Abstract
The gut microbiome is increasingly recognized as a second genome that contributes to the health and diseases of the host. A major function of the gut microbiota is to convert primary bile acids (BAs) produced from cholesterol in the liver into secondary BAs that activate distinct host receptors to modulate xenobiotic metabolism and energy homeostasis. The goal of this study was to investigate to what extent oral exposure to an environmentally relevant polychlorinated biphenyl (PCBs mixture), namely the Fox River mixture, impacts gut microbiome and BA homeostasis. Ninety-day-old adult female conventional (CV) and germ-free (GF) C57BL/6 mice were orally exposed to corn oil (vehicle), or the Fox River mixture at 6 or 30 mg/kg once daily for 3 consecutive days. The PCB low dose profoundly increased BA metabolism related bacteria Akkermansia (A.) muciniphila, Clostridium (C.) scindens, and Enterococcus in the large intestinal pellet (LIP) of CV mice (16S rRNA sequencing/qPCR). This correlated with a PCB low dose-mediated increase in multiple BAs in serum and small intestinal content (SIP) in a gut microbiota-dependent manner (UPLC-MS/MS). Conversely, at PCB high dose, BA levels remained stable in CV mice correlated with an increase in hepatic efflux transporters and ileal Fgf15. Interestingly, lack of gut microbiota potentiated the PCB-mediated increase in taurine conjugated α and β muricholic acids in liver, SIP, and LIP. Pearson’s correlation identified positive correlations between 5 taxa and most secondary BAs. In conclusion, PCBs dose-dependently altered BA homeostasis through a joint effort between host gut-liver axis and intestinal bacteria.
Keywords: PCBs, gut microbiota, bile acids, liver
Polychlorinated biphenyls (PCBs) were formerly used in industrial and consumer products before their production was banned in the United States in 1979 followed by a world-wide ban years later (EPA press release April 19, 1979). However, due to their lipophilic and bio-accumulative nature, these persistent organic pollutants are still a major health concern and negatively impact the health of humans and wildlife (Byard et al., 2015; Quinete et al., 2014). PCBs are frequently detected in serum samples of the general U.S. population (Patterson et al., 2009), suggesting ongoing exposures to PCBs via inhalation (Grimm et al., 2015; Jamshidi et al., 2007) and diet (Schecter et al., 2010; Shin et al., 2015). In breast milk, World Health Organization (WHO) surveys showed that global levels of PCBs were significantly above those considered toxicologically safe for breastfed infants (van den Berg et al., 2017). The multifaceted toxicities of PCBs and their metabolites observed in humans and laboratory animals include carcinogenicity (Freeman and Kohles, 2012; Ludewig and Robertson, 2013; Pesatori et al., 2013; Quinete et al., 2014; Zani et al., 2013), reproductive disorders (Bell, 2014; Eskenazi et al., 2017; Grandjean et al., 2012; Meeker et al., 2011; Toft, 2014; Yao et al., 2017), developmental neurotoxicity (Bell, 2014; Kania-Korwel et al., 2017a), cardiovascular inflammation (Gupta et al., 2017; Petriello et al., 2014), thyroid toxicity (Duntas and Stathatos, 2015), abdominal discomfort (Kim et al., 2017), as well as obesity, type-II diabetes, and fatty liver disease (Ghosh et al., 2014; Kania-Korwel et al., 2017b; Tang et al., 2014; Wahlang et al., 2014b).
The gut microbiome is increasingly recognized as an organ for xenobiotic biotransformation in pharmacology (Carmody and Turnbaugh, 2014; Haiser and Turnbaugh, 2013; Spanogiannopoulos et al., 2016); however, little is known regarding the importance of the gut microbiome in environmental toxicology in general and PCB-mediated toxicities in particular. The gut microbiome can directly metabolize drugs via reduction, hydrolysis (deconjugation) and other types of reactions, or remotely modulate the host xenobiotic biotransformation by releasing distinct microbial metabolites into the systemic circulation (Fu and Cui, 2017; Klaassen and Cui, 2015). Our previous work has shown that in livers and various intestinal sections of germ-free (GF) mice, the absence of gut microbiota profoundly altered the expression and enzyme activities of many Cyps and other xenobiotic biotransformation related genes (Fu et al., 2017; Selwyn et al., 2015a,c, 2016). These findings in unexposed mice are important because PCB exposure alters the composition of gut microbiome in mice. Specifically, oral exposure to a simplified PCB mixture consisting of PCB 153, PCB 138, and PCB 180 induced changes in the gut microbiome primarily by decreasing Proteobacteria (16S rRNA microarray), whereas exercise attenuated such change (Choi et al., 2013; Potera, 2013). Larval exposure to PCB 126 also persistently altered the amphibian gut microbiota (16S rRNA sequencing) (Kohl et al., 2015). Another dioxin-like persistent organic pollutant, namely 2, 3, 7, 8-tetrachlorodibenzofuran (TCDF), modified gut microbiome and host metabolic homeostasis in mice through activation of the aryl hydrocarbon receptor (AhR) (Zhang et al., 2015). However, very little is known regarding how changes in the gut microbiome caused by exposure to environmental PCB mixtures alter the functions of the gut microbiome including the microbial metabolite formation.
One of the major functions of gut microbiota is to convert primary BAs, which are cholesterol metabolites produced in the liver, to secondary BAs via dehydroxylation, deconjugation, and epimerization reactions (Jia et al., 2017; Kang et al., 2008; Ridlon et al., 2006, 2014). In addition to their classic functions in modulating lipid emulsification and absorption, BAs can activate various host receptors, including the farnesoid X receptor (FXR) and the pregnane X receptor (PXR) in liver and intestine, as well as G protein-coupled bile acid (BA) receptor (TGR-5) in brown adipose tissue and muscle (Chiang, 2013; Li and Chiang, 2015; Watanabe et al., 2006). It has been shown that the secondary BA lithocholic acid (LCA) was an activator of PXR that protects against liver toxicity (Staudinger et al., 2001), whereas the secondary BAs LCA and deoxycholic acid (DCA) were more potent activators of TGR-5 than primary BAs and contribute to thermogenesis and energy expenditure (Chiang, 2013). Therefore, it is important to understand the contribution of microbial-derived BAs in modulating the physiological and pathological responses of the host. Moreover, depending on their three-dimensional structure, individual PCB congeners can be activators of xenobiotic-sensing transcription factors, such as AhR, PXR, constitutive androstane receptor (CAR), or the lipid sensors peroxisome proliferator-activated receptors (PPARs) (Wahlang et al., 2014a). Previously the effect of PCBs on the BA precursor, namely cholesterol, has been reported in humans and animal models. For example, mono-ortho substituted PCBs 74 and 156, di-ortho PCBs 172 and 194, and tri- and tetra-ortho PCBs 199, 196 to 203, 206, and 209 each were significantly associated with total lipids, total cholesterol and triglycerides from a study in residents of Aniston Alabama (Aminov et al., 2013). In rodents, oral PCB exposure causes hypercholesterolemia by increasing hepatic cholesterol synthesis without modifying the fecal total steroid excretion (Nagaoka et al., 1990; Quazi et al., 1983). However, relatively little is known regarding the effect of PCBs on BAs. Since BAs and PCBs can target the same nuclear transcription factors, PCBs and BAs likely interact in the liver to influence the expression of xenobiotic response genes in a microbiome dependent manner.
The goal of the present study was to use conventional mice (CV, ie, specific pathogen-free mice with a normal gut microbiome) and GF mice (ie, mice without a gut microbiome) to test our hypothesis that oral exposure to an environmentally relevant synthetic PCB mixture alters the gut microbiome and subsequent functions in BA metabolism in a PCB-dependent manner. The Fox River mixture is a PCB mixture that mimics the complex PCB congener profile in fish from the Fox River, and thus the oral exposure of a human population in Green Bay, Wisconsin that consume these fish as their major source of dietary protein. The Fox River mixture has been widely used in laboratory studies to recapitulate the PCB-mediated toxicities in humans and consists of 35% Aroclor 1242, 35% Aroclor 1248, 15% Aroclor 1254, and 15% Aroclor 1260. The PCB congeners in this mixture resemble 71% of those reported for the Fox River fish (Kostyniak et al., 2005a). In Long-Evan rats, developmental exposure to the Fox River mixture (6 mg/kg PCBs) decreased the amplitudes and elevated the thresholds of distortion product otoacoustic emissions, which is a potential assessment of cochlear function (Poon et al., 2011). Developmental exposure to Fox River mixture also decreased serum thyroxine levels (Poon et al., 2011), altered the interceptive cues of psycho-stimulants (Sable et al., 2011), and delayed the propagation of electrical kindling from the amygdala in rats (Bandara et al., 2017). However, very little is known regarding the effect of different doses of Fox River mixture (6 mg/kg and 30 mg/kg) on gut microbiota. These doses used in this study were lower than the dose used by Choi et al., which used 43 mg/kg (150 µmol/kg) that resulted in plasma levels of PCBs representative of an acutely exposed human population (Jensen et al., 1989). Here we used a comprehensive systems biology approach, including 16S rRNA sequencing, QIIME (Kuczynski et al., 2012), PICRUSt, LefSe (Segata et al., 2011), RNA-Seq, and targeted proteomics to investigate the effect of PCBs on the gut-liver axis.
