Abstract
Objective
To develop an open-source information extraction system called Eligibility Criteria Information Extraction (EliIE) for parsing and formalizing free-text clinical research eligibility criteria (EC) following Observational Medical Outcomes Partnership Common Data Model (OMOP CDM) version 5.0.
Materials and Methods
EliIE parses EC in 4 steps: (1) clinical entity and attribute recognition, (2) negation detection, (3) relation extraction, and (4) concept normalization and output structuring. Informaticians and domain experts were recruited to design an annotation guideline and generate a training corpus of annotated EC for 230 Alzheimer’s clinical trials, which were represented as queries against the OMOP CDM and included 8008 entities, 3550 attributes, and 3529 relations. A sequence labeling–based method was developed for automatic entity and attribute recognition. Negation detection was supported by NegEx and a set of predefined rules. Relation extraction was achieved by a support vector machine classifier. We further performed terminology-based concept normalization and output structuring.
Results
In task-specific evaluations, the best F1 score for entity recognition was 0.79, and for relation extraction was 0.89. The accuracy of negation detection was 0.94. The overall accuracy for query formalization was 0.71 in an end-to-end evaluation.
Conclusions
This study presents EliIE, an OMOP CDM–based information extraction system for automatic structuring and formalization of free-text EC. According to our evaluation, machine learning-based EliIE outperforms existing systems and shows promise to improve.
Keywords: natural language processing, machine learning, clinical trials, patient selection, common data model, named entity recognition
INTRODUCTION
Clinical trial eligibility criteria and formalization
As the gold standard for generating medical evidence, randomized controlled trials are fundamental for advancing medical science and improving public health. However, recruitment for clinical trials remains a major barrier.1,2 Recruitment follows eligibility criteria (EC), whose free-text format and lack of standardization have inhibited their effective use for automatic identification of eligible patients in the electronic health record (EHR). Formal representation of EC has been pursued by the biomedical informatics research community for nearly 3 decades3 to optimize cohort selection and EC knowledge reuse4 and to support large-scale aggregative analytics5–7 and collaborative clinical research.8,9 Notable systems include Evaluation of Ontology (EON)10 – ontology for intervention protocols and guidelines, agreement on standardized protocol inclusion requirements for eligibility,11 eligibility criteria extraction and representation,12 and an eligibility rule of grammar and ontology.13 Unfortunately, often these formalizations not only require laborious manual interpretation of the syntactic rules and semantic concepts in EC, but also largely lack semantic interoperability with EHR data.
Use of a common data model intended for EHR data to represent EC shows promise in bridging this interoperability gap. The Observational Medical Outcomes Partnership (OMOP) Common Data Model (CDM) (http://www.ohdsi.org/data-standardization/) is ideal for this purpose because it enables interoperability among disparate observational databases by standardizing data using a common information model and multiple standard terminologies for pertinent clinical entities, such as condition, observation, and medication.14 All data analytical tools based on the OMOP CDM can be easily shared among data owners. For example, an open-source tool made available by the Observational Health Data Sciences and Informatics community15 called ATLAS (http://www.ohdsi.org/web/atlas) allows researchers to manually define sharable and structured EC rules as cohort queries against EHR data, formatted using the OMOP CDM and standard terminologies such as Systematized Nomenclature of Medicine – Clinical Terms (SNOMED-CT) and Logical Observation Identifiers Names and Codes (LOINC) to achieve interoperability between clinical research EC and EHR data. However, this manual approach is not scalable. An information extraction system is desired to automate the transformation from EC text to structured EHR data queries. In this study, we contribute such a system to make EC computable and interoperable with the rapidly growing observational databases enabled by the wide adoption of EHRs.16
Natural language processing for biomedical information extraction
Information extraction (IE) refers to the task of automatically extracting structured semantics (eg, entities, relations, and events) from unstructured text.17 Biomedical IE (bioIE) is often performed on unstructured scientific literature or clinical narratives in EHRs to prepare structured information input needed by clinical decision support systems.18–30 BioIE includes 3 major subtasks17: (1) named entity recognition (NER)25,31; (2) extraction of binary relationships between named entities, such as problem-treatment relationships25 or protein-protein interactions32; and (3) event identification, which identifies highly complex relations among extracted entities, such as gene regulation.33 Methodologies for bioIE fall into 5 categories: rule-based, knowledge-based, statistics-based, learning-based, and hybrid.21,34 One of the earliest and most advanced rule-based systems for clinical text processing is Medical Language Extraction and Encoding System (MedLEE).18 Learning-based methods have rapidly advanced in the past 5 years.21 The representative learning-based methods include conditional random fields (CRFs) and structured support vector machines (SVMs).35 For example, the best system reported in the Informatics for Integrating Biology and the Bedside (i2b2) named entity recognition (NER) challenge from de Bruijn et al.36 used semi-Markov (F1 score 0.85), followed by the system from Jiang et al.37 using CRF (F1 score 0.84). Relation extraction has evolved from simple co-occurrence statistics to syntactic analysis and dependency parsing.17,38 The best-performing system for the i2b2 relation extraction challenge achieved an F1 score of 0.74. Meanwhile, deep neural networks have also been increasingly applied to both general IE and bioIE, represented by the emerging open IE (openIE) system39 and deep learning for NER40–42 and relation extraction.43–45 Some recent works on word embedding show learning of word vectors via neural networks. One of the most popular embedding techniques is called word2vec.46 Instead of using each word as a feature, words are represented as vectors that encode rich contextual information. Word embeddings and deep learning techniques have shown great promise for the NER task for clinical text.47,48
Unfortunately, the above systems were designed for clinical notes in EHRs or text in the literature. Their adaptability to free-text clinical research EC remains untested. To date, the most specialized natural language processing (NLP) parser for automatically structuring EC free text is EliXR, developed by Weng et al.12 in 2011. Through a rule-based approach, EliXR recognizes Unified Medical Language System (UMLS) concepts based on dictionary matching and encodes clinical entities and relations using OMOP CDM v.4.49 Like most other rule-based bioIE systems, EliXR exhibits high precision but poor recall due to the existence of morphological variants50 or poor coverage of concepts or rules.51
Contributions
This study makes 2 primary contributions. First, we designed a novel annotation guideline for clinical research EC following OMOP CDM v.5 and constructed a new annotated corpus using this guideline for training or validating various clinical research EC parsers. Second, we developed and validated a machine learning–based IE system to automatically formalize clinical research EC. We named this system Eligibility Criteria IE (EliIE). EliIE takes 4 steps to structure EC free text: (1) named entity recognition and attribute recognition, (2) negation detection, (3) relation extraction, and (4) standardization by concept normalization and output structuring. We validated EliIE’s accuracy for formalizing EC using the OMOP CDM, which can be used to perform patient screening in the EHR or enable development of a large knowledge base of structured clinical research EC for knowledge reuse or aggregate analyses. (In this study, we selected Alzheimer’s disease for methodology illustration. EliIE is able to parse all free-text eligibility criteria text, though the performance of trials in other domains may be weakened. In the future, we plan to extend the use case to more domains.) To the best of our knowledge, EliIE is the first open-source machine learning–based IE system specifically designed for clinical research EC.
