Abstract
DNA mismatch repair was proposed to play a pivotal role in the development and prognosis of colorectal cancer. However, the prognostic value of mismatch repair on colorectal cancer is still unknown. The PubMed, EMBASE, and Cochrane Central Register of Controlled Trials databases were searched. The articles about mismatch repair (including hMLH1, hMSH2, hMSH3, hMSH6, hPMSH1, and hPMSH2) deficiency for the prognosis of patients with colorectal cancer were included in the study. The hazard ratio and its 95% confidence interval were used to measure the impact of mismatch repair deficiency on survival time. Twenty-one articles were included. The combined hazard ratio for mismatch repair deficiency on overall survival was 0.59 (95% confidence interval: 0.50-0.69) and that on disease-free survival was 0.57 (95% confidence interval: 0.43-0.75). In subgroup analysis, there were a significant association between overall survival and mismatch repair deficiency in Asian studies (hazard ratio: 0.67; 95% confidence interval: 0.50-0.91) and Western studies (hazard ratio: 0.56; 95% confidence interval: 0.46-0.67). For disease-free survival, the hazard ratios in Asian studies and Western studies were 0.55 (95% confidence interval: 0.38-0.81) and 0.62 (95% confidence interval: 0.50-0.78), respectively. Our meta-analysis indicated that mismatch repair could be used to evaluate the prognosis of patients with colorectal cancer.
Keywords: DNA mismatch repair (MMR), prognosis, colorectal cancer, meta-analysis, overall survival, disease-free survival
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer in males and the second in females, with an estimated 1.4 million cases and 693 900 deaths occurring in 2012.1 Molecular markers for the biological behavior and prognosis of CRC were extensively studied; among these markers, DNA mismatch repair (MMR) was proposed to play a pivotal role in the development and prognosis of CRC.2
Approximately 10% to 20% of sporadic CRC is associated with impaired function of DNA MMR genes.3 Mismatch repair genes encode corresponding enzymes that can recognize and repair mismatched base pairs during DNA replication. Deficient MMR leads to genetic instability and accounts for the accumulation of widespread alterations in the length of short repeated DNA sequences, known as microsatellite instability (MSI). The accelerating accumulation of gene mutations in proto-oncogenes and cancer suppressor genes because of aberrant MMR can affect the proliferation of normal cells and promote the development of carcinoma.4-6 A meta-analysis by Guastadisegni, including 31 eligible studies reporting survival in 12 782 patients with CRC, showed that MSI predicted favorable prognosis, with both longer overall survival (OS) and disease-free survival (DFS).7 However, the detection of MSI by polymerase chain reaction (PCR) in 5 highly monomorphic mononucleotidic microsatellite markers (BAT26, BAT25, NR21, NR24, NR27) is complex and costly, limiting its application in the clinic. Currently, different MMR genes (hMLH1, hMSH2, hMSH3, hMSH6, hPMSH1, and hPMSH2) have been identified to cause the development of MSI.8 The hMLH1 and hMSH2 account for more than 90% of MSI development. Thus, their relationship with the development and prognosis of CRC has been extensively studied.9-18 However, the results were controversial. Thus, the aim of this meta-analysis is to identify the association between deficient MMR and the prognosis of CRC.
Materials and Methods
Search Strategy
A meta-analysis was conducted according to the guidelines of the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement.19 Two reviewers (JTH and XLC) independently searched the following databases from their inceptions to June 1st, 2017: PubMed, EMBASE, and the Cochrane Central Register of Controlled Trials. The search terms included the following:
“colorectal cancer” OR “colon cancer” OR “rectal cancer”
“mismatch repair gene” OR “hMLH1” OR “hMSH2” OR “hMSH3” OR “hMSH6” OR “hPMSH1” OR “hPMSH2” OR MMR
“prognosis” OR “prognoses” OR “prognostic” OR “predictive” OR “biomarker” OR “marker” OR “survival” OR “survive” OR “Cox” OR “Logrank” OR “Kaplan-Meier”
The search was not limited by language. The potentially relevant studies were manually reviewed in the relevant systematic reviews and meta-analysis. The relevant studies were also obtained by searching Google scholar with the search terms “colorectal cancer, colon cancer, or rectal cancer,” “mismatch repair gene,” and “prognosis, predictive, or survive.”
Inclusion Criteria
The inclusion criteria included the following: (1) patients who were diagnosed with primary CRC (including colon cancer, or rectal cancer) were included. Patients exhibiting different clinical stages, histological types, or treatment methods were all included; (2) the expression of MMR genes was measured using PCR, immunohistochemistry (IHC), or enzyme-linked immunosorbent assay in the CRC tissue; (3) the association between MMR with patient prognosis was investigated, and the hazard ratio (HR), its 95% confidence interval (CI), or the relevant information were provided; and (4) a full paper was published. When the same team reported several studies from the same patients, the most recent study was included. Studies published in the abstract were excluded.
Study Selection
The same studies from the different databases were identified. The titles and abstracts were read for eligibility by 2 of the 3 authors (DJZ, PRL, or LZC). The full texts of potentially eligible studies were retrieved and reviewed independently by 2 authors (HBL and DJZ). Any disagreements were recorded and resolved by consensus under the guidance of another author (XLC).
Data Collection
The data in the eligible studies were extracted by 2 authors (JTH and DJZ). The study information (the first author, the year of publication), study participants (the type of patients, gender, mean age, and sample size), the characteristics of treatment (surgery, chemotherapy, radiotherapy), the characteristics of MMR (gene subtype, test sample, test content, test method), and the prognostic outcomes of interest (OS, DFS, and/or relapse-free survival) were extracted. If the relevant data were not reported in the study, the item was recorded as “NR (not reported).”