MATERIALS AND METHODS
Chemicals and preparation of the Fox River mixture
Aroclor 1248 (lot #: 106-248) and Aroclor 1260 (lot #: 021-020-1A) were purchased from Accustandard (New Haven, Connecticut). Aroclor 1242 (lot # KB 05-415) and Aroclor 1254 (lot #: KB 05-612) were a generous gift from Dr Larry Hansen and have been used previously for the preparation of PCB mixtures (Kostyniak et al., 2005b; Zhao et al., 2010). Pesticide grade acetone was purchased from Fisher Scientific (Fair Lawn, New Jersey). Isotope labeled internal standards, 13C12-2, 5-dichlorbiphenyl (13C12-PCB 9, purity 100%, 13C12, 99%) and 13C12-2, 2′, 3, 3′, 4, 4′, 5, 5′-octachlorobiphenyl (13C12-PCB 194, purity 98%, 13C12, 98%) were obtained from Cambridge Isotope Laboratories (Tewksbury, Massachusetts). Analytical standard solutions containing all 209 PCB congeners were purchased from AccuStandard (New Haven, Connecticut).
Solutions of each Aroclor (50 mg/ml in acetone) were combined in pre-weighted headspace vials in a ratio of Aroclor 1242: Aroclor 1248: Aroclor 1254: Aroclor 1260 ratio of 35:35:15:15% by weight (Kostyniak et al., 2005b). The mixture was vortexed and inverted for 30 min. Aliquots of the PCB mixture in acetone or acetone alone (for the preparation of the vehicle control) were placed into pre-weighted hypo vials, and the solvent was evaporated under a gentle stream of nitrogen. Traces of acetone were subsequently removed under vacuum for 1 hour and the final mass of the PCB mixture in each vial was determined gravimetrically. Preliminary experiments demonstrated that this procedure did not alter the PCB congener profiles of the Fox River mixture before and after evaporation of the solvent.
The Fox River mixture and the original four Aroclors were analyzed on an Agilent 7890A GC system coupled with an Agilent 5975C Inert Mass Selective Detector operated in electron ionization mode and equipped with a SLB-5MS capillary column (30 m length, 250 µm inner diameter, 0.25 µm film thickness, Supelco, Bellefonte, Pennsylvania) using a published congener specific analysis method (Hu et al., 2015). The following temperature program was used for the congener-specific analysis: hold at 80°C for 1 min, then 2°C/min to 160°C, 1°C/min to 170°C and hold for 15 min, 1°C/min to 180°C and hold for 15 min, 1°C/min to 245°C, then 10°C/min to 300°C and hold for 15 min. The operating temperatures of the mass selective detector were 280°C, 230, and 250°C for the transfer line, source, and quadrupole, respectively. Helium was used as the carrier gas with a flow rate of 1.09 ml/min. All PCB congener profiles were determined in triplicate using a reference standard containing all 209 PCB congeners. 13C12-PCB 9 and 13C12-PCB 194 (80 ng each) were added to each sample prior to analysis as internal standards (volume corrector). The PCB congener profile of the Fox River mixture was highly similar to the theoretical profile calculated based on the analysis of the individual Aroclors, with both correlation and similarity coefficients >0.99 (Kostyniak et al., 2005b; Zhao et al., 2010).
Animals and PCB exposure
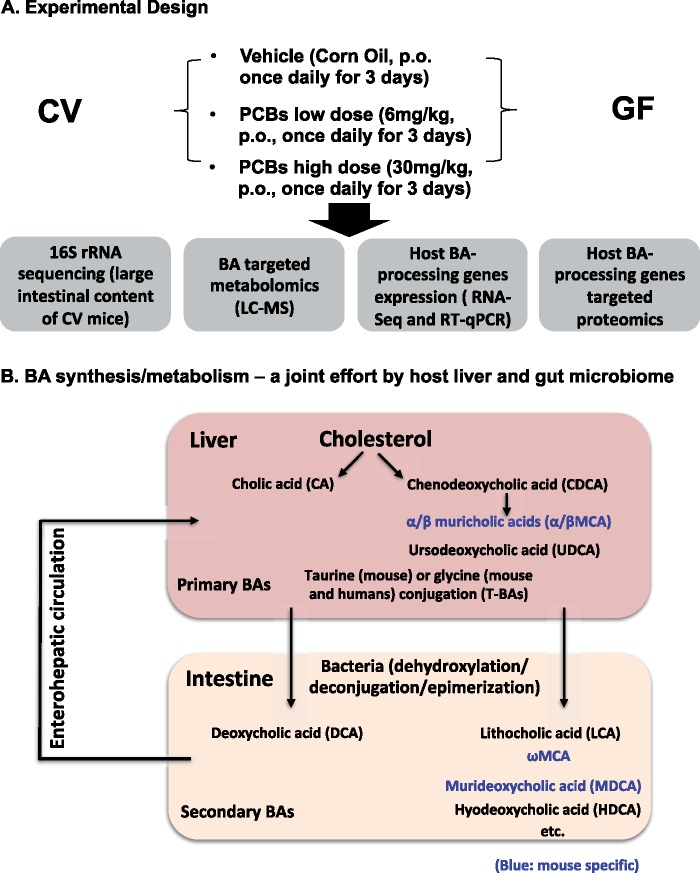
Three-month-old female C57BL/6J CV mice (which were specific pathogen free) were purchased from the Jackson Laboratory (Bar Harbor, Maine), and acclimated to the animal Facility at the University of Washington for one week before beginning PCB exposure. The initial GF breeders of C57BL/6 background were obtained from the National Gnotobiotic Rodent Resource Center (University of North Carolina, Chapel Hill). Age-matched female GF pups were born in the Gnotobiotic Animal Core (GNAC) Facility at University of Washington. Female mice were used in this study to lay the foundation for subsequent studies of the developmental neurotoxicity in mice exposed to PCBs via the maternal diet, and to compare the results from this study with our previous work using the females (Kania-Korwel et al., 2008a, 2008b, 2008c, 2010; Wu et al., 2013, 2015). All mice were housed according to the Association for Assessment and Accreditation of Laboratory Animal Care International Guidelines. Both CV and GF mice were exposed to the same Laboratory Autoclavable Rodent Diet 5010 (LabDiet, St. Louis, Missouri), water (nonacidified autoclaved water), and bedding (autoclaved Enrich-N’Pure). All chemical solutions were sterilized using the Steriflip Vacuum-driven Filtration System with a 0.22 μM Millipore Express Plus Membrane (EMD Millipore, Temecula, California). All gavage needles and syringes were sterilized by autoclave. As described in Figure 1A, mice were exposed to corn oil (p.o., 10 ml/kg b.w., n = 5), or Fox River mixture low dose (p.o., 6 mg/kg b.w. dissolved in corn oil, n = 5), or Fox River mixture high dose (p.o., 30 mg/kg b.w. dissolved in corn oil, n = 5), once daily between 8 and 10 am for 3 consecutive days. Analogous dosing paradigms are frequently used to study dose-dependent effect of xenobiotics on the expression of hepatic drug metabolizing enzymes (Guengerich, 2001). At the same time, the comparatively high doses allow us to detect effects of the Fox River mixture on microbiome composition and function in a short time frame, thus following a fundamental concept used in chemical risk assessment (Doull, 2003). Importantly, we have used the lower PCB dose in an earlier disposition study (Kania-Korwel et al., 2012), and several other groups have used this dose in developmental neurotoxicity studies (Poon et al., 2013; Sable et al., 2006; Yang et al., 2009). Tissues were harvested 24 h after the final dose. Whole blood was collected via cardiac puncture and transferred to a MiniCollect serum separator (Greiner Bio-One, Kremsmunster, Austria), and was kept on ice for at least 30 min to allow sufficient time for coagulation. Serum was collected by centrifugation at 4000 rpm (1503 g) at 4°C for 20 min, and was stored at −80°C until further analysis. Small and large intestinal content was flushed out in 15 ml of ice-cold phosphate-buffered saline (PBS) that contains 0.1% dithiothreitol (DTT) (Sigma Aldrich, St. Louis, Missouri), and immediately frozen on dry ice. Intestine tissue was then divided into 4 sections, namely duodenum, jejunum, ileum, and colon. All tissues were immediately frozen in liquid nitrogen and stored at −80°C until further analysis. The downstream experiments, including 16S rRNA sequencing, BA targeted profiling, RNA-Seq of BA-processing genes in host liver and intestine (Figure 1B) are described in the following method sections.
Figure 1.