METHODS
Dataset and annotations
We randomly selected 230 Alzheimer’s disease (AD) trials from ClinicalTrials.gov.52 AD trials were chosen for methodology illustration, given that AD is one of the most well-studied diseases in the United States.53 Next, we extracted the EC text from the “eligibility criteria” section of each trial for annotation. The EC text varies in size from around 100 words to over 1000 words. Example free-text EC is provided below:
– No evidence of major depression.
– Normal B12, rapid plasma reagin and Thyroid Function Tests or without any clinically significant abnormalities that would be expected to interfere with the study.
One clinician (AR) and 2 informatics students (TK, GH) designed the annotation guideline for entity and attribution annotation using an iterative process. First, they studied OMOP CDM v.5.0 (https://github.com/OHDSI/CommonDataModel/blob/master/OMOP%20CDM%20v5.pdf) and focused on 4 classes of entities: condition, observation, drug/substance, and procedure or device. The context for each entity consists of 4 types of attributes: modifiers/qualifiers, temporal constraints, measurements, and anatomic location. (Anatomic location is not commonly defined in eligibility criteria for AD, and thus is not evaluated in this paper. However, this attribute is included, as it is an important component of eligibility criteria for trials on other diseases [eg, myocardial infarction].) The team first independently annotated 5 trials based on each person’s interpretation of the CDM definition. Next, the team compared the annotation results and discussed the discrepancies until a consensus was reached, resulting in revised annotation guidelines. Using the updated guidelines, the team independently annotated 5 new trials and repeated the process until there was no discrepancy among annotators. In this manner, the guidelines were refined using at least 5 iterations of annotation, discussion, and amendment until eventually the team reached a stable state of consensus, finalizing the annotation guidelines. One of the challenges in the annotation guideline design was reaching consensus regarding what constitutes a “qualifier/modifier”; eg, in the phrase “unstable major depression,” is there only 1 modifier, “unstable,” or 2, “unstable” and “major”? We followed the “longest concept” rule, which is to choose the UMLS concept with the longest length.54 For this example, the longest concept found in UMLS is “major depression (CUI: C1269683),” therefore the entity would be “major depression,” and “unstable” is its modifier.
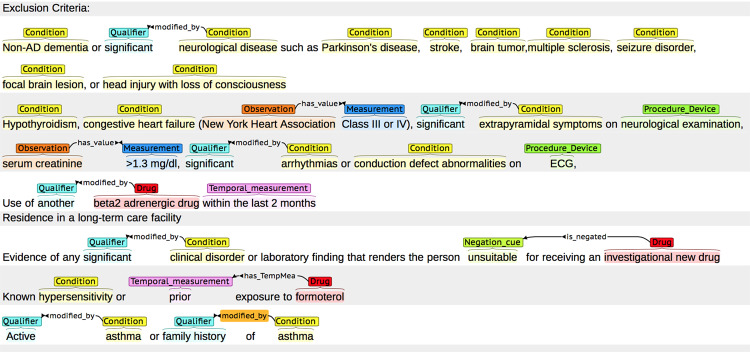
Our guidelines also defined relations between the entities and the corresponding attributes “modified by,” “has value,” and “has TempMea” (see relation annotation guideline in supplementary materials online for details, https://github.com/Tian312/EliIE/tree/master/Supp%20Materials). All relations are directional, pointing from the entity to its attribute. Entity annotation was completed by 1 clinician (YT) and 1 informatics student (TK). Relation annotation was completed by 1 informatics student (TK) and verified by the clinician (YT). All annotation was completed in Brat, a web-based annotation tool.55 Example annotations in Brat are shown in Figure 1 . The annotated corpus includes 8008 entities, 3550 attributes, and 3529 relations.
Figure 1.
Example annotations using the Brat tool (http://brat.nlplab.org). Entities and attributes from different classes are distinguished by colors; relations are annotated as arrows between entities and attributes. It is able to generate structured annotation result files using Brat.
System modules of EliIE
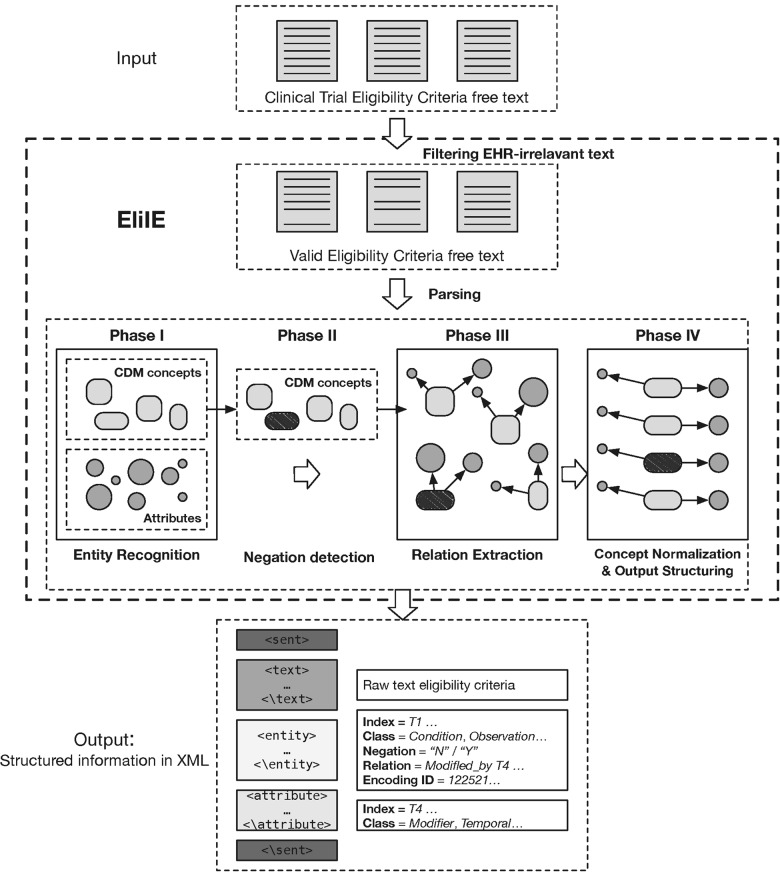
The workflow of EliIE is presented in Figure 2 . It includes preprocessing and 4-phase parsing. These steps are described in detail below.
Figure 2.
General workflow of EliIE. It includes a filtering step and 4-phase parsing. The final outputs are stored in an XML file.
Preprocessing
The EC text was preprocessed by normalizing the punctuation and removing criteria not available in the EHR, such as criteria for informed consent or patients’ willingness to participate. To exclude these criteria, we defined a list of keywords, eg, “informed consent” and “willing to,” to filter out nonapplicable EC for EHR settings. The concept annotation guidelines provide further details about our excluded EC categories.