Data Analysis
The MMR genes were classified as either “deficiency” (weak or negative) or “proficiency” (strong or positive). If the HRs and their 95% CI were reported explicitly in the study, the data were collected. If these data were not reported explicitly, they were calculated from the available numerical data or survival curves using the methods reported by Tierney et al.20 The meta-analysis was conducted according to 2 types of indexes, OS, and DFS.
To measure the impact of deficient MMR on survival time, the combined HR and its 95% CI were calculated. The heterogeneity of the individual HR was calculated with a χ2 test. The heterogeneity test with the inconsistency index (I 2) statistic and Q statistic was performed. For the Q statistic, a P value of less than 0.1 was considered representative of statistically significant heterogeneity. The I 2 is the proportion of total variation contributed by between-study variation. An I 2 index of approximately 25% was considered to demonstrate low levels of heterogeneity, 50% was considered medium, and 75% was considered high. The sensitivity analysis were conducted by reestimating the pooled HR and omitting each study in turn to investigate the influence of each individual study on the overall meta-analysis summary estimate. Furthermore, subgroup analysis based on geographical regions (Asia, West [Europe and America]), different subtypes of gene (MMR, hMSH2, hMLH1), and different methods for detection of MMR (IHC, PCR) were performed to clarify the source of heterogeneity. In addition, the heterogeneity of the effect was discovered using meta-regression, including country, type of patient, sample size (≤200, and >200), test content, analysis method, subtype of gene, and therapy methods as covariates. Heterogeneity was defined as a P ≤ .05. An observed HR >1 implied a worse prognosis in the high expression of MMR compared to the low expression of MMR. Publication bias and small study effects were assessed by Begg test and Egger test, with P < .05 considered to show significant publication bias. All of the calculations were performed by STATA version 12.0.
Results
The Characteristics of the Studies
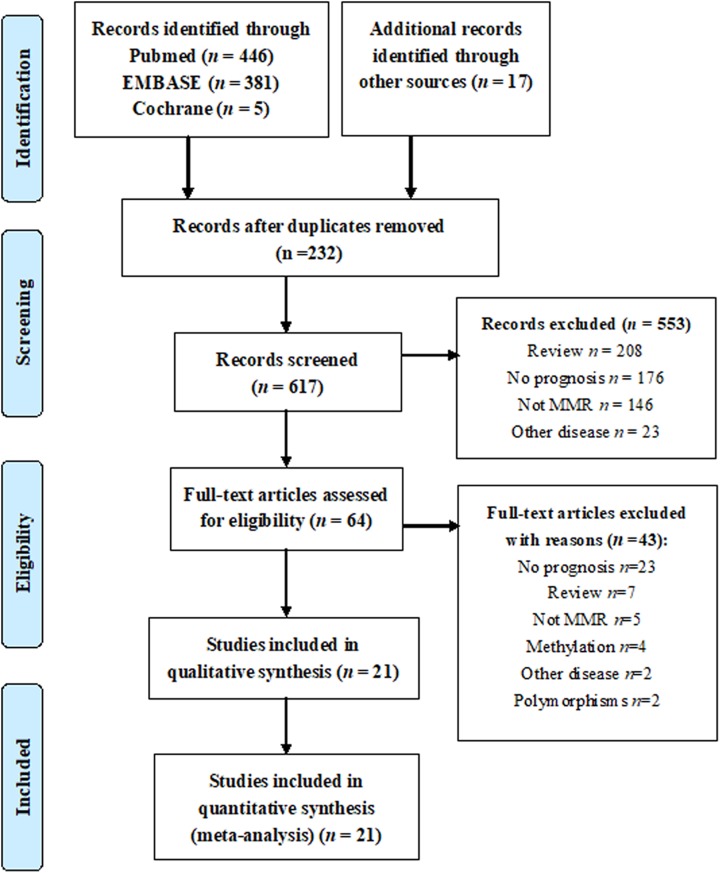
A total of 849 studies met the inclusion criteria (Figure 1), of which 232 studies were excluded for duplicates, and 553 studies were excluded by reviewing the titles and abstracts. The full texts of the remaining 64 studies were obtained for review. Eventually, 21 studies were included in the meta-analysis.9-18,21-31
Figure 1.
Flow chart of the search strategy.
The characteristics of the included studies are shown in Table 1. Twenty-one studies contained a total of 5340 patients with CRC. All the studies were published from 1999 to 2016. Of the 21 studies, 15 studies reported OS and 10 reported DFS (Table 2).
Table 1.
The Main Characteristics of the Included Studies.
