As a general conclusion, the outcome of prion coinfection is strongly dependent on the strain combination and the model utilized and is therefore difficult to predict. The coexistence of several prion strains may remain undetected if one of the strains has more favorable conditions to replicate in the host. The use of several models (such as a transgenic mouse expressing PrP from different species) to analyze field prion isolates is recommended to avoid this situation. The inference effect exerted by nonreplicative prion strains should be considered an interesting tool to advance in new therapeutic strategies for treating prion diseases; it may even be a proper therapeutic strategy.
KEYWORDS: coexistence, coinfection, prion interference, prion propagation, prion replication, prion strain, TSEs
ABSTRACT
Co-occurrence of different prion strains into the same host has been recognized as a natural phenomenon for several sporadic Creutzfeldt-Jakob disease (sCJD) patients and natural scrapie cases. The final outcome of prion coinfection is not easily predictable. In addition to the usual factors that influence prion conversion, the replication of one strain may entail positive or negative consequences to the other. The main aim of this study was to gain insights into the prion coinfection and interference concepts and their potential therapeutic implications. Here, different mouse models were challenged with several combinations of prion strains coupled on the basis of the lengths of their incubation periods and the existence/absence of a species barrier in the tested animal model. We found that nontransmissible strains can interfere the replication of fully transmissible strains when there is a species transmission barrier involved, as happened with the combination of a mouse (22L) and a human (sCJD) strain. However, this phenomenon seems to be strain dependent, since no interference was observed when the human strain coinoculated was vCJD. For the other combinations tested in this study, the results suggest that both strains replicate independently without effect on the replication of one over the other. It is common that the strain with more favorable conditions (e.g., a higher speed of disease development or the absence of a species barrier) ends being the only one detectable at the terminal stage of the disease. However, this does not exclude the replication of the least favored strain, leading to situations of the coexistence of prion strains.
IMPORTANCE As a general conclusion, the outcome of prion coinfection is strongly dependent on the strain combination and the model utilized and is therefore difficult to predict. The coexistence of several prion strains may remain undetected if one of the strains has more favorable conditions to replicate in the host. The use of several models (such as a transgenic mouse expressing PrP from different species) to analyze field prion isolates is recommended to avoid this situation. The inference effect exerted by nonreplicative prion strains should be considered an interesting tool to advance in new therapeutic strategies for treating prion diseases; it may even be a proper therapeutic strategy.
INTRODUCTION
Prion diseases, also known as transmissible spongiform encephalopathies (TSEs), are a group of slow progressive and invariably fatal neurodegenerative diseases that affect a wide range of mammal species, including humans. The key molecular event in the pathogenesis of TSEs is the conversion of the host-encoded cellular prion protein PrPC into a disease-associated isoform, PrPSc, which is considered to be, if not entirely, at least the main component of the prion agent (1). PrPSc self-catalyzes its formation by recruiting and transforming PrPC into PrPSc (2). For that, PrPSc aggregates act as a template that promote the PrPC conformational change into new PrPSc particles, which are characterized by an increase in their β-sheet content. Prion conversion results in a change in the protein biochemical features. Whereas PrPC is monomeric, protease sensitive, and soluble in nonionic detergents (3), PrPSc has a high tendency to aggregate, is partially resistant to protease digestion, and is insoluble in nonionic detergents (1).
Early experiments using PrP transgenic mice showed that the efficacy of PrPC conversion into PrPSc mainly depends on the identity between the primary sequence of exogenous PrPSc and host PrPC (4). This concept is known as a “species barrier.” PrPC-PrPSc interaction between identical primary sequences is homologous, and the efficiency of conversion is fairly high. However, when differences in the amino acid sequences of both proteins exist, such a mismatch promotes heterologous interaction, and this is associated with reduced transformation efficiency (5). Prion strains also affect prion conversion. Prion strains are defined conformational variants which show distinct prion disease phenotypes (differences in incubation times, the distribution of prion deposits in the brain, clinical symptoms, and biochemical features) when transmitted to congenic hosts (6). Hence, the efficiency of prion replication is also affected by the prion strain being this replication favorable only when the structure of exogenous PrPSc is one of the possible tridimensional structures that the host PrPC can adopt, as stated by the conformational selection model (7).
Taking into account both concepts, the species barrier and the prion strains, the use of a heterologous PrPC protein to hamper the efficient conversion between homologous PrPs has been proposed in recent years. Either peptides (8, 9) or whole heterologous proteins (10) have been used with promising results in cellular and animal models. However, the use of a second PrPSc protein to stop homologous PrP conversion could be another therapeutic strategy to treat prion diseases. Further understanding of the factors governing prion coinfection and the possible outcomes of this phenomenon are first necessary in order to advance the development of these therapies.
Co-occurrence of two different prion strains into the same host is a naturally existing phenomenon, since it has been reported for certain sporadic Creutzfeldt-Jakob disease (sCJD) patients (11, 12) and natural scrapie isolates (13, 14). PrP posttranslational modifications are a natural source of prion strain emergence (15). However, whether the host is coinfected by two different prion conformations or a distinct conformation of prions emerges within the same host is completely unknown. Experimentally, prion coinfection has been studied using several animal and cellular models (16–24). These experiments mainly conclude that the final outcome of the coinfection process is not easily predictable and is highly dependent on the combination of the strains used.
The main aim of this study was to gain insights into prion coinfection and interference concepts. For that purpose, several transgenic mouse models for the PrP protein were challenged with different combinations of prion strain pairs. The length of the incubation period and the existence of species barrier for one of the strains were the basic features considered for the selection of the prion strain pairs used in the experiments.
RESULTS
In this study, several mouse models were challenged by the intracranial route with different combinations of prion strain pairs. The selection of each strain pair was performed attending to the length of the incubation period (fast and slow strains) and the existence of a species barrier for one of the strains (transmissible and nontransmissible strains). All of the prion strains used in this study have been previously characterized and adapted to replication in the corresponding mouse models by at least two previous passages. Detailed information about prion strain origins, the strain combinations, and the inoculated animal models can be found in (Tables 1 and 2).
TABLE 1.