Phase 1: CRF-based sequence labeling
The tasks of entity recognition and attribute recognition were cast jointly as a sequence-labeling problem. Seven categories, including 4 types of entities (condition, observation, drug/substance, procedure/device) and 3 types of attributes (measurement, temporal constraint, qualifier/modifier), were considered as labels. CRF, an established method for numerous sequence-labeling problems, has previously demonstrated outstanding performance for such tasks in the clinical domain.32 Thus, in our study, CRF was adopted to perform entity and attribute recognition by its publicly available implementation, CRF++.56 We used classical “BIO” tags to represent the boundary of terms of interest, ie, entities and attributes. “O” means it is outside the target terms. “B” represents the beginning word, and “I” tags all the inside words. Example tagging output is provided below:
– Subject/Ohas/Obleeding/B-Conditiondiathesis/I-Condition.
– Subject/Owith/Oblood/B-Observationpressure/I-Observation higher/B-Measurementthan/I–Measurement180/-Measurement-/I-Measurement110/I-MeasurementmmHg/I–Measurement.
We experimented with different types of features as CRF labels to identify the ones with the best performance. For each labeler, the input was a sentence and the output was a BIO-type sequence. The first system in this study was a CRF labeler, which relied on the basic set of word-level features, ie, the input words in the sentence and bigrams. For each word to be labeled, a 1-hot feature vector, which is a 1 × N vector used to distinguish each term in a vocabulary from every other word, where N is the count of terms in the vocabulary, the vector consisted of N−1 0 s and one 1 (to represent the target word). The system based on the “bag of words” as the only features was the baseline for CRF performance in our study. We then added other surface syntactic features to the CRF baseline system, such as part-of-speech tags and lemmas using the Natural Language Toolkit (NLTK) package.57 To include domain knowledge to recognize entities, a UMLS-based feature was generated for the system. For each input criterion, we used MetaMap58 to identify the UMLS concepts in it and its corresponding sematic types. “BI” (semantic types) and “O” (outside) labels were used to represent such features. For example, the MetaMap result of the criterion “History of myocardial infarction or cerebrovascular disease” is labeled as follows:
– History/B-[dsyn]of/I-[dsyn]myocardial/I-[dsyn]infarction/I-[dsyn]or/Ocerebrovascular/B-[dsyn]disease/I-[dsyn]
Here, “dsyn” is the shortcut of the semantic type “T047 Disease and Syndrome.”
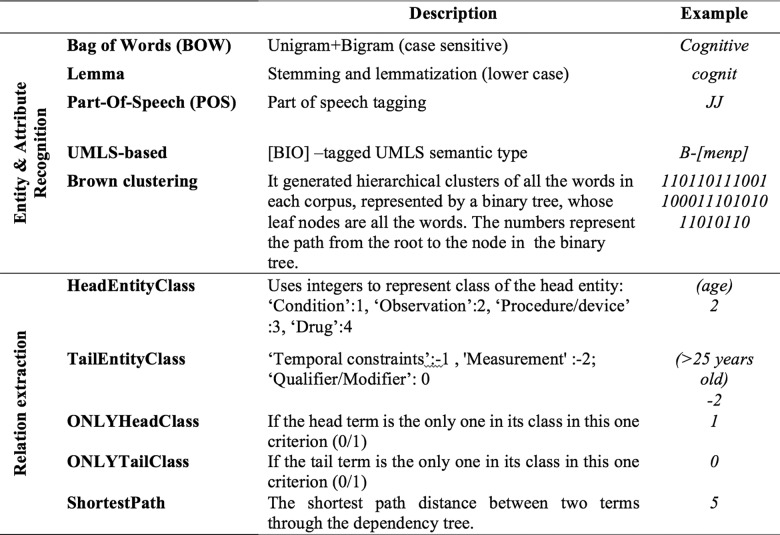
We also implemented a feature for each word to generate semantic information for the tokens learned from a large similar context. A word representation is often a vector. Each dimension’s value corresponds to a feature and might even have a semantic or grammatical interpretation, so we call it a word representation.59 Traditional 1-hot representations like bag of words (BOW) suffer from data sparsity. Words that are rare in the labeled training corpus will be poorly estimated. Typically, there exist 3 major kinds of word semantic representation approaches59: distributional representation (eg, latent Dirichlet allocation [LDA],60 latent semantic analysis [LSA]61), clustering-based word representation (eg, Brown-clustering [BC]62), and word embedding (eg, Word2vec46). In our study, we selected 2 approaches to test on EC texts, BC and Word2Vec. BC is a hierarchical clustering algorithm that clusters words to maximize the mutual information of bigrams. It generates hierarchical clusters of all the words in each corpus, represented by a binary tree whose leaf nodes are all the words. Word2vec performs vectorized embeddings to get richer representations of linguistic units, such as words.63 Tang et al.64 showed that when the 3 kinds of word representation were evaluated independently, BC achieved the best results in a clinical NER system. We retrieved the EC text of all the clinical trials from ClinicalTrials.gov. BC and word2vec were trained on this large unlabeled corpus to generate word representation. We ran these 2 features on a subset of trials and found that BC outperformed word2vec in EC texts. Thus, we chose BC as the word representation feature in our system. Figure 3 gives a detailed description of features we included to recognize entities and attributes. We then ran CRF models on the annotated corpus and used 5-fold cross-validation to evaluate the results.
Figure 3.
Detailed feature description for entity/attribute recognition and relation extraction.
Phase 2: negation detection
Negation detection is important in determining whether a criterion is used for inclusion or exclusion purposes. In EliIE, each recognized clinical entity is assessed for its negation by implementing the NegEx algorithm65 followed by a set of rules designed for EC text and invented in EliXR49 (https://github.com/Tian312/EliIE/blob/master/bin/EC_triggers.txt). To evaluate EliIE’s negation detection accuracy, each entity in our annotated corpus was also labeled “affirmed” or “negated” by annotators.
Phase 3: relation extraction
Once the entity and attribute recognition was completed, we ran the relation classifier on those recognized terms to identify relations between entities and their related attributes. To predict relations, all possible pairwise relations between entities and attributes in 1 criterion were enumerated and classified using SVM with the radial basis function. The directions of all the relations were predefined from each entity to its attributes. We implemented the SVM classifier through LibSVM.66 The features selected to predict relations were accommodated from.67 The detailed description is shown in Figure 3, including the class of the head entity, the class of the attribute, the shortest path between the 2 terms in the dependency tree, and whether the entity was the only one in its class in this criterion. Head term and tail term were determined by their classes. We used the multiclassification mode in LibSVM, using “0” as a label to represent that there was no relation between this entity and the attribute, and “1/2/3” to represent 3 kinds of relations. We ran the SVM classifier using 5-fold cross-validation to select the best parameters.
Phase 4: concept normalization and output structuring
After recognition and extraction of entities and their attributes and identification of the relations between them, the entities were encoded by the standard terminologies in OMOP CDM (http://www.ohdsi.org/web/wiki/doku.php?id=documentation:vocabulary). For example, the concept “AD” is encoded by CDM concept id: 378419. To support downstream large-scale analysis, we standardized the output results in the XML format. An example output format is illustrated in Figure 5.
