Short abstract
Objective
To investigate whether the mechanism by which a microRNA, miR-520a, suppresses the replication of hepatitis B virus (HBV) involves the regulation of the serine/threonine kinase (AKT) gene.
Methods
The effects of miR-520a on the proliferation, mitotic index and apoptosis of the HBV-replicating human hepatocellular carcinoma cell line HepG2.2.15 were measured using standard laboratory methods including flow cytometry. The effects of miR-520a on HBV transcription and replication were assessed using methods including immunoassays and reverse transcription–polymerase chain reaction. The effect of small interfering RNA (siRNA) to AKT on the levels of AKT mRNA and protein were also evaluated.
Results
In HepG2.2.15 cells, miRNA-520a reduced HBV transcription and replication by reducing AKT levels. MiRNA-520a decreased cell proliferation and mitosis entry of cells and increased apoptosis in HepG2.2.15 cells. AKT levels were reduced significantly by its siRNA, which resulted in suppression of HBV replication in HepG2.2.15 cells.
Conclusions
MiRNA-520a inhibited AKT gene expression and suppressed HBV transcription and replication. These findings suggest that miRNA-520a may be a novel target for the treatment of HBV infection because miRNA-520a reduced HepG2.2.15 cell survival and inhibited HBV replication associated with the AKT signalling pathway.
Keywords: MicroRNA, hepatitis B virus (HBV), replication, AKT
Introduction
Hepatitis B virus (HBV) infection can result in both acute and chronic liver disease, which in most patients will develop into cirrhosis and liver cancer.1 Approximately 240–350 million people worldwide live with chronic HBV infection.2 Each year, more than 750 000 people die of diseases associated with HBV infection.2 As one of the most common primary malignant carcinomas, the mortality rate for hepatocellular carcinoma (HCC) is one of the highest among all of the cancers.3 The major risk factor for HCC is chronic HBV infection in 55% of cases worldwide.3 Evidence suggests that HBV X protein (HBx) plays a crucial multifunctional role in the regulation of hepatocarcinogenesis.1
There is considerable evidence that microRNAs (miRNAs) are involved in the life cycle and infectivity of HBV.4 In addition, HBV can regulate the expression of host miRNAs, thus providing a beneficial environment for the survival and replication of HBV.4 Researching the complex relationship between HBV and miRNAs may help in the development of effective treatment for the liver diseases associated with HBV infection.3–5
The genes encoding the peroxisome proliferator-activated receptors (PPARs) have been identified as the targets of miR-141.6 The replication levels of HBV were inhibited via the downregulation of PPAR gene expression using small interfering RNA (siRNA);6 and similar results were found for miR-520a by our previous unpublished research. In addition, our previous unpublished research demonstrated that miR-520a siRNA or mimics of miR-520a could mediate the suppression of HBV replication by obstructing the promoter functions of HBV. These previous findings were consistent with research that indicated that PPARs can regulate the gene expression of HBV by activating the promoter regulatory areas of HBV genes.7 This previous study demonstrated that miR-520a could inhibit the replication of HBV by inactivating the promoter regulatory areas of HBV genes by the knock-down of the expression of the PPAR gene.7 There is considerable interest in exploring the molecular mechanisms associated with the relationship between the host genes and HBV replication because this might lead to the development of new therapeutic strategies against HBV replication.7
Glabridin (GLA) is a new antitumour drug that can suppress inflammation, proliferation and oxidization in cancer cells.8 MiR-520a promoted the antitumour activities of GLA by inhibiting the nuclear factor (NF)-κB/interleukin (IL)-6/signal transducer and activator of transcription (STAT)-3 signalling pathway.9 In summary, GLA could upregulate the expression of miR-520a, which targets the 3ʹ untranslated region (UTR) of the NF-κB/RELA mRNA, thus inhibiting the production and function of NF-Κb.9
Previous preliminary research by the current authors demonstrated that miR-520a suppressed HBV replication in the HBV-replicating human HCC cell line HepG2.2.15, but the mechanism remains unknown.10 The serine/threonine kinase 1 (AKT) gene was predicted to be the target of miR-520a with online software by our team. HBx may play a crucial role in the replication of HBV.11 HBx could inhibit the apoptosis of hepatocytes and the replication of HBV by activating the AKT gene.12 HBV replication is regulated by inhibiting the activity of the transcription factor hepatocyte nuclear factor 4α by AKT.13 HBx activation of AKT may contribute to the tumorigenesis of HCC by promoting the replication of HBV.12 AKT might be an important therapeutic target for treatment of HBV replication and thus the prevention of HBV-associated HCC.
This current study aimed to investigate whether the mechanism by which miR-520a suppresses the replication of HBV involves the regulation of the AKT gene.
Materials and methods
Cell culture
The human hepatoblastoma cell line HepG2 was obtained from American Type Culture Collection (Manassas, VA, USA) and cultured in Dulbecco's modified Eagle medium (Gibco BRL, Grand Island, NJ, USA) with 380 µg/ml G418 (Thermo Fisher Scientific, Waltham, MA, USA), 100 µg/ml penicillin and 100 µg/ml streptomycin antibiotics (Thermo Fisher Scientific) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Thermo Fisher Scientific) in a humidified incubator (SANYO, Osaka, Japan) containing 5% CO2 at 37 °C.
