ABSTRACT
Circular RNAs (circRNAs) are a large class of endogenously expressed non-coding RNAs formed by covalently closed loops through back-splicing. High throughput sequencing technologies have identified thousands of circRNAs with high sequence conservation and cell type specific expression in eukaryotes. CircRNAs play multiple important roles in cellular physiology functioning as miRNA sponges, transcriptional regulators, RBP binding molecules, templates for protein translation, and immune regulators. In a clinical context, circRNAs expression is correlated with patient’s clinicopathological features in cancers including breast, liver, gastric, colorectal, and lung cancer. Additionally, distinct properties of circRNAs, such as high stability, exonuclease resistance, and existence in body fluids, show promising role for circRNAs as molecular biomarkers for tumor diagnosis, non-invasive monitoring, prognosis, and therapeutic intervention. Therefore, it is critical to further understand the molecular mechanism underlying circRNAs interaction in tumors and the recent progress of this RNA species in cancer development. In this review, we provide a detailed description of biological functions, molecular role of circRNAs in different cancers, and its potential role as biomarkers in a clinical context.
KEYWORDS: Circrnas, circRNAs functions, miRNA sponge, carcinogenesis, cancer, tumor
1. Background: origin of circRNAs as functional non-coding RNAs
Circular RNAs (circRNAs) are a large group of non-coding RNAs formed by back-splicing. Initial microscopy studies provided the early evidence of circRNAs under denaturing conditions primarily in viral genetic materials [1–3]. Following the observation of circular molecules in viral genome, the first circRNAs in eukaryotic cells were confirmed in the cytoplasmic region of HeLa cells [4], and yeast mitochondrial RNA [5]. Subsequent evidence on individual circRNAs was then continually reported in different human and rat tissues, for example circRNAs derived from ETS-1, SRY, P450, ABP, DYSTROPHIN, MBL, and AML [6–12]. Though multiple experiments prove the existence of circRNAs, the identified circRNAs were generally less abundant than the linear products from the same parental genes. Therefore, circRNAs were considered as rare events with unclear biological functions before the advent of genome-wide sequencing technologies. The development of high throughput sequencing has enabled in-depth characterization of circRNAs for identification, abundance, and potential functions. Among the major features of high throughput sequencing includes longer read lengths, improved circRNA mining bioinformatics algorithms, and more importantly, ribosomal RNA (rRNA)-depleted non-polyadenylated RNA sequencing [13–15]. Since then, the focus of circRNA research has shifted to elucidate the biogenesis and the functional roles of circRNAs. Several biogenesis models have been proposed, including direct back-splicing with ALU and inverted repeats complementation [14,16,17], exon lariat [18], and RNA binding protein (ADAR, MBL, DHX9, FUS, RBM20, and QKI) mediated models [16,19–23]. Furthermore, several biological functions such as microRNA (miRNA) sponges, transcriptional regulator, protein interaction, protein translation, regulator of cancer progression, and immune responses have been implicated (Figure 1). Remarkably, cancer development and progression has been shown to be associated with deregulated circRNA expression. Regulation of gene expression by circRNAs through sponging disease-related miRNAs, forming a complex network of miRNA-circRNA-ncRNA and circRNA-protein interaction will be further discussed in this review. As a result of these diverse functions, aberrant expression of circRNAs impacts the development of a wide array of human diseases, particularly cancer. Furthermore, the distinct characteristics of circRNAs, including long half-life, resistance to exonuclease, expression in tissue, serum, urine, blood, and saliva, make it a promising biomarker with tremendous potentials in a clinical setting. In this review, we have summarised the key functions of circRNAs in the specific context of cancer development and progression and its putative biomarker potential.
Figure 1.
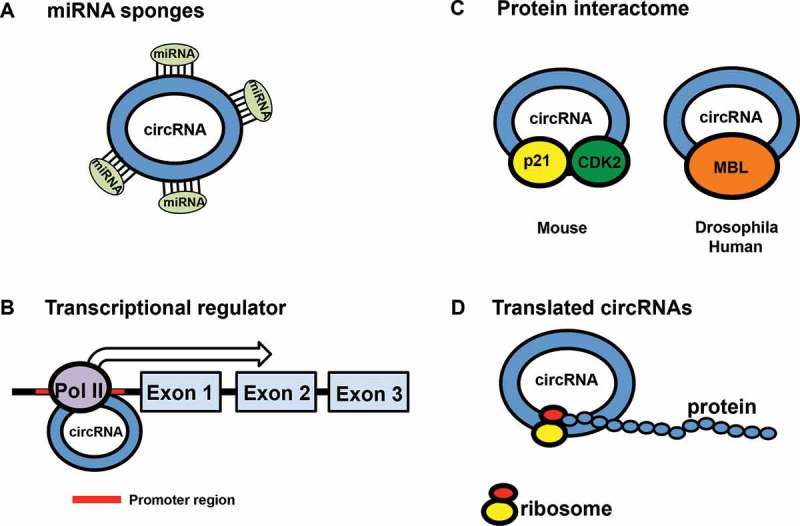
Key functions of circRNAs. (A) CircRNA serves as miRNA sponge that harbors multiple miRNA binding sites that indirectly controls gene expression; (B) CircRNA functions as a transcriptional regulator via binding to RNA polymerase II; (C) CircRNAs act as a protein interactome. In human and Drosophila, RBP (MBL) binds to circRNA to compete with linear alternative splicing; In mouse, cell cycle related proteins bind to circRNAs to strengthen p21/CDK2 interaction and block cell cycle progression; (D) CircRNA that contains open reading frame (ORF) and in-frame stop codon is translated into proteins in a splicing-dependent, cap-independent manner.
2. Known molecular mechanism of circular RNAs
2.1. Microrna sponge
Growing evidence for the functional roles of circRNAs has been discovered in different cellular physiologies (Figure 1). The mechanism of action of circRNAs that received the most attention is their miRNA sponging activities (Figure 1(A)). CircRNAs sequester miRNAs via complementary RNA base-pairing and thus prevent miRNAs from binding their mRNA targets. The first line of evidence for miRNA sponging activity is proven both in vitro and in vivo in CDR1-as circRNAs [15,24]. Both experiments show that CDR1-as is densely bound by AGO and harbor seed regions for miR-7. In vitro assay shows that CDR1-as binds to miR-7 [24]. In addition, over-expression of CDR1-as reduces the miR-7 levels, which then enhances the expression of miR-7 target genes. Using zebrafish and mouse as model systems, in vivo analysis demonstrates the role of CDR1-as in brain development through regulating the activity of miR-7 [15]. Furthermore, the outcome of over-expression of CDR1-as mimics the phenotypes of miR-7 knockdown with morpholinos, which causes the reduction of mid-brain size in zebrafish embryo [15]. In addition, miR-671 has also been proposed to directly cleave CDR1-as due to its almost perfect match, suggesting additional functions of CDR1-as beside miR-7 inhibition [25,26]. In mouse, CDR1-as KO mouse showed impaired sensorimotor gating [25]. The loss of CDR1-as demonstrated a critical interaction between circRNA and miRNA in brain development of zebrafish embryo and brain function of mouse through affecting miR-7 and miR-671 expression [25]. Following the description of circRNAs as miRNA sponges, numerous circRNAs have been implicated to bind disease-associated miRNAs, suggesting the involvement of circRNAs in disease development as discussed in later sections. Nonetheless, genome-wide sequencing analysis reveals that miRNA sponging activity cannot be widely applied across all circRNAs [14,15,27,28] since very few of circRNAs harbor more than 10 seed regions for a single miRNA [28].