Figure 5.
Example results from ELIIE, i2b2-based CliNER, and EliXR (EliXR output format: identified UMLS CUI {concept; Negation; Uncertain; Temporal; Measurement; Dosage}).
EVALUATION METRICS
To measure how well EliIE performs overall and for specific tasks, we designed an evaluation framework including both task-specific and end-to-end evaluations. In the task-specific evaluation, we used standard classification metrics: precision, recall, and F1-score, which are defined below (TP: true positive; FP: false positive; FN: false negative).
| (Eq. (1)) |
Since our goal was to generate structured queries in EHR, where clinical entities (eg, condition, drug) were the primary content, we conducted our end-to-end evaluations focused on clinical entities (eg, a simple criterion, “Current treatment with antipsychotics and antidepressants,” includes 2 queries by our entity-based definition: presence of drug “antipsychotics” with the temporal constraint “current” and presence of drug “antidepressants” with the temporal constraint “current”). Therefore, a “true positive query” in the end-to-end evaluation is counted if and only if a correctly recognized entity (condition, drug, observation, procedure, or device) is correctly assigned right relation(s) to its correctly recognized attribute(s) (if it has any), and at the same time, the result of negation detection is accurate. Then we report a final number for accuracy according to the following definition:
| (Eq. (2)) |
Comparison with related systems
Due to the scarce literature on parsing EC text and discrepant data models used by related systems, we had difficulty identifying comparable systems for EliIE and eventually selected 2 systems for extrinsic evaluation. One is an open-source clinical NER system recently developed and tested in the i2b2 dataset, CliNER.68 Its best F1 score for the i2b2 corpus is 0.8. The other system is EliXR,12,49 the only dedicated rule-based EC parser. Both define different entity classes of varied granularities, introducing difficulty for comparison.
CliNER extracts 3 entity classes defined in the i2b2 challenge: test, problem, and treatment. Among the 3 classes, the definition of “problem” is the most similar to the entity class “Condition” in EliIE. Thus, here we chose the performance of “problem” class in CliNER for NER baseline evaluation and compared it to the performance of “Condition” in EliIE. EliXR implements dictionary matching to recognize UMLS concepts and their attributes and constraints. We selected a group of semantic types in UMLS describing disorders, such as “T047: Disease or Syndrome” and “T048: Mental or Behavioral Dysfunction” (for full list, see Table 2). We used EliXR to identify concepts that belong to this group of semantic types to compare with the “Condition” entities recognized by EliIE as the baseline. We further contrasted the raw output from the 3 systems for more qualitative comparison.
Table 2.
Performance of all CRF systems for entity and attribute recognition
| Feature seta | Step 1: Boundary detection |
Steps 1 + 2: Boundary detection + Classification |
|||||
|---|---|---|---|---|---|---|---|
| Precision | Recall | F1-score | Precision | Recall | F1-score | ||
| BOW | Exact | 0.8284 | 0.6661 | 0.7384 | 0.7917 | 0.6363 | 0.7054 |
| Inexact | 0.9411 | 0.8137 | 0.8728 | 0.8715 | 0.7536 | 0.8083 | |
| BOW + POS + Lemma | Exact | 0.8687 | 0.7393 | 0.7988 | 0.8342 | 0.7100 | 0.7671 |
| Inexact | 0.9480 | 0.8325 | 0.8865 | 0.8894 | 0.7811 | 0.8317 | |
| BOW + POS + Lemma + UMLS | Exact | 0.8644 | 0.7574 | 0.8073 | 0.8341 | 0.7309 | 0.7791 |
| Inexact | 0.9445 | 0.8541 | 0.8970 | 0.8836 | 0.7991 | 0.8392 | |
| BOW + POS + Lemma + UMLS + BC | Exact | 0.8682 | 0.7661 | 0.8137 | 0.8382 | 0.7400 | 0.7861 |
| Inexact | 0.9491 | 0.8558 | 0.8978 | 0.8866 | 0.8037 | 0.8432 | |
| Entity classes | Precision |
Recall |
F1 score |
|||
|---|---|---|---|---|---|---|
| Inexact | Exact | Inexact | Exact | Inexact | Exact | |
| *Baseline – CliNER (Problem class) | 0.3692 | 0.3421 | 0.4809 | 0.4140 | 0.4177 | 0.3746 |
| *Baseline – EliXR (Disorder group) | 0.6402 | 0.4289 | 0.8138 | 0.7089 | 0.7176 | 0.5345 |
| Condition | 0.9071 | 0.8566 | 0.8788 | 0.8209 | 0.8927 | 0.8384 |
| Observation | 0.83.97 | 0.8169 | 0.7378 | 0.6760 | 0.7855 | 0.7398 |
| Procedure/Device | 0.8817 | 0.7951 | 0.6581 | 0.6110 | 0.7537 | 0.6910 |
| Drug/Substance | 0.9027 | 0.8573 | 0.7287 | 0.7179 | 0.8064 | 0.7814 |
| Qualifier/Modifier | 0.8807 | 0.8505 | 0.7412 | 0.7253 | 0.8049 | 0.7829 |
| Temporal Constraints | 0.8808 | 0.8045 | 0.8239 | 0.7254 | 0.8514 | 0.7629 |
| Measurement | 0.8984 | 0.8101 | 0.8401 | 0.7168 | 0.8683 | 0.7606 |
|
Overall |
0.8866 |
0.8382 |
0.8037 |
0.7400 |
0.8432 |
0.7861 |
aFeature notation: BOW: bag of words; POS: part of speech; BC: brown clustering. The upper table describes the general performance with different feature sets. The lower table shows the detailed results of each class using the best feature set (BOW + POS + Lemma + UMLS + BC.
*Here we choose the performance of “problem” entity class in CliNER and concepts that belong to UMLS disorder semantic types identified by EliXR as 2 baselines. We compare 2 baselines with the performance of the “Condition” entity class by EliIE. The full list of semantic types we include is: T020, T190, T049, T019, T047, T050, T033, T037, T048, T191, T046, T184. The bold values in feature set (BOW + POS + Lemma + UMLS + BC) correspond to the overall best performance was achieved using the combination of all the features. The bold values in Entity classes (Procedure/Device) due to the less occurrence in the trials, Procedure/Device has the worst performance with F1 score of 0.69 among all the entity classes. The bold values in Entity classes (Overall) indicate by implementing the system with the best setting (BOW+POS+Lemma+UMLS), the overall performance achieves precision, recall and F1 score with 0.84, 0.74, and 0.79 respectively.
RESULTS
All our software source codes and annotation guidelines for EliIE are available at https://github.com/Tian312/ELIIE.
Annotation
Descriptive statistics of our annotated EC corpus are provided in Table 1. To assess whether the training corpus was large enough, we drew a learning curve against the different sizes of the training corpus using the system with the best performance in our study (Figure 4 ). It has been observed that when the size gets close to 200 trials, the results stabilize. Therefore, our training set with 230 trials was sufficient to develop a bioIE system and achieve stable performance.