A, A diagram illustrating the experimental design. Three-month-old female CV and GF mice (n = 5 per group) were exposed to vehicle (corn oil, 10 ml/kg b.w., p.o.), the low PCB dose (6 mg/kg b.w., p.o.), or the high PCB dose (30 mg/kg b.w., p.o.), once daily for 3 consecutive days. Tissues were removed 24 h after the final dose. Microbial DNA from large intestinal content (LIP) of CV mice was subjected to 16S rRNA sequencing. BAs were extracted from serum, liver, SIP, and LIP of CV and GF mice and quantified by LC/MS-MS-based targeted metabolomics. The expression of BA-processing genes was determined by RNA-Seq (the mRNAs of selected genes were validated by RT-qPCR) and as well as LC-MS-based targeted proteomics. Detailed procedures are described in the Materials and Methods section. B, A diagram illustrating the BA synthesis and metabolism as a joint effort between host liver and gut microbiome. Briefly, primary BAs are synthesized in liver from cholesterol. The major primary BAs are CA and CDCA in humans. In mice, CDCA is further metabolized to αMCA and βMCA. UDCA is another primary BA produced by mouse liver although it was thought to be solely produced by intestinal bacteria (Selwyn et al., 2015b). Most of the BAs are conjugated in human liver with taurine (T) and glycine, whereas taurine-conjugation is more predominant in mouse liver. In intestine, primary BAs are metabolized by bacteria via dehydroxylation, epimerization, and deconjugation reactions, generating DCA, LCA, ωMCA, murideoxycholic acid (MDCA), and hyodeoxycholic acid (HDCA), as well as various deconjugated primary BAs. Blue: mouse-specific BAs. (For interpretation of the reference to color in this figure legend, the reader is referred to the web version of this article.)
Bacterial 16S rRNA sequencing, qPCR analysis, and bioinformatics
The large intestinal content in PBS/DTT solution from CV mice was thawed overnight at 4°C, and the large intestinal pellet (LIP) was obtained by centrifugation at 10 000 g at 4°C for 1 h. Bacterial DNA was isolated from the LIP using an OMEGA E.Z.N.A. Stool DNA Kit (OMEGA Biotech Inc., Norcross, Georgia). The concentration of DNA was determined using Qubit (Thermo Fisher Scientific, Waltham, Massachusetts). The integrity and quantity of all DNA samples were confirmed using an Agilent 2100 Bioanalyzer (Agilent Technologies Inc., Santa Clara, California). Bacterial 16S rRNA V4 amplicon sequencing was performed using a HiSeq 2500 second-generation sequencer (250 bp paired-end) (Beijing Genome Institute Americans Corporation, Cambridge, Massachusetts) (n = 3 per group). Raw data in fastq format were analyzed using various python scripts in QIIME (Caporaso et al., 2010) including de-multiplexing, quality filtering, operational taxonomy unit (OTU) picking, alpha- and beta-diversity determinations, as well as format conversions and visualizations. Predictive functional profiling of 16S rRNA data was performed using PICRUSt (Langille et al., 2013) and differentially regulated KEGG pathways were visualized in 2-way hierarchical clustering dendrograms. The sequencing data have been deposited into the Sequence Read Archive (SRA) of the National Center for Biotechnology Information (NCBI). Selected differentially regulated bacteria were validated by quantitative polymerase chain reaction (qPCR) using a BioRad CFX384 Real-Time PCR Detection System (Bio-Rad, Hercules, California). The qPCR primers targeting the 16S rRNA sequences are shown in Supplementary Table 1.
BA-targeted profiling using ultra-performance liquid chromatography-tandem mass spectrometry (UPLC/MS-MS)
BAs from liver, serum, small intestinal pellet (SIP), and LIP compartments were extracted with internal standards (2H4-GCDCA [40 μg/ml] and 2H4-CDCA [20 μg/ml]) using a similar method as described previously (Zhang and Klaassen, 2010). Nineteen major BAs were quantified, namely taurine-conjugated cholic acid (T-CA), T-α muricholic acid (T-αMCA), T-β muricholic acid (T-βMCA), T-chenodeoxycholic acid (T-CDCA), T-ursodeoxycholic acid (T-UDCA), T-murideoxycholic acid (T-MDCA), T-hyodeoxycholic acid (T-HDCA), T-deoxycholic acid (T-DCA), T-lithocholic acid (T-LCA), αMCA, βMCA, CA, CDCA, UDCA, ωMCA, MDCA, HDCA, DCA, and LCA. CA, CDCA, DCA, and LCA were purchased from Sigma Aldrich (St. Louis, Missouri); αMCA, βMCA, ωMCA were purchased from Steraloids (Newport, Rhode Island). Other BAs were kindly obtained from University of Kansas Medical Center. ACQUITY UPLC BEH C18 columns and VanGuard BEH C18 pre-columns were purchased from Waters Corporation (Milford, Massachusetts). The samples were eluted using gradient mobile phases of A (10 mM ammonium acetate in 20% acetonitrile) and B (10 mM ammonium acetate in 80% acetonitrile). The UPLC/MS-MS operating parameters are shown in Supplementary Table 2.
For serum BA extraction, 100 µl of serum spiked with 10 µl of the internal standard solution was extracted. The samples were vortexed for 30 s and then allowed to stand for 10 min on ice. The mixture was centrifuged at 12 000 g for 10 min and then the supernatant was collected and evaporated under vacuum. The residue was reconstituted in 50 μl of 50% methanol water solution. Samples were centrifuged at 20 000 g for 10 min before injection.
For liver BA extraction, approximately 50 mg accurately weighted frozen liver were homogenized in 5 volumes of water, from which 600 µl of homogenate was taken and mixed with 10 µl of internal standard solution. After 10 min equilibration on ice, the homogenate was mixed with 3 ml of ice-cold acetonitrile containing 5% ammonia, vortexed vigorously, and shaken for 1 h at room temperature. The mixture was centrifuged at 12 000 g for 10 min and the supernatant was collected. The pellet was extracted with 1 ml of methanol, sonicated for 5 min, and centrifuged at 12 000 g for 10 min. The two supernatants were pooled and evaporated under vacuum, and reconstituted in 100 µl of 50% methanol. The supernatant was transferred into 0.2 µm Costar Spin-X HPLC microcentrifuge filter (Corning Inc., Corning, New York), and centrifuged at 20 000 g for 10 min before injection.
For BA analyses in intestinal content, samples were centrifuged at 10 000 g under vacuum for 1 h at 4°C to collect pellets. One hundred milligram of the pellet was weighted accurately and extracted twice with 3 ml of methanol. After shaking for 30 min in room temperature, the mixture was centrifuged at 12 000 g for 20 min to collect the supernatant. The two supernatants were combined, evaporated under vacuum, and reconstituted in 100 µl of 50% methanol water solution. The suspension was filtered by centrifugation using a 0.22 μm Spin-X filter polypropylene tube (Corning, New York) before injection.
RNA isolation, library preparation, and RNA-Seq
Total RNA was isolated from liver and ileum using RNA-Bee reagent per the manufacturer’s protocol (Tel-Test Inc., Friendswood, Texas). RNA concentration was quantified using a NanoDrop 1000 Spectrophotometer (Thermo Scientific, Waltham, Massachusetts) at a wavelength of 260 nm. Integrity of total RNA samples was evaluated by formaldehyde-agarose gel electrophoresis with visualization of 18S and 28S rRNA bands under ultraviolet light, and was confirmed by an Agilent 2100 Bioanalyzer (Agilent Technologies Inc. Santa Clara, California). Samples with RNA integrity (RIN) above 8.0 were used for RNA-Seq using a similar method as we described previously (Cheng et al., 2017). The raw and analyzed RNA-sequencing data will be deposited into NCBI GEO database.
Statistical analyses
Statistically significant differences were determined using analysis of variance (ANOVA) followed by Ducan’s post-hoc test (p < .05). Two-way hierarchical clustering dendrograms were generated using JMP v. 13 (SAS Institute Inc., Cary, North Carolina). Linear discriminant analysis effect size (LefSe) was used for high-dimensional biomarker discovery for the 16S rRNA data at the species level (α = 0.1; threshold on the logarithmic LDA score for discriminative features = 2.0; strategy for multi-class analysis: one-against all) (MicrobiomeAnalyst, Marker Data Profiling [MDP]). Pearson’s correlation analysis between BAs and bacteria was performed using R (ggplot2 and reshape2 packages).