2.2. Transcriptional regulators
CircRNAs function as transcriptional regulators (Figure 1(B)). Additionally, nuclear residing exon-intron circRNAs (ElciRNAs), associate with RNA polymerase II, interact with U1 snRNP and Pol II transcription complex at the promoter of their parental genes, thereby control the expression of parental genes [29]. Taken together, circRNAs act as transcriptional regulators to control host gene expression.
2.3. Protein interactome
Multiple evidence demonstrated the role of non-coding RNA in control of gene expression both at the transcription and post-transcriptional levels via physical interaction with RNA binding proteins or other non-coding RNAs [30]. It raised the possibility that circRNAs might have similar roles in mediating cellular functions through serving as a platform for protein interaction (Figure 1(C)). For instance, circRNAs are associated with transcriptional related proteins AGO2 and RNA Pol II [29]. In addition, circ-Mbl competes for binding to splicing related protein Mbl [19]. Furthermore, circFoxo3 forms a ternary complex with cell cycle related proteins, such as CDK2 and p21 [31]. Besides, a subfamily of circRNA is also associated with IMP3, a known oncofetal and tumor marker with post-transcriptional regulation roles [32]. Furthermore, circPABPN1 binds to HuR, and thus prevents HuR from binding to PABPN1 mRNA, thereby reduces PABPN1 translation [33]. The biological functions of related circRNAs and protein interaction are listed in Table 1.
Table 1.
CircRNA and protein interacting partners.
| CircRNAs | Protein involved | Functions | References |
|---|---|---|---|
| circEIF3J, circPAIP2 | RNA Pol II, U1 | Regulate parental host gene expression | Li et al. [29] |
| circMbl | MBL | Competes for alternative splicing with Mbl transcript | Ashwal-Fluss et al. [19] |
| circFoxo3 | CDK2, p21 | Blocks cell cycle progression | Du et al. [31] |
| circNFATC3, circANKRD17 | IMP3 | Subclass of RNA binding protein potentially linked to circRNA biogenesis | Schneider et al. [32] |
| circPABPN1 | HuR | Binds to HuR RBP, serves as a competition between circRNA and cognate mRNA, affecting PABPN1 protein translation | Abdelmohsen et al. [33] |
2.4. Translation of circRNAs
In vitro introduction of internal ribosome entry site (IRES) and reading frames result in translation of engineered circRNAs [34–37]. Therefore, it is possible that AUG-containing exon within circRNAs could be translated into mini proteins (Figure 1(D)). Guo et al showed that majority of the circRNAs provides no evidence for translation [28]. However, the first evidence of protein coding circRNA in eukaryotes is described for circ-ZNF609 [38]. This circRNA contains a start codon with an in-frame stop codon created upon circularization. Translated circ-ZNF609 controls myoblast proliferation, associates with heavy polysomes, and is translated through splicing-dependent, cap-independent mechanism [38]. In addition, a number of circRNAs, for example, circMBL, circDMD, and circFMN, behave as ‘mRNA trap’ which binds to cognate linear mRNA and sequesters the mRNA from undergoing translation, ultimately leading to reduced protein expression level [10,19,39].
3. Additional roles of circRNAs
3.1. Cancer association and potential as a biomarker
The expression of circRNAs is associated with several cancers, which will be reviewed in the following sections (Figure 2(A)). There are two major experimental readouts in measuring the expression of circRNAs in cancer tissues or cell lines: global expression changes (circRNA microarray or RNA-sequencing) and specific circRNA expression measurement.
Figure 2.
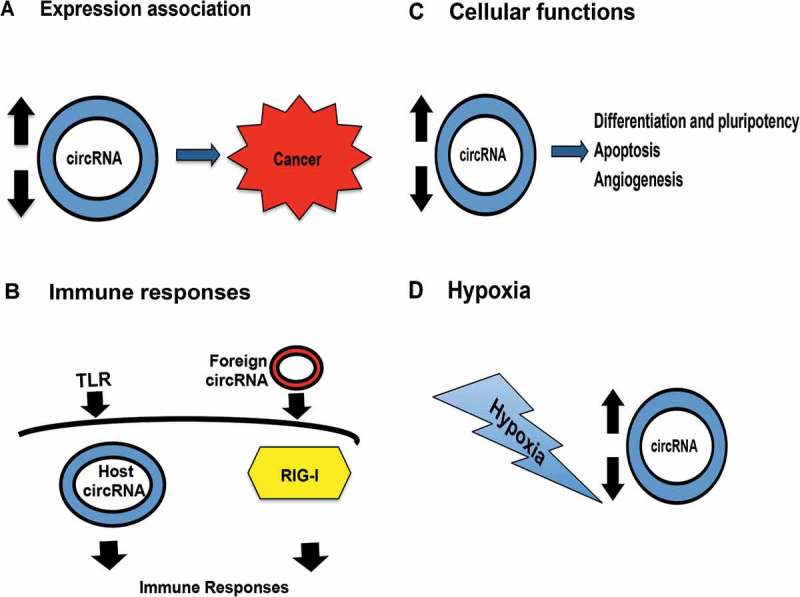
Roles of circRNAs. (A) Aberrant circRNAs expression associates with different cancer outcome. (B) Immune signaling, such as Toll-like receptor, induces the production of host circRNAs (left). Foreign circRNAs evoke RIG-I mediated immune responses via intron identity (right). (C) CircRNAs expression associates with cellular functions. (D) Hypoxia induces circRNAs expression.
3.2. Immune responses
CircRNAs have been implicated in mediating immune responses (Figure 2(B)). For example, circRasGEF1B positively regulates the expression of TLR4/LPS- induced ICAM-1 mature mRNA stability, thereby controlling the innate immune responses [40]. Furthermore, cellular mechanism recognises foreign circRNAs through intron identity, which enables the sensing of self and foreign circRNAs within the cytoplasm of the host [41]. Intriguingly, the mechanism of the lack of 5ʹ triphosphate group in circRNAs in activating the RIG-I pathway remains unknown.
3.3. Cellular differentiation and pluripotency
CircRNAs also play additional roles in cellular functions (Figure 2(C)). From a developmental context, circRNAs are found to be abundantly expressed and upregulated in epidermal stem cell differentiation and largely independent of host gene expression. In particular, circHECTD1 and circZNF91 are predicted to have multiple miRNA and AGO2 binding sites. For example, circZNF91 harbors 24 sites for miR-23b-3p, a key player in keratinocyte differentiation [42]. In addition, Yu et al. showed the involvement of circRNAs in embryonic stem cells for maintaining pluripotency. They showed that circBIRC6 (AGO2-associated) and circCORO1C (non AGO2-associated) act as regulators in pluripotent state. However, only circBIRC6 acts as a ‘miRNA sponge’ to miR-34a and miR-145 to modulate human pluripotency and differentiation. Its biogenesis is promoted by ESRP1 splicing factor, whose expression is controlled by OCT4 and NANOG [43].