Two copies of the HBV genome were stably transfected into HepG2 cells as described below, which then became HBV-replicating HepG2.2.15 cells. These cells were used for the subsequent experiments. One day before transfection, 5 × 105 cells were seeded per well in 500 µl of growth medium without antibiotics to attain 90–95% confluence at the time of transfection. For each transfection sample, DNA-Lipofectamine® 3000 (Thermo Fisher Scientific) complexes were prepared as follows: dilute 0.8 µg DNA in 50 µl Opti-MEM I (Thermo Fisher Scientific), then mix gently and let it stand for 6 h at room temperature. Incubate the DNA-Lipofectamine® 3000 complexes with the cells at 37 °C in a humidified incubator with 5% CO2 for 48 h until the cells were ready to assay.
Luciferase reporter assays
Online software TargetScanHuman (version 7.2) was used for miRNA-target prediction.14 The software predicted that miR-520a would target the 3ʹUTR of the AKT mRNA and this was amplified using polymerase chain reaction (PCR) from genomic DNA from HepG2.2.15 cells. The 3ʹUTR of the AKT mRNA was then cloned into pmirGLO-NULL plasmid (Life Technologies, Grand Island, NJ, USA). Then the pmirGLO-AKT 3′UTR construct plasmid and its negative control (pmirGLO-NULL vector) plasmid were transfected into HepG2.2.15 cells using Lipofectamine™ 2000 following the manufacturer’s instructions (Life Technologies). A dual-luciferase reporter assay system (Promega Corporation, Madison, WI, USA) was used to analyse the activity of luciferase.
Quantitative reverse transcription–polymerase chain reaction
Total RNA was extracted from 6 × 107 HepG2.2.15 cells using TRIzol® reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. Total RNA (5 µg) was reverse transcribed to cDNA using a SMART-cDNA synthesis kit (Clontech Laboratories, Mountainview, CA, USA). Reverse transcription–polymerase chain reaction (RT–PCR) assays were performed using an Applied Biosystems® 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the internal control to normalize the data. The intrinsic miR-520a was determined using the quantitative RT–PCR method and RNU6B was used to normalize the data. Quantitative RT–PCR was performed with SYBR® Green Master PCR Mix (Merck, Darmstadt, Germany). The primer sequences used were as follows: AKT, forward 5ʹ-ATGAGCGACGTGGCTATTGT-3ʹ, reverse 5ʹ-ACAATAGCCACGTcGCTCAT-3ʹ; GAPDH, forward 5ʹ-TGAAGGTGCCATCATTCTTG-3ʹ, reverse 5ʹ-CAAGAATGATGGCACCTTCA-3ʹ; miR-520a, forward 5ʹ-CUCAGGCUGUGACCCUCCAGAGGGAAGUACUUUCUGUUGUCUG-3ʹ, reverse 5ʹ-GAGUUUGGCUUUGUCAGGUUUCCCUUCGUGAAAGAAAAGAGAG-3ʹ; and RNU6B, forward 5ʹ-CTCGCTTCGGCAGCACA-3ʹ, reverse 5ʹ-TGTGCTGCCGAAGCGAG-3ʹ. These primers were purchased from Thermo Fisher Scientific (Shanghai, China).
The thermocycling conditions for the quantitative PCR were as follows: denaturation at 95°C for 30 s, followed by 40 cycles of denaturation at 94°C for 30 s and annealing at 60°C for 30 s. The 2–ΔΔCT method was used to calculate relative expression and quantify the results.
The cycling programme for the RT–PCR involved preliminary denaturation at 95°C for 1 min, followed by 40 cycles of denaturation at 95°C for 6 s, annealing at 58°C for 30 s, and elongation at 72°C for 45 s, followed by a final elongation step at 72°C for 7 min. The PCR products were separated on 1% agarose gels and visualized using ethidium bromide staining and ultraviolet light. The sizes (in base pairs [bp]) of the PCR amplification products were 20 bp for AKT, 20 bp for GAPDH, 43 bp for miR-520a, and 16 bp for U6.