RESULTS
Effect of PCBs on Gut Microbiota Diversities
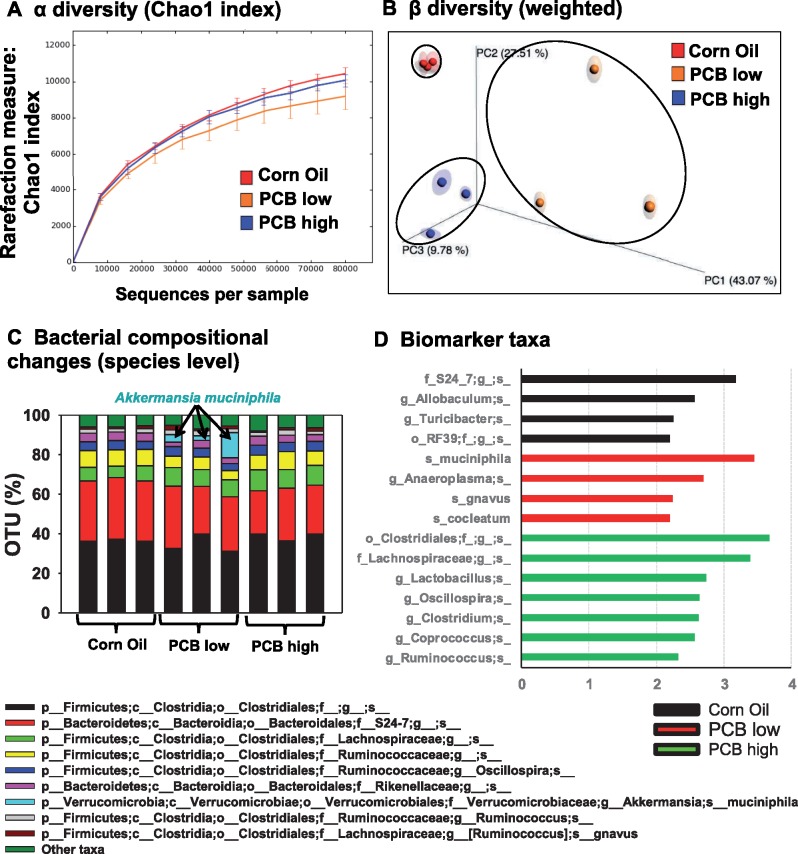
The 16S rRNA sequencing targeting the V4 hyper-variable region was performed on the content from the large intestine of CV mice exposed to corn oil, the low PCB dose, or the high PCB dose to determine the effect of the Fox River mixture on gut microbiota of CV mice (n = 3 per group). Approximately 109 000−140 000 reads were generated among these samples, with a median sequence length of 253 bp (Supplementary Table 3). In silico rarefraction was performed for all exposure groups from 0 to 80 000 reads with an incrementing step size of 10 000 (Figure 2A). To determine the effect of PCBs on the α diversity, which is a measure of species richness or diversity within an individual sample, alpha_diversity.py (QIIME) was used to calculate the α diversity metrics for all the rarefied OTU tables in three ways: chao1 index (Figure 2A), observed OTUs, and Faith’s Phylogenetic Diversity (ie, PD whole tree) (Supplementary Figure 1A). The α diversity quantifications demonstrated that there were no marked differences between corn oil and high PCB exposure groups; however, the low PCB exposure group tended to have moderately lower observed OTUs as the sampling depth increased. This observation suggests that lower PCB exposure appears to be more effective in reducing the diversity/enrichment of bacterial species.
Figure 2.
Alpha (A) and beta (B) diversities of gut microbiota in CV mice exposed to corn oil, the low PCB dose, and the high PCB dose (n = 3 per group). The 16S rRNA sequencing data were analyzed using QIIME as described in the Materials and Methods section. (C) Bacterial compositional changes at the species level in corn oil, low PCB and high PCB dose exposure groups. The top 9 species are shown, whereas the sum of the remaining taxa is displayed as “other taxa” as the 10th category. (D) Microbial biomarker prediction in each exposure group using LDA effect size (LefSe) analysis of the QIIME-generated 16S rRNA OTU table at the species level. Data were analyzed using MicrobiomeAnalyst, MDP. Biomarker bacteria with a LDA score above 2 and p < .05 (indicating a significantly enriched biomarker) were plotted in each exposure group.
Regarding the β diversity, which compares the diversity change among different exposure groups, both the weighted unifrac, a quantitative measure that reveals the community differences due to changes in relative taxon abundance (Figure 2B), as well as the unweighted unifrac, which is a qualitative measure that assess the community differences that are due to changes in the overall environment (Supplementary Figure 1B), were considered. Principal coordinate analysis revealed a distinct separation among the three exposure groups for both the weighted and unweighted unifrac (analysis of similarity [ANOSIM] p = .007 for weighted, and p = .006 for unweighted unifrac method), suggesting that the PCBs at low and high doses uniquely modified the gut microbiota configuration compared to the control group.
A total of 36 classes including 154 species of bacteria were identified by QIIME across all three exposure groups, and the top 10 most abundant taxa are shown in Figure 2C (note: the remaining taxa were summed and labeled “other taxa” as the 10th category). Interestingly, the most profound change among all taxa was a PCB dose mediated increase in A. muciniphila, which is known to positively correlate with circulating primary BAs during high fat diet induced obesity (Pierre et al., 2016). As shown in Figure 2D, the microbial response to PCB exposure displayed high dose specificity. The linear discriminant analysis (LDA) effect size (LefSe) method, which considers both statistical significance and effect relevance, was applied to the bacterial OTUs data (MicrobiomeAnalyst, MDP) to identify intestinal bacteria that are representative biomarkers for each exposure group. Biomarker bacteria associated with control conditions are the S24-7 family, the Allobaculum genus, the Turicibacter genus, and the RF 39 order (Figure 4A). In contrast, the biomarker bacteria associated with the low PCB dose are A. muciniphila, the Anaeroplasma genus, Ruminococcus gnavus, and C. cocleatum; whereas the biomarker bacteria associated with the high PCB dose are the Clostridiales order, the Lachnospiraceae family, the Lactobacillus genus, as well as Oscillospira, Clostridium, the Coprococcus, and Ruminococcus genera.
Figure 4.
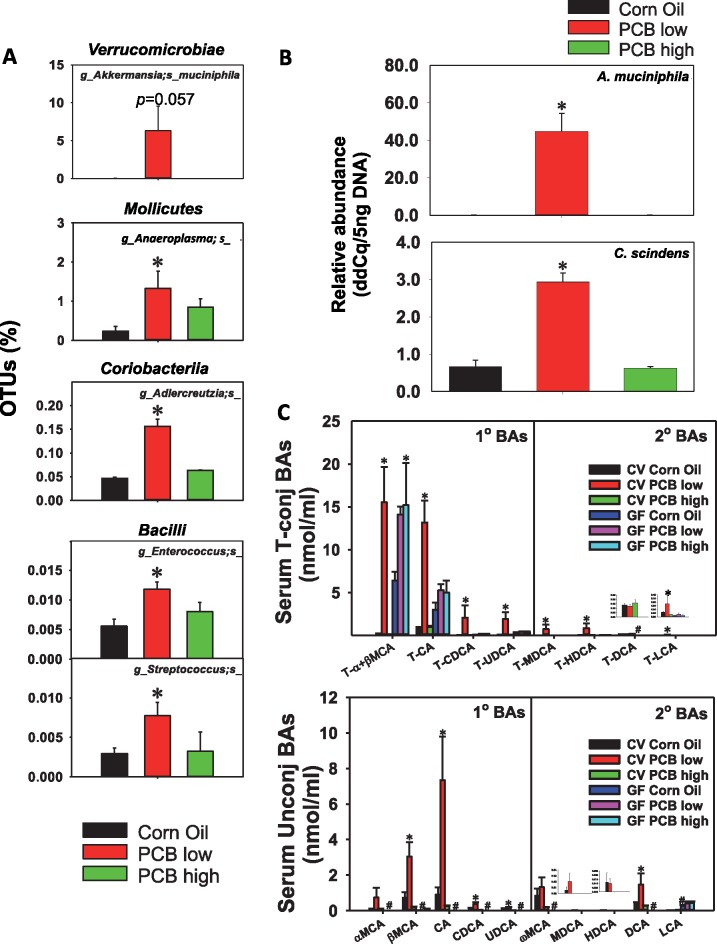
Intestinal bacteria and serum BAs that were uniquely up-regulated by PCB low dose in CV conditions. A, PCB low dose-induced intestinal bacteria at the species level in the content of the large intestine of CV mice (n = 3 per group). Asterisks represent statistically significant differences as compared with corn oil exposure group (1-way ANOVA, Duncan’s post-hoc test, p < .05). B, qPCR validation of Akkermansia muciniphila and C. scindens in LIC of CV mice exposed to corn oil, a low PCB or the high PCB dose (n = 5 per group). Asterisks represent statistically significant differences as compared with corn oil exposure group (1-way ANOVA, Duncan’s post-hoc test, p < .05). C, Individual BA profiles (T-BAs: top; unconjugated BAs: bottom) that were predominantly up-regulated by PCB low dose in serum in a gut microbiota-dependent manner (n = 5 per group). Asterisks represent statistically significant differences as compared to corn oil exposure group; pounds (#) represent statistically significant differences between CV and GF mice exposed to the same chemical (2-way ANOVA, Duncan’s post-hoc test, p < .05).