3.4. Apoptosis
Several circRNAs are involved in regulating apoptosis. Over-expression of circFoxo3 releases Foxo3 from MDM2-dependent degradation and leads to Puma and Bax-mediated apoptosis [44]. In contrast, unlike circFoxo3, circUBAP2 is shown to inhibit apoptosis [45].
3.5. Angiogenesis
CircFoxo3 has been shown in the inhibition of angiogenesis [46], while hypoxia- induced circZNF292 shows proangiogenic activities [47]. Additionally, circMYLK promotes angiogenesis via VEGFA/VEGFR2 pathway [48].
3.6. Hypoxia
Hypoxia levels positively correlate with the prognosis of cancer patients and serves as a key feature of most solid tumors [49]. Cancer cells increase HIF1α transcription factor expression and promote angiogenesis, proliferation, and metastasis [50]. After hypoxia induction, several hypoxia-associated circRNAs were induced in endothelial cells including circZNF292, circAFF1, circTHSD1, circDENND4C, circSRSF4, and circFOXJ3 [47],(Figure 2(D,). Further experiment on silencing HIF1α in breast cancer cell line MDA-MB-231 showed that hypoxia-induced circDENND4C is a HIF1α associated circRNA. CircDENND4C mediates breast cancer cells proliferation in hypoxic environment and the expression of this circRNA positively correlates with HIF1α level and the tumor size in 30 paired breast cancer and adjacent normal tissues [51].
3.7. Growing number of databases of circRNAs
Over thousands of circRNAs have been identified and documented by different research groups. Hence, the up to date compilation of circRNAs databases and resources is necessary for better navigation and experimental design. Existing databases include circBase [52], circInteractome [53], CircNet [54], circPedia [55], circRNABase [56], circRNADb [57], Circ2Traits [58], CSCD [59], deepBase v2.0 [60], plant-specific PlantcircBase [61] and SomamiR v2.0 [62],(Table 2). In particular, CSCD [59] serves as a comprehensive database for cancer-specific circRNAs navigation.
Table 2.
List of available up-to-date circRNA databases.
| Database | Cell types | Highlights | Link |
|---|---|---|---|
| CircBase |
H. sapiens (hg19) M.musculus (mm9) C.elegans (ce6) L. chalumnae (latCha1) L. menadoensis (latCha1) D.melanogaster (dm3) |
One of the earliest and most comprehensive databases. The custom python scripts can be downloaded | http://www.circbase.org |
| circInteractome | 109 datasets of RNA binding proteins (RBP) and circRNAs for RNA binding sites [39]. | Searches RBP binding to a circRNA and sequences upstream or downstream of circRNAs Identifies RBPs binding to circRNA junctions Identifies miRNAs targeting circRNAs Designs divergent primers Designs siRNAs specific for circRNAs |
https://circinteractome.nia.nih.gov |
| CircNet | 464 RNA-seq samples without PolyA selection from 26 different human tissue samples | Provides a total of 212,950 circRNAs Provides a total of 34,000 circRNAs with junction sites > 3, as highly expressed circRNAs. Provides circRNA expression profiles across 26 different human tissues Predicts circRNA-miRNA interactions and regulatory networks Provides genomic annotation of circRNAs using integrated genome browser |
http://circnet.mbc.nctu.edu.tw |
| CircPedia | 31 human 26 mouse 30 fly 12 worm |
Annotate alternative back-splicing in circRNAs across different cell lines with circRNA characterization pipeline CIRCexplorer2 | http://www.picb.ac.cn/rnomics/circpedia/ |
| circRNABase |
H. sapiens (hg19) M.musculus (mm9) |
Predicts miRNA-circRNA interactions by overlapping circRNA sequence with CLIP-seq peaks from miRNA targets | http://starbase.sysu.edu.cn/mirCircRNA.php |
| circRNADb | CD_19 CD_34 HEK293 Neutrophils Hs68 H9 hESCs Leukemia Leukocyte Normal brain tissue Glioblastoma Oligodendroma |
Contains 32,914 exonic circRNAs with 16,328 protein-coding annotation A total of 46 circRNAs from 37 genes have corresponding proteins expressed based on mass-spectrometry |
http://reprod.njmu.edu.cn/circrnadb |
| Circ2Traits | Data sources taken from 1953 predicted human circRNAs [15], and miR2Disease (174 different human disease) [49] | Measures the likelihood of a circRNA and disease association using hypergeomtric test, p < 0.01 Visualizes circRNA-miRNA-mRNA-lncRNA interactome network for individual disease Information about disease associated SNP in circRNA loci |
http://gyanxet-beta.com/circdb/ |
| CSCD | Human cancer 228 total RNA or polyA(-) RNA seq samples from cancer and normal cell lines |
Identifies 272,152 cancer-specific circRNA Predicts miRNA response element sites Predicts ORF for translatable circRNAs Predicts RBP sites of each circRNA |
http://gb.whu.edu.cn/CSCD |
| deepBase v2.0 |
H. sapiens M.musculus Drosophila C. elegans |
Predicts 101,739 circRNAs Provides a distribution of circRNAs in each species: Caenorhabditis (0.5%), Fly (5.6%), Mouse (1.6%), Human (92.3%) |
http://biocenter.sysu.edu.cn/deepBase/ |
| PlantcircBase |
Arabidopsis thalina (TAIR10) Hordeum vulgare (ASM32608v1.26) Oryza sativa (RGAPv7) Solanum lycopersicum (SL2.40.25) Zea mays (AGPv3.22) |
Predicts circRNAs as miRNA sponges. Provides plant circRNAs with related information (sequence, host genes, expression, experimental validation) |
http://ibi.zju.edu.cn/plantcircbase/ |
| SomaMIR v2.0 | H. sapiens (Contains 620 types of cancer) | Predicts somatic mutations in miRNA target sites and displays somatic mutations within 3ʹ UTR of genes, lncRNAs, and circRNAs which creates or disrupts the target sites | http://compbio.uthsc.edu/SomamiR |
4. Role of circular RNA in cancer progression
4.1. Aberrant expression of circRNAs in cancer
CircRNA expression has been confirmed in various human cell types [15,63], where they play important physiological roles such as cell proliferation and hematopoiesis [64,65]. Deregulated expression of circRNAs and its clinical significance has been reported in several cancers. Global circular RNA abundance negatively correlates with cellular proliferation and a global reduction of circular RNA abundance has been reported in colorectal cell lines, ovarian cancer and idiopathic lung fibrosis compared to normal human tissues [64]. Besides, a genome-wide circRNA array study in breast cancer detected 1155 differentially expressed circRNAs (715 up- and 440 down- regulated) in breast cancer tissues [66]. In blood cancer, circRNA microarray identified 464 differentially expressed circRNAs (147 up and 317 down- regulated) in CN-AML patients compared to healthy controls, in which 12 dysregulated circRNAs were expressed 10 fold more than healthy controls [67]. In bladder carcinoma, two independent microarray studies detected 469 (285 up- and 184 down- regulated) [68], and 571 (47 up- and 524 down- regulated) dysregulated circular transcripts respectively [69]. In pancreatic ductal adenocarcinoma (PDAC), using circRNA microarray, a total of 351 (209 up- and 142 down- regulated) circRNAs were aberrantly expressed between 6 PDAC cancer samples and paired adjacent tissues [70]. Interestingly, more circRNAs were upregulated rather than downregulated in liver cancer. A microarray study identified 127 (113 up- and 14 down- regulated) differentially expressed circRNAs in liver cancer while another independent study reported a total of 226 (189 up- and 37- down regulated) differentially expressed circRNAs [71,72].