| Author | Year of Publication | Country | Time | Patients | Sample Size | Male | Mean of Age (range) | Stage | Surgery Treatment | Chemotherapy | Radiotherapy | Median Follow-Up Month (Range) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bendardaf R | 2008 | Finland | 1996-2003 | CRC | 73 | 46 | 57.9 (NR) | II-IV | Partial | All | No patient | 32.6 (NR) |
| Cawkwell L | 1999 | UK | NR | CRC | 101 | NR | >50 (NR) | NR | NR | NR | NR | 60.0 (NR) |
| Huh JW | 2016 | Korea | NR | Rectal cancer | 209 | 136 | 56 (27-81) | II, III | All | All | All | 44 (2-87) |
| Ide T | 2008 | Japan | 1999-2005 | CRC | 94 | 60 | 68.2 (40-87) | I-IV | All | All | NR | 26.1 (NR) |
| Jansson A | 2003 | Sweden | 1972-1996 | CRC | 301 | NR | 70 (34-94) | I-IV | All | NR | NR | NR (NR) |
| Jensen LH | 2007 | Denmark | NR | CRC | 28 | 13 | 61 (49-74) | III-IV | NR | All | NR | 12.2 (6.7-20.2) |
| Jensen SA | 2009 | Denmark | 1996-2003 | CRC | 340 | 159 | NR (NR) | II-IV | NR | All | NR | 6.1 (4.1-11.3) |
| Langner E | 2010 | Poland | NR | CRC | 75 | 45 | NR (NR) | I-IV | All | NR | NR | NR (NR) |
| Lanza G | 2006 | Italy | 1986-1995 | CRC | 718 | 359 | 65 (27-85) | II, III | Partial | Most | Partial | 90.5 (63-144) |
| Ma J | 2015 | China | 2008-2011 | Colon cancer | 184 | 111 | NR (NR) | NR | NR | All | NR | 17.6 (7-36) |
| Park JW | 2010 | Korea | 2001-2003 | CRC | 318 | 191 | 60.5 (27-87) | I-IV | All | Partial | NR | 24.0 (NR) |
| Pu C | 2015 | China | 2005-2008 | CRC | 327 | 201 | NR (NR) | I-IV | NR | Partial | NR | 24.0 (NR) |
| Rau B | 2003 | Germany | 1993-1999 | CRC | 66 | 41 | 59 (39-74) | NR | NR | All | All | 39.3 (11.3-83.4) |
| Russo A | 2009 | Italy | NR | CRC | 526 | 288 | 48 (20-88) | I-IV | NR | NR | NR | 64 (6-383) |
| Sinicrope FA | 2006 | USA | NR | Colon cancer | 528 | 274 | NR (NR) | II, III | NR | All | NR | NR (NR) |
| Smyth EF | 2004 | UK | 1995-1998 | Colon cancer | 111 | NR | 72 (3990) | I-IV | All | NR | NR | NR (NR) |
| Sun Z | 2014 | China | 2009-2012 | CRC | 404 | 233 | NR (NR) | I-IV | NR | NR | NR | >36 (NR) |
| Wang H | 2014 | China | 2005-2008 | CRC | 327 | 201 | 58.7 (2581) | I-IV | NR | NR | NR | 60.0 (NR) |
| Wang JB | 2016 | China | 2011-2012 | Colon cancer | 90 | 58 | NR (NR) | II, III | All | Partial | NR | 27 (5-35) |
| Wang Y | 2014 | China | NR | CRC | 433 | 254 | 58.6 (2482) | I-IV | All | Partial | Partial | 52 (1-87) |
| Wu HW | 2013 | China | 2004-2006 | CRC | 87 | 56 | 59 (35-82) | I-III | All | NR | NR | 60.0 (NR) |
Abbreviations: CRC, colorectal cancer; NR, not report.
Table 2.
The Gene and Results of the Included Studies.
| Author | Subtypes of Gene | Test Sample | Test Content | Test Method | Analysis Method | Survival Type |
|---|---|---|---|---|---|---|
| Bendardaf R | MMR | Tissue | Protein | IHC | Univariate | DFS |
| Cawkwell L | MMR | Tissue | Protein | IHC | Univariate | OS |
| Huh JW | hMSH2 | Tissue | Protein | IHC | Multivariate | DFS |
| Ide T | hMLH1 | Tissue | mRNA | PCR | Univariate | DFS |
| Jansson A | hMSH2 | Tissue | Protein | IHC | Univariate | OS |
| Jensen LH | hMSH2 | Tissue | RNA | PCR | Univariate | OS |
| Jensen SA | MMR | Tissue | Protein | IHC | Multivariate | OS, RFS |
| Langner E | hMSH2 | Tissue | RNA | PCR | Univariate | OS |
| Lanza G | MMR | Tissue | Protein | IHC | Multivariate | OS |
| Ma J | MMR | Tissue | Protein | IHC | Multivariate | OS, DFS |
| Park JW | MMR | Tissue | Protein | IHC | Multivariate | OS |
| Pu C | hMLH1 | Tissue | Protein | IHC | Multivariate | DFS |
| Rau B | hMSH2 | Tissue | Protein | IHC | Univariate | OS, DFS |
| Russo A | MMR | Blood | mRNA | PCR | Multivariate | OS |
| Sinicrope FA | MMR | Tissue | Protein | IHC | Univariate | OS, DFS |
| Smyth EF | hMLH1 | Tissue | Protein | IHC | Multivariate | OS |
| Sun Z | MMR | Tissue | Protein | IHC | Univariate | DFS |
| Wang H | hMSH2 | Tissue | Protein | IHC | Multivariate | OS |
| Wang JB | MMR* | Tissue | Protein | IHC | Univariate | OS |
| Wang Y | MMR | Blood | DNA | PCR | Multivariate | OS |
| Wu HW | hMSH2 | Tissue | Protein | IHC | Multivariate | DFS |
Abbreviations: DFS, disease-free survival; DNA, deoxyribonucleic acid; IHC, immunohistochemistry; MMR, contain hMLH1, hMSH2; MMR*, contain hMLH1, hMSH2, hMSH6, and hPMS2; mRNA, messenger RNA; Multivariate, multivariate survival analysis; OS, overall survival; PCR, polymerase chain reaction; RFS, recurrence-free survival (which was used as DFS); RNA, ribonucleic acid; Univariate, univariate survival analysis.