Western blotting
Total protein was extracted from 7 × 1011 cells using RIPA buffer (50 mM Tris-HCl pH 8.0, 1 mM ethylenediaminetetra-acetic acid, 500 mM NaCl, 0.1% (w/v) sodium dodecyl sulphate, 1% (v/v) Triton X-100; Pierce Biotechnology, Rockford, IL, USA), containing protease inhibitors (Roche Diagnostics, Basel, Switzerland), then boiled for 5 min. The protein concentration was quantified using the BCA method (Pierce Biotechnology). Proteins (50 µg) were separated by sodium dodecyl sulphate–polyacrylamide gel electrophoresis, and then transferred to a 0.45-µm polyvinylidene fluoride membrane (Roche Diagnostics), which was blocked with 1 × Tris-buffered saline-Tween-20 (0.05%; pH 7.4; TBST) containing 5% nonfat dry milk and agitated for 1 h at room temperature.15 The membranes were incubated with primary antibodies to β-actin (1:2000 dilution; CST 4970; Cell Signaling Technology, Inc., Danvers, MA, USA) and AKT (SC-5298; 1:3000 dilution; Santa Cruz Biotechnology, Santa Cruz, CA, USA) at 4°C overnight. Then they were washed with 1 × TBST three times at 37°C. The horseradish peroxidase (HRP)-conjugated secondary antibodies were goat anti-rabbit IgG-HRP (SC-2004; 1:5000 dilution; Santa Cruz Biotechnology) or goat anti-mouse IgG-HRP (SC-2005; 1:5000 dilution; Santa Cruz Biotechnology). The membranes were incubated with the HRP-conjugated secondary antibodies for 1 h at room temperature. Then they were washed with 1 × TBST three times at 37°C. The protein bands were detected and visualized using an enhanced chemiluminescence kit (GE Healthcare, Chicago, IL, USA).
Determination of the replication of HBV
After the HepG2.2.15 cells had been transfected with either the inhibitor (5ʹ-AAAGUGCUUCCCUUUGGACUGU-3′) or the mimic (5′-CUCAGGCUGUGACCCUCCAGAGGGAAGUACUUUCUGUUGUCUG-3′) of miR-520a, the supernatant above the cultured HepG2.2.15 cells was collected at 12, 24, 36 and 48 h. In addition, the sequence of the negative control used was as follows: 5′-ACGUGACACGUUCGGAGAAUU-3′. The levels of the surface antigen of the hepatitis B virus (HBsAg) and hepatitis B e-antigen (HBeAg) in the supernatant were detected using diagnostic kits (Shanghai Kehua Biotech Company, Shanghai, China). At the same time-points, intracellular HBV DNA was extracted from virus core particles as described previously.8,9 In brief, the nuclei and cellular debris were removed from HepG2.2.15 cell lysates and then the supernatants were digested with micrococcal nuclease at 37°C for 2 h. Viral DNA was separated and then used in the RT–PCR method described above.9
The levels of HBsAg and HBeAg in the supernatant and intracellular HBV-DNA were also detected in HepG2.2.15 cells treated with the control siRNA (sc-37077) and siRNA of the AKT gene (sc-38910), which were obtained from Santa Cruz Biotechnology.
BrdU incorporation assay and mitotic index analysis
To examine effect of miR-520a on HepG2.2.15 cell proliferation, cells transfected with the mimic or inhibitor of miR-520a and its negative control were plated into 96-well plates at 2 × 103 cells/well and cultured at 37°C in a humidified atmosphere containing 5% CO2. Cells were synchronized using the block method with double thymidine (10 µM) for 4 h. After synchronizing for 16 h, cells were released and then collected or fixed for assays. DNA synthesis or mitotic entry analysis were performed using the 5-bromo-2-deoxyuidine (BrdU) incorporation assay (BD Biosciences, San Jose, CA, USA).16 Cells were counted every day at fixed time-points. Cells that were BrdU-positive were scored using fluorescence microscope (IX71; Olympus Corporation, Tokyo, Japan). Data from three independent experiments were analysed.16
Mitotic events were determined with DNA staining and time-lapse videomicroscopy (Medium Throughput Long Term Time Lapse IncuCyte; Solent Scientific, Segensworth, UK). After synchronizing the cells as described above, the real-time images of cells were obtained at 10-min intervals with a fluorescence microscope at 200 × magnification (IX71; Olympus Corporation). Mitotic events in cells were detected through change of morphology. Mitotic cells were counted based on DNA condensation and nuclear morphology using a DNA dye (Hoechst 33258; Shimadzu Corporation, Kyoto, Japan).
Detection of apoptotic cells
To examine effect of miR-520a on HepG2.2.15 cell apoptosis, cells transfected with the mimic or inhibitor of miR-520a and its negative control were plated into 6-well plates at 3 × 106 cells/well and cultured at 37°C in a humidified atmosphere containing 5% CO2. Using an Annexin V-FITC Apoptosis Detection Kit (BD Biosciences), the cells were subjected to flow cytometric analysis at 12, 24, 36 and 48 h after being transfected with the inhibitor or mimic of miR-520a using a FACScan flow cytometer (BD Biosciences) and Flowjo software (BD Biosciences).17
Statistical analyses
All statistical analyses were performed using the SPSS® statistical package, version 18.0 (SPSS Inc., Chicago, IL, USA) for Windows®. The normally distributed continuous variables are presented as mean ± SD. Between-group comparisons were undertaken using Student’s t-test or analysis of variance (ANOVA). Data that were not normally distributed were analysed using the Kruskal–Wallis ANOVA method. A P-value < 0.05 was considered statistically significant.