The expression levels of specific circRNAs such as hsa_circRNA_103809, hsa_circRNA_104700 and hsa_circ_001988 are downregulated in cancerous tissue compared to the normal adjacent tissue [73,74]. This is significantly associated with clinicopathological features in colorectal cancer (CRC) patients such as metastasis, differentiation and perineural invasion respectively [73,74]. On the other hand, circ_BANP and hsa_circ_0000069, and hsa_circ_001569 have been shown to be overexpressed in CRC cancerous tissue and proposed to be a prognostic and therapeutic marker because of their role in cancer progression [75–77]. In addition, a microarray analysis was performed by one group to study chemoradiation resistance (CRR) in response to 5-fluorouracil in CRC patients [78]. This study had identified 71 circRNAs that are differentially expressed in chemoradiation-resistant CRC cells. Out of these expression of 5 circRNAs was validated with quantitative reverse transcription PCR (qRT-PCR) and interacts with 355 miRNAs according to bioinformatics predictions [78]. Several other circular RNAs have been shown to play a role in CRC progression by acting as miRNA sponges or through transcriptional regulation and have been reviewed elsewhere [79]. Additionally, downregulation of circRNA abundance at a global level has been shown in several colon cancer cell lines having a mutant KRAS indicating a role of circRNA in tumorigenesis [80].
In gastric cancer (GC), hsa_circRNA_002059, hsa_circRNA_0000181 and hsa_circ_0000190 expression has been shown to be downregulated in cancerous tissue compared to adjacent normal tissue with significant correlation to metastasis [81–83]. Based on the stability of hsa_circRNA_0000181 and hsa_circ_0000190 expression in tissue and plasma samples, they had been proposed to be novel biomarkers for prognosis and diagnosis of gastric cancer [82,83]. At the same time, another study reported that the expression of circPVT1 that was derived from PVT1 gene is upregulated in GC tissue and acts as a sponge for miR-125 family [84]. The levels of circPVT1 has been proposed to be an independent prognostic biomarker for disease-free and overall survival in GC patients [84]. Moreover, in a cohort of 51 patients, hsa_circRNA_0067934 was significantly overexpressed in esophageal squamous cell carcinoma (ESCC) compared to the adjacent normal tissue and was associated with poor cellular differentiation [85].
Additionally, by combining results from 5 gastric cancer (GC) cell lines and 257 tumor tissue specimens it was found that hsa_circRNA_0001895 expression levels were significantly correlated with patient’s clinicopathological factors suggesting its crucial role in gastric cancerogenesis and therefore is a potential biomarker for disease prognosis prediction [86]. In a similar study, hsa_circRNA_0003159 was shown to be downregulated in GC tissue compared to the adjacent normal tissue which was of diagnostic significance [87]. Global circRNA expression profiling in GC found a list of top 10 upregulated and downregulated circRNAs that have a significant aberrant expression in GC, amongst which hsa_circ_ 0014717 was downregulated in 77.2% gastric cancer tissues [88].
In prostate cancer, circSMARCA5 is an androgen-induced circRNA. In a study by Kong et al, circSMARCA5 is upregulated in 5 prostate cancer cell lines and 21 paired prostate cancer tissue samples compared with match normal tissues [89].
In hepatocellular carcinoma (HCC), hsa_circ_0001649 expression was downregulated in 89 paired samples of HCC and adjacent tissues, and its expression was correlated with tumor size and tumor embolus [90]. In addition, it has also been shown that hsa_circ_0004018 expression is lower in HCC and correlates with serum alpha-fetoprotein (AFP) level tumor diameter, differentiation, and tumor-node-metastasis stage [91]. However, another observational study showed that hsa_circ_0005075 expression was associated solely with the tumor size but not with other clinicopathological characteristics [92]. Interestingly, a detailed study on both ZKSCAN1 mRNA and circZKSCAN1 showed that the expression level of both transcript species was downregulated in HCC [93]. Additionally, knockdown of both species accelerated cell proliferation, migration, and invasion, while over-expression repressed the progression of HCC [93].
4.2. Network of circRNA-miRNA-mRNA in cancer
Recently, a study on gastric cancer (GC) tissues identified a network of circRNA, miRNA and mRNAs that are differentially expressed between the GC tissues and adjacent normal tissue samples [94]. This raises a possibility of a direct linkage between the regulatory properties among the network of circRNA-miRNA-mRNA expressions.
For instance, a regulatory network of circ_0006528-miR7–5p-Raf1 has been proposed in breast cancer. There is a negative correlation between circ_0006528 and miR-7–5p expression in Adriamycin (ADM) resistant cancer tissues. Raf1 is a direct target of miR-7–5p [95]. Knocking down circ_0006528 in MCF-7/ADM and MDA-MB-231/ADM cell lines reduced Raf1 mRNA and protein levels, suggesting a regulatory role of circRNA-miRNA-mRNA axis in ADM resistant breast cancer [96].
In addition, a network of circHIAT1/miR-195-5p/29a-3p/29c-3p/CDC42 is involved in clear cell renal cell carcinoma (ccRCC) development [97]. Androgen receptor suppressed circHIAT1 expression resulting in deregulated miR-195-5p/29a-3p/29c-3p expression, ultimately leading to increased CDC42 expression to enhance ccRCC cell migration and invasion. CircHIAT1 possibly acts as a ‘miRNA reservoir’ to stabilise miRNAs in this context.
Moreover, a circRNA and miRNA expression study in hepatocellular carcinoma (HCC) demonstrated a potential regulatory role of circRNA_000839/miR-200b/RhoA in the pathogenesis of HCC. This study indicated an inverse correlation between miR-200b with RhoA and circ_000839, suggesting a role of this interplay for miR-200b in suppression of the invasion and migration in HCC [98].
Furthermore, a microarray expression study in bladder carcinoma demonstrated a differential expression profile of circTCF25/miR-103a-3p/miR-107/CDK6 regulatory pathway. Over-expression of circTCF25 promotes proliferation and migration of EJ and T24 bladder cancer cell lines [68].
4.3. Oncogenic properties of circRNAs
CircRNAs also have oncogenic and proto-oncogenic roles to promote cancer formation. Fusion circRNAs (f-circRNA) are formed as a result of cancer associated genomic translocations and have tumor-promoting properties such as increased cell viability, resistance to therapy and cellular transformation in leukemic cells in vivo [99]. For example, cZNF292 has been shown to promote tube formation in gliomas and its inhibition is associated with suppression of tube formation by inhibition of cell cycle progression [100]. In non-small cell lung cancer, over expression of circRNA_100876 in tumor tissues compared to adjacent non-tumorous tissue has been correlated to lymph node metastasis, tumor staging and overall survival which is of prognostic and therapeutic significance [101].