Meta-Analysis for OS
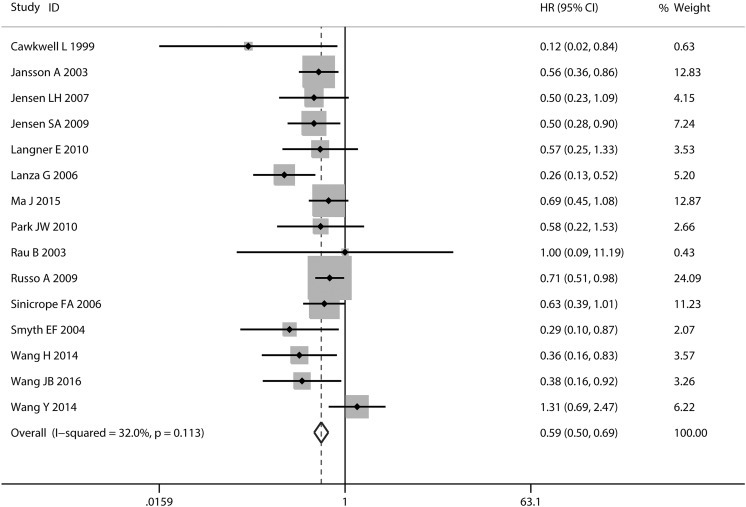
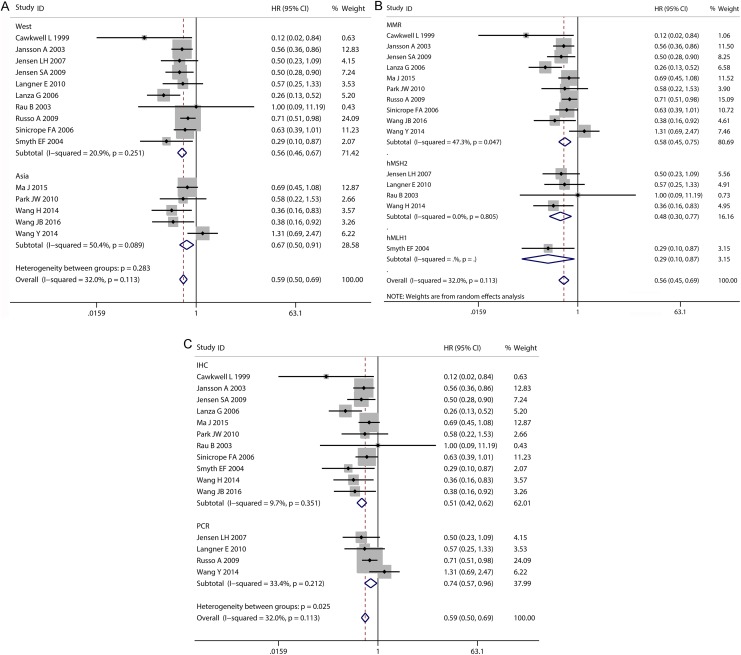
Fifteen studies of MMR for OS in patients with CRC were included in the meta-analysis. There was no significant heterogeneity across 15 studies with OS (I 2 = 32.0%, P = 0.113; Figure 2). The combined HR of the 15 studies was 0.59 (95% CI: 0.50-0.69; Figure 2). Mismatch ratio was significantly associated with improved prognosis in Asian studies (HR: 0.67; 95% CI: 0.50-0.91; Figure 3) and western studies (HR: 0.56; 95% CI: 0.46-0.67; Figure 3). The HR was 0.58 (95% CI: 0.45-0.75) for MMR as a marker in 10 studies, 0.29 (95% CI: 0.10-0.87) for hMLH1 as a marker in one study, and 0.48 (95% CI: 0.30-0.77) for hMSH2 as a marker in 4 studies (Table 3; Figure 3). When the subgroups were analyzed based on the test method, the combined HRs for IHC and PCR were 0.51 (95% CI: 0.42-0.62) and 0.74 (95% CI: 0.57-0.96), respectively (Figure 3).
Figure 2.
Forest plot for the association between MMR expression and OS in CRC.
CRC indicates colorectal cancer; MMR, mismatch repair; OS, overall survival.
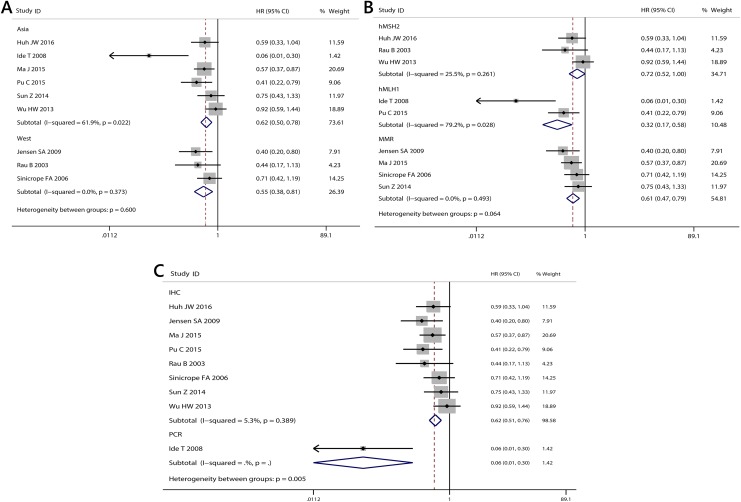
Figure 3.
Subgroup analysis for the association between MMR expression and OS in CRC. A, different geographical regions (Asia, west); (B) different subtypes of gene (MMR, hMSH2, hMLH1); (C) different methods for detection of MMR (immunohistochemistry [IHC], polymerase chain reaction [PCR]). CRC indicates colorectal cancer; MMR, mismatch repair; OS, overall survival.
Table 3.