Results
MiR-520a was identified as possibly targeting the 3′ UTR of AKT mRNA. Whether miR-520a bound to the 3′ UTR of AKT mRNA was determined using luciferase reporter assays. The relative activity of 3′ UTR AKT luciferase was significantly reduced in HepG2.2.15 transfected with miR-520a compared with the control group (Figure 1A) (P = 0.001). These data confirmed that AKT mRNAs were bound to miR-520a. Western blot analysis was then undertaken to identify whether miR-520a regulates the expression of the AKT gene in HepG2.2.15 cells (Figure 1B). The results indicated that miR-520a decreased the levels of AKT protein in HepG2.2.15 cells.
Figure 1.
Results of luciferase reporter assays to determine if miR-520a bound to the 3ʹ untranslated regions (UTRs) of the AKT gene in HepG2.2.15 cells (A). The data are presented as mean ± SD from three independent experiments; Student’s t-test; **P < 0.01; pmirGLO-NULL, negative control vector; pmirGLO-AKT 3ʹUTR, vector cloned with the 3′UTR of AKT mRNA. Western blot analysis was undertaken to determine the effect of miR-520a on the levels of AKT protein in HepG2.2.15 cells (B). NC, negative control. The loading control for the Western blot was β-actin.
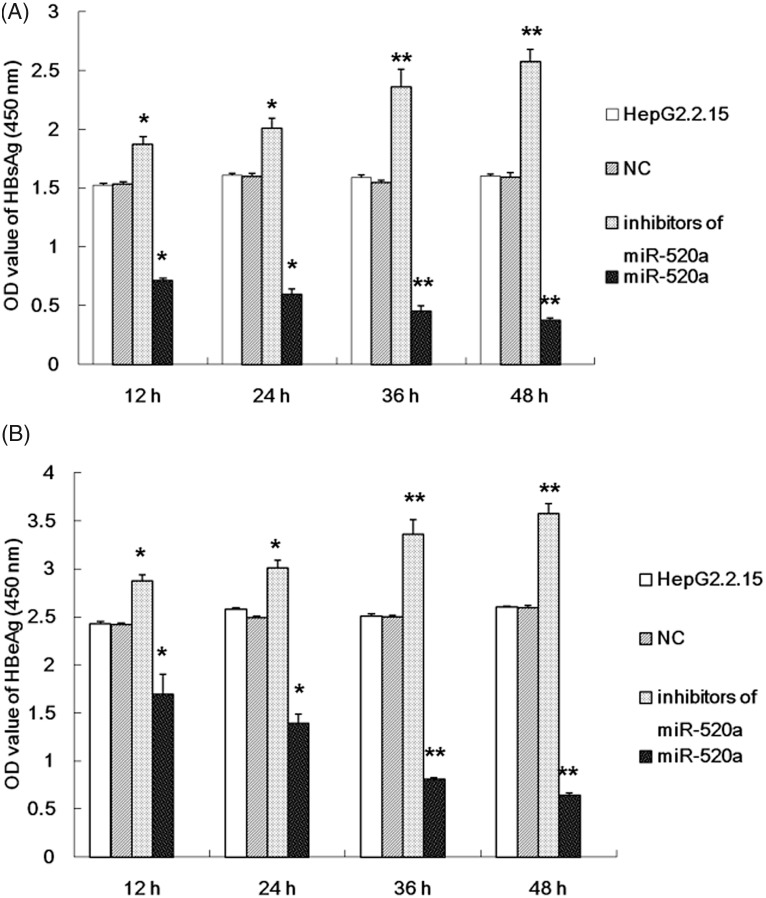
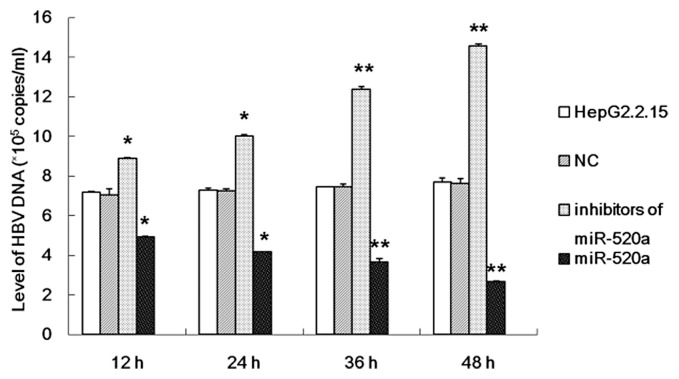
The possible role of miR-520a in the replication of HBV was investigated using an inhibitor or a mimic of miR-520a transfected into HepG2.2.15 cells. The HBsAg and HBeAg levels in the supernatants from HepG2.2.15 cells were examined at different time-points. The results demonstrated that miR-520a significantly suppressed the replication of HBV as demonstrated by a significant reduction in the levels of HBsAg and HBeAg compared with the untreated HepG2.2.15 cells (Figure 2). Furthermore, miR-520a inhibited the replicative ability of HBV in a time-dependent manner as determined by measuring HBV DNA load (Figure 3). The inhibitor of miR-520a was able to effectively activate HBV replication, resulting in a significant increase in the levels of HBsAg and HBeAg (Figure 2) and HBV DNA (Figure 3) compared with the untreated HepG2.2.15 cells in a time-dependent manner (P < 0.05 for all comparisons).
