Abstract
Background: It is critical to integrate and analyze data from biological, translational, and clinical studies with data from health systems; however, electronic artifacts are stored in thousands of disparate systems that are often unable to readily exchange data.
Objective: To facilitate meaningful data exchange, a model that presents a common understanding of biomedical research concepts and their relationships with health care semantics is required. The Biomedical Research Integrated Domain Group (BRIDG) domain information model fulfills this need. Software systems created from BRIDG have shared meaning “baked in,” enabling interoperability among disparate systems. For nearly 10 years, the Clinical Data Standards Interchange Consortium, the National Cancer Institute, the US Food and Drug Administration, and Health Level 7 International have been key stakeholders in developing BRIDG.
Methods: BRIDG is an open-source Unified Modeling Language–class model developed through use cases and harmonization with other models.
Results: With its 4+ releases, BRIDG includes clinical and now translational research concepts in its Common, Protocol Representation, Study Conduct, Adverse Events, Regulatory, Statistical Analysis, Experiment, Biospecimen, and Molecular Biology subdomains.
Interpretation: The model is a Clinical Data Standards Interchange Consortium, Health Level 7 International, and International Standards Organization standard that has been utilized in national and international standards-based software development projects. It will continue to mature and evolve in the areas of clinical imaging, pathology, ontology, and vocabulary support. BRIDG 4.1.1 and prior releases are freely available at https://bridgmodel.nci.nih.gov.
Keywords: data sharing, data modeling, clinical trials, translational medical research, pharmacogenetics
BACKGROUND AND SIGNIFICANCE
Meaningful exchange of electronic data has long been a challenge in biomedical research. Semantic and syntactic standards promote interoperability, where data can be more readily exchanged between computational systems in unambiguous, meaningful ways. Many standards now exist for health care and research, and it can be difficult to understand how diverse standards from the Clinical Data Interchange Standards Consortium (CDISC), Health Level 7 (HL7), the National Cancer Institute (NCI), and others fit together in a holistic picture of biomedicine.
In 2004, to harmonize global CDISC standards among one another and link with health care concepts, CDISC initiated the Biomedical Research Integrated Domain Group (BRIDG) model. Shortly thereafter, the NCI, the Food and Drug Administration (FDA), and HL7’s Regulated Clinical Research Information Management Technical Committee joined this collaborative effort. The BRIDG model is now a CDISC, HL7, and International Standards Organization (ISO) standard domain information model (DIM). It provides a Unified Modeling Language–class diagram representation of the protocol-driven biomedical research domain, “bridging” biomedicine and health care concepts. BRIDG’s biomedical semantics include molecular biology, biospecimens, experiments, study conduct, protocols, and other key concepts. Its health care concepts range across patients, health care sites, providers, diagnoses, procedures, observations, drugs, devices, etc., where each of these concepts can also participate in or be part of clinical research. BRIDG includes semantics from CDISC standards, including those required for new submissions to the FDA1,2 and Japan’s Pharmaceuticals and Medical Device Agency3,4 beginning in 2016 and from HL7.
DIMs represent semantics of an entire domain using language that subject matter experts (SMEs) understand. Applications, programming interfaces, enterprise applications, and other electronic systems can be developed using DIMs. Even though those systems can be implemented using different programming languages, all systems using a DIM share the same semantics. Common semantics provides a critical foundation for software interoperability. To facilitate meaningful health research data exchange, a DIM that presents a common understanding of the concepts and relationships among biomedical research domains and key health care semantics and datatypes is required for shared understanding of health-related research. BRIDG fulfills this need, such that models, software, and systems created using BRIDG have shared meaning “baked in” to support interoperability among such disparate systems.
MATERIALS AND METHODS
UML model development
BRIDG is developed through stakeholders’ use cases and harmonization with other models. BRIDG analysts work directly with SMEs to harmonize the semantics of stakeholders’ projects with concepts in BRIDG. Use cases, processes, and/or external models are analyzed, mapping and design artifacts such as spreadsheets listing data elements or physical models are created, and changes are made in BRIDG. Where gaps exist, concepts from the external model are added to BRIDG as new classes and/or attributes. Where the same or similar concepts are present in both models, their nontechnical descriptions, data types, and relationships are reviewed and harmonized to BRIDG. This activity may include adding new attributes to BRIDG classes, updating BRIDG descriptions to improve definition clarity, etc. All modeling is performed in Enterprise Architect by Sparx Systems.
Data Types
BRIDG attributes use data types common to the health care messaging standard HL7 Abstract Data Types Release 2. These are complex, combining integer, Boolean, and other data types to express the increasingly complicated semantics of biomedical research.
Model representations
Three model representations are available within BRIDG: (1) the UML representation, which includes packages for each subdomain; (2) the HL7 reference information model (RIM) representation in Visio files as HL7 restricted message information models, an HL7 v3 graphical style; and (3) a Web Ontology Language (OWL) representation that can be used for semantic validation and inferencing. The UML representation is intended for domain experts, analysts, and architects. The RIM is intended for HL7 v3 modelers who can leverage BRIDG-to-RIM mappings to build v3 messages. The OWL representation is intended for analysts. The HL7 RIM and OWL models are not yet available for BRIDG 4+, but are available for release 3.2.
BRIDG modeling group
The BRIDG analysts and modelers are referred to as the Modeling Group. They are responsible for developing and maintaining the BRIDG model, working with all stakeholders during harmonization of project semantics, developing all BRIDG-related documentation, and publishing BRIDG releases. The Modeling Group also takes the model through the HL7/CDISC/ISO balloting. Modeling Group tasks are governed by the BRIDG Steering Committee.