Detailed information about the prion agents used in this study
| Prion strain | Prion isolate generation | Rate of disease development |
PrPres MWa
(unglycosylated band [kDa]) |
Infectious titer (ID50/g)c |
||
|---|---|---|---|---|---|---|
| Classification | Host | Mean survival time ± the SD (attack rate [%]) |
||||
| 22L | 22L adapted to 129 Ola mice | Fast | 129 Ola mice | 128 ± 10 (100) | 21 | 1.5 × 107* |
| NTb | HuTg340 | >700 (0) | 21 | 1.5 × 107* | ||
| RML | Rocky Mountain Laboratory (RML) adapted to 129 Ola mice | Fast | 129 Ola mice | 147 ± 4 (100) | 21 | 1.6 × 107* |
| BSEm | Bovine spongiform encephalopathy (BSE) adapted to 129 Ola mice | Slow | 129 Ola mice | 142 ± 5 (100) | 19 | 1.2 × 106* |
| Fast-sCJD | Sporadic Creutzfeldt-Jakob disease type 1 from a 129MM individual adapted to 129M-HuTg340 mice | Fast | HuTg340 | 201 ± 15 (100) | 21 | 4.1 × 107† |
| NTb | 129 Ola mice | >700 (0) | 21 | 4.1 × 107† | ||
| Slow-sCJD | Sporadic Creutzfeldt-Jakob disease type 2 from a 129VV individual adapted to 129M-HuTg340 mice | Slow | HuTg340 | 507 ± 85 (100) | 21 | 1.3 × 107† |
| vCJD | Variant Creutzfeldt-Jakob disease adapted to 129M-HuTg340 mice | Slow | HuTg340 | 650 ± 60 (100) | 19 | 4.2 × 107‡ |
| C-BSE | Epidemic bovine spongiform encephalopathy (BSE) adapted to BoTg110 mice | Slow | BoTg110 | 241 ± 7 (100) | 19 | 1.9 × 107‡ |
| H-BSE | H type atypical bovine spongiform encephalopathy (BSE) adapted to BoTg110 mice | Slow | BoTg110 | 278 ± 17 (100) | 21 | NA |
MW, molecular weight.
NT, nontransmissible.
*, Infectious titer in Tga20 mice (transgenic mice overexpressing mouse PrP at ∼10-fold the levels expressed in wild-type mouse brain); †, infectious titer in HuTg340 mice; ‡, infectious titer in BoTg110 mice; NA, not available.
TABLE 2.
Summary of the coinfection experiments
| Animal model | Prion strain 1 | Prion strain 2 | Experimental design | Mean survival time ± the SD (attack rate [%]) |
PrPres type | Coinfection outcome | ||
|---|---|---|---|---|---|---|---|---|
| Prion strain 1 | Prion strain 2 | Coinoculation | ||||||
| 129 Ola mice | 22L | RML | Fast strain vs fast strain | 128 ± 10 (100) | 147 ± 4 (100) | 136 ± 15 (100) | 22L/RMLa | No effect |
| 22L | BSEm | Fast strain vs slow strain | 128 ± 10 (100) | 142 ± 5 (100) | 135 ± 1 (100) | 22L | No effect; imposition of the most favorable strain | |
| 22L | Fast-sCJD | Transmissible strain vs nontransmissible strain | 128 ± 10 (100) | >700 (0) | 164 ± 31 (100) | 22L | Interference | |
| HuTg340 | Fast-sCJD | vCJD | Fast strain vs slow strain | 201 ± 15 (100) | 650 ± 60 (100) | 197 ± 36 (100) | Fast-sCJD | No effect; imposition of the most favorable strain |
| Fast-sCJD | Slow-sCJD | Fast strain vs slow strain | 201 ± 15 (100) | 507 ± 85 (100) | 204 ± 7 (100) | Fast-sCJD | No effect; imposition of the most favorable strain | |
| vCJD | Slow-sCJD | Slow strain vs slow strain | 650 ± 60 (100) | 507 ± 85 (100) | 546 ± 32 (100) | vCJD/Slow-sCJD | No effect; coexistence of both strains | |
| Fast-sCJD | 22L | Transmissible strain vs nontransmissible strain | 201 ± 15 (100) | >700 (0) | 247 ± 27 (100) | Fast-sCJD | Interference | |
| vCJD | 22L | Transmissible strain vs nontransmissible strain | 650 ± 60 (100) | >700 (0) | 632 ± 63 (100) | vCJD | No effect; imposition of the most favorable strain | |
| BoTg110 | C-BSE | H-BSE | Slow strain vs slow strain | 241 ± 7 (100) | 278 ± 17 (100) | 253 ± 6 (100) | C-BSE/H-BSE | No effect; coexistence of both strains |
Impossible to distinguish between strains.
Coinfection experiments in wild-type 129 Ola mice.
Three different experiments were conducted in wild-type 129 Ola mice: (i) coinoculation of 22L and RML mouse-adapted prions (fast strain versus fast strain); (ii) coinoculation of 22L and BSE mouse-adapted prions (fast strain versus slow strain); and (iii) coinoculation of 22L mouse-adapted prions and fast-sCJD (transmissible strain versus nontransmissible strain).