Effect of Interactions Between PCBs and Gut Microbiota on BA Homeostasis
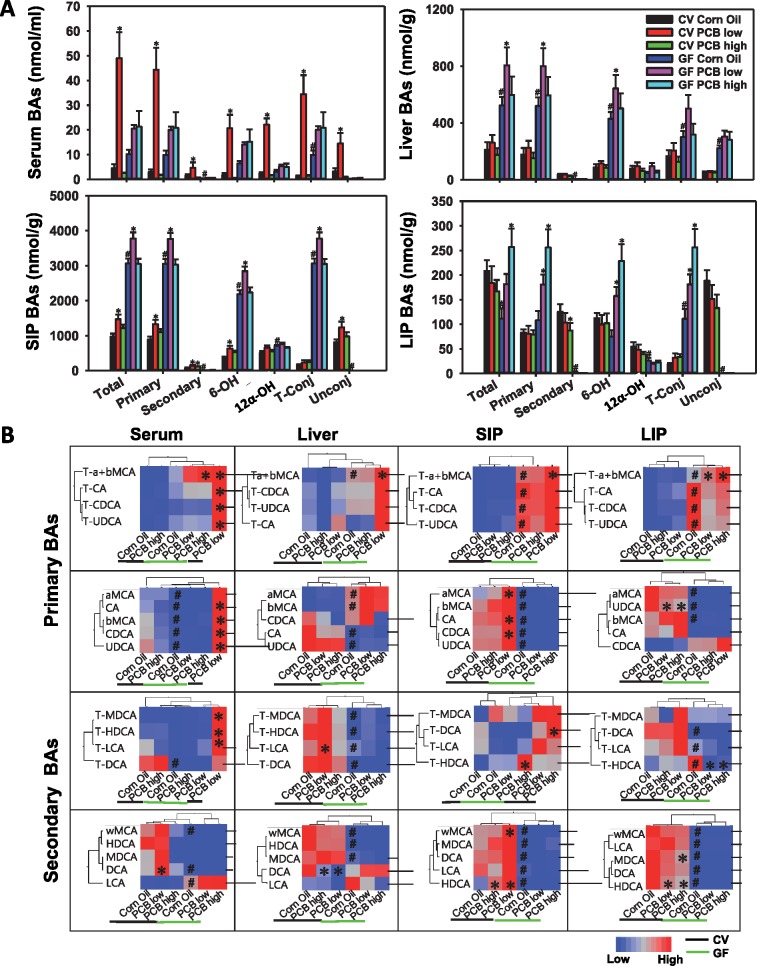
As shown in Figure 3A, in serum of CV mice, only PCB low dose, but not PCB high dose, markedly increased serum total BAs (9.69-fold increase). Such increase was attributed to an increase in both primary and secondary BAs (13.64-fold and 1.96-fold increase, respectively), as well as an increase in both conjugated and unconjugated BAs (24.74-fold and 3.47-fold increase, respectively). In addition, BAs from both the classic pathway (12-OH) and alternative pathway (6-OH) of host hepatic synthesis were increased at the low PCB dose in CV mice (8.73-fold and 10.33-fold increase, respectively). Interestingly, in the serum of GF mice, the PCB-mediated increase in BAs was completely abolished, indicating that the PCB low dose-mediated increase in these BAs are gut microbiota dependent. As expected, the basal levels of total, primary, and conjugated BAs tended to be higher in serum of GF mice as compared with CV mice exposed to the same dose of corn oil, due to lack of microbial metabolism in GF mice.
Figure 3.
A, LC-MS/MS analysis of BAs in serum, liver, SIP, and LIP of CV and GF mice (n = 5 per group). A total of 19 major BAs were quantified as described in the Materials and Methods section. BAs in each of the following categories are displayed: total BA; primary BA (a sum of T-αMCA, T-βMCA, T-CDCA, T-UDCA, CA, αMCA, βMCA, CDCA, and UDCA); secondary BAs (a sum of T-DCA, T-LCA, T-HDCA, T-MDCA, ωMCA, MDCA, HDCA, DCA, and LCA); 6-OH BAs derived from alternative pathway of host hepatic BA synthesis (a sum of T-αMCA, T-βMCA, αMCA, βMCA, and ωMCA), 12α-OH BAs derived from classic pathway of host hepatic BA synthesis (T-CA, T-DCA, CA, and DCA), taurine-conjugated BAs (a sum of T-αMCA, T-βMCA, T-CDCA, T-UDCA, T-DCA, T-LCA, T-HDCA, T-MDCA), and unconjugated BAs (a sum of CA, αMCA, βMCA, CDCA, UDCA, ωMCA, MDCA, HDCA, DCA, and LCA). Asterisks (*) indicate the PCB exposure effect, which represents statistically significant differences as compared to corn oil-treated group of the same enterotype; pounds (#) indicate the enterotype effect, which represents statistically significant differences between CV and GF mice exposed to the same chemical (p < .05). B. Hierarchical clustering dendrograms of individual BAs in serum, liver, SIP, and LIP of CV (black line) and GF (green line) mice (average values of n = 5 were used). Red represents relatively high levels, whereas blue represents relatively low levels. Asterisks represent statistically significant differences as compared with corn oil-treated control group; pounds (#) represent statistically significant differences between CV and GF mice exposed to the same chemical (p < .05). (For interpretation of the reference to color in this figure legend, the reader is referred to the web version of this article.)
In SIP of CV mice, PCB low dose uniquely increased total (51.37% increase), primary (49.01% increase), secondary (77.64% increase), 6-OH (70.26% increase), and unconjugated BAs (53.04% increase), whereas both PCB low and high doses increased SIP secondary BAs (Figure 3A). Except for secondary and unconjugated BAs, which were extremely low in SIP of GF mice as expected, all of the PCB-mediated increase in SIP BAs appeared to be gut microbiota independent, because a similar up-regulation pattern was observed in SIP of GF mice. As expected, the basal total, primary, and taurine-conjugated BAs were higher in SIP of GF than CV mice due to lack of microbial metabolism, and the increased BA levels were mainly contributed by the alternative pathway (evidenced by increased 6-OH-BAs) but not through the primary pathway of BA de novo synthesis (evidenced by no change in 12-OH-BAs). At the individual BA level, PCB low dose increased αMCA, CA, CDCA, and ωMCA, and HDCA in a gut microbiota-dependent manner (HDCA was also increased by PCB high dose in CV conditions); whereas lack of gut microbiota sensitized PCB low dose-mediated increase in T-α/βMCA in GF mice (Supplementary Figure 3).
In liver and LIP compartments of CV mice, BA profiles remained relatively stable following PCB exposure (Figure 3A, Supplementary Figure 4). In GF mice, lack of gut microbiota sensitized liver to PCB low dose mediated increase in total, primary, and 6-OH BAs, and sensitized LIP to PCB high dose-mediated increase in total, primary, 6-OH, and taurine-conjugated BAs (T-α/βMCA) (Figure 3A, Supplementary Figure 5).
In summary, oral exposure to PCBs altered BA profiles in a bio-compartment specific manner, and the majority of the changes were observed at PCB low dose. In serum, most of the increased BAs by PCB low dose were dependent on gut microbiota, and so were the PCB-mediated increases in secondary and unconjugated BAs in SIP. Conversely, in GF mice, lack of gut microbiota sensitized liver to PCB low dose-mediated increase in primary BAs from alternative pathway of host synthesis, but sensitized LIP to PCB high dose-mediated increase in primary BAs in primary pathway of host synthesis as well as taurine-conjugated BAs. Individual BA changes in all four bio-compartments are summarized as two-way hierarchical clustering dendrograms as shown in Figure 3B.
Corresponding to an increase in circulating BAs in serum of CV mice, PCB at the low dose uniquely increased the percentage of OTUs of A. muciniphila (345.77-fold, p = .057) from the Verrucomicrobiae class, Anaeroplasma from the Mollicutes class, Adlercreutzia from the Coriobacteriia class, as well as Enterococcus and Streptococcus from the Bacilli class (16S rRNA sequencing) (Figure 4A). The PCB low dose-mediated increase in A. muciniphila was confirmed by qPCR; in addition, C. scindens was also increased by PCB low dose only as shown by qPCR (Figure 4B). To note, A. muciniphila is known to positively associate with increased circulating primary BAs (Pierre et al., 2016), whereas C. scindens is known to carry BA-metabolizing enzymes for secondary BA synthesis (Ridlon and Hylemon, 2012; Wells and Hylemon, 2000). In addition, Enterococcus is also known to have BA metabolism activities (Ridlon et al., 2016). The DNA abundance of the BA-metabolizing enzymes bsh and baiJ were increased by both PCB doses, although PCB low dose tended to have slightly higher abundance; whereas baiCD, which is another BA-metabolizing enzyme, as well as universal bacteria, remained unchanged (Supplementary Figure 2).
At individual BA level, the PCB low dose-specific increase in these taxa correlated with an increase in primary BAs α/βMCA, CA, CDCA, and UDCA in both conjugated and unconjugated forms; conjugated secondary BAs T-MDCA, T-HDCA, T-LCA, as well as the unconjugated secondary BA DCA (Figure 4C).