The Results of the Meta-Analysis.
| Number of Studies | Patients | HR (95% CI) | Heterogeneity (I 2, P) | |
|---|---|---|---|---|
| Overall survival | ||||
| All | 15 | 4146 | 0.59 (0.50-0.69) | 32.0%, 0.113 |
| Asian | 5 | 1352 | 0.67 (0.50-0.91) | 50.4%, 0.089 |
| Western | 10 | 2794 | 0.56 (0.46-0.67) | 20.9%, 0.251 |
| Gene | ||||
| MMR | 10 | 3539 | 0.58 (0.45-0.75)a | 47.3%, 0.047 |
| hMSH2 | 4 | 496 | 0.48 (0.30-0.77) | 0.0%, 0.805 |
| hMLH1 | 1 | 111 | 0.29 (0.10-0.87) | NR |
| Test method | ||||
| IHC | 11 | 3084 | 0.51 (0.42-0.62) | 9.7%, 0.351 |
| PCR | 4 | 1062 | 0.74 (0.57-0.96) | 33.4%, 0.212 |
| Disease-free survival | ||||
| All | 10 | 2312 | 0.62 (0.44-0.88) | 66.5%, 0.001 |
| Allb | 9 | 2239 | 0.57 (0.43-0.75) | 48.0%, 0.052 |
| Asian | 6 | 1305 | 0.62 (0.50-0.78)a | 61.9%, 0.022 |
| Western | 3 | 934 | 0.55 (0.38-0.81) | 0.0%, 0.373 |
| Gene | ||||
| MMR | 4 | 1456 | 0.61 (0.47-0.79) | 0.0%, 0.493 |
| hMLH1 | 2 | 421 | 0.32 (0.17-0.58)a | 79.2%, 0.028 |
| hMSH2 | 3 | 362 | 0.72 (0.52-1.00) | 25.5%, 0.261 |
| Test method | ||||
| IHC | 8 | 2145 | 0.62 (0.51-0.76) | 5.3%, 0.389 |
| PCR | 1 | 94 | 0.06 (0.01-0.30) | NR |
Abbreviations: IHC, immunohistochemistry; NR: not report; MMR, mismatch repair; PCR, polymerase chain reaction.
a Results from random-effect model.
b Bendardaf study was excluded, which was also excluded in subgroup analysis.
Meta-Analysis for DFS
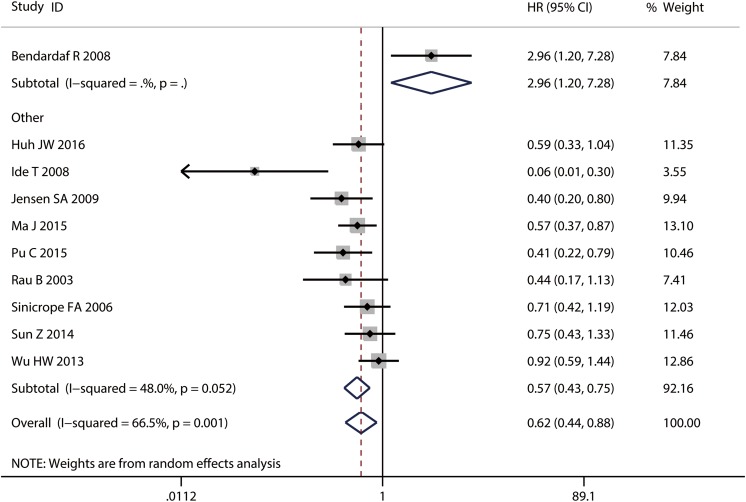
In a pooled analysis of 10 DFS studies, deficient MMR was associated with a better prognosis for CRC (HR: 0.62; 95% CI: 0.44-0.88; Figure 4). There was significant heterogeneity across 10 studies with DFS (I 2 = 66.5%, P = .001). A meta-regression was performed to explore the source of heterogeneity for DFS. The results showed that all the variables were not related with the heterogeneity (Table 4). The tested content was nearly significant (P = .077). In addition, Bendardaf study included 73 patients, and thymidylate synthase (TS) and MMR expressions were assessed for each patient.27 Its HR was calculated by comparing 18 patients with both high TS and MMR expression with 27 patients with both low TS and MMR, while other patients with low MMR and high TS or high MMR and low TS were excluded. This may result in significant heterogeneity between Bendardaf study and other studies (P = .001). Thus, Bendardaf study was excluded in subgroup analysis.
Figure 4.
Forest plot for the association between MMR expression and DFS in CRC. CRC indicates colorectal cancer; DFS, disease-free survival; MMR; mismatch repair.
Table 4.
The results of meta-regression for DFS.
| Coefficient | 95% CI | P | |
|---|---|---|---|
| Patients | −0.149 | (−1.196 to 0.898) | .681 |
| Country | −0.124 | (−1.812 to 1.563) | .830 |
| Sample size | −0.723 | (−2.387 to 0.940) | .260 |
| Subtypes of gene | −0.220 | (−1.140 to 0.699) | .501 |
| Test content | 3.157 | (−0.626 to 6.940) | .077 |
| Analysis method | −0.501 | (−2.196 to 1.195) | .417 |
Abbreviation: CI, confidence interval.
The combined HRs for studies without Bendardaf study were 0.57 (95% CI: 0.50-0.73; I 2 = 48.0%, P = .052). The HRs of deficient MMR on DFS in Asian studies or western studies were 0.62 (95% CI: 0.50-0.78; Figure 5) and 0.55 (95% CI: 0.38-0.81; Figure 5). The HR was 0.61 (95% CI: 0.47-0.79) for MMR as a marker in 5 studies, 0.32 (95% CI: 0.17-0.58) for hMLH1 as a marker in 2 studies, and 0.72 (95% CI: 0.52-1.00) for hMSH2 as a marker in 3 studies (Table 3; Figure 5). When the subgroups were analyzed based on the test method, the combined HRs for IHC and PCR were 0.62 (95% CI: 0.51-0.76) and 0.06 (95% CI: 0.01-0.30), respectively (Figure 5).
Figure 5.