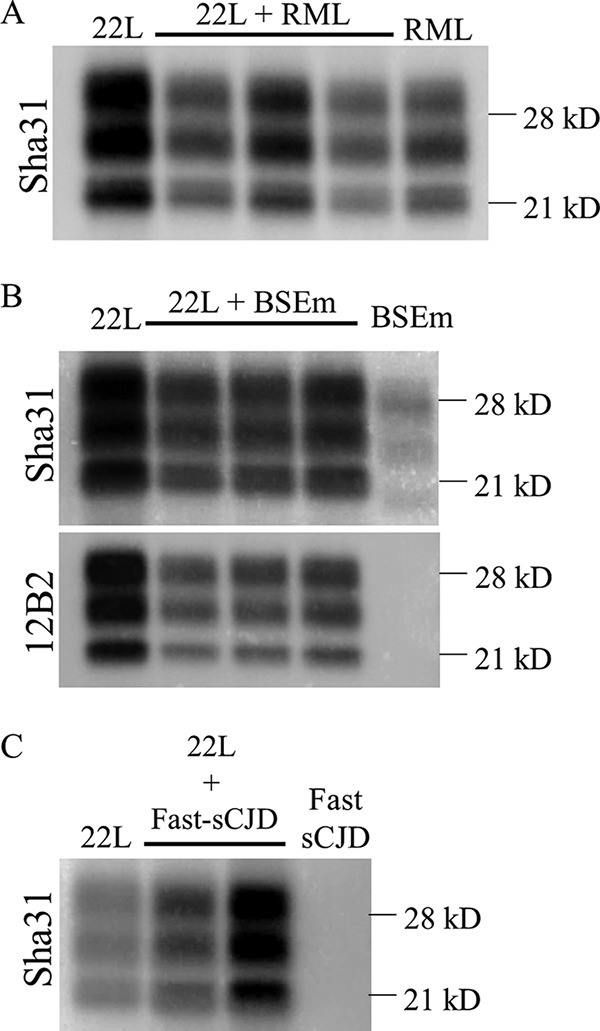
All mice coinoculated with 22L and RML developed prion disease with a mean survival time of 136 ± 15 days postinoculation (dpi). Their control counterparts, inoculated with either 22L or RML, showed mean survival times of 128 ± 10 dpi and 147 ± 4 dpi, respectively (Table 2). Therefore, the coinoculated animals showed an intermediate incubation time between both controls. Regarding the propagated PrPSc on coinoculated animals, both strains used in this experiment typically showed an indistinguishable 21-kDa PrPres signature in Western blotting (WB) and, unfortunately, we do not possess any antibody able to discriminate between these two strains (Fig. 1A). In wild-type mice, differentiation of both strains by histological analysis is also difficult due to their similarity (25), and paraffin-embedded tissue blotting (PET-blot) analyses did not clarify the results (see Fig. 4). Overall, these results suggest a no-effect outcome of the coinoculation with an impossibility to determine whether one of the strains has succeeded in imposing against the other.
FIG 1.
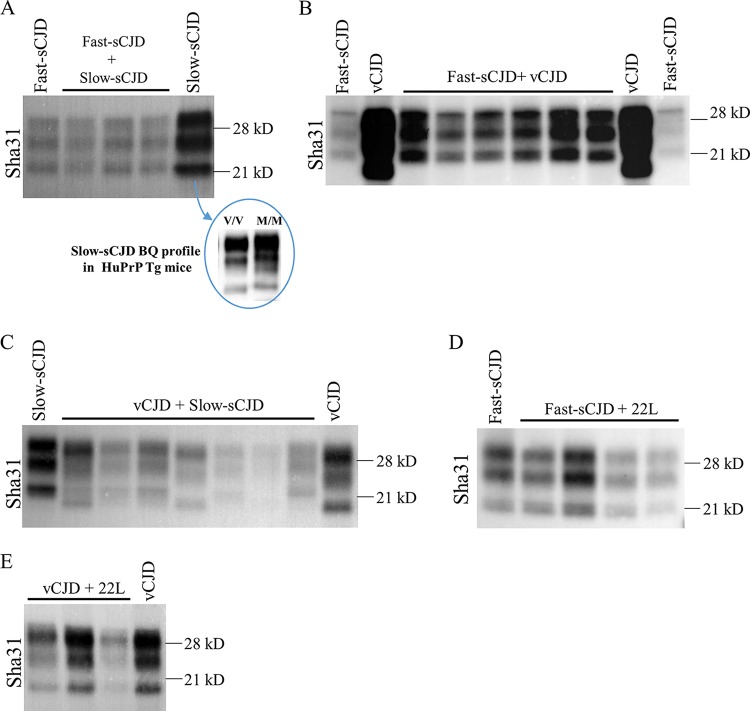
Brain PrPres signature obtained from coinfection experiments in 129 Ola mice. Western blot images of the PrPres signature displayed by the coinfected wild-type 129 Ola mice and the corresponding controls. (A) Coinfection of 22L and RML. Both strains show an undistinguishable 21-kDa pattern. (B) Coinfection of 22L and BSEm. The only PrPres component in coinoculated animals is the one given by 22L strain, since the Sha31 antibody does not reveal any BSEm pattern (19-kDa band) in the samples. (C) Coinfection of 22L and fast-sCJD. Fast-sCJD is nontransmissible in this animal model, so the PrPres displayed is for the 22L strain.
FIG 4.
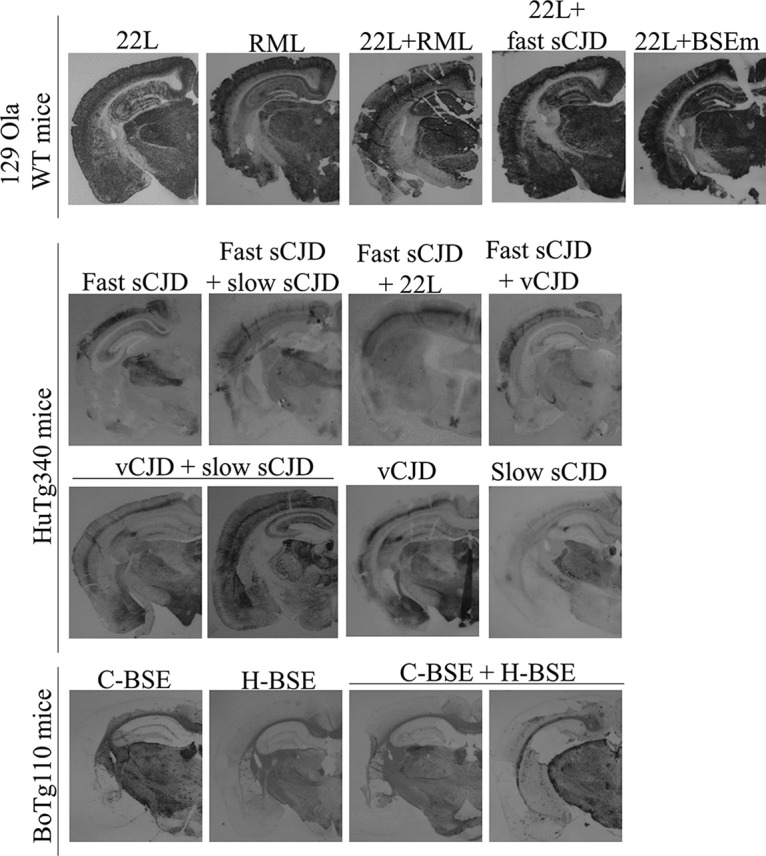
PrPres deposition patterns in the brains of different coinoculated and control animals. Sha31 immunoblotted PET-blot images of the PrPres deposits in coinfected animals and the corresponding controls are shown.