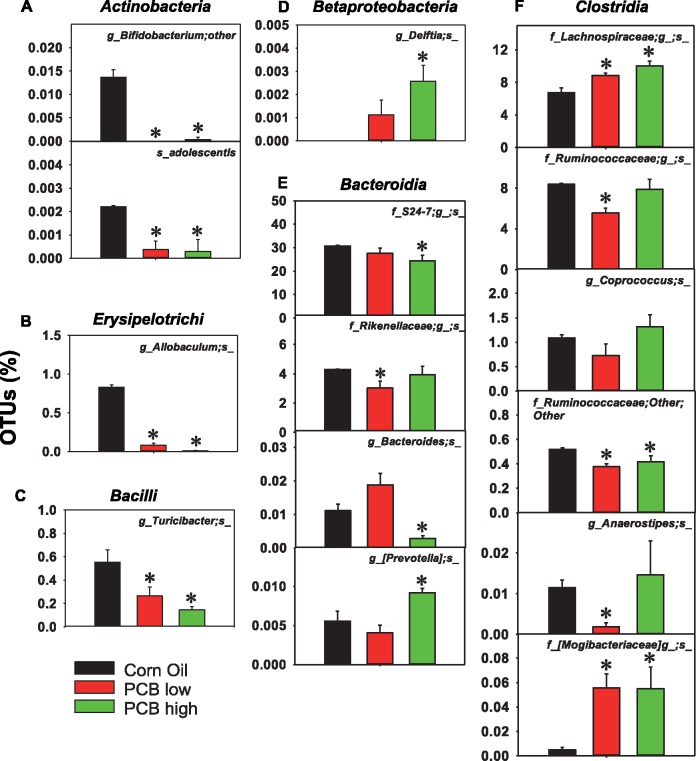
Oral exposure to PCBs also dose-dependently altered other taxa in LIP of CV mice, including a decrease in Bifidobacterium and adolescentis of the Actinobacteria class, in Allobaculum of the Erysipelotrichi class, Turicibacter in the Bacilli class, and Ruminococcaceae other in the Clostridia class, by both PCB low and high doses (Figs. 5A–C and 5F). Conversely, both PCB low and high doses increased Lachnospiraceae and Mogibacteriaceae in the Clostridia class (Figure 5F). PCB low dose decreased Rikenellaceae in the Bacteroidia class, as well as Ruminococcaceae and Anaerostipes in the Clostridia class (Figs. 5E and 5F); whereas PCB high dose increased Delftia in the Betaproteobacteria class and Prevotella in the Bacteroidia class, but decreased S24-7 and Bacteroides in the Bacteroidia class.
Figure 5.
Other differentially taxa by PCB exposure in LIC of CV mice (n = 3 per group). (A), Taxa in the Actinobacteria class; (B) taxa in the Erysipelotrichi class; (C) taxa in the Bacilli class; (D) taxa in the Betaproteobacteria class; (E) taxa in the Bacteroidia class; (F) taxa in the Clostridia class. Data were analyzed using QIIME. Asterisks represent statistically significant differences as compared with corn oil exposure group (1-way ANOVA, Duncan’s post-hoc test, p < .05).
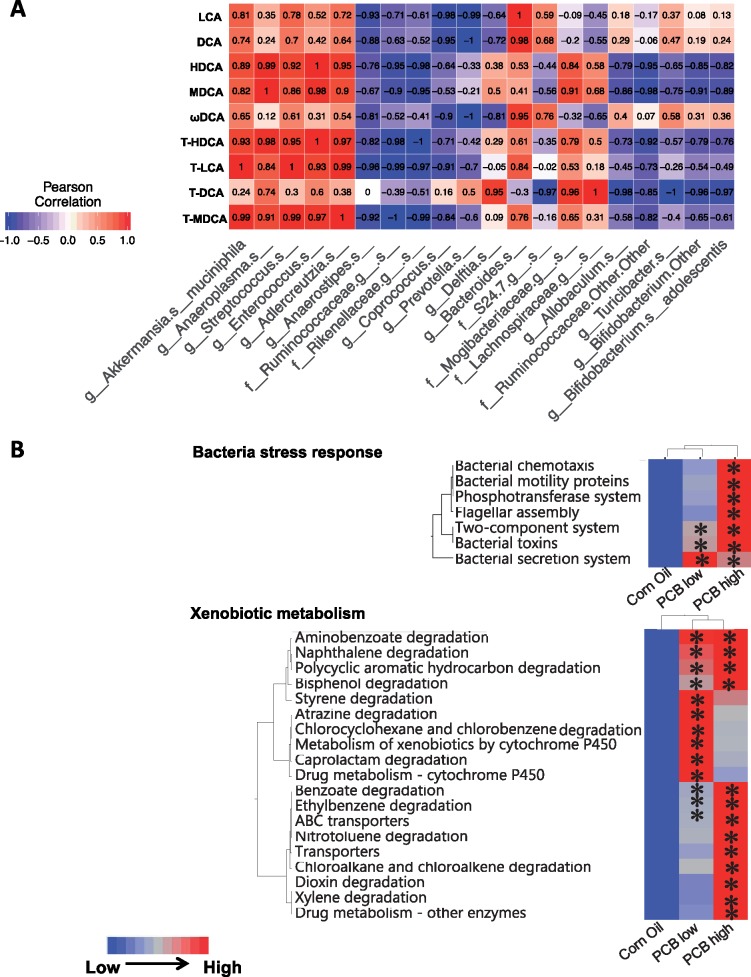
As shown in Figure 6a, because very little is known regarding what microbial species are responsible for PCB-mediated changes in BAs in various bio-compartments, as a first step to explore such mechanistic connections, Pearson’s correlation analysis was performed between bacteria that were differentially regulated by PCBs in large intestinal content and total secondary BA pool (Figure 7). To obtain the total secondary BA pool, levels of an individual secondary BA (either conjugated or conjugated) from the 4 biological compartments, namely serum, liver, SIP, and LIP, were summed together as one value. The reason for using the total pool of each BA in all 4 compartments to correlate with gut microbiota was to focus on the microbial contribution rather than the other confounding factors such as dispositional changes of BAs mediated by host transporters among various compartments. A total of 9 secondary BAs were considered, namely the unconjugated BAs LCA, DCA, HDCA, MDCA, and ωMCA, as well as the conjugated BAs T-HDCA, T-CA, T-DCA, and T-MDCA. A total of 20 taxa that were differentially regulated by PCBs at either low or high dose were considered. The r-values above 0.7 or below −0.7 were considered positive or negative correlations. For example, for the taxa that were up-regulated by PCB low dose, A. muciniphila was positively associated with most secondary BAs except for T-DCA. Anaeroplasma was positively associated with most secondary BAs including HDCA, MDCA, T-HDCA, T-LCA, T-DCA, and TMDCA. Streptococcus was positively associated with most secondary BAs except for ωMCA and T-DCA. Enterococcus was positively associated with most secondary BAs including HDCA, MDCA, T-HDCA, T-LCA, and T-MDCA. Adlercreutzia was positively associated with most secondary BAs except for DCA, ωMCA, and T-DCA. Conversely, for the taxa that were down-regulated by the low PCB dose, Anaerostipes was negatively correlated with most secondary BAs except for T-DCA and MDCA. Ruminococcaceae was negatively correlated with most secondary BAs including HDCA, MDCA, T-HDCA, T-LCA, and T-MDCA. Rikenellaceae was negatively correlated with HDCA, MDCA, T-HDCA, T-LCA, and T-MDCA.
Figure 6.
A, Pearson’s correlation analysis of differentially regulated bacterial species and total secondary BA pool (sum of serum, liver, SIP, and LIP). Values toward 1 mean positive correlation, whereas values toward −1 mean negative correlation. Data were analyzed using R. B, Hierarchical clustering dendrograms of selected KEGG pathways predicted from 16S rRNA sequencing data using PICRUSt as described in the Materials and Methods section. Red represents relatively high abundance, whereas blue represents relatively low abundance. Asterisks represent statistically significant differences as compared to control group (1-way ANOVA, Duncan’s post-hoc test, p < .05). (For interpretation of the reference to color in this figure legend, the reader is referred to the web version of this article.)
Figure 7.