HL7 BRIDG working group
Members of the BRIDG working group represent various stakeholders of BRIDG, the BRIDG Modeling Group, and other community members who are interested in contributing to the development of the DIM.
BRIDG Steering Committee
The Steering Committee includes 1 or 2 members from each stakeholder group (CDISC, NCI, FDA, and HL7), 1 member from ISO, and 2 to 4 at large academic centers and national government agencies. This group provides input on priorities for BRIDG development and maintenance and outreach opportunities within the biomedical field, and gives feedback on modeling efforts.
Community comment period
BRIDG is balloted or vetted through CDISC, HL7, and ISO standards development processes. BRIDG releases are made available on the BRIDG website for comment during a specified interval of time through CDISC, HL7, and ISO. Formal balloting through these stakeholder groups is put in place to encourage the community to provide feedback and comments on the BRIDG model. Any member of the public can create an account in the CDISC SharePoint-based information management site and mark up candidate release documents with comments. Comments are then compiled and reviewed, and changes are made to the model where persuasive. The final model, its documentation, and all comments (along with their disposition/resolution) are made available to the public.
RESULTS
BRIDG model subdomains
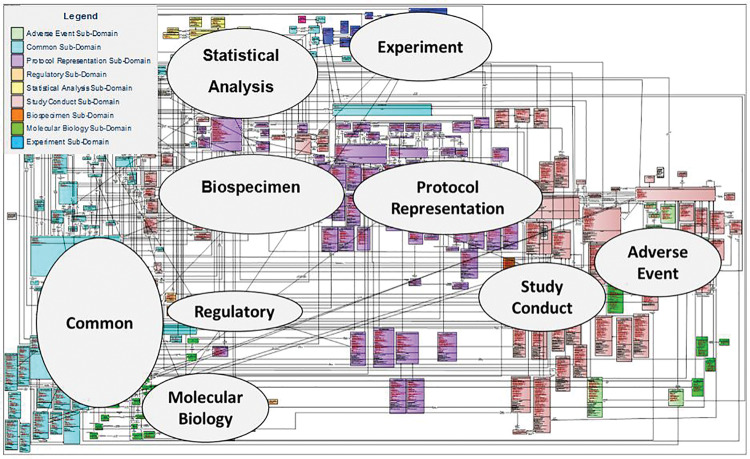
BRIDG concepts are organized into UML classes, attributes, and relationships(Figure 1 ). For a detailed description of how to interpret these UML artifacts, please see the Supplemental Materials and BRIDG User Guide, part of every BRIDG release package. The BRIDG UML representation contains 9 subdomain packages for exploring subsets of the model. Classes are color-coded to show which subdomain each belongs to. The subdomains are Common, Protocol Representation, Study Conduct, Adverse Events, Regulatory, Statistical Analysis, Experiment, Biospecimen, and Molecular Biology (Figure 2 ). In addition to being represented in the BRIDG model itself, each subdomain has its own view. Views show the classes for a given subdomain plus classes from any other subdomain where direct relationships exist. These views provide more focused “slices” of BRIDG while retaining greater semantic context.
Figure 1.
BRIDG’s common subdomain semantics. Example objects, attributes, and interobject relationships from the UML perspective of the common BRIDG subdomain are shown.
Figure 2.
The scope of BRIDG 4.0’s domains and classes. Classes for Protocol Representation (purple), Study Conduct (pink), Adverse Events (lime green), Regulatory (light orange), Statistical Analysis (yellow), Experiment (dark blue), Biospecimen (dark orange), Molecular Biology (dark green), and Common (light blue) domains are shown. For more details on classes within these domains, see the online model.
Common
This subdomain represents concepts and semantics shared across different types of protocol-driven research. Common semantic representations include people (eg, Subject or HealthcareProvider), organizations (eg, Organization or BiologicEntityGroup), places (eg, ServiceDeliveryLocation or TreatingSite), and materials (eg, Biologic or Cosmetic). The Common subdomain was developed through the use cases and various harmonization projects described below. As a consequence of their general nature, Common concepts are referenced as linked classes in the views of all other BRIDG subdomains.
Protocol representation
This subdomain represents concepts involved in the planning and design of a clinical research protocol. It focuses on the characteristics of a study and its activities as defined within study protocol(s) (eg, StudyObjective, StudyOutcomeMeasure, StudyActivity, or DefinedInclusionCriteria). It links these activities to study arms and epochs. The subdomain provides semantics for protocol versioning and amendments (eg, StudyProtocolDocumentVersion and AmendmentChangeSummaryVersion). Use cases for this subdomain came from various NCI, CDISC, and HL7 projects, such as CDISC’s Trial Design and Protocol Representation, NCI’s Clinical Participant Registry (C3PR,5 Patient Study Calendar6) and Clinical Trials Reporting Program, and HL7’s Clinical Trials Registration and Results (CTR&R) message. Concepts within the Protocol Representation subdomain are linked predominantly to those from the Common and Study Conduct subdomains, with additional touchpoints to the Regulatory and Biospecimen subdomains.
Study conduct
This subdomain represents the concepts of executing a research protocol. Semantics include personnel (eg, StudySiteInvestigator or StudySiteResearchCoordinator), funding sources (eg, GovernmentFunding), resources (eg, StudySiteResource or MaterialResource), locations (eg, StudySite or Laboratory), a range of activities (eg, PerformedSpecimenCollection, ScheduledSubstanceAdministration, or PerformedSubjectMilestone), and results (eg, ReferenceResult, PerformedClinicalResult or PerformedDiagnosis). Many Study Conduct classes have relationships with concepts from the Common and Protocol Representation classes. Additional relationships also exist between activities performed on biological material concepts in Study Conduct and the biomaterials included within the Biospecimen subdomain.