4.4. CircRNA and miRNA sponging activity in cancer
MiRNA and circRNA interaction have also been reported in multiple cancer types (Table 3). For example in bladder cancer, upregulation of circTCF25 has been shown to downregulate miR-103a-3p and miR-107 and at the same time increase CDK6 expression [68]. This interaction has been linked to increased proliferation and migration both in vitro and in vivo. Interestingly, analysis of differentially expressed mRNA, lncRNA and circRNA in bladder cancer revealed regulatory networks whose role in pathogenesis of bladder cancer has been demonstrated [102]. Besides, in bladder carcinoma, circMYLK sponges miR-29a and relieve the repression of VEGFA, leading to activation of VEGFA/VEGFR2 and downstream Ras/ERK pathway [48]. In addition, a circularised exon 2 from HIPK3 gene, circHIPK3, binds to miR-558, and suppresses the HPSE expression. Over-expression of circHIPK3 inhibits migration and invasion in vitro, while suppresses the growth, metastasis, and angiogenesis of bladder cancer cells in vivo [69].
Table 3.
miRNAs sponge activity of circRNAs in cancer and clinical outcomes.
| Cancer type | circRNA | miRNA | miRNA targets | Outcome | References |
|---|---|---|---|---|---|
| Bladder | circTCF25 | miR-103a-3p, miR-107 | CDK6 | Increase proliferation and migration | Zhong et al. [68] |
| Bladder | circMYLK | miR-29a | VEGFA | Relieve the repression and activate Ras/ERK pathway | Zhong et al. [48] |
| Bladder | circHIPK3 | miR-558 | HPSE | Suppress HPSE expression | Li et al. [69] |
| Breast | circABCB10 | miR-1271 | - | Promote proliferation and progression | Liang et al. [103] |
| Breast | circFoxo3 | miR-22 miR-136 miR-138 miR-149 miR-433 miR-762 miR-3614-5p miR-3622-5p |
- | Inhibit tumor growth and angiogenesis | Yang et al. [46] |
| Colorectal | hsa_circ001569 | miR-145 | E2F5, BAG4, FMNL2 | Proliferation and invasion | Xie et al. [77] |
| Esophageal | circITCH | miR-7 miR-17 miR-214 |
ITCH | Promote phosphorylated Dvl2, inhibit Wnt/b-catenin pathway | Li et al. [104] |
| Gastric | hsa_circLARP4 | miR-424 | LATS1 | Inhibit proliferation | Zhang et al. [105] |
| Gastric | hsa_circ100269 | miR-630 | - | Proliferation | Zhang et al. [106] |
| Hepatocellular carcinoma | ciRS-7 | miR-7 | - | Microvascular invasion | Xu et al. [107] |
| Hepatocellular carcinoma | circRNA-100338 | miR-141-3p | MTSS1 (in silico) | Promote metastatic progression | Huang et al. [72] |
| Hepatocellular carcinoma | circMTO1 | miR-9 | p21 | Suppress tumor development, correlate with poor prognosis | Han et al. [109] |
| Liver | circFUT8 | miR-570-3p miR-17-3p miR-552-3p |
Ren et al. [71] | ||
| Liver | circZFR | miR-511-5p miR-130b-5p miR-642a-5p miR-532-3p mir-329-5p |
|||
| Liver | circIPO11 | miR-659-3p miR-424-5p miR-106a-3p |
|||
Additionally, circABCB10 is abnormally upregulated in breast cancer tissue. The sponge activity of circABCB10 on miR-1271 resulted in breast cancer proliferation and progression [103]. Additionally, a mechanistic study on all three forms of Foxo3; circFoxo3, Foxo3 mRNA, and pseudogene (Foxo3P), showed that all three forms suppress tumor growth and cell proliferation, and circFoxo3, Foxo3P, and Foxo3 are targets of miRNAs [46].
In colorectal cancer (CRC), in vitro over-expression of hsa_circ_001569 in SW480 and HCT116 cells, and silencing of hsa_circ_001569 in SW620 and LOVO cells showed that this circRNA is a positive regulator in mediating cell proliferation and invasion through sponging the miR-145 expression levels, thereby upregulating its targets such as E2F5, BAG4, and FMNL2 [77].
In esophageal squamous cell carcinoma (ESCC), circITCH sponges multiple miRNAs (miR-7, miR-17, and miR-214), which subsequently increase the ITCH expression. Increased expression of ITCH promotes Dvl2 degradation, and thus inhibits Wnt/β-catenin pathway in ESCC [104].
In gastric cancer (GC), hsa_circRNA_LARP4 has been shown to sponge miR-424 level which inhibits tumor proliferation and regulates its target gene LATS1 gene in GC [105]. Hsa_circRNA_LARP4 has therefore been proposed to be a novel tumor suppressor and a potential biomarker in GC [105]. Moreover, hsa_circRNA_100269 downregulates the expression of miR-630 and the expression of hsa_circRNA_100269 negatively correlated with that of miR-630 in GC tissues [106]. The interaction of hsa_circRNA_100269 and miR-630 has been shown to regulate tumor cell proliferation in GC [106].
In liver cancer, circular RNA regulates expression of miRNAs by acting as a miRNA sponge and have clinical consequences, especially in hepatocellular carcinoma (HCC) [107]. CiRS-7 acts as a miRNA sponge and inhibitor of miR-7, a tumor suppressor. CiRS-7 may be considered as a risk factor in HCC as it is significantly correlated with hepatic microvascular invasion [107,108]. Besides, another study reported that three circRNAs, namely, circFUT8, circZFR, and circIPO11 were upregulated in 40 clinical samples. However, the sponging activities of the predicted circFUT8-miR-570-3p/miR-17-3p/miR-552-3p, circZFR-miR-511-5p/miR-130b-5p/miR-642a-5p/miR-532-3p/mir-329-5p, and circIPO11- miR-659-3p/miR-424-5p/miR-106a-3p remains to be tested [71]. Another study in hepatitis B-related HCC revealed that circRNA-100338 acts as an endogenous sponge for miR-141-3p in regulating invasion potential of liver cancer [72]. A functional analysis in HCC revealed the role of circMTO1 in inhibiting HCC growth through sponging miR-9 resulted in the increased expression of p21 [109].
Role of circRNAs in molecular pathogenesis of basal cell carcinoma has also been demonstrated through the identification of 71 deregulated circRNAs and 354 potential miRNA response elements among these circRNAs in basal cell carcinoma [110]. Similarly, deregulation of circRNA expression has been demonstrated in malignancies such as oral, gastric, breast and cervical cancers [58].
4.5. CircRNA responses to radiation therapy and chemotherapy
Deregulation of circular RNA has also been linked to the acquired resistance to radiation therapy. For instance, 57 upregulated and 17 downregulated circRNAs (fold change ≥ 2.0 and P < 0.05) were found in radioresistant esophageal cancer cell line KYSE-150R compared to the parental KYSE-150 cell line [111]. An interaction analysis revealed correlations between circRNA-microRNA-mRNA as distinct interaction nodes [111]. Moreover, a comparison between seven acute myeloid leukemia (AML) patients undergoing standard induction chemotherapy at both without prior treatment and complete remission (CR) stage showed increased hsa_circ_0004277 [67]. This implies a potential restoration of dysregulated hsa_circ_0004277 expression after chemotherapy.