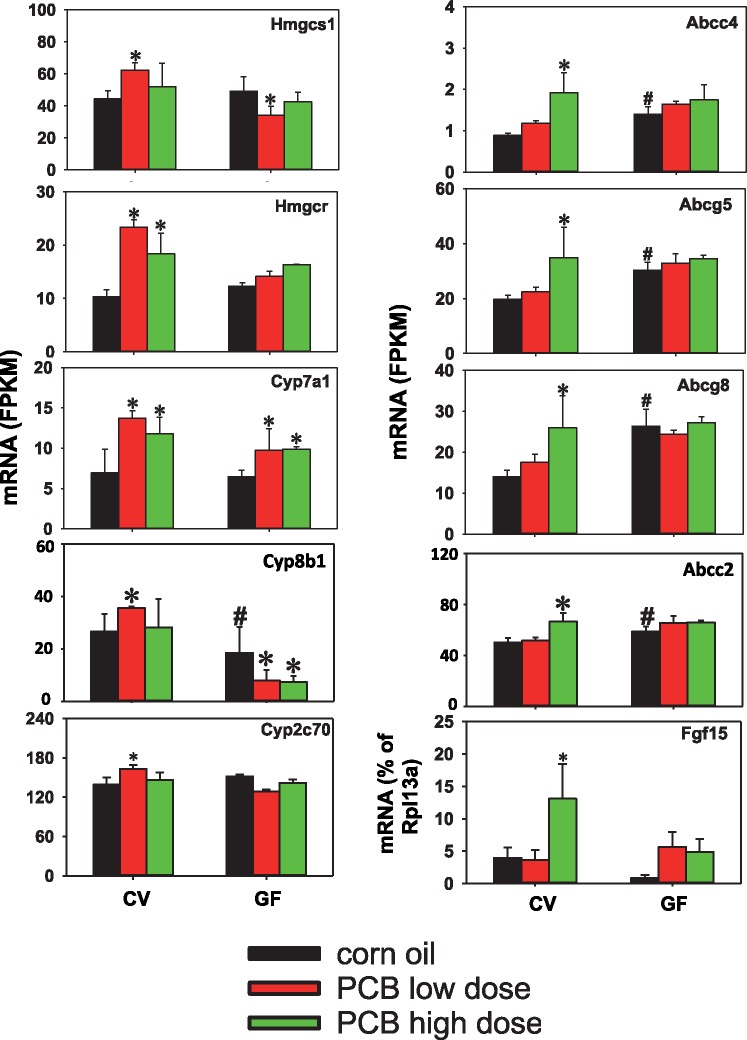
mRNA expression of host BA-processing genes (Hmgcs1, Hmgcr, Cyp7a1, Cyp8b, Cyp2c70, Abcc4, Abcg5, Abcg8, and Abcc2) in liver and ileum (Fgf15) following PCB exposure in CV and GF mice. The liver gene expression was determined using RNA-Seq (n = 3 per group), and the ileal Fgf15 mRNA was determined using RT-qPCR (n = 5 per group). Asterisks represent statistically significant differences as compared with corn oil-treated group of the same enterotype (p < .05); pounds (#) represent statistically significant differences between CV and GF mice exposed to the same chemical (p < .05).
As shown in Figure 6B, phylogenetic investigation of communities by reconstruction of unobserved states (PICRUSt) (Langille et al., 2013) was applied to the 16S rRNA sequencing data to predict the functional composition of the gut microbiota following oral exposure to the Fox River mixture. Of the total 328 KEGG pathways identified, 188 were differentially regulated by PCB exposure (ANOVA, p < .05). Among these pathways, 8 bacteria-specific pathways and 19 xenobiotic metabolism-specific pathways were identified. Oral PCB exposure dose-dependently increased the following bacteria-specific pathways: bacterial chemotaxis, bacterial motility proteins, phosphotransferase system (which is a direct method used by bacteria for sugar uptake) (Clore and Venditti, 2013), flagellar assembly, 2-component system (which is most commonly used in gram-negative and cyanobacteria) (Capra and Laub, 2012), and bacterial toxins, and bacterial secretion systems. The only exception was the bacterial secretion system, which increased more at the low compared to the high PCB dose. PCB exposure increased the microbial cytochrome P450s-mediated drug metabolism pathways at the low PCB dose and the ATP-binding cassette (ABC) transporters-mediated efflux pathways at the high PCB dose. In addition, many metabolic pathways involved in xenobiotic degradation including PCB-like compounds, were also increased by PCB exposure. Degradation pathways for aminobenozoate, naphthalene, polycyclic aromatic hydrocarbon, and bisphenol were enriched in both PCB exposure groups; degradation pathways of styrene, atrazine, chlorocyclohexane, chlorobenzene, and caprolactam were enriched at low PCB dose; whereas degradation pathways of benzoate, ethylbenzene, nitrotoluene, chloroalkane and chloroalkene, dioxin and xylene were enriched at high PCB dose.
In summary, whereas PCBs at low dose preferably increased BAs in a gut microbiota dependently manner, PCBs at high dose had the most predicted functional changes related to xenobiotic biotransformation.
Regulation of Host BA-Processing Genes in Liver and Ileum of CV and GF Mice by PCB Exposure
As shown in Figure 7 left panel, in livers of CV mice, PCB low dose increased the mRNA expression of the cholesterol bio-synthesis enzymes Hmgcs1 and Hmgcr1, as well as the BA-synthetic enzymes Cyp7a1 and Cyp8b1 from the classic pathway of BA synthesis. The BA-synthetic enzymes in alternative pathway of BA synthesis remained unchanged following PCB exposure in either CV or GF conditions (Supplementary Figure 6A). The mRNA of Cyp2c70, which is important in converting CDCA to MCA (Takahashi et al., 2016), was also up-regulated by PCB low dose. To note, similar to the PCB low dose, the PCB high dose also increased the mRNAs of Hmgcr and Cyp7a1 in livers of CV mice; however, the fold increase was less as compared with the PCB low dose-exposed CV mice. In livers of GF mice, the PCB-mediated increase in these mRNAs was all abolished except for Cyp7a1 mRNA, whereas PCBs at low dose decreased Hmgcs1 mRNA, and PCBs at both doses decreased Cyp8b1 mRNA. In addition, the basal level of Cyp8b1 mRNA was lower in GF mice.
As shown in Figure 7 right panel, in liver of CV mice, PCB at high dose uniquely increased the mRNA of the basolacteral efflux transporter Mrp4 (which can transport BAs), as well as the canalicular efflux transporter dimers Abcg5/g8 (which transport cholesterol) and Abcc2 (which contributes to bile flow). PCB at high dose also uniquely increased the mRNA of the ileum-derived hormone Fgf15, which suppress BA synthesis in liver. The PCB high dose mediated increase in the mRNAs of these genes was completed abolished in GF conditions. In addition, there was a basal mRNA increase in Abcc4, Abcg5/8, and Abcc2 in livers of GF mice. Other BA-processing genes in liver, including the basolateral BA uptake transporters Nctp Oatp1b2, the canalicular efflux transporters Bsep, Mdr2, Atp8b1, and the BA conjugate enzymes Slc27a1 and Baat, as well as the BA transporters in ileum (Slc10a2, Slc51a/b), remained unchanged by PCB exposure or lack of gut microbiota (Supplementary Figure 6).
DISCUSSION
In the present study, we showed that acute PCB exposure modulates BA levels in a dose and compartment-specific manner, associated with changes in BA-metabolizing gut microbiota and host BA-processing genes.
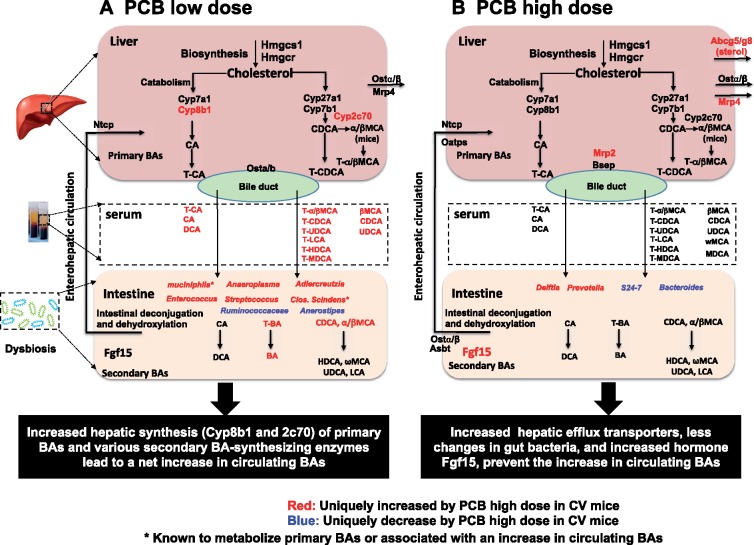
It is important to note that not all PCB-mediated changes in BAs are dependent on gut microbiota. For example, BA regulatory patterns by PCBs are similar between CV and GF mice SIP, although as expected the basal levels of BAs are higher in SIP of GF mice (Figure 3A). However, in all 3 other bio-compartments (serum, liver, and LIP), the presence of gut microbiota profoundly modified the PCB-mediated changes in BAs. Notably, the PCB low dose mediated a marked increase in multiple BA categories that was only present in serum of CV mice but not in GF mice. Moreover, the lack of gut microbiota sensitized the liver and LIP to PCB-mediated increase in BAs. These observations suggest that gut microbiota modulates changes of BAs following oral PCB exposure. As summarized in Figure 8A, we hypothesize that the PCB-mediated regulation of BAs is through a combination of both host factors and the gut microbiota.
Figure 8.
Schematic summary of mechanism of PCB exposure influencing BA biotransformation through modulation of intestinal microbiota, host BA synthesizing/metabolizing enzymes, and transporters in CV mice. (A) PCB low dose. (B) PCB high dose.