Adverse events
This subdomain represents safety-related activities, such as detection (CausalAssessment), evaluation (PerformedProductInvestigation or EvaluatedResultRelationship), and follow-up reporting (SafetyReportVersion) of adverse events (AEs) or serious AEs (AdverseEventSeriousness). These activities can involve people and materials such as drugs, devices, or biologics from the Common subdomain. AE concepts are also related to the procedures and observations that occur during the course of biomedical research or after a research protocol has been completed, and those relationships to Study Conduct semantics are also captured in the AE view.
Regulatory
This subdomain represents the creation, submission, and review of protocol artifacts to regulatory authorities. As such, Regulatory concepts include the activities (eg, Submission or RegulatoryAssessment), documents (eg, RegulatoryApplication or RelevantRegulation), and organizations (eg, RegulatoryAuthority or RegulatoryApplicationSponsor) involved with this subdomain, which was developed with an early version of HL7/FDA–regulated product submission message specification requirements (see http://www.hl7.org/implement/standards/rim.cfm). Regulatory concepts are related to study protocol versions from the Protocol Representation subdomain, as well as documents, materials, people, and organizations from the Common subdomain.
Statistical analysis
This subdomain represents concepts that describe the planning and performance of the statistical analysis of data collected during execution of the protocol. It includes semantics on the analysis plan, its considerations (GeneralStatisticalConsiderations or SampleSizeConsideration), its versions, and modifications (StatisticalAnalysisPlanModificationSummary). Concepts for the data and safety monitoring committee (DataSafetyCommitteeCharterVersion) are also captured. All the content for this subdomain came from the CDISC Analysis Data Model7 (ADaM) standard’s community volunteer SMEs. The Statistical Analysis subdomain’s concepts are related to documents and oversight committee semantics from the Common subdomain, as well as study protocol and objectives from the Protocol Representation subdomain.
Experiment
This subdomain represents concepts related to the design, planning, resourcing, and execution of biomedical experiments. These include semantics for the experiment (Experiment) and its purpose, the devices and parameters required to carry out the experiment (eg, a hot water bath set to 42°C), variables that can be manipulated by an experimentalist (eg, a gene knocked into a mouse as ExperimentalFactor and ExperimentalFactorValue), and subsequent capture of in vitro, in vivo, and physicochemical characterizations for data collection. The semantics of the Experiment subdomain were derived from NCI’s Life Sciences Domain Analysis Model (LS DAM8) and harmonized within the BRIDG 4.0 release. Experiment classes have relationships with animal, material, and activity classes from the Common subdomain, with defined processes and activities from Protocol Representation, performed observations and procedures from Study Conduct, and concepts from Molecular Biology.
Biospecimen
This subdomain represents concepts related to the collection and management of biospecimens. It includes specimen protocols (eg, SpecimenCollectionProtocol or SpecimenProcessingProtocol), the subject from whom the specimen was collected (eg, SpecimenCollectionProtocolSubject), the set of specimens that can be collected from a subject’s case (eg, SpecimenCollectionGroup), and the biological specimen (eg, BiologicSpecimen or Specimen). It also includes semantics on specimen processing, transfers within the inventory, and distributions to other groups (eg, PerformedSpecimenMove). These concepts were harmonized within the BRIDG 4.0 release primarily from the LS DAM to replace BRIDG 3+ Specimen classes and from the CDISC Study Data Tabulation Model’s (SDTM9) Pharmacogenomics and Pharmacokinetics domains. Because biospecimens are related to both clinical and basic research, Biospecimen semantics have relationships with other subdomains, including Common, Experiment, Protocol Representation, and Study Conduct.
Molecular biology
This subdomain, also new in BRIDG 4.0, represents molecular biology and performed observations to determine their presence, variations, and features. It includes genomics (eg, NucleicAcidSequence, Genome, Intron, SingleNucleotidePolymorphism), transcriptomics (eg, RNA, MessengerRNA), proteomics (eg, AminoAcidSequence, Protein), pathways, biomarkers (eg, MolecularBiomarker, StudiedMolecularBiomarkerGroup), and other concepts. Central concepts from popular bioinformatics databases are also included, such as gene, mRNA, protein, and pathway identifiers, molecular sequence annotations, and reference sequences. Broad concepts are present in this subdomain, but specific details for some areas still need to be filled in. This subdomain was created through harmonization mainly with the LS DAM’s molecular semantics. Some additional attributes arose from harmonization with CDISC’s SDTM Pharmacogenomics/genetics Implementation Guide v1.0 standard.10 Classes in this subdomain have relationships to materials, drugs, and organizations in Common; to protocols in Protocol Representation; and to performed observations and procedures in Study Conduct.