Figure 2.
The results of investigations into the effect of miR-520a on the levels of the surface antigen of the hepatitis B virus (HBsAg) and hepatitis B e-antigen (HBeAg) in the supernatant from HepG2.2.15 cells at different time-points after transfection with a miR-520a mimic or a miR-520a inhibitor. The data are presented as mean ± SD from three independent experiments; Student’s t-test; *P < 0.05, **P < 0.01 compared with negative control (NC) group.
Figure 3.
The results of investigations into the effect of miR-520a on the levels of hepatitis B virus (HBV) DNA in HepG2.2.15 cells at different time-points after transfection with a miR-520a mimic or a miR-520a inhibitor. The data are presented as mean ± SD from three independent experiments; Student’s t-test; *P < 0.05, **P < 0.01 compared with the negative control (NC) group.
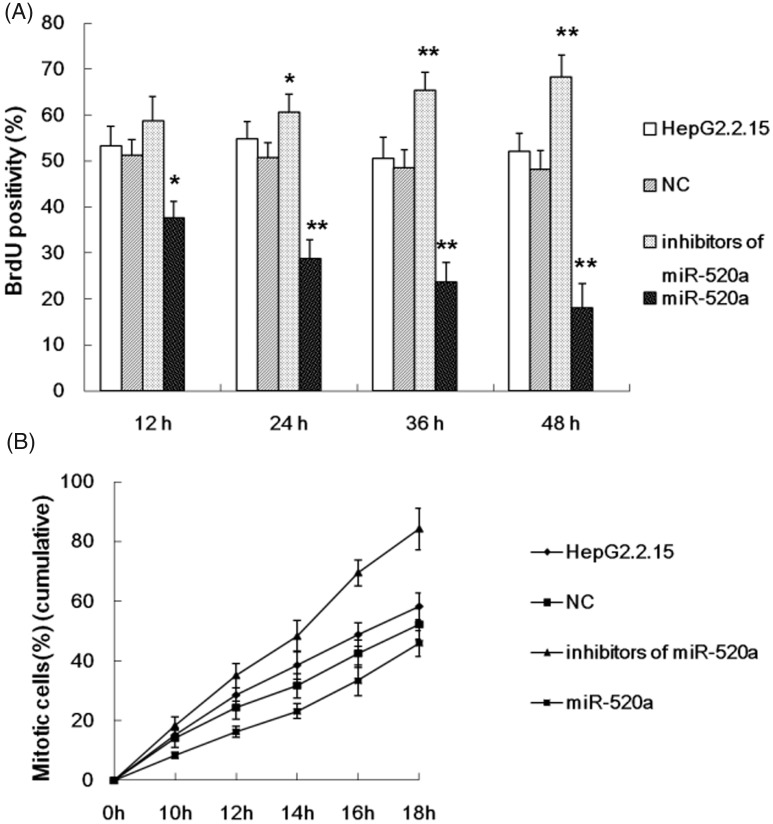
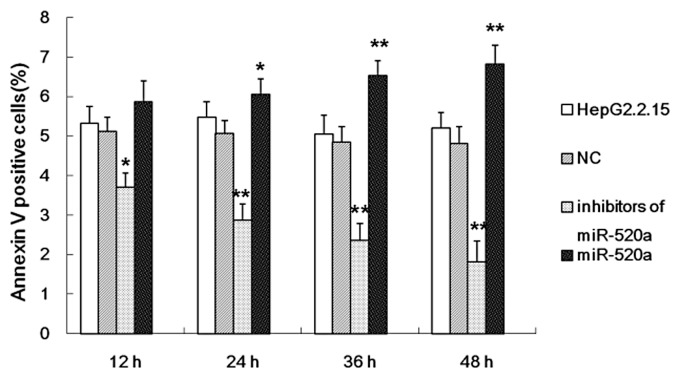
A BrdU incorporation assay and flow cytometry were used to study cell proliferation and apoptosis of HepG2.2.15 cells in response to an miR-520a mimic and inhibitor. The results demonstrated that miR-520a significantly suppressed HepG2.2.15 cell proliferation (Figure 4A) and mitosis entry (Figure 4B) compared with the untreated HepG2.2.15 cells. The mimic of miR-520a induced apoptosis of HepG2.2.15 cells in a time-dependent manner (Figure 5). The inhibitor of miR-520a promoted HepG2.2.15 cell proliferation (Figure 4A) and mitosis entry (Figure 4B) in a time-dependent manner; and repressed apoptosis of HepG2.2.15 cells (Figure 5).
Figure 4.
The results of investigations into the effect of miR-520a on HepG2.2.15 cell proliferation (A) and mitosis entry (B) at different time-points after transfection with a miR-520a mimic or a miR-520a inhibitor. The data are presented as mean ± SD from three independent experiments; Student’s t-test; *P < 0.05, **P < 0.01 compared with the negative control (NC) group.