Table 1.
Descriptive statistics of the annotated eligibility criteria corpus
| Measures | Counts |
|---|---|
| Corpus overview | |
| Total no. of trials | 230 |
| Total no. of sentences | 5634 |
| Average sentence length | 12 |
| Entity class | |
| Condition | 4136 |
| Observation | 1756 |
| Drug/substance | 1464 |
| Procedure/device | 652 |
| Attribute class | |
| Qualifier/modifier | 1715 |
| Temporal constraints | 811 |
| Measurement | 1025 |
| Relation | |
| Modified by | 1096 |
| Has temporal measure | 882 |
| Has measurement | 1551 |
Figure 4.
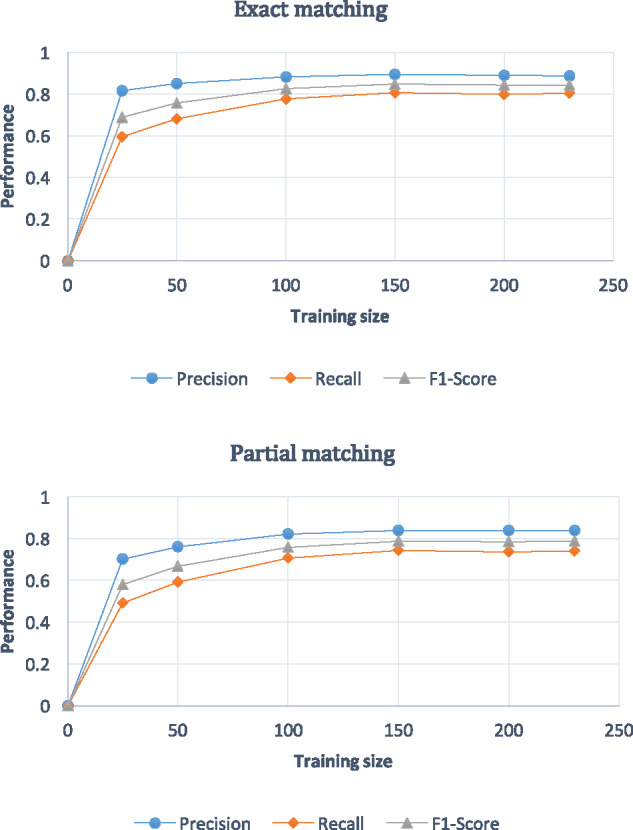
Learning curves for recognition tasks by different sizes of training sizes. The graph on the top describes the learning curves from exact matching evaluation, while the other is partial matching evaluation. Both results show that when the number of the training data is over 150, the performance reaches stable status. In the last version of revision, here the legend F-score should be F1-score.
Entity and attribute recognition
Interannotator partial agreement for the recognition task was 0.90 by F1, which was the upper bound of the named entity and attribute recognition task. For each strategy using additional features, we carried out 5-fold cross-validation and reported average performance in precision, recall, and F1 score. Detailed performance for all strategies is shown in Table 2. We used both exact matching and partial matching to generate evaluation for the 2 steps in NER, boundary detection and classification. The best performance of each entity and attribute class for EliIE is shown in Table 2. The intrinsic baseline performance was based on BOW features only. The best performance was achieved by using a combination of the features including BOW, POS, lemma, UMLS-based features, and BC learned from the entire ClinicalTrials.gov, with precision, recall, and F1 score of 0.84, 0.74, and 0.79, respectively. When using partial match evaluation, F1 score reached 0.84. The “Condition” class achieved the best F1 scores, 0.84 for exact matching and 0.89 for partial matching.
The results of 2 baseline systems and the detailed performance of our best system are shown in the lower part of Table 2. According to the “Condition” class alone, EliIE (F1 score 0.84) largely outperformed both CliNER (F1 score 0.37) and EliXR (F1 score 0.53). Because of different granularity levels predefined for each tool, we also examined the partial evaluation, in which EliIE still outperformed the 2 baselines.
Negation detection
EliIE employed 192 rules in addition to implementing NegEx for negation detection and achieved an accuracy of 0.94 for all the gold standard entities (= #correctly predicted entities/8008). Among those mistakenly predicted, 0.24 (N = 98) were FNs (“negated” predicted as “affirmed”) and 0.76 (N = 302) were FPs.
Relation extraction
We evaluated the relation extraction task independently by using gold standard entities and attributes (entities and attributes defined in annotation texts). Performance is reported in Table 3. The F1 score of all 3 types of relations, “modified by,” “has temporal measurement,” and “has measurement,” were 0.96, 0.76, and 0.92, respectively. The best overall performance (combining all 3 types of relations) by 5-fold cross-validation achieved a precision of 0.87, a recall of 0.92, and an F1 score of 0.89. The corresponding precision, recall, and F1 score for the most complex “has temporal measurement” relation were 0.75, 0.77, and 0.76, respectively.
Table 3.
Performance of clinical relation extraction
| Relation class | Precision | Recall | F1 score |
|---|---|---|---|
| Modified_By | 0.90 | 0.98 | 0.94 |
| Has_TempMea | 0.75 | 0.77 | 0.76 |
| Has_Measurement | 0.88 | 0.95 | 0.92 |
| Overall | 0.87 | 0.92 | 0.89 |
When using the gold standard entities and attributes from annotations, the relation extraction is able to achieve F1 score of 0.89.
The end-to-end evaluation
Using Equation 2 in the Methods section, we measured the general performance of EliIE by reporting an overall accuracy of the TP queries. Exact matching (the output of each step is exactly the same as that from the annotation) was used. The final results for both task-specific and end-to-end evaluation are reported in Table 4. The overall accuracy of EliIE was 0.71, which means EliIE was able to correctly formalize 71% of the queries from our test corpus.
Table 4.
EliIE performance summary
DISCUSSION
Performance analysis
Task-specific evaluations indicate that entity and attribute recognition has room to improve from an F1 score of 0.79. As shown in Table 2, adding valid features helps improve the overall performance of the CRF model but also increases the computing complexity and decreases the efficiency. In the relation extraction step, the performance is satisfactory for a simple set of features (with F1 score being 0.90). Since the definition of each type of relation is unambiguous, there is no misclassification across different relation classes. Errors are FNs (classes 1, 2, and 3 misclassified into class 0) or FP (class 0 misclassified into 1, 2, or 3). Unlike EHR data that contain many nuances of uncertainty, EC texts are relatively explicit regarding positive/negative contexts. EliIE achieves an accuracy of 0.94 in negation detection by simply implementing NegEx and further improves this by adding rules.
Since EliIE is a system composed of multiple subtasks, errors are propagated from task to task along the parsing pipeline. As reported in Table 4, the overall accuracy was 0.71, which means that among all the potential queries in the test free-text EC corpus, EliIE was able to correctly identify and formalize 71% of them. More work is still needed to improve EliIE to meet the needs of practices with higher accuracy requirements, but the results of this study show a promising start.