Subgroup analysis for the association between MMR expression and DFS in CRC. A, different geographical regions (Asia, west); (B) different subtypes of gene (MMR, hMSH2, hMLH1); (C) different methods for detection of MMR (immunohistochemistry [IHC], polymerase chain reaction [PCR]). CRC indicates colorectal cancer; DFS, disease-free survival; MMR; mismatch repair.
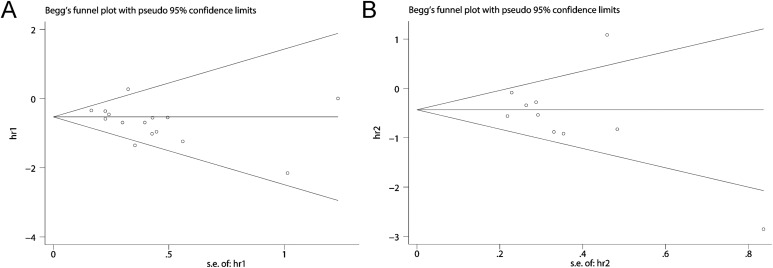
The publication bias was not significant (OS, P = .113; DFS, P = .210). However, one study was out of the reference line in the DFS group indicated that there might be publication bias for DFS (Figure 6).
Figure 6.
Begg funnel plot (A: OS, P = .113; B: DFS, P = .210). DFS indicates disease-free survival; OS, overall survival.
Discussion
Our pooled results from all the eligible studies showed that the HR was 0.59 for OS and 0.62 for DFS, with all showing statistically significant associations between MMR and CRC. The results indicated that deficient MMR was associated with better OS and DFS in the patients with CRC. Meanwhile, subgroup analysis of the different regions (Western and Asia) MMR showed consistent results. In addition, no obvious publication bias was determined by Begg test. These analyses enhanced the reliability of this meta-analysis.
Mismatch repairs are a group of enzymes that can recognize and repair mismatched base pairs during DNA replication, including hMLH1, hMSH2, hMSH3, hMSH6, hPMSH1, and hPMSH2, which are considered critical proteins for the formation of MSI.9-18,21-31 Mismatch repairs, along with MSI, are proposed to be useful markers in CRC. Approximately 90% hereditary nonpolyposis CRC and 10% to 20% of sporadic CRC demonstrate MSI.32 The vast majority of CRC with MSI is caused by aberrant hMLH1 expression (70% to 95%), while others primarily result from the inactivation of hMSH2 and hMSH6. Additionally, in sporadic CRC, approximately 95% deficient hMLH1 expression is due to hypermethylation of the hMLH1 promoter.33,34
In this meta-analysis, some studies defined negative hMLH1 or hMSH2 expression as MMR deficiency, and they demonstrated a significantly longer OS for patients with deficient MMR.7,14,16,17,23,28,35 Some studies defined deficient hMLH1 (or hMSH2) expression as deficient MMR and demonstrated the same result for OS. 11-13,24,26 Four studies showed longer DFS in the patients with CRC with deficient hMLH1 or hMSH2.14-16,28 However, Bendardaf et al suggested that deficient MMR demonstrated a shorter DFS. This inconsistency could be explained by combining with other genes (TS) and distinctive clinic features indicating that CRC with deficient MMR is apt to display marked peritumoral and intratumoral lymphocytic infiltration.36-39 To summarize, these results suggest that detection of both hMLH1 and hMSH2 could be used to identify most MMR and could be useful methods to evaluate OS for patients with CRC. However, further studies are required to confirm the relationship between MMR and DFS because of significant heterogeneity.
The prognosis of CRC is strongly associated with tumor stage, tumor site, and treatment. MMR-deficient CRC exhibit distinct clinical and pathological features, including proximal location and early tumor stage.38,40 Further, Scarpa et al reported that CRC with MMR deficiency exhibited a higher CD80 expression and CD8+ and Th1 T-cell infiltration.41 In vitro silencing of hMSH2, hMLH1, and hMSH6 significantly increased the CD80+ cell rate. These results suggest an enhanced immune surveillance mechanism in the presence of MMR deficiency,41 which may explain the improved OS and DFS for patients with CRC with MMR deficiency. These reports might explain our observations that deficient MMR was associated with better prognosis of patients with CRC. However, MMR-deficient CRC shows poor differentiation and mucinous histology, and strong preclinical and clinical evidence suggests a possible resistance to 5-FU in these tumors. Thus, further studies are required to explore the underlying mechanism of MMR deficiency and better CRC prognosis.
There are 2 broadly accepted methods for aberrant MMR detection including MSI testing by PCR and MMR protein expression analysis by IHC. It has been shown that the results of MMR protein expression by IHC are concordant with DNA-based MSI testing, with a favorable sensitivity and a dramatic specificity.42 Immunohistochemistry is commonly used as an alternative test when a molecular laboratory is not available.43 In addition, IHC for MMR is cost-effective and simple, which can determine the specific protein of MMR.43 In this meta-analysis, most studies adopted the methods of IHC for the detection of aberrant MMR. The subgroup of IHC demonstrated that deficient MMR was a protective factor for the prognosis of CRC, which could predict a longer OS and DFS. As a result, the detection of aberrant MMR by IHC could be a promising method to assess the prognosis of CRC.
There were several limitations in our study. First, the detection of MMR is different among the studies; however, most studies adopted IHC to determine the expression of MMR, and the results were consistent. Second, the definition of aberrant MMR is inconsistent; some studies considered aberrant MMR as negative MMR expression, some selected negative hMLH1 expression alone or negative hMSH2 expression alone, and 2 studies selected negative expression of hMLH1, hMSH2, hMSH6, and hPMS2. This may result in significant heterogeneity among these studies. Third, there is significant heterogeneity among the DFS studies. Although we investigated the influence of each individual study on the overall estimate and conducted a meta-regression and subgroup analysis according to geographical regions and different detection methods, the heterogeneity remained significant in some subgroup analysis and could not be clearly classified. Fourth, some studies did not mention certain vital data, especially the follow-up information, such as studies by Langner E, Jansson A, and Smyth EF, which may influence the reliability of the statistical analysis.