Figure 5.
The results of investigations into the effect of miR-520a on HepG2.2.15 apoptosis as assessed using the flow cytometric analysis of annexin V-positive cells at different time-points after transfection with a miR-520a mimic or a miR-520a inhibitor. The data are presented as mean ± SD from three independent experiments; Student’s t-test; *P < 0.05, **P < 0.01 compared with the negative control (NC) group.
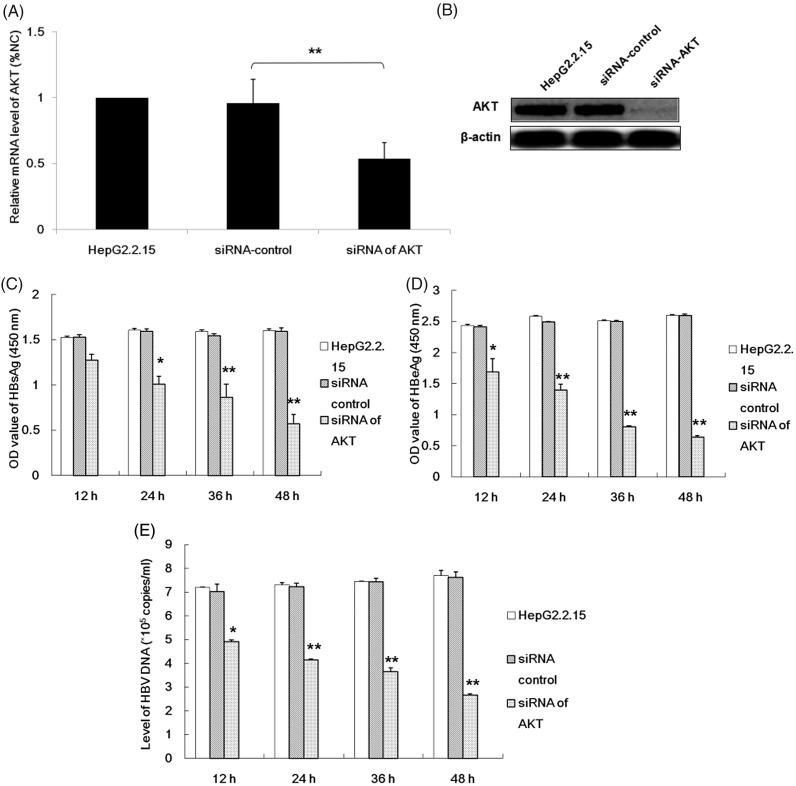
In order to investigate the role played by the AKT gene in the regulation of HBV replication in HepG2.2.15 cells, siRNAs of the AKT gene were transfected into HepG2.2.15 cells and the levels of AKT mRNA and protein were determined using RT–PCR and Western blot analysis. These data confirmed that a specific siRNA for AKT could significantly reduce AKT mRNA and protein levels compared with the untreated HepG2.2.15 cells (Figures 6A and 6B). Analysis of the levels of the HBsAg and HBeAg in cell culture supernatants from transfected HepG2.2.15 cells, as well as analysis of HBV DNA loads, demonstrated that decreased AKT protein levels in HepG2.2.15 cells were associated with a decrease in HBV replication in a time-dependent manner (Figures 6C–6E).
Figure 6.
The effects of silencing AKT gene expression using small interfering RNA (siRNA) on hepatitis B replication in HepG2.2.15 cells. Initially, the effect of siRNA on AKT gene expression was determined by measuring the levels of AKT mRNA (A) and protein (B). Glyceraldehyde 3-phosphate dehydrogenase and β-actin were used as internal controls, respectively. Relative levels were normalized as a percentage of the negative siRNA control. The effect of silencing AKT gene expression on hepatitis B virus (HBV) replication was then determined by measuring the levels of the surface antigen of the hepatitis B virus (HBsAg) and hepatitis B e-antigen (HBeAg) in the cell culture supernatant from HepG2.2.15 cells and the HBV DNA load at different time-points after transfection. The data are presented as mean ± SD from three independent experiments; Student’s t-test; *P < 0.05, **P < 0.01 compared with the HepG2.2.15 cells treated with siRNA-control.
Discussion
Hepatitis B virus is closely associated with the development and progression of HCC.3 As a transactivator, HBx regulates the activity of nuclear factor NF-κB and transcription factor AP-2.18 In addition, HBx affects the biological functions of non-coding RNAs, such as long ncRNAs and microRNAs (e.g. miRNA-205).18 HBx is also associated with epigenetic modification, such as acetylation and methylation, while it interacts with various signal transduction pathways, including activator of transcription NF-κB or signal transducer pathways Wnt/β-catenin, and protein kinase B/Akt.18 Furthermore, HBx influences cell survival by changing the balance between cell growth and apoptosis, thereby promoting the progression of HCC.19–21 HBV-related hepatocarcinogenesis is associated with HBV replication and chronic infection of HBV.22,23 Moreover, the evidence that HBV replication induces HCC is lacking.