Concerning 22L and BSE (BSEm) mouse-adapted prion coinoculation, the mean survival times of the coinoculated animals were similar to the that obtained for 22L-inoculated controls (135 ± 1 versus 128 ± 10 dpi) and significantly shorter than the result found for BSEm-inoculated controls (142 ± 5 dpi) (Table 2). WB assay of the brains belonging to coinoculated animals showed the same PrPres profile than 22L-inoculated controls (Fig. 1B), which is in consonance with the mean survival times and suggests a no-effect outcome of the coinoculation with the clear imposition of the strain under more favorable conditions, in this case 22L, probably due to its faster replication and/or short incubation period. In addition, the PET-blot analysis of the brains from coinoculated mice showed a result compatible with the replication of 22L alone (see Fig. 4) similar to that previously described (25).
We then tested the role of the species barrier in the coinfection outcome inoculating 22L and fast-sCJD, being the last nontransmissible strain in wild-type 129 Ola mice. Surprisingly, an interference outcome was obtained, since the survival times of the coinoculated animals were delayed compared to the 22L control inoculated animals (164 ± 31 versus 128 ± 10 dpi) (Table 2). All coinoculated animals propagated prions with the same PrPres signature as the 22L control inoculated animals (Fig. 1C). Moreover, the PrPSc deposition patterns in the brains of coinoculated mice were similar to those previously reported for animals inoculated with 22L alone (see Fig. 4) (25).
Coinfection experiments in HuTg340 transgenic mice.
Five different experiments were conducted in HuTg340 mice: (i) coinoculation of fast-sCJD with vCJD (fast strain versus slow strain); (ii) coinoculation of fast-sCJD with slow-sCJD (fast strain versus slow strain); (iii) coinoculation of vCJD with slow-sCJD (slow strain versus slow strain); (iv) coinoculation of fast-sCJD and 22L (fast strain versus nontransmissible strain); and (v) coinoculation of vCJD and 22L (slow strain versus nontransmissible strain).
As the fast strain, fast-sCJD was chosen. Slow-sCJD and vCJD were chosen as slow strains. The slow-sCJD isolate used in this study was previously adapted to HuTg340 mice to avoid any possible transmission barrier derived from the polymorphism present in human PrP codon 129 (see Table 1 for more information about the slow-sCJD isolate origin). In addition, the mean survival time of the second passage of this isolate in HuTg340 is 428 ± 37 dpi, with a 100% attack rate, which definitely proves that this agent behaves as a slow strain with a long incubation period in this animal model (Table 2).
All animals coinoculated with fast and slow-sCJD developed prion disease with a mean survival time similar to the one obtained for the control animals inoculated only with fast-sCJD (204 ± 7 versus 201 ± 15 dpi) (Table 2). The brain PrPres signature in coinoculated animals was also the same as the one observed in fast-sCJD (type 1) control inoculated animals (Fig. 2A). However, the transmission of slow-sCJD prions in HuTg340 mice yielded the same brain PrPres signature (type 1) as the transmission of fast-sCJD in these mice (Fig. 2A). Therefore, the contribution of slow-sCJD to the brain PrPres signature could not be discarded only by WB. When the slow-sCJD agent was changed by vCJD, similar results were observed. Coinoculated animals coincided with fast-sCJD control animals in terms of the mean survival time (197 ± 36 versus 201 ± 15 dpi) (Table 2) and brain PrPres signature (Fig. 2B). In both coinoculation experiments, PET-blot analyses revealed a typical fast-sCJD PrPSc deposition pattern (see Fig. 4), similar to that previously reported in HuTg340 mice inoculated with fast-sCJD alone (26) but different from that observed in these mice inoculated with vCJD (27). In summary, the outcome of both inoculations could be described as a no-effect outcome with the clear imposition of fast-sCJD, the strain in more favorable conditions due to its higher rate of replication and/or shorter incubation period.
FIG 2.
Brain PrPres signature obtained from coinfection experiments in HuTg340 transgenic mice. Western blot images of the PrPres signature displayed by the coinfected HuTg340 transgenic mice and the corresponding controls. (A) Coinfection of fast-sCJD and slow-sCJD. The biochemical profile of slow-sCJD (a type 2 isolate from a 129VV individual) is indistinguishable from that of fast-sCJD in these animal models (M/M in the blue circle) but is different in human PrP transgenic mouse lines valine homozygous at codon 129 (V/V in the blue circle). (B) Coinfection of fast-sCJD and vCJD. The PrPres pattern corresponds to fast-sCJD. (C) Coinfection of vCJD and slow-sCJD. Both patterns can be found in coinoculated animals. (D) Coinfection of fast-sCJD and 22L mouse-adapted prions. Coinoculated animals show a fast-sCJD pattern, since 22L is nontransmissible in HuTg340 mice. (E) Coinoculation of vCJD and 22L mouse-adapted prions. The biochemical PrPres signature corresponds to vCJD, since 22L is nontransmissible in HuTg340 mice.
Coinoculation of the two slow strains, slow-sCJD and vCJD, resulted in a mean survival time of coinoculated animals that was not very different from that for the control animals inoculated with either vCJD (546 ± 32 versus 650 ± 60 dpi) or slow-sCJD (546 ± 32 versus 507 ± 85 dpi) (Table 2). Strikingly, the brain PrPres signature obtained in some coinoculated animals was the one of vCJD, while in other animals a slow-sCJD-PrPres (type 1) signature or even a mixture of the two PrPres signatures in the same mice was detected (Fig. 2C). Similarly, PrPSc deposition patterns compatible with either vCJD (27) or slow-sCJD (26) replication were detected by PET-blot analyses in the brains of coinoculated animals (see Fig. 4). These results suggest that for each animal one prion has imposed against the other but without a general clear supremacy of one of the strains. However, in other animals, the coexistence of both strains can be observed.