5. Role of circular RNA as biomarkers
The stability due to their cyclic structure and tissue specific expression of circRNAs make them a suitable candidate for biomarker studies [13,15]. CircRNAs are secreted and transported in human exosomes where they are abundant and stable [112]. In colorectal cells (CRC), expression of circRNAs has been detected in cells as well as in exosomes secreted by three CRC cell lines differing in KRAS mutation status [80]. Mammalian brain has been shown to have abundant circRNAs which are conserved in sequence and expression [113]. Analysis of RNA-seq data of human whole blood samples has revealed reproducible and abundant expression of circRNAs making them amenable for routine blood sample testing [114]. Similarly, RNA-seq with in depth bioinformatics analysis in human cell-free saliva samples validated the expression of circRNAs [115]. These biological properties make circRNAs promising biomarker candidates for diagnosis, prognosis and detection of cancer from blood and saliva samples.
In gastric cancer for example, the expression of hsa_circRNA_002059 has been correlated with distal metastasis, TNM stage, gender and age and therefore it has been proposed to be a biomarker for the diagnosis [81]. Similarly, in esophageal squamous cell carcinoma, hsa_circRNA_0067934 has been associated with poor differentiation and promote proliferation [85]. It can plausibly serve as a novel biomarker for disease progression or as a therapeutic target [85]. In bladder carcinoma, circTCF25 has been suggested to be a promising biomarker [68]. Research to validate the expression of these biomarkers in easily available specimens such as blood and saliva will increase the utility of these biomarkers.
6. Current limitations and challenges
Despite the major progress made this far, several limitations and challenges need to be addressed. A standard naming system is required to standardise circRNAs. Annotation of each circRNAs would be useful for further research on specific circRNA species [116]. In addition, one of the major limitations in circRNA studies is that most publicly available RNA-seq data sets related to cancer were prepared using a poly (A) purification step to enrich mRNA [116]. This might reduce the possibility of identifying circRNAs, which naturally lack poly (A) tails. Furthermore, genome wide studies were performed using microarrays or limited number of tissue samples, partly due to the lack of experienced pathologist in tumor tissue examination [116]. In most circRNAs discoveries involving cancer, unbiased RNA-seq is employed. However, it should be noted that the RNA quality and sample handling during RNA-seq library preparation step affects the discovery of circRNAs of different types and sizes. Variation in RNA-seq preparation will alter circRNAs detection, for example, size selection excludes small circRNA while oligo dT favors linear mRNA and biases against circRNAs [116,117]. Another challenge for the discovery of circRNA involves the detection and quantification of circRNA. The study of circRNAs may be hampered by template switching and rolling circle amplification during reverse transcription and by amplification bias during PCR [117,118].
7. Concluding remark
In conclusion, recent investigations in circRNAs and their roles in cancer development and progression are still limited. However, constant improvement of circRNA bioinformatics pipelines and application of new techniques will eventually pinpoint the roles of circRNAs beyond acting as miRNA sponges. Better understanding of the regulation of circRNAs’ aberrant expression, circRNA/miRNA/mRNA networks and sponging activity would lead to robust biomarker development for cancer detection and clinical prognosis. Finally, further studies on this ancient RNA will provide a significant new perspective in cancer biology.
Biography
Ng. W.L., Mohidin. T.B.M., and Shukla. K., perceived the ideas, collected the related papers and wrote the manuscript.
Funding Statement
This work was supported by University of Malaya BKP grant [BK044-2016].
Abbreviations
- ADM
adriamycin
- AML
acute myeloid leukemia
- ccRCC
clear cell renal cell carcinoma
- circRNA
circular RNA
- CR
complete remission
- CRC
colorectal cancer
- CRR
chemoradiation resistance
- ESCC
esophageal squamous cell carcinoma
- f-circRNA
fusion circRNA
- GC
gastric cancer
- HCC
hepatocellular carcinoma
- KO
knockout
- miRNA
micro RNA
- PDAC
pancreatic ductal adenocarcinoma
- RBP: RNA
binding protein
Acknowledgments
The work was supported by research funding from University of Malaya BKP grant [BK044-2016] and School of Science, Monash University Malaysia. We thank Dr. Ea Chee Kwee for critically reading the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- 1.Kolakofsky D. Isolation and characterization of Sendai virus DI-RNAs. Cell. 1976;8:547–555. [DOI] [PubMed] [Google Scholar]
- 2.Hewlett MJ, Pettersson RF, Baltimore D. Circular forms of Uukuniemi virion RNA: an electron microscopic study. J Virol. 1977;21:1085–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kos A, Dijkema R, Arnberg AC, et al. The hepatitis delta (delta) virus possesses a circular RNA. Nature. 1986;323:558–560. [DOI] [PubMed] [Google Scholar]
- 4.Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280:339–340. [DOI] [PubMed] [Google Scholar]
- 5.Arnberg AC, Vanommen GJB, Grivell LA, et al. Some yeast mitochondrial Rnas are circular. Cell. 1980;19:313–319. [DOI] [PubMed] [Google Scholar]
- 6.Cocquerelle C, Mascrez B, Hetuin D, et al. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7:155–160. [DOI] [PubMed] [Google Scholar]
- 7.Capel B, Swain A, Nicolis S, et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73:1019–1030. [DOI] [PubMed] [Google Scholar]
- 8.Zaphiropoulos PG. Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: correlation with exon skipping. Proc Natl Acad Sci U S A. 1996;93:6536–6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zaphiropoulos PG. Exon skipping and circular RNA formation in transcripts of the human cytochrome P-450 2C18 gene in epidermis and of the rat androgen binding protein gene in testis. Mol Cell Biol. 1997;17:2985–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gualandi F, Trabanelli C, Rimessi P, et al. Multiple exon skipping and RNA circularisation contribute to the severe phenotypic expression of exon 5 dystrophin deletion. J Med Genet. 2003;40:e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu AN, Chen XH, Tan YH, et al. Identification of a novel circularized transcript of the AML1 gene. BMB Rep. 2013;46:163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Houseley JM, Garcia-Casado Z, Pascual M, et al. Noncanonical RNAs from transcripts of the Drosophila muscleblind gene. J Hered. 2006;97:253–260. [DOI] [PubMed] [Google Scholar]
- 13.Salzman J, Gawad C, Wang PL, et al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLOS ONE. 2012;7:e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. [DOI] [PubMed] [Google Scholar]
- 16.Ivanov A, Memczak S, Wyler E, et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Reports. 2015;10:170–177. [DOI] [PubMed] [Google Scholar]
- 17.Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28:2233–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrett SP, Wang PL, Circular SJ. RNA biogenesis can proceed through an exon-containing lariat precursor. Elife. 2015;4:e07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66. [DOI] [PubMed] [Google Scholar]
- 20.Aktas T, Avsar Ilik I, Maticzka D, et al. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature. 2017;544:115–119. [DOI] [PubMed] [Google Scholar]
- 21.Errichelli L, Dini Modigliani S, Laneve P, et al. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat Commun. 2017;8:14741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khan MA, Reckman YJ, Aufiero S, et al. RBM20 regulates circular RNA production from the titin gene. Circ Res. 2016;119:996–1003. [DOI] [PubMed] [Google Scholar]
- 23.Conn SJ, Pillman KA, Toubia J, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. [DOI] [PubMed] [Google Scholar]
- 24.Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. [DOI] [PubMed] [Google Scholar]