Alpha-fetoprotein (AFP) activates the phosphatidylinositol 3-kinase/protein kinase B (PI3K/AKT) signalling pathway and then up-regulates the expression of certain oncogenes, thus promoting the proliferation of HCC cells.24 Moreover, HBx increases the levels of AFP and AFP receptors (AFPR) in the normal liver cell line L-02.25 The expression of Src is stimulated by the AFPR signalling pathway.25 Inhibitors of PI3K, such as GDC0941 and Ly294002, effectively suppressed the AFPR-mediated over-expression of Src in AFPR-positive hepatoma cell lines.25 HBx induced the upregulation of Src associated with AFP and AFPR in HCC cell lines and normal liver specimens.25 The crucial role of HBV in the carcinogenesis of HCC is related to AFP and AFPR.26 Moreover, the serum levels of miR-181b are associated with the progression of chronic hepatitis B, especially the serum level of HBV DNA or HBV DNA levels in hepatocytes.26
A previous study demonstrated that HBx could activate the autophagy related to the PI3K-Akt-mTOR pathway in HepG2 and HepG2.2.15 cells.27 In the present study, the miRNAs that may regulate AKT gene expression in the HepG2.2.15 cell lines were predicted using TargetScanHuman (version 7.2) software and miR-520a was selected for the current experiments. The results of the present study showed that miR-520a markedly reduced the luciferase activities of the AKT 3′-UTR in HepG2.2.15 cells, which confirmed that AKT mRNA was the direct target of miR-520a.
MicroRNAs are short non-coding RNAs that regulate gene expression in animals, which influence gene expression by mediating posttranscriptional gene silencing by suppressing the mRNA translation of its target gene or by degrading mRNAs according to the mRNA sequence.28 Recent research has demonstrated that miRNAs take part in a variety of cellular processes, including differentiation, development, proliferation and tumorigenesis.29 A variety of expression models of miRNAs have been identified in different carcinomas,30 but evidence of the role of miRNAs in HBV-associated HCC is limited.
This current study investigated the role of miR-520a in the regulation of HBV replication. The present findings showed that miR-520a could directly regulate the expression of the AKT gene. In order to confirm the role of miR-520a in inhibiting HBV replication, a mimic or an inhibitor of miR-520a were transfected into HepG2.2.15 cells. The current results demonstrated that the levels of HBsAg and HBeAg and the replicative ability of HBV were significantly inhibited by miR-520a in a time-dependent manner. Furthermore, miRNA-520a decreased cell proliferation and mitosis entry and increased apoptosis of HepG2.2.15 cells. These current findings suggest that the AKT gene plays a key role in HBV replication. This was confirmed by experiments that demonstrated that AKT gene expression in HepG2.2.15 cells was suppressed by a specific siRNA, which also inhibited HBV replication.
In conclusion, these current results demonstrated that miRNA-520a reduced the expression of the AKT gene, which suppressed HBV transcription and replication. These findings suggest that miRNA-520a may be a novel target for anti-viral therapy of HBV because miRNA-520a suppressed the replicative ability of HBV and cell survival of HepG2.2.15 associated with the AKT signalling pathway.
Declaration of conflicting interests
The authors declare that there are no conflicts of interest.
Funding
This work was supported by grants from the National Natural Science Foundation of China (no. 30600524 and no. 81341067), the National Natural Science Foundation of Guangdong Province, China (no. 2017A030313510), Introduction of Talent Fund of Guangdong Second Provincial General Hospital (no. YY2016-006), and Capital Clinical Featured Applied Research and Results Promotion projects (no. Z161100000516141). The study sponsors had no involvement in the work.