We next tested the role of the species barrier in prion infection interference by coinoculation of two different transmissible agents in HuTg340, fast-sCJD (short incubation period) and vCJD (long incubation period), with the nontransmissible agent 22L. An interference outcome was obtained for fast-sCJD and 22L coinoculation, since coinoculated animals showed a mean survival time of 247 ± 27 dpi versus 201 ± 15 dpi obtained for the fast-sCJD control inoculated mice (Table 2). Such a delay in the mean survival time was statically significant (4.7 × 10−5; ***, P value [Student t test] with respect to fast-sCJD control animals). The brain PrPres signature in these animals was identical to that observed in fast-sCJD control mice (Fig. 2D). In this case, PET-blot analyses revealed a PrPSc deposition pattern characteristic of fast-sCJD (see Fig. 4) (26). However, the interference disappeared, and a no-effect outcome was obtained when the transmissible strain was changed by vCJD, since no significant delay in the mean survival time was reported compared to the vCJD control inoculated animals (632 ± 63 versus 650 ± 60 dpi) (Table 2). The brain PrPres signature was identical to the one for the vCJD control mice (Fig. 2E), and the PrPres deposition pattern (see Fig. 4) was similar to that previously described for these mice inoculated with vCJD (27).
Coinfection experiments in BoTg110 transgenic mice.
Coinoculation of classic BSE (C-BSE) and H-BSE (slow strain versus slow strain) was conducted in BoTg110 mouse line.
Coinoculation of both strains gave mean survival times that did not differ from those for both strains when inoculated alone, which showed few differences in their survival times (Table 2). In addition, the PrPres isoforms of both strains can be found in the brains of all coinoculated mice by differential WB using two different antibodies: 12B2, which detects just the H-BSE PrPres, and Sha31, which detects both H-BSE and C-BSE PrPres (Fig. 3). PET-blot analysis (Fig. 4) revealed that the PrPSc deposition pattern observed in some coinoculated animals has a greater resemblance to PrPSc deposition pattern described in BoTg110 inoculated with C-BSE alone (28), whereas in other coinoculated animals the PrPres deposition pattern was similar to that previously reported in these mice inoculated with H-BSE (28).
FIG 3.
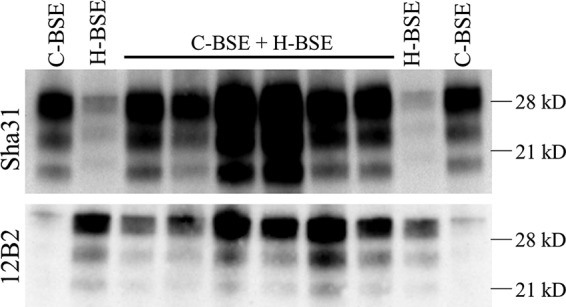
Brain PrPres signature obtained from coinfection experiments in BoTg110 transgenic mice. Western blot images of the PrPres signature displayed by the coinfected BoTg110 transgenic mice and the corresponding controls are presented. Coinfection results for C-BSE and H-BSE are shown. The C-BSE and H-BSE strains can be detected in the propagated PrPSc by antibodies Sha31 and 12B2, respectively.
DISCUSSION
In this study, different coinoculation experiments in three different animal models were performed to gain insights into the prion coinoculation and interference concepts. To summarize, three theoretical possible outcomes can result from prion coinfection: (i) interference, when the replication of one prion strain delays or blocks the replication of the other; (ii) synergy, when both strains replicate in a collaborative way accelerating the disease; and (iii) no effect, when both strains replicate without affecting each other. Our studies show that most coinfective strains can replicate without affecting its strain partner. As expected, the prion strain in more favorable conditions, either due to its shorter incubation period or to the absence of a species barrier, usually imposes above the other strain. This phenomenon could lead to the wrong conclusion that only one strain is present in the host brain, something that could have been happening in natural sCJD or scrapie cases (11–14). However, when coinoculated strains shared similar incubation times and replicated without a species barrier, such as vCJD plus slow-sCJD in HuTg340 or C-BSE plus H-BSE in BoTg110, no clear imposition of one of the strain could be detected. Survival times suffered no changes with respect to the controls, and both brain PrPres isoforms can be detected by WB either in separated animals or even in the same animal.
Furthermore, when we used nontransmissible strains in our experiments, these strains were sometimes able to interfere the replication process of the fully transmissible one, as happened in a reciprocal way with the combinations of 22L plus fast-sCJD in wild-type 129 Ola mice and of fast-sCJD plus 22L in HuTg340 mice. However, this phenomenon seems to be strain dependent, since the coinoculation of vCJD plus 22L in HuTg340 did not yield an interference result. Indeed, other cases of nontransmissible strains unable to interfere with prion replication have been reported (29), something totally expected since the outcome of a certain prion coinfection experiment is extremely difficult to predict and strongly dependent on the prion strain combination tested, the model used, and other experiment parameters such as the interval between the inoculations of the strains. It was previously suggested that the interfering strain must be able to replicate in the model of study in order to exert its effect above the other strain (19). Nevertheless, our results and those of others (22) postulate that this is not a necessary condition for producing interference. These different results could be due to discrepancies between the animal models and prion strains used in the different studies. Although the most likely explanation for our results is that the interfering strains are not replicating, agent replication and the contribution of this process to the interference results cannot be ruled out. As a matter of fact, it has been reported that prion replication can occur in the absence of disease development (30, 31). Thus, the phenomenon observed here should be referred to as prion interference mediated by nonpathogenic prion strains. Our results show that prion interference, as a result of coinfection, can be exploited as a possible therapeutic approach for treating TSEs. In light of our results, the use of a nontransmissible or nonpathogenic prion strain in humans could be an interesting choice for interfering with or blocking prion replication. Taking into account that this is a daring proposal, at least the elements involved in the interference should be identified in order to increase our knowledge about prion replication and also to be used as possible therapeutic targets to treat prion diseases. How exactly prion replication is interfered with by a nonreplicative, nonpathogenic, or a deficient-replicative prion strain is not known. Hypothetically, the interfering strain could act as a PrPC blocker, binding but not transforming PrPC molecules, hindering the access of the interfered strain to its substrate and retarding PrPSc self-catalysis. In the case of totally nonreplicative/nonpathogenic strains, this effect would not be perpetual, and a delay in the apparition of the disease would be observed, as happened in our experiments. In contrast, deficient replicative prion strains that sustain a poor but constant rate of replication could extend the blocking effect, even achieving a total absence of disease, as reported elsewhere (19, 20).