Conclusion
In summary, our pooled results indicated that deficient MMR was associated with a better OS and DFS for the CRC patients. However, large well-designed studies with a uniform method of MMR detection are required to confirm these results.
Abbreviations
- CI
confidence interval
- CRC
colorectal cancer
- DNA
deoxyribonucleic acid
- DFS
disease-free survival
- HR
hazard ratio
- IHC
immunohistochemistry
- MMR
mismatch repair
- MSI
microsatellite instability
- OS
overall survival
- PCR
polymerase chain reaction
- TS
thymidylate synthase.
Footnotes
Authors’ Note: Jiang-tao Hou and Li-na Zhao equally contributed to the work. The study was approved by the ethics committees of Guangzhou University of Chinese Medicine.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was funded by the National Natural Science Foundation of China (81774451 and 81503532), the Natural Science Foundation of Guangdong Province (2017A030313827 and 2015A030313036), the Outstanding Youth Foundation of Guangdong Province Colleges and Universities (YQ2015041), the Young Talents Foundation of Guangzhou University of Chinese Medicine (QNYC20140101), and Guangdong high level universities program of Guangzhou University of Chinese Medicine.
ORCID iD: Xin-lin Chen  https://orcid.org/0000-0002-2650-8051
https://orcid.org/0000-0002-2650-8051
References
- 1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. [DOI] [PubMed] [Google Scholar]
- 2. Gelsomino F, Barbolini M, Spallanzani A, Pugliese G, Cascinu S. The evolving role of microsatellite instability in colorectal cancer: a review. Cancer Treat Rev. 2016;51:19–26. [DOI] [PubMed] [Google Scholar]
- 3. Zhang CM, Lv JF, Gong L, et al. Role of deficient mismatch repair in the personalized management of colorectal cancer. Int J Environ Res Public Health. 2016;13(9):892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol. 2005;23(3):609–618. [DOI] [PubMed] [Google Scholar]
- 5. Kerr DJ, Midgley R. Defective mismatch repair in colon cancer: a prognostic or predictive biomarker? J Clin Oncol. 2010;28(20):3210–3212. [DOI] [PubMed] [Google Scholar]
- 6. Sargent DJ, Marsoni S, Monges G, et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J Clin Oncol. 2010;28(20):3219–3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guastadisegni C, Colafranceschi M, Ottini L, Dogliotti E. Microsatellite instability as a marker of prognosis and response to therapy: a meta-analysis of colorectal cancer survival data. Eur J Cancer. 2010;46(15):2788–2798. [DOI] [PubMed] [Google Scholar]
- 8. Ogino S, Goel A. Molecular classification and correlates in colorectal cancer. J Mol Diagn. 2008;10(1):13–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aparicio T, Schischmanoff O, Poupardin C, et al. Deficient mismatch repair phenotype is a prognostic factor for colorectal cancer in elderly patients. Dig Liver Dis. 2013;45(3):245–250. [DOI] [PubMed] [Google Scholar]
- 10. Russo A, Sala P, Alberici P, et al. Prognostic relevance of MLH1 and MSH2 mutations in hereditary non-polyposis colorectal cancer patients. Tumori. 2009;95(6):731–738. [DOI] [PubMed] [Google Scholar]
- 11. Wu HW, Gao LD, Wei GH. hMSH2 and nm23 expression in sporadic colorectal cancer and its clinical significance. Asian Pac J Cancer Prev. 2013;14(3):1995–1998. [DOI] [PubMed] [Google Scholar]
- 12. Rau B, Sturm I, Lage H, et al. Dynamic expression profile of p21WAF1/CIP1 and Ki-67 predicts survival in rectal carcinoma treated with preoperative radiochemotherapy. J Clin Oncol. 2003;21(18):3391–3401. [DOI] [PubMed] [Google Scholar]
- 13. Langner E, Przybylowska K, Trzcinski R, et al. Loss of hMSH2 gene expression correlates with improved survival in patients with sporadic colorectal cancer. J Genet. 2010;89(1):101–104. [DOI] [PubMed] [Google Scholar]
- 14. Ma J, Zhang Y, Shen H, et al. Association between mismatch repair gene and irinotecan-based chemotherapy in metastatic colon cancer. Tumour Biol. 2015;36(12):9599–9609. [DOI] [PubMed] [Google Scholar]
- 15. Sun Z, Yu X, Wang H, Zhang S, Zhao Z, Xu R. Clinical significance of mismatch repair gene expression in sporadic colorectal cancer. Exp Ther Med. 2014;8(5):1416–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jensen SA, Vainer B, Kruhoffer M, Sorensen JB. Microsatellite instability in colorectal cancer and association with thymidylate synthase and dihydropyrimidine dehydrogenase expression. BMC Cancer. 2009;9:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cawkwell L, Gray S, Murgatroyd H, et al. Choice of management strategy for colorectal cancer based on a diagnostic immunohistochemical test for defective mismatch repair. Gut. 1999;45(3):409–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang H, Sun Z, Ye L, et al. , [hMSH2 aberrant expression in patients with sporadic colorectal cancer in Xinjiang]. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2014;39(6):552–557. [DOI] [PubMed] [Google Scholar]
- 19. Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Bmj. 2009;339:b2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pu C, Ren W, Sun Z, et al. Human mutL homolog 1 expression characteristic and prognostic effect on patients with sporadic colorectal cancer. Int J Clin Exp Med. 2015;8(10):19652–19661. [PMC free article] [PubMed] [Google Scholar]
