar]
- 59.Glatzer T, Killig M, Meisig J, Ommert I, Luetke-Eversloh M, Babic M, Paclik D, Blüthgen N, Seidl R, Seifarth C, Gröne J, et al. RORγt+ Innate Lymphoid Cells Acquire a Proinflammatory Program upon Engagement of the Activating Receptor NKp44. Immunity. 2013;38:1223–1235. doi: 10.1016/j.immuni.2013.05.013. [DOI] [PubMed] [Google Scholar]
- 60.Killig M, Glatzer T, Romagnani C. Recognition strategies of group 3 innate lymphoid cells. Front Immunol. 2014;5:142. doi: 10.3389/fimmu.2014.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mebius RE, Rennert P, Weissman IL. Developing lymph nodes collect CD4+CD3- LTbeta+ cells that can differentiate to APC, NK cells, and follicular cells but not T or B cells. Immunity. 1997;7:493–504. doi: 10.1016/s1074-7613(00)80371-4. [DOI] [PubMed] [Google Scholar]
- 62.Brugman S. The zebrafish as a model to study intestinal inflammation. Dev Comp Immunol. 2016;64:82–92. doi: 10.1016/j.dci.2016.02.020. [DOI] [PubMed] [Google Scholar]
- 63.Aggad D, Mazel M, Boudinot P, Mogensen KE, Hamming OJ, Hartmann R, Kotenko S, Herbomel P, Lutfalla G, Levraud JP. The two groups of zebrafish virus-induced interferons signal via distinct receptors with specific and shared chains. J Immunol. 2009;183:3924–31. doi: 10.4049/jimmunol.0901495. [DOI] [PubMed] [Google Scholar]
- 64.Gury-BenAri M, Thaiss CA, Serafini N, Winter DR, Giladi A, Lara-Astiaso D, Levy M, Salame TM, Weiner A, David E, Shapiro H, et al. The Spectrum and Regulatory Landscape of Intestinal Innate Lymphoid Cells Are Shaped by the Microbiome. Cell. 2016;166:1231–1246.e13. doi: 10.1016/j.cell.2016.07.043. [DOI] [PubMed] [Google Scholar]
- 65.Picelli S, Björklund ÅK, Faridani OR, Sagasser S, Winberg G, Sandberg R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods. 2013;10:1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- 66.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lun ATL, Bach K, Marioni JC. Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol. 2016;17:75. doi: 10.1186/s13059-016-0947-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brennecke P, Anders S, Kim JK, Kołodziejczyk AA, Zhang X, Proserpio V, Baying B, Benes V, Teichmann SA, Marioni JC, Heisler MG. Accounting for technical noise in single-cell RNA-seq experiments. Nat Methods. 2013;10:1093–1095. doi: 10.1038/nmeth.2645. [DOI] [PubMed] [Google Scholar]
- 69.Lefranc MP, Giudicelli V, Duroux P, Jabado-Michaloud J, Folch G, Aouinti S, Carillon E, Duvergey H, Houles A, Paysan-Lafosse T, Hadi-Saljoqi S, et al. IMGT®, the international ImMunoGeneTics information system® 25 years on. Nucleic Acids Res. 2015;43:D413–D422. doi: 10.1093/nar/gku1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Obenchain V, Lawrence M, Carey V, Gogarten S, Shannon P, Morgan M. VariantAnnotation: a Bioconductor package for exploration and annotation of genetic variants. Bioinformatics. 2014;30:2076–2078. doi: 10.1093/bioinformatics/btu168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sole X, Guino E, Valls J, Iniesta R, Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22:1928–1929. doi: 10.1093/bioinformatics/btl268. [DOI] [PubMed] [Google Scholar]
- 72.Zhao J, Yu PLH, Kwok JT. Bilinear probabilistic principal component analysis. IEEE Trans neural networks Learn Syst. 2012;23:492–503. doi: 10.1109/TNNLS.2012.2183006. [DOI] [PubMed] [Google Scholar]
- 73.Stacklies W, Redestig H, Scholz M, Walther D, Selbig J. pcaMethods--a bioconductor package providing PCA methods for incomplete data. Bioinformatics. 2007;23:1164–7. doi: 10.1093/bioinformatics/btm069. [DOI] [PubMed] [Google Scholar]
- 74.Scrucca L, Fop M, Murphy TB, Raftery AE. mclust 5: Clustering, Classification and Density Estimation Using Gaussian Finite Mixture Models. R J. 2016;8:289–317. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.