Other aspects which seem to be relevant for prion interference are the window time between the inoculation of both strains and the spatial distribution of both strains in the brain during the replication (20). Both aspects were not extensively studied in our experiments, since all isolates were coinoculated at the same time with a single injection, but we could not rule out that different outcomes could have been obtained if such variables were introduced into the experiments since well-established interference results have been reported to be strongly dependent on these parameters (20).
Natural cases of prion co-occurrence in scrapie and sCJD are surprisingly common (11–14). Misclassification of sCJD cases due to the sampling process for diagnosis can occur easily if PrPSc is only analyzed in the frontal cortex, as is usually done (12). Indeed, it has been proposed that the different combinations of mixed phenotypes be incorporated into the current sCJD classification (12). Whether these cases are due to a proper coinfection by two different prion strains or whether the emergence of a second strain is determined by posttranslational modification remains unanswered. In these cases, both strains would not have to deal with any species barrier. Regarding their speed of disease development and according to our results, if it is similar for both strains then none of them would be able to impose against the other, and both of them would be detectable and easily diagnosed, at least in cases of vCJD plus slow-sCJD (type 2) and C-BSE plus H-BSE coinfections. However, if one of the strains is faster or if the second strain appears when the replication of the first one is already extended, this strain would impose and kill the host and the existence of the second strain could be ruled out, missing important information about the prion replication process.
Finally, one of the main conclusions determined here are that the outcome of prion coinfection is strongly dependent on the prion strain pair used and is difficult to predict, as previously noted by other authors (16–24). Nevertheless, we highlight here two main factors involved in this phenomenon regarding the prion strain combination: (i) their incubation period length and (ii) their transmissibility in the model of study can deeply impact the result of the process, ranging from no effect to prion interference. Interference with prion infection mediated by nonreplicating/nonpathogenic prion strains could constitute an interesting tool for developing new therapeutic strategies for treating prion diseases, or even be a therapeutic strategy itself.
MATERIALS AND METHODS
Ethics statements.
Animal experiments were carried out in strict accordance with the recommendations of the Code for Methods and Welfare Considerations in Behavioural Research with Animals (Directive 86/609EC and 2010/63/EU), and all efforts were made to minimize suffering. Experiments were approved by the Committee on the Ethics of Animal Experiments (CEEA) of the Spanish Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria (INIA; permit CEEA2011/050).
Animal models and prion strain pair selection.
Three different animal models were used in these studies: wild-type 129 Ola mice, HuTg340 mice (methionine homozygous at codon 129) (32), and BoTg110 transgenic mice (33) which overexpress 4- and 8-fold levels of human and bovine PrP proteins in human and cow brains, respectively.
In each experiment, two prion strains were coinoculated by the intracranial route in the same host. The selection of each strain pair was performed attending to two distinct criteria: the length of the incubation period (fast and slow strains) and the existence of a species barrier for one of the strains (transmissible and nontransmissible strains). All of the prion isolates used in this work were extensively characterized and adapted to replication in the corresponding mouse models by at least two passages of intracranial inoculation (Table 1). Detailed information about prion strain origin, strain combination, and animal models can be found in Tables 1 and 2. All combinations were inoculated in a 1:1 proportion, which results in a 5% brain homogenate inoculation of each strain. Corresponding controls were developed by inoculating 5% brain homogenate of each strain separately.
Two different sCJD isolates have been utilized on this study: a sCJD catalogued as a type 1 isolate coming from a 129-methionine homozygous individual (referred to here as fast-sCJD) and a sCJD isolate classified as a type 2 coming from a 129-valine homozygous individual (referred to here as slow-sCJD). The latter isolate, like other sCJD type 2 isolates, undergoes a change in its electrophoretic mobility when transmitted in human PrP transgenic mice homozygous for the amino acid 129 polymorphism. Its PrPres profile changes from a typical type 2 pattern to a common “21-kDa” profile that is indistinguishable from the one expected from a sCJD type 1 isolate (Fig. 2).
Prion transmission studies.
Groups of 6 to 9 individually identified animals (6 to 7 weeks old) were anesthetized and intracranially inoculated with 20 µl of prion-infected brain homogenate or a mixture of two prion-infected brain homogenates in the right parietal lobe using a 25-gauge disposable hypodermic needle, as previously detailed (33). The mouse neurologic status was assessed twice a week. Animals were killed for ethical reasons when progression of the disease was evident or at the set endpoint of the experiment, 700 dpi. Necropsy was then performed, and each brain was harvested. Half of the brain was collected for histological analysis, and the remaining brain tissue was used for PrPres detection by WB. In all cases, the survival time and attack rate were calculated for each coinfection experiment. The survival time was expressed as the mean survival dpi for all mice scoring positive for PrPres, along with the corresponding standard deviation. The attack rate was determined as the proportion of mice scoring positive for brain PrPres among the total numbers of inoculated mice. The mean survival times obtained for coinoculated and control groups were subjected to a Student t test in order to find statistical differences among them (*, P < 0.01; ** P < 0.05; ***, P < 0.001).
Western blotting.