BRIDG 3.2 Release
BRIDG releases 1 and 211 were critical in providing a framework for core concepts in clinical trials research. BRIDG 3.2 included harmonization with projects such as the FDA Janus Clinical Trials Repository, HL7’s CTR&R project, and NCI’s Clinical Trials Reporting Program, a regularly updated database of trial information and accruals on all NCI-supported clinical trials. HL7’s CTR&R semantics have since been absorbed into CDISC’s CTR.xml technical standard12 that populates ClinicalTrials.gov,13 the European Clinical Trials Database (EudraCT14), and the World Health Organization’s International Clinical Trials Registry Platform.15
A full listing of BRIDG 3.2 harmonization projects is available at https://bridgmodel.nci.nih.gov/download-model/bridg-releases/release-3-2. Project harmonization is not limited to the addition of new classes, as is, from external models. Rather, it is a careful review process in which modelers and SMEs assess BRIDG and the external project to determine which content is appropriate to add to BRIDG, whether alterations and extra information are needed, and where existing content within BRIDG needs to be changed or deleted. The 3.2 release added 26 classes and 77 attributes for statistical analysis plans, anatomic sites, donor registries, study activities, and other areas (Table 1). Many of the new attributes were associated with new classes, but some were added to existing classes, such as BiologicEntity and DefinedCompositionRelationship. Twenty-one existing classes and 58 attributes were updated to broaden and correct names, definitions, examples, and notes. Many new relationships were added between new and existing classes, and existing relationships were updated to amend cardinality, relationship name, destination class, or endpoint class. Constraints include rules on how classes and attributes should be interpreted or used. Roughly equal numbers of new constraints and changes to existing constraints’ descriptions were implemented in this release. A summary of changes for each BRIDG release package is available as an Excel file and is included in every BRIDG release package, published on the bridgmodel.org website.
Table 1.
Change list summary of recent BRIDG releases
| Release | Asset Type | Additions | Deletions | Updates |
|---|---|---|---|---|
| 3.2 | Classes | 26 | 0 | 21 |
| Attributes | 77 | 3 | 58 | |
| Relationships | 51 | 9 | 12 | |
| Constraints | 28 | 6 | 24 | |
| 4.0 | Classes | 76 | 0 | 66 |
| Attributes | 153 | 8 | 97 | |
| Relationships | 155 | 5 | 15 | |
| Constraints | 23 | 3 | 4 | |
| 4.1 | Classes | 3 | 0 | 34 |
| Attributes | 2 | 9 | 15 | |
| Relationships | 9 | 7 | 2 | |
| Constraints | 0 | 2 | 0 |
BRIDG 4.0 through 4.1.1
Although the clinical research domains of BRIDG matured from release 1 through release 3.2, there were calls for BRIDG to support molecular-based medicine, in which treatment of disease is informed by the patient’s genomic and other molecular characteristics.16 This change represented a major scope increase to include semantics from life sciences, where BRIDG 3.2 was harmonized with the multiple external standards described above for the Biospecimen, Experiment, and Molecular Biology domains. The majority of changes made to BRIDG 3.2 in the creation of BRIDG 4.0 included work to harmonize semantics from the NCI’s LS DAM and CDISC PGx 1.0. These harmonization projects included analyzing source materials, getting input from SMEs from cancer centers, and compiling examples and references from trusted community sources.
NCI’s LS DAM provided a shared view of translational and life science semantics to cover basic and preclinical research protocols.8 It was informed in part by the NCI Life Sciences Business Architecture Model.17 After harmonizing with BRIDG as part of its 4.0 release, the LS DAM was no longer supported. The FDA launched its Voluntary Genomic Data Submission program18 to provide guidance on submission of genetic data for FDA review. In turn, CDISC developed PGx 1.0, which includes information on genotyping, gene expression profiling, cytogenetics, and proteomics. By harmonizing with PGx 1.0, BRIDG 4.0 incorporated those semantics for human, microbiological, and proteogenomic studies. The BRIDG 4.0 release included, for the first time, semantics covering the full spectrum of biomedical protocol–driven research, including life sciences.
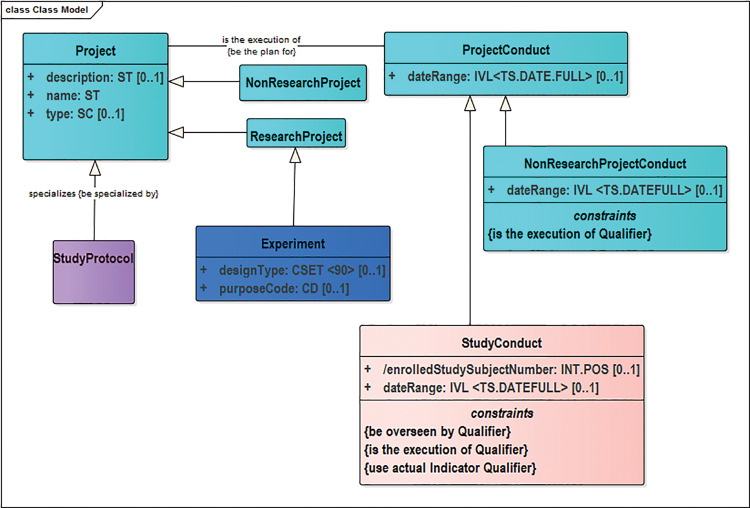
As a consequence of the broad scope increase, nearly 400 modifications (eg, classes, attributes, relationships, definitions, etc.) were added for biospecimens and their management and storage, cell cultures, molecular biology, research projects, experimental factors, etc., in BRIDG 4.0 (Table 1). The 66 updated classes include changes made as part of routine maintenance to the model and also to broaden existing classes, their definitions, and examples to encompass biological research. The 97 updated attributes were changed to ensure that their definitions, notes, and examples were accurate for all of biomedicine. As a consequence of the influx of new classes, 155 relationships and 28 constraints were added, with a smaller number of deletions and updates made as part of model maintenance based upon input from SMEs. In turn, 3 new subdomains were created (Biospecimen, Experiment, and Molecular Biology) and a new BRIDG 4.0 Project Backbone (Figure 3 ) emerged as a cross-subdomain core supporting both wet bench and clinical research, with classes such as Project, ResearchProject, NonResearchProject, and ProjectRelationship. This new backbone provides support to nonregulated research efforts.