Comparison with 2 baseline systems
In the end-to-end evaluation for EliIE against the 2 counterpart systems, EliXR12 and CliNER,68 we first compared the performance of the 3 systems in recognizing the concepts in the class “Condition/disorder.” As shown in Table 2, for both exact matching and partial matching, EliIE (F1 score 0.84/0.90) outperformed the 2 counterpart systems (CliNER, 0.37/0.42; EliXR, 0.53/0.72). Even though EC text is known to be syntactically simpler compared to other types of biomedical texts (some are just bullet lists of items),69 it is still semantically complex and has its own style characteristics. The low performance of CliNER shows that it is impractical to adapt existing NLP tools trained in clinical notes to parse EC text; specially designed tools for EC text are needed.
Further, we randomly selected some EC text to input into the 3 systems, whose parsing results for 3 example criteria are shown in Figure 5 and analyzed from the following aspects:
Ability to extract concepts not covered by selected vocabularies. One of the major limitations of all UMLS-based bioIE systems, including EliXR, is their inability to recognize concepts not defined in UMLS. For example, example 2 in Figure 4 is a simple criterion defining 1 standard diagnosis scoring for AD: “GDS-15 score <6.” However, since there is no concept for “GDS-15” in the UMLS Metathesaurus, EliXR recognized “GDS” instead of “GDS-15.” In contrast, EliIE correctly identified the latter based on its frequency of use.
Ability to extract constraints and assign clinical relations from more complex syntax. EliXR is a rule-based system and predefines a set of rules to extract measurements and temporal constraints for identified concepts. However, when the syntactic structure is beyond those rules, EliXR fails. In example 2, since EliXR failed to identify “GDS-15” as 1 concept, subsequently it failed to retrieve the measurement constraint “<6” using its predefined rules. Also, in example 3, EliXR recognized “unstable” as an independent concept for this criterion only because it is a UMLS concept, and incorrectly assigned the temporal constraint “within the previous 2 years” to “unable.” However, EliIE, as a learning-based system, is capable of parsing these constraints correctly.
Ability to define modifiers to represent granularity. In both EliXR and CliNER, there is no class for modifier. In the i2b2 corpus, it includes all modifiers with concepts (https://www.i2b2.org/NLP/Relations/assets/Concept%20Annotation%20Guideline.pdf). Thus, as we can see in Figure 4, CliNER recognizes “severe post-treatment hypersensitivity reaction” and “clinically significant cardiovascular disease” as single concepts. However, from examples 1 and 3, when several modifiers paratactically modify the same concept, CliNER fails to extract this kind of information. Thus, in the EliIE annotation guideline, it defines a “modifier/qualifier” attribute class to constrain the clinical concepts and achieves good performance.
Interoperability with EHR data. In example 2, “GDS-15” is a common diagnosis score for AD. However, since there was no proper class defined in the i2b2 corpus for “GDS-15,” CliNER recognized “GDS-15” and misclassified it as a problem. In comparison, our annotation guideline clearly defines it as an instance of “Observation,” according to OMOP CDM, thus EliIE correctly extracts “GDS-15” and classifies it in the “Observation” table. Generally, OMOP CDM is a suitable data model for EC to achieve interoperability with EHR.
Multitask comprehensiveness. EliIE performs entity recognition, relation extraction, and concept normalization and can be called a comprehensive bioIE system. In contrast, many existing machine learning–based NLP tools in the biomedical field, such as CliNER, can only perform part of the functions and are not comprehensive bioIE systems.
Overall, rule-based systems are largely constrained by the rules and vocabularies on which they depend, while existing machine learning systems trained on public data are not accurate and comprehensive enough and lack interoperability with EHRs. As the first machine learning–based bioIE system specifically designed for EC text and based on a suitable EHR data standard, EliIE achieves promising performance.
LIMITATIONS
This study has a couple of limitations. First, as previously pointed out, the lack of annotator agreement was primarily caused by different concept granularity. We hope the publicly shared annotation guideline can be used to generate more annotated EC corpora in the future to enable continued improvement of EliIE. Second, the generalizability of EliIE outside AD and other neuropsychological diseases is untested. Though different diseases share common EC and similar syntactic structures help with parsing, diseases in different fields have their own distinct characteristics. For example, “anatomical location” is not common in AD but is frequently used in diseases such as cancers, which need pathology and radiology reports. So is genetic information, as we expect there will be more genetic information because of the need for precision medicine. Another limitation is that the “query” defined in our study was simplified and each query contained only 1 clinical entity with its attributes and relations. Queries containing interactions among multiple entities (eg, cohort with disease A only taking drug B) are identified as several independent “unit queries” with “AND” logic relation (query “disease A” AND query “drug B”) in EliIE at present. This limitation will be addressed in our future plan.
Future work
Future work will focus on improving the accuracy and portability of EliIE. First of all, there needs to be clinical relation recognition between clinical entities. For example, 1 criterion is:
Uncontrolled hypertension with systolic BP ≥160 and/or diastolic ≥95 mmHg.
(ClinicalTrials.gov ID: NCT00675090)
EliIE can recognize 1 condition, “hypertension” modified by “uncontrolled,” and 2 observation entities, “systolic BP” and “diastolic (BP)” constrained by 2 measurements, “≥160 (mmHg)” and “≥95 mmHg,” respectively. However, it does not parse how those 3 clinical entities correlate with one another. Similarly for database query, we need information about the logical operator among 3 concepts (eg, AND/OR) to further define a cohort. Thus, to improve EliIE, in addition to understanding relations between entities and attributes, predicting relations among entities will be our next task. Moreover, we will apply EliIE to parse trials in other diseases beyond AD. Finally, further studies are warranted to evaluate EliIE’s portability to other text.
CONCLUSIONS
Our study proposes the first machine learning–based open-source bioIE system, called EliIE, to structure and formalize clinical research EC into OMOP CDM. It achieves competent performance when compared to baseline systems. Our study demonstrates the effectiveness of machine learning methods and the need for a corpus specifically constructed for EC text. We also share an annotation guideline to enable the development of more shared annotated corpora for clinical research of EC in the future.
COMPETING INTERESTS
None.
FUNDING
This work was supported by R01 LM009886 (bridging the semantic gap between research eligibility criteria and clinical data; principal investigator CW).