- 25.Piwecka M, Glazar P, Hernandez-Miranda LR, et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017. Sep 22;357(6357). pii: eaam8526 [DOI] [PubMed] [Google Scholar]
- 26.Hansen TB, Wiklund ED, Bramsen JB, et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414–4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.You X, Vlatkovic I, Babic A, et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci. 2015;18:603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo JU, Agarwal V, Guo H, et al. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–264. [DOI] [PubMed] [Google Scholar]
- 30.Turner M, Galloway A, Vigorito E. Noncoding RNA and its associated proteins as regulatory elements of the immune system. Nat Immunol. 2014;15:484–491. [DOI] [PubMed] [Google Scholar]
- 31.Du WW, Yang W, Liu E, et al. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schneider T, Hung LH, Schreiner S, et al. CircRNA-protein complexes: IMP3 protein component defines subfamily of circRNPs. Sci Rep. 2016;6:31313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abdelmohsen K, Panda AC, Munk R, et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14:361–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abe N, Matsumoto K, Nishihara M, et al. Rolling circle translation of circular RNA in living human cells. Sci Rep. 2015;5:16435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen CY, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. 1995;268:415–417. [DOI] [PubMed] [Google Scholar]
- 36.Kramer MC, Liang D, Tatomer DC, et al. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 2015;29:2168–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA. 2015;21:172–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Legnini I, Di Timoteo G, Rossi F, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66:22–37 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chao CW, Chan DC, Kuo A, et al. The mouse formin (Fmn) gene: abundant circular RNA transcripts and gene-targeted deletion analysis. Mol Med. 1998;4:614–628. [PMC free article] [PubMed] [Google Scholar]
- 40.Ng WL, Marinov GK, Liau ES, et al. Inducible RasGEF1B circular RNA is a positive regulator of ICAM-1 in the TLR4/LPS pathway. RNA Biol. 2016;13:861–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen YG, Kim MV, Chen X, et al. Sensing Self and Foreign Circular RNAs by Intron Identity. Mol Cell. 2017;67:228–38 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kristensen LS, Okholm TLH, Veno MT, et al. RNAs are abundantly expressed and upregulated during human epidermal stem cell differentiation. RNA Biol. 2018;15:280–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu CY, Li TC, Wu YY, et al. The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nat Commun. 2017;8:1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Du WW, Fang L, Yang W, et al. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24:357–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang H, Wang G, Ding C, et al. Increased circular RNA UBAP2 acts as a sponge of miR-143 to promote osteosarcoma progression. Oncotarget. 2017;8:61687–61697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang W, Du WW, Li X, et al. Foxo3 activity promoted by non-coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene. 2016;35:3919–3931. [DOI] [PubMed] [Google Scholar]
- 47.Boeckel JN, Jae N, Heumuller AW, et al. Identification and Characterization of Hypoxia-Regulated Endothelial Circular RNA. Circ Res. 2015;117:884–890. [DOI] [PubMed] [Google Scholar]
- 48.Zhong Z, Huang M, Lv M, et al. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305–317. [DOI] [PubMed] [Google Scholar]
- 49.Rundqvist H, Johnson RS. Tumour oxygenation: implications for breast cancer prognosis. J Intern Med. 2013;274:105–112. [DOI] [PubMed] [Google Scholar]
- 50.Pawlus MR, Wang L, Hu CJ. STAT3 and HIF1alpha cooperatively activate HIF1 target genes in MDA-MB-231 and RCC4 cells. Oncogene. 2014;33:1670–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liang G, Liu Z, Tan L, et al. HIF1alpha-associated circDENND4C Promotes Proliferation of Breast Cancer Cells in Hypoxic Environment. Anticancer Res. 2017;37:4337–4343. [DOI] [PubMed] [Google Scholar]
- 52.Glazar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20:1666–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dudekula DB, Panda AC, Grammatikakis I, et al. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13:34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu YC, Li JR, Sun CH, et al. CircNet: a database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res. 2016;44:D209–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang XO, Dong R, Zhang Y, et al. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016;26:1277–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li JH, Liu S, Zhou H, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen X, Han P, Zhou T, et al. circRNADb: A comprehensive database for human circular RNAs with protein-coding annotations. Sci Rep. 2016;6:34985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ghosal S, Das S, Sen R, et al. Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits. Front Genet. 2013;4:283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xia S, Feng J, Chen K, et al. CSCD: a database for cancer-specific circular RNAs. Nucleic Acids Res. 2018. Jan 4;46(D1):D925–D929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zheng LL, Li JH, Wu J, et al. deepBase v2.0: identification, expression, evolution and function of small RNAs, LncRNAs and circular RNAs from deep-sequencing data. Nucleic Acids Res. 2016;44:D196–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chu Q, Zhang X, Zhu X, et al. PlantcircBase: A database for plant circular RNAs. Mol Plant. 2017;10:1126–1128. [DOI] [PubMed] [Google Scholar]
- 62.Bhattacharya A, Cui Y. Somami R. 2.0: a database of cancer somatic mutations altering microRNA-ceRNA interactions. Nucleic Acids Res. 2016;44:D1005–D1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Salzman J, Chen RE, Olsen MN, et al. Cell-type specific features of circular RNA expression. PLOS Genetics. 2013;9:e1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bachmayr-Heyda A, Reiner AT, Auer K, et al. Correlation of circular RNA abundance with proliferation–exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. 2015;5:8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bonizzato A, Gaffo E, Te Kronnie G, et al. CircRNAs in hematopoiesis and hematological malignancies. Blood Cancer J. 2016;6:e483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lu L, Sun J, Shi P, et al. Identification of circular RNAs as a promising new class of diagnostic biomarkers for human breast cancer. Oncotarget. 2017;8:44096–44107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li W, Zhong C, Jiao J, et al. Characterization of hsa_circ_0004277 as a new biomarker for acute myeloid leukemia via circular RNA profile and bioinformatics analysis. Int J Mol Sci. 2017. Mar 9;18(3). pii: E597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhong Z, Lv M, Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci Rep. 2016;6:30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li Y, Zheng F, Xiao X, et al. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Reports. 2017;18:1646–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li H, Hao X, Wang H, et al. Circular RNA expression profile of pancreatic ductal adenocarcinoma revealed by microarray. Cell Physiol Biochem. 2016;40:1334–1344. [DOI] [PubMed] [Google Scholar]
- 71.Ren S, Xin Z, Xu Y, et al. Construction and analysis of circular RNA molecular regulatory networks in liver cancer. Cell Cycle. 2017;16(22):2204–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang XY, Huang ZL, Xu YH, et al. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-100338/miR-141-3p pathway in hepatitis B-related hepatocellular carcinoma. Sci Rep. 2017;7:5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang P, Zuo Z, Shang W, et al. Identification of differentially expressed circular RNAs in human colorectal cancer. Tumour Biol. 2017;39:1010428317694546. [DOI] [PubMed] [Google Scholar]
- 74.Wang X, Zhang Y, Huang L, et al. Decreased expression of hsa_circ_001988 in colorectal cancer and its clinical significances. Int J Clin Exp Pathol. 2015;8:16020–16025. [PMC free article] [PubMed] [Google Scholar]