References
- 1.Kew MC. Epidemiology of chronic hepatitis B virus infection, hepatocellular carcinoma, and hepatitis B virus-induced hepatocellular carcinoma. Pathol Biol (Paris) 2010; 58: 273–277. [DOI] [PubMed] [Google Scholar]
- 2.Omata M, Cheng AL, Kokudo N, et al. Asia-Pacific clinical practice guidelines on the management of hepatocellular carcinoma: a 2017 update. Hepatol Int 2017; 11: 317–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petruzziello A. Epidemiology of Hepatitis B Virus (HBV) and Hepatitis C Virus (HCV) Related Hepatocellular Carcinoma. Open Virol J 2018; 12: 26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet 2011; 12: 861–874. [DOI] [PubMed] [Google Scholar]
- 5.Chen Y, Verfaillie CM. MicroRNAs: the fine modulators of liver development and function. Liver Int 2014; 34: 976–990. [DOI] [PubMed] [Google Scholar]
- 6.Hu W, Wang X, Ding X, et al. MicroRNA-141 represses HBV replication by targeting PPARA. PLoS One 2012; 7: e34165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giordano S, Columbano A. . MicroRNAs: new tools for diagnosis, prognosis, and therapy in hepatocellular carcinoma? Hepatology 2013; 57: 840–847. [DOI] [PubMed] [Google Scholar]
- 8.Chen CT, Chen YT, Hsieh YH, et al. Glabridin induces apoptosis and cell cycle arrest in oral cancer cells through the JNK1/2 signaling pathway. Environ Toxicol 2018; 33: 679–685. [DOI] [PubMed] [Google Scholar]
- 9.Mu J, Ning S, Wang X, et al. The repressive effect of miR-520a on NF-κB/IL-6/STAT-3 signal involved in the glabridin-induced anti-angiogenesis in human breast cancer cells. RSC Adv 2015; 5: 34257–34264. [Google Scholar]
- 10.Huang J, Yang J, Lei Y, et al. An ANCCA/PRO2000-miR-520a-E2F2 regulatory loop as a driving force for the development of hepatocellular carcinoma. Oncogenesis 2016; 5: e229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hensel KO, Cantner F, Bangert F, et al. Episomal HBV persistence within transcribed host nuclear chromatin compartments involves HBx. Epigenetics Chromatin 2018; 11: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee WP, Lan KH, Li CP, et al. Oncogenic circuit constituted by Ser31-HBx and Akt increases risks of chronic hepatitis and hepatocellular carcinoma. Biochim Biophys Acta 2016; 1862: 837–849. [DOI] [PubMed] [Google Scholar]
- 13.Tao NN, Gong R, Chen X, et al. Interleukin-35 stimulates hepatitis B virus transcription and replication by targeting transcription factor HNF4α. J Gen Virol 2018; 99: 645–654. [DOI] [PubMed] [Google Scholar]
- 14.Whitehead Institute for Biomedical Research. To predict biological targets of miRNAs by searching for the presence of conserved 8mer, 7mer, and 6mer sites that match the seed region of each miRNA, http://www.targetscan.org/vert_72/ (2015, accessed March 2018).
- 15.Mao Y, Wu S, Zhao R, et al. MiR-205 promotes proliferation, migration and invasion of nasopharyngeal carcinoma cells by activation of AKT signalling. J Int Med Res 2016; 44: 231–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J, Feng W, Chen L, et al. Downregulation of SMC1A inhibits growth and increases apoptosis and chemosensitivity of colorectal cancer cells. J Int Med Res 2016; 44: 67–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li B, Chen X, Yang W, et al. Single-walled carbon nanohorn aggregates promotes mitochondrial dysfunction-induced apoptosis in hepatoblastoma cells by targeting SIRT3. Int J Oncol 2018; 53: 1129.–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang XD, Wang Y, Ye LH. Hepatitis B virus X protein accelerates the development of hepatoma. Cancer Biol Med 2014; 11: 182–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiu X, Dong S, Qiao F, et al. HBx-mediated miR-21 upregulation represses tumor-suppressor function of PDCD4 in hepatocellular carcinoma. Oncogene 2013; 32: 3296–3305. [DOI] [PubMed] [Google Scholar]
- 20.Xu X, Fan Z, Kang L, et al. Hepatitis B virus X protein represses miRNA-148a to enhance tumorigenesis. J Clin Invest 2013; 123: 630–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu CS, Yen CJ, Chou RH, et al. Downregulation of microRNA-15b by hepatitis B virus X enhances hepatocellular carcinoma proliferation via fucosyltransferase 2-induced Globo H expression. Int J Cancer 2014; 134: 1638–1647. [DOI] [PubMed] [Google Scholar]
- 22.Zhang T, Zhang J, Cui M, et al. Hepatitis B virus X protein inhibits tumor suppressor miR-205 through inducing hypermethylation of miR-205 promoter to enhance carcinogenesis. Neoplasia 2013; 15: 1282–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lan SH, Wu SY, Zuchini R, et al. Autophagy suppresses tumorigenesis of hepatitis B virus-associated hepatocellular carcinoma through degradation of microRNA-224. Hepatology 2014; 59: 505–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng L, Gong W, Liang P, et al. Effects of AFP-activated PI3K/Akt signaling pathway on cell proliferation of liver cancer. Tumour Biol 2014; 35: 4095–4099. [DOI] [PubMed] [Google Scholar]
- 25.Zhu M, Guo J, Li W, et al. HBx induced AFP receptor expressed to activate PI3K/AKT signal to promote expression of Src in liver cells and hepatoma cells. BMC Cancer 2015; 15: 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu F, Zhou G, Li G, et al. Serum miR-181b is correlated with hepatitis B virus replication and disease progression in chronic hepatitis B patients. Dig Dis Sci 2015; 60: 2346–2352. [DOI] [PubMed] [Google Scholar]
- 27.Wang P, Guo QS, Wang ZW, et al. HBx induces HepG-2 cells autophagy through PI3K/Akt-mTOR pathway. Mol Cell Biochem 2013; 372: 161–168. [DOI] [PubMed] [Google Scholar]
- 28.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 2008; 9: 102–114. [DOI] [PubMed] [Google Scholar]
- 29.Chen CZ. MicroRNAs as oncogenes and tumor suppressors. N Engl J Med 2005; 353: 1768–1771. [DOI] [PubMed] [Google Scholar]
- 30.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature 2005; 435: 834–838. [DOI] [PubMed] [Google Scholar]