CONTRIBUTORS
TK proposed methods, designed and carried out the experiments, and drafted the manuscript. SZ participated in the study design and manuscript writing. YT, GH, and AR provided data annotations and guidelines. NE participated in study design and manuscript review. CW supervised the research, participated in study design, and edited the manuscript.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Supplementary Material
References
- 1. Lovato LC, Hill K, Hertert S, Hunninghake DB, Probstfield JL. Recruitment for controlled clinical trials: literature summary and annotated bibliography. Controlled Clinical Trials. 1997;184:328–52. [DOI] [PubMed] [Google Scholar]
- 2. McDonald AM, Knight RC, Campbell MK. et al. What influences recruitment to randomised controlled trials? A review of trials funded by two UK funding agencies. Trials. 2006;71:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Weng C, Tu SW, Sim I, Richesson R. Formal representation of eligibility criteria: a literature review. J Biomed Inform. 2010;433:451–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Weng C, Yaman A, Lin K, He Z. Trend and network analysis of common eligibility features for cancer trials in ClinicalTrials.gov. Smart Health. 2014;8549:130–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. He Z, Carini S, Sim I, Weng C. Visual aggregate analysis of eligibility features of clinical trials. J Biomed Inform. 2015;54:241–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. He Z, Wang S, Borhanian E, Weng C. Assessing the collective population representativeness of related type 2 diabetes trials by combining public data from ClinicalTrials.gov and NHANES. Stud Health Technol Inform. 2015;216:569–73. [PMC free article] [PubMed] [Google Scholar]
- 7. Weng C, Li Y, Ryan P. et al. Distribution-based method for assessing the differences between clinical trial target populations and patient populations in electronic health records. Appl Clin Inform. 2014;52:463–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hernandez AF, Fleurence RL, Rothman RL. The ADAPTABLE Trial and PCORnet: Shining Light on a New Research Paradigm. Ann Intern Med. 2015;1638:635–36. [DOI] [PubMed] [Google Scholar]
- 9. Hripcsak G, Ryan PB, Duke JD. et al. Characterizing treatment pathways at scale using the OHDSI network. Proc Natl Acad Sci USA. 2016;11327:7329–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tu SW, Musen MA. The EON model of intervention protocols and guidelines. In: Cimino JJ, ed. Proc AMIA Annu Fall Symp. Philadelphia: Hanley & Belfus; 1996:587–91. [PMC free article] [PubMed] [Google Scholar]
- 11. Niland J. ASPIRE: Agreement on Standardized Protocol Inclusion Requirements for Eligibility; 2007. Disponible sur: (Consulté le 03/02/2010); 2007. [Google Scholar]
- 12. Weng C, Wu X, Luo Z, Boland MR, Theodoratos D, Johnson SB. EliXR: an approach to eligibility criteria extraction and representation. J Am Med Inform Assoc. 2011;18(Suppl 1): i116–i124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tu S, Peleg M, Carini S, Rubin D, Sim I. Ergo: A Template-based Expression Language for Encoding Eligibility Criteria. 2009, Technical report. [Google Scholar]
- 14. Overhage JM, Ryan PB, Reich CG, Hartzema AG, Stang PE. Validation of a common data model for active safety surveillance research. J Am Med Inform Assoc. 2012;191:54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hripcsak G, Duke JD, Shah NH. et al. Observational Health Data Sciences and Informatics (OHDSI): Opportunities for Observational Researchers. Stud Health Technol Inform. 2015;216:574–78. [PMC free article] [PubMed] [Google Scholar]
- 16. Adler-Milstein J, DesRoches CM, Kralovec P. et al. Electronic health record adoption in US hospitals: progress continues, but challenges persist. Health Aff (Millwood). 2015;3412:2174–80. [DOI] [PubMed] [Google Scholar]
- 17. Cohen AM, Hersh WR. A survey of current work in biomedical text mining. Briefings Bioinformatics. 2005;61:57–71. [DOI] [PubMed] [Google Scholar]
- 18. Friedman C, Hripcsak G, Shagina L, Liu H. Representing information in patient reports using natural language processing and the extensible markup language. J Am Med Inform Assoc. 1999;61:76–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cao Y, Liu F, Simpson P. et al. AskHERMES: An online question answering system for complex clinical questions. J Biomed Inform. 2011;442:277–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Harpaz R, Callahan A, Tamang S. et al. Text mining for adverse drug events: the promise, challenges, and state of the art. Drug Safety. 2014;3710:777–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu F, Chen J, Jagannatha A, Yu H. Learning for Biomedical Information Extraction: Methodological Review of Recent Advances. arXiv preprint arXiv:1606.07993, 2016. [Google Scholar]
- 22. Kim J-D, Ohta T, Tsuruoka Y, Tateisi Y, Collier N. Introduction to the bio-entity recognition task at JNLPBA. In Proc International Joint Workshop on Natural Language Processing in Biomedicine and Its Applications. Geneva, Switzerland: Association for Computational Linguistics; 2004:70–75. [Google Scholar]
- 23. Kim J-D, Ohta T, Pyysalo S, Kano Y, Tsujii JI. Overview of BioNLP’09 shared task on event extraction. In Proceedings of the Workshop on Current Trends in Biomedical Natural Language Processing: Shared Task. Boulder, Colorado: Association for Computational Linguistics; 2009:1–9. [Google Scholar]
- 24. Kim J-D, Pyysalo S, Ohta T, Bossy R, Nguyen N, Tsujii JI. Overview of BioNLP shared task 2011. In Proceedings of the BioNLP Shared Task 2011 Workshop. Association for Computational Linguistics; 2011:1–6. [Google Scholar]
- 25. Uzuner Ö, South BR, Shen S, DuVall SL. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc. 2011;185:552–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Uzuner Ö, Solti I, Cadag E. Extracting medication information from clinical text. J Am Med Inform Assoc. 2010;175:514–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Suominen H, Salanterä S, Velupillai S. et al. Overview of the ShARe/CLEF eHealth evaluation lab 2013. In International Conference of the Cross-Language Evaluation Forum for European Languages. Springer; 2013:212–31. [Google Scholar]
- 28. Pradhan S, Elhadad N, South BR. et al. Evaluating the state of the art in disorder recognition and normalization of the clinical narrative. J Am Med Inform Assoc. 2015;221:143–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Elhadad N, Pradhan S, Chapman W, Manandhar S, Savova G. SemEval-2015 task 14: analysis of clinical text. In Proc of Workshop on Semantic Evaluation. Denver: Association for Computational Linguistics; 2015:303–10. [Google Scholar]
- 30. Pradhan S, Elhadad N, Chapman W, Manandhar S, Savova G. SemEval-2014 task 7: analysis of clinical text. SemEval. Dublin 2014;19999:54. [Google Scholar]
- 31. Smith L, Tanabe LK, nee Ando RJ. et al. Overview of BioCreative II gene mention recognition. Genome Biol. 2008;92:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Krallinger M, Leitner F, Rodriguez-Penagos C, Valencia A. Overview of the protein-protein interaction annotation extraction task of BioCreative II. Genome Biology. 2008;92:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Van Landeghem S, Björne J, Wei C-H. et al. Large-scale event extraction from literature with multi-level gene normalization. PloS One. 2013;84:e55814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Piskorski J, Yangarber R. Information extraction: past, present and future. In Multi-source, Multilingual Information Extraction and Summarization. Berlin: Springer; 2013:23–49. [Google Scholar]