Figure 3.
The BRIDG 4.0+ project backbone. These classes form the new higher-level backbone of BRIDG as a translational model for nonregulated research.
BRIDG 4.1 refined BRIDG 4.0 semantics based on comments from the HL7 balloting process and requests by SMEs to improve representation of the conduct of multinational studies. In contrast with the addition-heavy BRIDG 4.0 release, 4.1 included only 14 new assets. The majority of changes in 4.1 were updates (51), largely in the Molecular Biology and Experiment subdomains, and deletions (18) from the DefinedImaging and PerformedImaging classes. BRIDG 4.1.1 was a minor release that contained a handful of changes in response to HL7 informative ballot comments in May 2016.
DISCUSSION
The impact of BRIDG
With the FDA’s interest in leveraging electronic health records, wearable devices, and other databases for regulated research, there is a pressing need for a tool that provides a common understanding of research with touchpoints to health care semantics. BRIDG is the only such standard model recognized by health care and clinical research standards development organizations (CDISC, HL7, and ISO) to fill this need and serve as a foundation for many systems and initiatives in a learning health system.
The FDA and Japan’s Pharmaceuticals and Medical Device Agency now mandate that submissions for new trials comply with CDISC’s SDTM and ADaM standards beginning December 2016. As a consequence, many pharmaceutical, medical device, biotechnology, and other companies as well as academic medical centers are overhauling their reporting systems to help ensure submission compliance.19 CDISC standards such as SDTM and Clinical Data Acquisition Standards Harmonization,20 which provides a minimal set of electronic case report form elements common to all clinical protocols, are modeled within or mapped to BRIDG. The model, therefore, provides a graphical view of CDISC’s interrelated semantics. Over 20 CDISC Therapeutic Area (TA21) and indication standards have been developed, with FDA naming dyslipidemia, chronic hepatitis C virus, diabetes, QT interval, and tuberculosis in technical conformance guides. All CDISC TA standards contain mappings to variables from SDTM and Controlled Terminologies. Work is ongoing to add ADaM and Clinical Data Acquisition Standards Harmonization for building core electronic case report forms with these standards. Because many concepts from TA standards are being mapped to BRIDG, systems that are designed using BRIDG will be traceable to these various standards, in order to help with interoperability and data analysis (eg, when aggregating data from related indications).
Increasingly, leaders at clinical research organizations, device manufacturers, and pharmaceutical companies understand the importance of establishing and utilizing standard information models and data schemata to manage and exchange clinical trial and other data generated from protocol-driven research. This is reflected in TransCelerate Biopharma Inc.’s Technical Council, which will recommend the use of BRIDG as part of a common data model for its biopharmaceutical member companies in its coming enterprise architecture guidance.
How to utilize BRIDG to develop semantically interoperable software
No usable software system could implement all objects within BRIDG; the model is simply too large. That completeness of coverage, however, allows end users to review BRIDG’s universe of semantics and select the subset of concepts that are required to implement a specific software solution. Because BRIDG uses concepts and examples that make sense to domain experts, such experts can work closely with software developers and BRIDG analysts to review the DIM and select classes appropriate for their project. Where no classes within BRIDG cover the necessary semantics of a new project, end users can work with BRIDG analysts to identify these gaps, provide use cases to describe them, and then fill the gaps with new semantics within BRIDG. The BRIDG-based information model can then be utilized by the development team to develop a logical model. Logical models from existing projects can also be informed by a DIM to improve interoperability. A physical model can be developed from a logical model, and it will include details specific to the system, such as programming language–specific data types, database tables, access constraints, etc. All such specific implementations would be readily traced back to the reference standard semantics within BRIDG.
Examples of BRIDG use
Multiple examples of BRIDG use exist in international settings. Genzyme utilized BRIDG to develop its rare-disease central registry, called RegistryNXT!, which won an internal innovation award from its parent company, Sanofi. The Innovative Medicines Initiative’s Security and Interoperability in Next Generation PPDR Infrastructures (SALUS) project is an interoperability framework for proactive postmarket safety studies in the European Union. The SALUS team developed a resource description framework (RDF) representation of BRIDG to provide semantics for its metadata repository (MDR). BRIDG was selected because it maps to various CDISC standards, HL7 information models, and Integrating the Healthcare Enterprise clinical document types.22,23 SALUS and its collaborators used this BRIDG-based MDR as part of a proof-of-concept mechanism to meaningfully exchange data between European EHRs. As detailed below, Enterprise Data Engineering Technology (EDETEK) and PAREXEL International both use BRIDG as their common data model, from which many different software systems have been developed.24
BRIDG’s value centers around more rapid design, increased semantic consistency and quality, and improved interoperability. EDETEK utilized BRIDG’s classes when designing its CONFORM informatics suite (personal communication with J Chen, 2016). This suite provides a “hub” that links “spoke” tools that had previously been stand-alone, noninteroperable tools. These spokes include MDR, pipelines for data standardization and informatics processes, and event management tools. EDETEK realized ∼50% cost savings when creating a logical model for the “hub” platform. A 70% time savings was achieved when mapping to the BRIDG-based common data model and harmonizing the spoke tools’ physical models with that model.25 These efficiencies translate to improved interoperability. Because BRIDG concepts have tags to CDISC, HL7, Observational Medical Outcomes Partnership (OMOP), and other standards, use of BRIDG provides stability and protection of the development investment through better alignment with industry and community standards. The tags that bind HL7 and other semantics to BRIDG concepts were utilized to design the Exchange Connector module for integration with EHRs’ HL7 V3 messaging.