Brain tissue was homogenized in 5% glucose in distilled water in grinding tubes (Bio-Rad) and adjusted to 10% (wt/vol) by using a TeSeE Precess 48TM homogenizer (Bio-Rad) according to the manufacturer’s instructions. To determine the presence of PrPres in mouse brains, 100 μl of 10% brain homogenate were analyzed by WB as previously described (32). Briefly, digestion was performed with 40 μg/ml of proteinase K using the reagents of the TeSeE (Bio-Rad) enzyme-linked immunosorbent assay at 37°C for 15 min. Samples were electrophoresed in 12% Criterion XT Bis-Tris gel (Bio-Rad). For immunoblotting, membranes were incubated with Sha31 (34) or 12B2 (35) PrP monoclonal antibody (epitopes 145-YEDRYYRE-152 and 89-WGQGG-93 of the human PrP sequence, respectively) at a final concentration of 1 μg/ml. Immunocomplexes were detected with horseradish peroxidase-conjugated anti-mouse IgG (Amersham Pharmacia Biotech) after incubation of the membranes for 1 h, and blots were developed with chemiluminescent substrate ECL Select (GE Healthcare Amersham Biosciences). Images were captured using the ChemiDoc XRS+ system and then processed using Image Lab 5.2.1 Software.
Histopathological analysis.
Brain samples were immediately fixed in neutral buffered 10% formalin (4% 2-formaldehyde) during mouse necropsy and paraffin embedded later. PET-blot analyses were conducted as previously described (26).
ACKNOWLEDGMENTS
This study was supported by grants from the Spanish Ministerio de Economía y Competitividad (AGL2016-78054-R [AEI/FEDER, UE], AGL2012-37988-C04-04, and RTA2012-00004-00-00). A.M.-M. was supported by a fellowship from the INIA (FPI-SGIT-2015-02), and P.A.-C. was supported by a fellowship from the Spanish Ministerio de Economía y Competitividad (BES-2010-040922). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. The authors declare no conflict of interest.
We thank Olivier Andreoletti for assistance in performing the PET-blot experiments. We thank Juan Píquer, Irene Prieto, Patricia Lorenzo, and Ana Villa for technical assistance and the staff of the biosafety level 3 animal facility and the Biosafety Office at the CISA-INIA (Valdeolmos-Madrid) for excellent animal care and work.
REFERENCES
- 1.Colby DW, Prusiner SB. 2011. Prions. Cold Spring Harb Perspect Biol 3:a006833. doi: 10.1101/cshperspect.a006833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prusiner SB. 1991. Molecular biology of prion diseases. Science 252:1515–1522. doi: 10.1126/science.1675487. [DOI] [PubMed] [Google Scholar]
- 3.Prusiner SB. 1998. Prions. Proc Natl Acad Sci U S A 95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prusiner SB, Scott M, Foster D, Pan KM, Groth D, Mirenda C, Torchia M, Yang SL, Serban D, Carlson GA, Hoppe PC, Westaway D, DeArmond SJ. 1990. Transgenetic studies implicate interactions between homologous PrP isoforms in scrapie prion replication. Cell 63:673–686. doi: 10.1016/0092-8674(90)90134-Z. [DOI] [PubMed] [Google Scholar]
- 5.Béringue V, Vilotte JL, Laude H. 2008. Prion agent diversity and species barrier. Vet Res 39:47. doi: 10.1051/vetres:2008024. [DOI] [PubMed] [Google Scholar]
- 6.Scott M, Peretz D, Ridley RM, Baker HF, DeArmond SJ, Prusiner SB. 2004. Transgenetic investigations of the species barrier and prion strains, p 435–482 In Prusiner SB. (ed), Prion biology and diseases, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 7.Collinge J, Clarke A. 2007. A general model of prion strains and their pathogenicity. Science 318:930–936. doi: 10.1126/science.1138718. [DOI] [PubMed] [Google Scholar]
- 8.Chabry J, Caughey B, Chesebro B. 1998. Specific inhibition of in vitro formation of protease-resistant prion protein by synthetic peptides. J Biol Chem 273:13203–13207. doi: 10.1074/jbc.273.21.13203. [DOI] [PubMed] [Google Scholar]
- 9.Horiuchi M, Baron GS, Xiong LW, Caughey B. 2001. Inhibition of interactions and interconversions of prion protein isoforms by peptide fragments from the C-terminal folded domain. J Biol Chem 276:15489–15497. doi: 10.1074/jbc.M100288200. [DOI] [PubMed] [Google Scholar]
- 10.Skinner PJ, Kim HO, Bryant D, Kinzel NJ, Reilly C, Priola SA, Ward AE, Goodman PA, Olson K, Seelig DM. 2015. Treatment of prion disease with heterologous prion proteins. PLoS One 10:e0131993. doi: 10.1371/journal.pone.0131993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Polymenidou M, Stoeck K, Glatzel M, Vey M, Bellon A, Aguzzi A. 2005. Coexistence of multiple PrPSc types in individuals with Creutzfeldt-Jakob disease. Lancet Neurol 4:805–814. doi: 10.1016/S1474-4422(05)70225-8. [DOI] [PubMed] [Google Scholar]
- 12.Parchi P, Strammiello R, Notari S, Giese A, Langeveld JP, Ladogana A, Zerr I, Roncaroli F, Cras P, Ghetti B, Pocchiari M, Kretzschmar H, Capellari S. 2009. Incidence and spectrum of sporadic Creutzfeldt-Jakob disease variants with mixed phenotype and co-occurrence of PrPSc types: an updated classification. Acta Neuropathol 118:659–671. doi: 10.1007/s00401-009-0585-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thackray AM, Lockey R, Beck KE, Spiropoulos J, Bujdoso R. 2012. Evidence for coinfection of ovine prion strains in classical scrapie isolates. J Comp Pathol 147:316–329. doi: 10.1016/j.jcpa.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 14.Chong A, Kennedy I, Goldmann W, Green A, González L, Jeffrey M, Hunter N. 2015. Archival search for historical atypical scrapie in sheep reveals evidence for mixed infections. J Gen Virol 96:3165–3178. doi: 10.1099/jgv.0.000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aguilar-Calvo P, Xiao X, Bett C, Eraña H, Soldau K, Castilla J, Nilsson KP, Surewicz WK, Sigurdson CJ. 2017. Posttranslational modifications in PrP expand the conformational diversity of prions in vivo. Sci Rep 8:43295. doi: 10.1038/srep43295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dickinson AG, Fraser H, Meikle VM, Outram GW. 1972. Competition between different scrapie agents in mice. Nat New Biol 237:244–245. doi: 10.1038/newbio237244a0. [DOI] [PubMed] [Google Scholar]