- 75.Zhu M, Xu Y, Chen Y, et al. Circular BANP, an upregulated circular RNA that modulates cell proliferation in colorectal cancer. Biomed Pharmacother. 2017;88:138–144. [DOI] [PubMed] [Google Scholar]
- 76.Guo JN, Li J, Zhu CL, et al. Comprehensive profile of differentially expressed circular RNAs reveals that hsa_circ_0000069 is upregulated and promotes cell proliferation, migration, and invasion in colorectal cancer. Onco Targets Ther. 2016;9:7451–7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xie H, Ren X, Xin S, et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7:26680–26691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xiong W, Ai YQ, Li YF, et al. Microarray analysis of circular RNA expression profile associated with 5-fluorouracil-based chemoradiation resistance in colorectal cancer cells. Biomed Res Int. 2017;2017:8421614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Taborda MI, Ramirez S, Bernal G. Circular RNAs in colorectal cancer: possible roles in regulation of cancer cells. World J Gastrointest Oncol. 2017;9:62–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dou Y, Cha DJ, Franklin JL, et al. Circular RNAs are down-regulated in KRAS mutant colon cancer cells and can be transferred to exosomes. Sci Rep. 2016;6:37982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li P, Chen S, Chen H, et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132–136. [DOI] [PubMed] [Google Scholar]
- 82.Zhao Q, Chen S, Li T, et al. Clinical values of circular RNA 0000181 in the screening of gastric cancer. J Clin Lab Anal. 2018. May;32(4):e22333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen S, Li T, Zhao Q, et al. Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clin Chim Acta. 2017;466:167–171. [DOI] [PubMed] [Google Scholar]
- 84.Chen J, Li Y, Zheng Q, et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208–219. [DOI] [PubMed] [Google Scholar]
- 85.Xia W, Qiu M, Chen R, et al. Circular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Sci Rep. 2016;6:35576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shao Y, Chen L, Lu R, et al. Decreased expression of hsa_circ_0001895 in human gastric cancer and its clinical significances. Tumour Biol. 2017;39:1010428317699125. [DOI] [PubMed] [Google Scholar]
- 87.Tian M, Chen R, Li T, et al. Reduced expression of circRNA hsa_circ_0003159 in gastric cancer and its clinical significance. J Clin Lab Anal. 2018. Mar;32(3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shao Y, Li J, Lu R, et al. Global circular RNA expression profile of human gastric cancer and its clinical significance. Cancer Med. 2017;6:1173–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kong Z, Wan X, Zhang Y, et al. Androgen-responsive circular RNA circSMARCA5 is up-regulated and promotes cell proliferation in prostate cancer. Biochem Biophys Res Commun. 2017;493:1217–1223. [DOI] [PubMed] [Google Scholar]
- 90.Qin M, Liu G, Huo X, et al. Hsa_circ_0001649: A circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016;16:161–169. [DOI] [PubMed] [Google Scholar]
- 91.Fu L, Yao T, Chen Q, et al. Screening differential circular RNA expression profiles reveals hsa_circ_0004018 is associated with hepatocellular carcinoma. Oncotarget. 2017;8:58405–58416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shang X, Li G, Liu H, et al. Comprehensive circular RNA profiling reveals that hsa_circ_0005075, a new circular RNA biomarker, is involved in hepatocellular crcinoma development. Medicine (Baltimore). 2016;95:e3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yao Z, Luo J, Hu K, et al. ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Mol Oncol. 2017;11:422–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sui W, Shi Z, Xue W, et al. Circular RNA and gene expression profiles in gastric cancer based on microarray chip technology. Oncol Rep. 2017;37:1804–1814. [DOI] [PubMed] [Google Scholar]
- 95.Hsiao YC, Yeh MH, Chen YJ, et al. Lapatinib increases motility of triple-negative breast cancer cells by decreasing miRNA-7 and inducing Raf-1/MAPK-dependent interleukin-6. Oncotarget. 2015;6:37965–37978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gao D, Zhang X, Liu B, et al. Screening circular RNA related to chemotherapeutic resistance in breast cancer. Epigenomics. 2017;9:1175–1188. [DOI] [PubMed] [Google Scholar]
- 97.Wang K, Sun Y, Tao W, et al. Androgen receptor (AR) promotes clear cell renal cell carcinoma (ccRCC) migration and invasion via altering the circHIAT1/miR-195-5p/29a-3p/29c-3p/CDC42 signals. Cancer Lett. 2017;394:1–12. [DOI] [PubMed] [Google Scholar]
- 98.Wang BG, Li JS, Liu YF, et al. MicroRNA-200b suppresses the invasion and migration of hepatocellular carcinoma by downregulating RhoA and circRNA_000839. Tumour Biol. 2017;39:1010428317719577. [DOI] [PubMed] [Google Scholar]
- 99.Guarnerio J, Bezzi M, Jeong JC, et al. Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell. 2016;165:289–302. [DOI] [PubMed] [Google Scholar]
- 100.Yang P, Qiu Z, Jiang Y, et al. Silencing of cZNF292 circular RNA suppresses human glioma tube formation via the Wnt/beta-catenin signaling pathway. Oncotarget. 2016;7:63449–63455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yao JT, Zhao SH, Liu QP, et al. Over-expression of CircRNA_100876 in non-small cell lung cancer and its prognostic value. Pathol Res Pract. 2017;213:453–456. [DOI] [PubMed] [Google Scholar]
- 102.Huang M, Zhong Z, Lv M, et al. Comprehensive analysis of differentially expressed profiles of lncRNAs and circRNAs with associated co-expression and ceRNA networks in bladder carcinoma. Oncotarget. 2016;7:47186–47200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liang HF, Zhang XZ, Liu BG, et al. Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am J Cancer Res. 2017;7:1566–1576. [PMC free article] [PubMed] [Google Scholar]
- 104.Li F, Zhang L, Li W, et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget. 2015;6:6001–6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang J, Liu H, Hou L, et al. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer. 2017;16:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang Y, Liu H, Li W, et al. CircRNA_100269 is downregulated in gastric cancer and suppresses tumor cell growth by targeting miR-630. Aging (Albany NY). 2017;9:1585–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xu L, Zhang M, Zheng X, et al. The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2017;143:17–27. [DOI] [PubMed] [Google Scholar]
- 108.Fang Y, Xue JL, Shen Q, et al. MicroRNA-7 inhibits tumor growth and metastasis by targeting the phosphoinositide 3-kinase/Akt pathway in hepatocellular carcinoma. Hepatology. 2012;55:1852–1862. [DOI] [PubMed] [Google Scholar]
- 109.Han D, Li J, Wang H, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151–1164. [DOI] [PubMed] [Google Scholar]
- 110.Sand M, Bechara FG, Sand D, et al. Circular RNA expression in basal cell carcinoma. Epigenomics. 2016;8:619–632. [DOI] [PubMed] [Google Scholar]
- 111.Su H, Lin F, Deng X, et al. Profiling and bioinformatics analyses reveal differential circular RNA expression in radioresistant esophageal cancer cells. J Transl Med. 2016;14:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rybak-Wolf A, Stottmeister C, Glazar P, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58:870–885. [DOI] [PubMed] [Google Scholar]
- 114.Memczak S, Papavasileiou P, Peters O, et al. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLOS ONE. 2015;10:e0141214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bahn JH, Zhang Q, Li F, et al. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem. 2015;61:221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kristensen LS, Hansen TB, Veno MT, et al. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37:555–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Szabo L, Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat Rev Genet. 2016;17:679–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chen D, Zhang LJ, Tan K, et al. Application of droplet digital PCR in quantitative detection of the cell-free circulating circRNAs. Biotechnol Biotechnol Equip. 2018;32:116–123. [Google Scholar]