In a second example, PAREXEL and PAREXEL Informatics have utilized BRIDG as the basis for a central common data model (CDM) to increase semantic consistency and quality across a diverse portfolio of products and services (personal communication with H Glover, J Jones, and B Egersdoerfer, 2016). BRIDG has provided (1) standard, well-defined terminologies, (2) consistent data types that support the use of those terminologies and provide metadata for structured data management, and (3) domain models.26 Together, these benefits create a “shared language” across disparate applications. BRIDG has been valuable in improving the quality of system integrations and new software development. Historically, PAREXEL Informatics created custom integration solutions for its customers. BRIDG is now being utilized to standardize integration models and semantics. This effort was launched to reduce the amount of time required to map customers’ applications, increase the ability to reuse existing code, and achieve greater consistency across projects. All new development projects will utilize the BRIDG-based CDM to have a common understanding of how business processes work, a single source of interoperable schemas, and greater consistency across those interoperable systems. This CDM will also make it easier to reuse existing code for disparate software implementations. Implementation of any standard requires an up-front investment for future value. BRIDG has already delivered quality and consistency, and PAREXEL and PAREXEL Informatics expect to achieve cost and time savings via their BRIDG-based CDM.
Within the United States, BRIDG has also been used for several national initiatives. The National Marrow Donor Program and the Center for International Blood and Marrow Transplant Research utilize BRIDG to place hematopoietic cell transplants in the context of health care and research for use as a central specification for data submissions and interoperability. Since BRIDG’s inception in 2005, the FDA has used it as the conceptual model for the Janus Clinical Trials Repository (CTR) warehouse. As part of a marketing application, clinical trial sponsors submit subject-level data from trials in CDISC SDTM format to the FDA for storage in the Janus CTR. The CTR is used by FDA staff to support regulatory review and cross-study analyses to address emergent public health questions.
Vocabulary binding in BRIDG
Members of the community routinely comment on the lack of specific controlled vocabularies that are implemented within the model. Historically, references to 1 or 2 exemplar vocabularies, terminologies, and identifier schemes have been included within descriptions of attributes as examples only. No basic research or medical vocabulary standard was ever formally bound to classes or attributes within BRIDG. The strategic decision was made, in part, because when multiple vocabularies exist in the same space, the selection of an appropriate vocabulary can depend on many factors. For example, software derived from BRIDG for AEs in hospitals or clinics would likely implement Systemized Nomenclature of Medicine – Clinical Terms (SNOMED CT)27 for AE capture, whereas regulated research systems would implement Medical Dictionary for Regulatory Activities (MedDRA)28 due to global regulatory agency reporting requirements. Predetermining which vocabularies are appropriate under which conditions was not seen as being within the scope of BRIDG. In addition, the BRIDG team was concerned about appearing to favor some vocabularies over others, accidentally leaving out any vocabularies or overrepresenting vocabularies used only by certain countries. This modeling decision is the major limitation of BRIDG’s supporting full semantic interoperability in the long run.
Future development of BRIDG
The long-term goal is for BRIDG to provide an evolving model for use with trans-health initiatives. Previously BRIDG 3.2 was mapped to the OMOP CDM,29,30 which represents concepts in active drug surveillance utilizing observational data. This exercise found that many OMOP CDM concepts were already present in BRIDG, and that future enhancements would improve support for retrospective surveillance studies modeled within the CDM.31 FDA Commissioner Dr Robert Califf has described a need to improve evidence gathering to more quickly identify effective treatments in a way that is not limited to slow and expensive clinical trials. Thus, he has called for leveraging prospective clinical research study data alongside claims databases, medical device/wearable data from patients, and federated databases from large national projects.32 Enhancements of BRIDG to include additional surveillance and safety signal concepts will provide a comprehensive model to support this goal.
Future work on controlled vocabularies is another key goal. Now that the model has matured, it is clear that the benefit of vocabulary binding outweighs the risks described above, and this is a critical future direction. As a global model, it must be ontology and terminology agnostic, but this agnosticism does not preclude the binding of BRIDG class attributes to 1 or more common terminologies/code sets from medicine (eg, SNOMED CT, International Classification of Diseases,33 Current Procedural Terminology,34 Logical Observation Identifiers Names and Codes [LOINC],35 RxNorm36) and/or from research such as CDISC Labs terminologies,37 PubChem,38 ChEMBL,39 Entrez Gene,40 or UBERON for cross-species anatomic sites.41 For example, most LOINC codes could be bound to the attributes within the Study Conduct class PerformedObservationResult. Special cases will exist, such as observation results for cancer pathology tests, where the subset of LOINC codes for histology (eg, 59847‐4 and 33732‐9) would be bound instead to Study Conduct’s PerformedHistopathologyResult. The Gene class in Molecular Biology could be bound to multiple terminology and identifier lists, such as those from Entrez Gene, Ensembl,42 Human Genome Nomenclature Committee,43,44 Rat Genome Database,45 and other common resources. By binding ontologies and terminologies to BRIDG concepts, interoperability will be greatly improved beyond the model’s current value.
Groups that use BRIDG to architect their systems will have “out-of-the-box” access to synonyms and “sister” terms or codes from one code list that are clearly mapped to other code systems. Working groups within HL7 are working to define how to best bind vocabularies and terminologies to diverse models. We will utilize these technical principles to “hook” external semantics to the model. In some cases, multiple vocabularies, terminologies, and identifier schemes will be hooked to some BRIDG classes. Some community groups are mapping or have mapped different vocabularies, terminologies, or identifiers to one another. Where such mapping exists, external terminology services will help BRIDG end users resolve synonyms and other relationships between terms or codes.