- 35. Tang B, Cao H, Wu Y, Jiang M, Xu H. Recognizing clinical entities in hospital discharge summaries using Structural Support Vector Machines with word representation features. BMC Med Inform Decis Mak. 2013;13(Suppl 1):S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. de Bruijn B, Cherry C, Kiritchenko S, Martin J, Zhu X. Machine-learned solutions for three stages of clinical information extraction: the state of the art at i2b2 2010. J Am Med Inform Assoc. 2011;185:557–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jiang M, Chen Y, Liu M. et al. A study of machine-learning-based approaches to extract clinical entities and their assertions from discharge summaries. J Am Med Inform Assoc. 2011;185:601–06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zweigenbaum P, Demner-Fushman D, Yu H, Cohen KB. Frontiers of biomedical text mining: current progress. Briefings Bioinformatics. 2007;85:358–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Banko M, Cafarella MJ, Soderland S, Broadhead M, Etzioni O. Open information extraction from the web. In IJCAI. 2007;2670–6. [Google Scholar]
- 40. Wu H, Gu Y, Sun S, Gu X. Aspect-based Opinion Summarization with Convolutional Neural Networks. In Neural Networks (IJCNN), Vancouver, Canada: International Joint Conference on; 2016:3157–63. IEEE. [Google Scholar]
- 41. Marujo L, Ling W, Ribeiro R. et al. Exploring events and distributed representations of text in multi-document summarization. Knowledge-Based Sys. 2016;94:33–42. [Google Scholar]
- 42. Huang H, Heck L, Ji H. Leveraging Deep Neural Networks and Knowledge Graphs for Entity Disambiguation. arXiv preprint arXiv:1504.07678, 2015. [Google Scholar]
- 43. Nguyen TH, Grishman R. Combining Neural Networks and Log-linear Models to Improve Relation Extraction. arXiv preprint arXiv:1511.05926, 2015. [Google Scholar]
- 44. Xu Y, Mou L, Li G, Chen Y, Peng H, Jin Z. Classifying relations via long short term memory networks along shortest dependency paths. In Proceedings of Conference on Empirical Methods in Natural Language Processing. 2015. [Google Scholar]
- 45. Miwa M, Bansal M. End-to-end Relation Extraction using LSTMs on Sequences and Tree Structures. In Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics. Berlin, Germany; 2016 (Volume 1: Long Papers):1105–16. [Google Scholar]
- 46. Mikolov T, Chen K, Corrado G, Dean J. Efficient Estimation of Word Representations in Vector Space. arXiv preprint arXiv:1301.3781, 2013. [Google Scholar]
- 47. Jagannatha AN, Yu H. Bidirectional RNN for medical event detection in electronic health records. In Proceedings of the conference. Association for Computational Linguistics. North American Chapter. Meeting; 2016:473. NIH Public Access. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang S, Kang T, Zhang X, Wen D, Elhadad N, Lei J. Speculation detection for Chinese clinical notes: Impacts of word segmentation and embedding models. J Biomed Inform. 2016;60:334–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Levy-Fix G, Yamin A, Weng C. Structuring clinical trial eligibility criteria with common data model. In Proc of 2015 AMIA Joint Summits for Translational Science. San Francisco, CA: AMIA; 2015. [Google Scholar]
- 50. Tuason O, Chen L, Liu H, Blake JA, Friedman C. Biological nomenclatures: a source of lexical knowledge and ambiguity. In Proceedings of the Pacific Symposium of Biocomputing. 2003;9:238. [DOI] [PubMed] [Google Scholar]
- 51. Hao T, Liu H, Weng C. Valx: a system for extracting and structuring numeric lab test comparison statements from text. Methods Inform Med. 2016;553:266–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. National Institutes of Health. ClinicalTrials.gov. http://clinicaltrials.gov. Accessed March 15, 2017. [Google Scholar]
- 53. Alzheimer’s Association. Alzheimer’s disease facts and figures. Alzheimer’s Dementia. 2015;113:332. [DOI] [PubMed] [Google Scholar]
- 54. Schuyler PL, Hole WT, Tuttle MS, Sherertz DD. The UMLS Metathesaurus: representing different views of biomedical concepts. Bull Med Library Assoc. 1993;812:217. [PMC free article] [PubMed] [Google Scholar]
- 55. Stenetorp P, Pyysalo S, Topić G, Ohta T, Ananiadou S, Tsujii JI. BRAT: a web-based tool for NLP-assisted text annotation. In Proceedings of the Demonstrations at the 13th Conference of the European Chapter of the Association for Computational Linguistics. Avignon, France: Association for Computational Linguistics; 2012:102–07. [Google Scholar]
- 56. Kudo T. CRF++: Yet another CRF toolkit. Software. http://crfpp/. Sourceforge. Net, 2005.
- 57. Bird S. NLTK: the natural language toolkit. In Proceedings of the COLING/ACL on Interactive Presentation Sessions. Association for Computational Linguistics; 2006:69–72. [Google Scholar]
- 58. Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. In Proceedings of the AMIA Symposium. American Medical Informatics Association; 2001:17. [PMC free article] [PubMed] [Google Scholar]
- 59. Turian J, Ratinov L, Bengio Y. Word representations: a simple and general method for semi-supervised learning. In Proceedings of the 48th Annual Meeting of the Association for Computational Linguistics. Association for Computational Linguistics; 2010:384–94. [Google Scholar]
- 60. Blei DM, Ng AY, Jordan MI. Latent dirichlet allocation. J Machine Learning Res. 2003;3:993–1022. [Google Scholar]
- 61. Hofmann T. Probabilistic latent semantic analysis. In Proceedings of the 15th Conference on Uncertainty in Artificial Intelligence. Morgan Kaufmann Publishers; 1999:289–96. [Google Scholar]
- 62. Brown DE, Huntley CL. A practical application of simulated annealing to clustering. Pattern Recognition. 1992;254:401–12. [Google Scholar]
- 63. Collobert R, Weston J, Bottou L, Karlen M, Kavukcuoglu K, Kuksa P. Natural language processing (almost) from scratch. J Machine Learning Res. 2011;12:2493–537. [Google Scholar]
- 64. Tang B, Cao H, Wang X, Chen Q, Xu H. Evaluating word representation features in biomedical named entity recognition tasks. BioMed Res Int. 2014;2014:240403. doi:10.1155/2014/240403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform. 2001;345:301–10. [DOI] [PubMed] [Google Scholar]
- 66. Chang C-C, Lin C-J. LIBSVM: a library for support vector machines. ACM Transact Intell Syst Technol. 2011;23:27. [Google Scholar]
- 67. Yim W-W, Denman T, Kwan S, Yestigen M. Tumor information extraction in radiology reports for hepatocellular carcinoma patients. In American Medical Informatics Association Summit on Clinical Research Informatics. San Francisco, CA; 2016:2016:455–64. [PMC free article] [PubMed] [Google Scholar]
- 68. Boag W, Wacome K, Tristan Naumann M, Rumshisky A.. CliNER: A Lightweight Tool for Clinical Named Entity Recognition. AMIA Joint Summits on Clinical Research Informatics (poster). 2015. [Google Scholar]
- 69. Kang T, Elhadad N, Weng C. Initial readability assessment of clinical trial eligibility criteria. AMIA Annu Symp Proc. 2015;2015:687–96. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.