- 17.Dickinson AG, Fraser H, McConnell I, Outram GW, Sales DI, Taylor DM. 1975. Extraneural competition between different scrapie agents leading to loss of infectivity. Nature 253:556. doi: 10.1038/253556a0. [DOI] [PubMed] [Google Scholar]
- 18.Baron TG, Biacabe AG. 2001. Molecular analysis of the abnormal prion protein during coinfection of mice by bovine spongiform encephalopathy and a scrapie agent. J Virol 75:107–114. doi: 10.1128/JVI.75.1.107-114.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartz JC, Aiken JM, Bessen RA. 2004. Delay in onset of prion disease for the HY strain of transmissible mink encephalopathy as a result of prior peripheral inoculation with the replication-deficient DY strain. J Gen Virol 85:265–273. doi: 10.1099/vir.0.19394-0. [DOI] [PubMed] [Google Scholar]
- 20.Bartz JC, Kramer ML, Sheehan MH, Hutter JA, Ayers JI, Bessen RA, Kincaid AE. 2007. Prion interference is due to a reduction in strain-specific PrPSc levels. J Virol 81:689–697. doi: 10.1128/JVI.01751-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manuelidis L, Lu ZY. 2003. Virus-like interference in the latency and prevention of Creutzfeldt-Jakob disease. Proc Natl Acad Sci U S A 100:5360–5365. doi: 10.1073/pnas.0931192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishida N, Katamine S, Manuelidis L. 2005. Reciprocal interference between specific CJD and scrapie agents in neural cell cultures. Science 310:493–496. doi: 10.1126/science.1118155. [DOI] [PubMed] [Google Scholar]
- 23.Nilsson KP, Joshi-Barr S, Winson O, Sigurdson CJ. 2010. Prion strain interactions are highly selective. J Neurosci 30:12094–12102. doi: 10.1523/JNEUROSCI.2417-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schutt CR, Bartz JC. 2008. Prion interference with multiple prion isolates. Prion 2:61–63. doi: 10.4161/pri.2.2.6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karapetyan YE, Saá P, Mahal SP, Sferrazza GF, Sherman A, Salès N, Weissmann C, Lasmézas CI. 2009. Prion strain discrimination based on rapid in vivo amplification and analysis by the cell panel assay. PLoS One 4:e5730. doi: 10.1371/journal.pone.0005730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cassard H, Torres JM, Lacroux C, Douet JY, Benestad SL, Lantier F, Lugan S, Lantier I, Costes P, Aron N, Reine F, Herzog L, Espinosa JC, Beringue V, Andréoletti O. 2014. Evidence for zoonotic potential of ovine scrapie prions. Nat Commun 5:5821. doi: 10.1038/ncomms6821. [DOI] [PubMed] [Google Scholar]
- 27.Fernández-Borges N, Espinosa JC, Marín-Moreno A, Aguilar-Calvo P, Asante EA, Kitamoto T, Mohri S, Andréoletti O, Torres JM. 2017. Protective effect of Val129-PrP against bovine spongiform encephalopathy but not variant Creutzfeldt-Jakob disease. Emerg Infect Dis 23:1522–1530. doi: 10.3201/eid2309.161948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Torres JM, Castilla J, Pintado B, Gutiérrez-Adan A, Andréoletti O, Aguilar-Calvo P, Arroba AI, Parra-Arrondo B, Ferrer I, Manzanares J, Espinosa JC. 2013. Spontaneous generation of infectious prion disease in transgenic mice. Emerg Infect Dis 19:1938–1947. doi: 10.3201/eid1912.130106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taylor DM, Dickinson AG, Fraser H, Marsh RF. 1986. Evidence that transmissible mink encephalopathy agent is biologically inactive in mice. Neuropathol Appl Neurobiol 12:207–215. doi: 10.1111/j.1365-2990.1986.tb00051.x. [DOI] [PubMed] [Google Scholar]
- 30.Dickinson AG, Fraser H, Outram GW. 1975. Scrapie incubation time can exceed natural lifespan. Nature 256:732–733. doi: 10.1038/256732a0. [DOI] [PubMed] [Google Scholar]
- 31.Race R, Raines A, Raymond GJ, Caughey B, Chesebro B. 2001. Long-term subclinical carrier state precedes scrapie replication and adaptation in a resistant species: analogies to bovine spongiform encephalopathy and variant Creutzfeldt-Jakob disease in humans. J Virol 75:10106–10112. doi: 10.1128/JVI.75.21.10106-10112.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Padilla D, Béringue V, Espinosa JC, Andreoletti O, Jaumain E, Reine F, Herzog L, Gutierrez-Adan A, Pintado B, Laude H, Torres JM. 2011. Sheep and goat BSE propagate more efficiently than cattle BSE in human PrP transgenic mice. PLoS Pathog 7:e1001319. doi: 10.1371/journal.ppat.1001319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Castilla J, Gutiérrez Adán A, Brun A, Pintado B, Ramírez MA, Parra B, Doyle D, Rogers M, Salguero FJ, Sánchez C, Sánchez-Vizcaíno JM, Torres JM. 2003. Early detection of PrPres in BSE-infected bovine PrP transgenic mice. Arch Virol 148:677–691. doi: 10.1007/s00705-002-0958-4. [DOI] [PubMed] [Google Scholar]
- 34.Féraudet C, Morel N, Simon S, Volland H, Frobert Y, Créminon C, Vilette D, Lehmann S, Grassi J. 2005. Screening of 145 anti-PrP monoclonal antibodies for their capacity to inhibit PrPSc replication in infected cells. J Biol Chem 280:11247–11258. doi: 10.1074/jbc.M407006200. [DOI] [PubMed] [Google Scholar]
- 35.Yull HM, Ritchie DL, Langeveld JP, van Zijderveld FG, Bruce ME, Ironside JW, Head MW. 2006. Detection of type 1 prion protein in variant Creutzfeldt-Jakob disease. Am J Pathol 168:151–157. doi: 10.2353/ajpath.2006.050766. [DOI] [PMC free article] [PubMed] [Google Scholar]