While the LS DAM was used as the primary project with which to harmonize for life science expansion, it has (as do many useful models) gaps that now also exist in BRIDG 4.1.1. For example, some Molecular Biology classes are concepts only, containing only a single attribute. This issue is common in any model that is driven by community use cases, many of which are required to mature the semantics over time to include additional attributes, constraints, and examples to more completely model a given domain. This model evolution is expected to occur through new use cases and harmonization with other models as part of BRIDG 4+ development. Overlap also exists with these concepts in BRIDG and the models under development by the HL7’s Clinical Genomics, Imaging with the Digital Imaging and Communications in Medicine standard, Orders and Observations, and Anatomic Pathology work groups. An important future goal will be to refine BRIDG, fleshing out these subdomains by abstraction via “modeling by reference,” where shared concepts between BRIDG and external models are identified with appropriate linkages to those external models included within the BRIDG UML. Other key goals include developing an oncology-specific view of BRIDG to benefit NCI stakeholders. This oncology view will leverage the existing CDISC Breast Cancer TA standard, the upcoming CDISC Prostate, Colorectal, and Lung Cancer TA standards, and other pertinent standards.
CONCLUSION
The current release of BRIDG 4.1.1, its documentation, and prior releases are all freely available at https://bridgmodel.nci.nih.gov/. Specific releases can be found under the page’s Download/View Model tab. Parties who are interested in harmonizing with BRIDG are encouraged to reach out to the authors.
CONTRIBUTORS
SH and WVH serve as BRIDG modelers, harmonizing project semantics, coordinating development, and working with SMEs such as LB, DW, and many others from NCI-designated cancer centers, CDISC, and the biopharmaceutical research community. EH has served as a BRIDG product owner within the NCI for many years. All save WVH and SH have served or actively serve on the BRIDG Steering Committee. LB drafted the manuscript and prepared the figures, with support from SH, RM, BB, MAS, RK, and EH and special consultation from DW. All authors reviewed and edited the manuscript.
FUNDING
LB and RM were supported through a subcontract with NCI and the Frederick National Laboratory for Cancer Research. LB also received funding from NCI Cancer Center support grant P30CA125123.
COMPETING INTERESTS
The authors have no competing interests to declare.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge Lisa Schick, Dr Baris E Suzek, Dr Robert R Freimuth, and Sharon Elcombe for subject matter expertise in the harmonization of BRIDG and LS DAM; Dr Juli Klemm for leadership in increasing BRIDG’s scope; Joyce Hernandez for PGx support; Dr Mitra Rocca for consultation; and Ann White for editorial support. We wish to especially acknowledge the discussions about the value of BRIDG to real-world software projects and thank Jian Chen and Peter Smilanskey from EDETEK and Hugh Glover, Julie James, and Benedikt Egersdoerfer from PAREXEL and PAREXEL Informatics.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
References
- 1. FDA. Providing Regulatory Submissions in Electronic Format—Standardized Study Data: Guidance for Industry. 2014http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/UCM292334.pdf. Accessed December 21, 2016.
- 2. FDA. Study Data Technical Conformance Guide V3.0. 2016http://www.fda.gov/downloads/ForIndustry/DataStandards/StudyDataStandards/UCM384744.pdf. Accessed December 21, 2016.
- 3. PMDA. Notification of Practical Operations of Electronic Study Data Submissions. 2015http://www.pmda.go.jp/files/000206451.pdf. Accessed December 21, 2016.
- 4. PMDA. Technical Conformance Guide on Electronic Study Data Submissions. 2015https://www.pmda.go.jp/files/000206449.pdf. Accessed December 21, 2016.
- 5. National Cancer Institute. caBIG(R) Central Clinical Participant Registry (C3PR). U.S. Department of Health and Human Services, National Institutes of Health. 2010;10:7433. [Google Scholar]
- 6. Payne PR, Kwok A, Greaves AW. Integrating web portlet technologies with caGrid to enable rapid application development: the CRC Patient Study Calendar. AMIA Annu Symp Proc. 2008:1087. [PubMed] [Google Scholar]
- 7. CDISC. Analysis Data Model (ADaM). 2016https://www.cdisc.org/standards/foundational/adam. Accessed December 21, 2016.
- 8. Freimuth RR, Freund ET, Schick L et al. Life sciences domain analysis model. J Am Med Inform Assoc. 2012;196:1095–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. CDISC. Study Data Tabulation Model (SDTM), 2016. 2016https://www.cdisc.org/standards/foundational/sdtm. Accessed December 21, 2016.
- 10. CDISC. Pharmacogenomics/genetics (PGx). 2016https://www.cdisc.org/standards/foundational/pgx. Accessed December 21, 2016.
- 11. Fridsma DB, Evans J, Hastak S, Mead CN. The BRIDG project: a technical report. J Am Med Inform. Assoc 2008;152:130–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. CDISC. CTR-XML. 2016https://www.cdisc.org/standards/foundational/ctr-xml. Accessed December 21, 2016.