- 22. Wang Y, Li D, Li X, et al. Prognostic significance of hMLH1/hMSH2 gene mutations and hMLH1 promoter methylation in sporadic colorectal cancer. Med Oncol. 2014;31(7):39. [DOI] [PubMed] [Google Scholar]
- 23. Park JW, Chang HJ, Park S, et al. Absence of hMLH1 or hMSH2 expression as a stage-dependent prognostic factor in sporadic colorectal cancers. Ann Surg Oncol. 2010;17(11):2839–2846. [DOI] [PubMed] [Google Scholar]
- 24. Smyth EF, Sharma A, Sivarajasingham N, Hartley J, Monson JR, Cawkwell L. Prognostic implications of hMLH1 and p53 immunohistochemical status in right-sided colon cancer. Dis Colon Rectum. 2004;47(12):2086–2091; discussion 2091-2. [DOI] [PubMed] [Google Scholar]
- 25. Ide T, Kitajima Y, Ohtaka K, Mitsuno M, Nakafusa Y, Miyazaki K. Expression of the hMLH1 gene is a possible predictor for the clinical response to 5-fluorouracil after a surgical resection in colorectal cancer. Oncol Rep. 2008;19(6):1571–1576. [PubMed] [Google Scholar]
- 26. Jansson A, Arbman G, Zhang H, Sun XF. Combined deficiency of hMLH1, hMSH2, hMSH3 and hMSH6 is an independent prognostic factor in colorectal cancer. Int J Oncol. 2003;22(1):41–49. [PubMed] [Google Scholar]
- 27. Bendardaf R, Lamlum H, Ristamaki R, Korkeila E, Syrjanen K, Pyrhonen S. Thymidylate synthase and microsatellite instability in colorectal cancer: implications for disease free survival, treatment response and survival with metastases. Acta Oncol. 2008;47(6):1046–1053. [DOI] [PubMed] [Google Scholar]
- 28. Sinicrope FA, Rego RL, Foster N, et al. Microsatellite instability accounts for tumor site-related differences in clinicopathologic variables and prognosis in human colon cancers. Am J Gastroenterol. 2006;101(12):2818–2825. [DOI] [PubMed] [Google Scholar]
- 29. Jensen LH, Danenberg KD, Danenberg PV, Jakobsen A. Predictive value of MSH2 gene expression in colorectal cancer treated with capecitabine. Clin Colorectal Cancer. 2007;6(6):433–435. [DOI] [PubMed] [Google Scholar]
- 30. Wang JB, Ma DL, Li JY, Sun QD, Liu YE. Association between expression of DNA mismatch repair genes and clinical features and prognosis of patients with radical resection of colon cancer. Genet Mol Res. 2016;15(3). [DOI] [PubMed] [Google Scholar]
- 31. Huh JW, Kim HC, Kim SH, et al. Mismatch repair gene expression as a predictor of tumor responses in patients with rectal cancer treated with preoperative chemoradiation. Medicine (Baltimore). 2016;95(3):e2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Strate LL, Syngal S. Hereditary colorectal cancer syndromes. Cancer Causes Control. 2005;16(3):201–213. [DOI] [PubMed] [Google Scholar]
- 33. Koopman M, Kortman GA, Mekenkamp L, et al. Deficient mismatch repair system in patients with sporadic advanced colorectal cancer. Br J Cancer. 2009;100(2):266–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Devaud N, Gallinger S. Chemotherapy of MMR-deficient colorectal cancer. Fam Cancer. 2013;12(2):301–306. [DOI] [PubMed] [Google Scholar]
- 35. Lanza G, Gafa R, Santini A, Maestri I, Guerzoni L, Cavazzini L. Immunohistochemical test for MLH1 and MSH2 expression predicts clinical outcome in stage II and III colorectal cancer patients. J Clin Oncol. 2006;24(15):2359–2367. [DOI] [PubMed] [Google Scholar]
- 36. Jass JR, Do KA, Simms LA, et al. Morphology of sporadic colorectal cancer with DNA replication errors. Gut. 1998;42(5):673–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gafa R, Maestri I, Matteuzzi M, et al. Sporadic colorectal adenocarcinomas with high-frequency microsatellite instability. Cancer. 2000;89(10):2025–2037. [PubMed] [Google Scholar]
- 38. Ward R, Meagher A, Tomlinson I, et al. Microsatellite instability and the clinicopathological features of sporadic colorectal cancer. Gut. 2001;48(6):821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hawkins NJ, Tomlinson I, Meagher A, Ward RL. Microsatellite-stable diploid carcinoma: a biologically distinct and aggressive subset of sporadic colorectal cancer. Br J Cancer. 2001;84(2):232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zeinalian M, Hashemzadeh-Chaleshtori M, Salehi R, Kazemi M, Emami MH. Tumor microsatellite instability and clinicopathologic features in Iranian colorectal cancer patients at risk for Lynch syndrome. J Res Med Sci. 2015;20(2):154–160. [PMC free article] [PubMed] [Google Scholar]
- 41. Scarpa M, Ruffolo C, Canal F, et al. Mismatch repair gene defects in sporadic colorectal cancer enhance immune surveillance. Oncotarget. 2015;6(41):43472–43482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lindor NM, Burgart LJ, Leontovich O, et al. Immunohistochemistry versus microsatellite instability testing in phenotyping colorectal tumors. J Clin Oncol. 2002;20(4):1043–1048. [DOI] [PubMed] [Google Scholar]
- 43. Zhang X, Li J. Era of universal testing of microsatellite instability in colorectal cancer. World J Gastrointest Oncol. 2013;5(2):12–19. [DOI] [PMC free article] [PubMed] [Google Scholar]