- 13. Zarin DA, Tse T, Williams RJ, Carr S. Trial Reporting in ClinicalTrials.gov: The Final Rule. N Engl J Med. 2016;375:1998–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Watson R. European clinical trials database gathers pace. BMJ. 2013;346:f3614. [DOI] [PubMed] [Google Scholar]
- 15. Stegemann H. [The International Clinical Trials Registry Platform: ICTRP]. Arch Latinoam Nutr. 2007;574:311–12. [PubMed] [Google Scholar]
- 16. Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015;3729:793–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Boyd LB, Hunicke-Smith SP, Stafford GA et al. The caBIG(R) Life Science Business Architecture Model. Bioinformatics. 2011;2710:1429–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Goodsaid FM, Amur S, Aubrecht J et al. Voluntary exploratory data submissions to the US FDA and the EMA: experience and impact. Nat Rev Drug Discovery. 2010;96:435–45. [DOI] [PubMed] [Google Scholar]
- 19. Rockhold FW, Bishop S. Extracting the value of standards: The role of CDISC in a pharmaceutical research strategy. CDISC Articles. 2012;401 http://www.cdisc.org/system/files/all/article/application/pdf/extracting_the_value_of_standards_rockhold__bishop.pdf. Accessed December 21, 2016. [Google Scholar]
- 20. CDISC. Clinical Data Acquisition Standards Harmonization (CDASH) Webpage. 2016https://www.cdisc.org/standards/foundational/cdash. Accessed December 21, 2016.
- 21. CDISC. Therapeutic Area Standards Webpage. 2016http://www.cdisc.org/therapeutic. Accessed December 21, 2016.
- 22. Laleci GB, Yuksel M, Dogac A. Providing semantic interoperability between clinical care and clinical research domains. IEEE J Biomed Health Inform. 2013;172:356–69. [DOI] [PubMed] [Google Scholar]
- 23. Daniel C, Sinaci A, Ouagne D et al. Standard-based EHR-enabled applications for clinical research and patient safety: CDISC - IHE QRPH - EHR4CR & SALUS collaboration. AMIA Jt Summits Transl Sci Proc. 2014;2014:19–25. [PMC free article] [PubMed] [Google Scholar]
- 24. Bache R, Daniel C, James J et al. An approach for utilizing clinical statements in HL7 RIM to evaluate eligibility criteria. Stud Health Technol Inform. 2014;205:273–77. [PubMed] [Google Scholar]
- 25. Chen J, Smilansky P. Personal communication. 2016. [Google Scholar]
- 26. Glover H, James J, Egersdoerfer B. Personal communication. 2016. [Google Scholar]
- 27. Millar J. The Need for a Global Language: SNOMED CT Introduction. Stud Health Technol Inform. 2016;225:683–85. [PubMed] [Google Scholar]
- 28. Brown EG, Wood L, Wood S. The medical dictionary for regulatory activities (MedDRA). Drug Saf. 1999;202:109–17. [DOI] [PubMed] [Google Scholar]
- 29. Matcho A, Ryan P, Fife D, Reich C. Fidelity assessment of a clinical practice research datalink conversion to the OMOP common data model. Drug Saf. 2014;3711:945–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rijnbeek PR. Converting to a common data model: what is lost in translation? Commentary on “fidelity assessment of a clinical practice research datalink conversion to the OMOP common data model.” Drug Saf. 2014;3711:893–96. [DOI] [PubMed] [Google Scholar]
- 31. Rocca M, Kubick W, Kush R, Haider S, Ryan P. Harmonization of the OMOP Common Data Model with the BRIDG Model. 2007http://omop.org/sites/default/files/07_Rocca_FDA_Harminization%20of%20the%20OMOP%20CDM%20with%20BRIDG%20Model.pdf. Accessed December 21, 2016.
- 32. Appel A. Califf Sees New Role for E-records in Drug, Device Approval. New York City, New York: Bloomberg BNA Snapshot, Bloomberg L.P; 2016. [Google Scholar]
- 33. WHO. ICD-10 Version: 2016. World Health Organization; 2016http://apps.who.int/classifications/icd10/browse/2016/en. Accessed December 21, 2016. [Google Scholar]
- 34. Hirsch JA, Leslie-Mazwi TM, Nicola GN et al. Current procedural terminology; a primer. J Neurointerv Surg. 2015;74:309–12. [DOI] [PubMed] [Google Scholar]
- 35. Vreeman DJ, McDonald CJ, Huff SM. LOINC(R): A universal catalog of individual clinical observations and uniform representation of enumerated collections. Int J Funct Inform Personal Med. 2010;34:273–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nelson SJ, Zeng K, Kilbourne J, Powell T, Moore R. Normalized names for clinical drugs: RxNorm at 6 years. J Am Med Inform Assoc. 2011;184:441–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. CDISC. Laboratory Data Model (LAB). 2004http://www.cdisc.org/standards/foundational/lab. Accessed December 21, 2016.
- 38. Kim S, Thiessen PA, Bolton EE et al. PubChem Substance and Compound databases. Nucleic Acids Res. 2016;44(D1):D1202–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Willighagen EL, Waagmeester A, Spjuth O et al. The ChEMBL database as linked open data. J Cheminform. 2013;51:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2011;39(Database issue):D52–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Haendel MA, Balhoff JP, Bastian FB et al. Unification of multi-species vertebrate anatomy ontologies for comparative biology in Uberon. J Biomed Semantics. 2014;5:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yates A, Akanni W, Amode MR et al. Ensembl 2016. Nucleic Acids Res. 2016;44(D1):D710–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gray KA, Seal RL, Tweedie S, Wright MW, Bruford EA. A review of the new HGNC gene family resource. Hum Genomics. 2016;10:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yates B, Braschi B, Gray KA, Seal RL, Tweedie S, Bruford EA. Genenames.org: the HGNC and VGNC resources in 2017. Nucleic Acids Res. 2016;45(D1):D619–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shimoyama M, De Pons J, Hayman GT et al. The Rat Genome Database 2015: genomic, phenotypic and environmental variations and disease. Nucleic Acids Res. 2015;43(Database issue):D743–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.