The majority of the BA changes were observed at PCB low dose, as evidenced by a marked increase in most circulating BAs. The appeared to be contributed by a PCB low dose-mediated increase in several taxa that are known to generate secondary BAs (eg, C. scindens and Enterococcus), or associated with high circulating primary BAs (A. muciniphila). Several other taxa that were also uniquely increased by PCB low dose, including Anaeroplasma, Adlercreutzia, and Streptococcus, have not been characterized regarding their relevance to BA metabolism, and our study was among the first to show a positive correlation between their abundance and BA levels during PCB exposure. Host BA-synthetic enzymes may also contribute to this process, in that the PCB low dose-specific up-regulation in the host BA-synthetic enzymes Cyp8b1 and Cyp2c70 as well as the cholesterol synthetic enzyme Hmgcs1 may jointly contribute to the rise in circulating BAs together with the microbial contributions. In contrast, as summarized in Figure 8B, at PCB high dose, much less bacteria were altered by PCBs, whereas S24-7 and Bacteroides, which are known to contribute to BA metabolism (according to our FishTaco analysis, data not shown), were down-regulated by PCB high dose. In addition, the PCB high dose-mediated increase in various efflux transporters in the liver, as well as the intestinal suppressive hormone Fgf15 for BA-synthesis, may contribute to higher elimination of BAs out of the body and counteracting the host de novo synthesis, together preventing the increase in BA levels in general.
Gut microbiota plays an essential role in deconjugation of primary BAs and secondary BAs synthesis (Ramirez-Perez et al., 2017) and can be affected by environmental pollutants such as PCBs (Choi et al., 2013). Prior to this work, very little was known regarding the dose-response relationship of the Fox River mixture and the functional changes of the gut microbiome. The present study has filled this critical knowledge gap and specifically unveiled a novel link between this PCB mixture and BAs, which are a well-known toxicological endpoint implicated in various human diseases such as inflammation, metabolic syndrome, and cancer.
A previous study also identified changes in gut microbiota composition in exercising and sedentary mice exposed to a simple PCB mixture containing environmentally relevant mixture of PCBs (PCB138, PCB153, and PCB180) (Choi et al., 2013). Therefore, we compared results from the present study with a subset of the previous work using sedentary mice. Both studies determined the acute effect of PCB oral exposure in adult C57BL/6 mice (3-days exposure in the present study versus 2 days exposure in the previous study). The PCB dose used by Choi et al. was 150 μmol/kg (43 mg/kg), which is expected to result in plasma levels of PCBs representative of an acutely exposed human population (Jensen et al., 1989), whereas the PCB doses used in the present study were below this level (20.53 μmol/kg for 6 mg/kg low PCB dose, and 102.67 μmol/kg for 30 mg/kg high PCB dose).
Consistent with the previous study, we also showed that PCB exposure did not alter the overall biodiversity of the gut microbiome, however, the low PCB dose tended to decrease the richness of the gut microbiota although a statistical difference was not achieved. The low PCB dose-mediated increase in A. muciniphila was consistent with the previously observed increase in Verrucomicrobiceae (Choi et al., 2013), which is the family that includes the A. muciniphila species. Differences in the regulation of gut microbiota were also observed, for example, the previous study showed that bacteria in the Proteobacteria phylum were differentially regulated by PCB exposure, whereas in the present study no such effect was observed at either dose. This difference may be due to differences in gender (Choi et al. used male mice), the use of a complex PCB mixture versus a mixture containing only 3 PCB congeners, different diet and microbial environment between the two animal facilities, and 16S rRNA quantification techniques (Choi et al. used microarrays).
In addition, the dosing frequencies were different between the present study and Choi et al. (2013), in that the present study administered the low (6 mg/kg) and the high PCB (30 mg/kg) doses once daily for three consecutive days, and samples were collected on the 4th day, whereas Choi et al. performed a single dose of a different PCB mixture at 43 mg/kg, and samples were collected over the next 2 days. Therefore, the cumulative dose of PCBs over 3 days in the present study is expected to be higher than the dose used by Choi et al. Last but not least, the differences in the animal husbandry (especially diet and bedding) between the two animal facilities may also contribute to the different observations in gut microbiome changes between the present study and by Choi et al. These differences mentioned above (eg, different PCB mixtures, gender of animals, dosing frequency and tissue collection regimen, and animal husbandry) may contribute to the differences in the taxa that were differentially regulated in the two studies. It is important to note that following the recently proposed “Anna Karenina Principle” regarding the stress and stability of the microbial communities (Zaneveld et al., 2017), the variation in the dysbiotic organisms is both a common and important response of animal microbiomes to stressors. These stressors from the external environment reduce the ability of the host and/or its microbiome to regulate community compositional changes. It could be argued that although the exact differentially regulated taxa may be dissimilar following similar xenobiotic insults, the common microbial pathways that were differentially regulated may be functionally overlapping to accomplish the same protective functions against xenobiotic insult.
The present study suggested that PCBs at the low dose contribute to both primary and secondary BA synthesis associated with an up-regulation in Bacteroides, Bifidobacterium and Enterococcus, C. scindens, and A. muciniphila. Bshs (Ridlon et al., 2006), known as conjugated bile salt hydrolases, can result in local BA deconjugation which expressed in several genera include Bacteroides, Bifidobacterium, and Enterococcus (Klaassen and Cui, 2015). These findings suggest that the increase of unconjugated BAs in SIP of CV mice by PCB low dose could be regulated by these bacteria. C. scindens is also well known for being responsible for secondary BA synthesis via 7α dehydroxylation via enzymes produced from the BA inducible operons (Ridlon and Hylemon, 2012). Due to current limitations in the reference database and high sequence homologies among various clostridia genes, C. scindens was not mapped in the 16S rRNA sequencing data, but its increase was demonstrated using qPCR. The low PCB dose mediated increase in C. scindens positively correlated with the increase in serum- and intestinal-secondary BAs. It should be noted that other microbial species displayed positive associations with individual BA levels from the total BA pool and may also contribute to BA homeostasis directly or indirectly. Due to limited information in their complete genomic sequences and technical limitations in culturing them under anaerobic conditions, future studies and technological advancements are needed to further elucidate the roles of these bacteria in PCB-mediated changes in BA signaling.
Interestingly, the PCB-mediated changes in BAs in CV mice is non-monotonic in that the PCB low dose produced a more prominent effect in increasing primary BAs in serum and SIP and appeared to have a moderate stimulatory effect on secondary BA synthesis. The up-regulated taxa may be responsible for increased secondary BA synthesis, whereas the down-regulated taxa may counteract the low PCB dose-mediated changes in secondary BA synthesis. The effects of PCB high dose on synthesis of unconjugated and secondary BAs was inconsistent with the increase in the predicted-bacterial BA metabolism as well as the apparent increase in DNA encoding microbial enzymes for secondary BA synthesis (bsh and baiJ). One potential explanation for this discrepancy is that the high PCB dose causes the microbial community to shift from performing intermediary metabolism (such as secondary BA synthesis) toward xenobiotic biotransformation as a compensatory mechanism to protect the microorganism and the host from high dose of toxic insults. This interpretation is consistent with the preferred enrichment of predicted functions in bacterial stress response and xenobiotic metabolism at the PCB high dose. Another possibility for this discrepancy is that, the apparent increase in microbial DNA does not necessarily translate into a proportional increase in functional microbial enzymes, or other bai genes not determined in this assay may play important roles. The involvement of host BA-processing genes, especially efflux transporters in liver and the intestinal inhibitory hormone Fgf15 for BA synthesis may also contribute to the nonmonotonic increase in BA levels.
The expression of the cholesterol-synthetic enzyme Hmgcs1, the BA-synthetic enzyme Cyp8b1, and the MCA-producing Cyp2c70 was increased only by the low PCB dose in a gut microbiota-dependent manner. To note, Cyp2c70 has been shown to be responsible for the production of α/βMCAs (Takahashi et al., 2016), which may explain the PCB low dose mediated increase in circulating α/βMCA. The increase in Cyp2c70, together with the increase in Hmgcs1 and Cyp8b1, may contribute to the increase in total BAs mediated by the PCB low dose. The expression of Fgf15 mRNA was preferably increased by the PCB high dose in ileum of CV mice. It remains elusive why hepatic Cyp7a1 mRNA was not decreased following the increase in Fgf15. One possible explanation is that the intestine-derived Fgf15 may prevent a further increase in Cyp7a1, but is not potent enough to counteract hepatic PCB-mediated increase in BAs. At the PCB high dose, the mRNA increase in efflux transporters (Abcg5/g8, Abcc2, and Abcc4) may lead to a rapid elimination of hepatic accumulation of the BA precursor cholesterol and other bile constituents. This may be responsible for the lack of increase in total BAs at the PCB high dose.
Another interesting observation in the present study is that the PCB-mediated changes in BA levels were compartment- and enterotype-specific. Regarding the compartment specificity, serum BA levels of CV mice were most impacted by PCBs (low dose), followed by SIP. In contrast, level of BAs in the liver and LIP were not significantly affected by PCB exposure (Figure 5B). One possible explanation for this observation is that PCBs modulate the expression of host genes that are involved in disposition of PCBs and their metabolites. Indeed, it has been reported that another PCB congener, namely PCB126, increased the hepatic expression of efflux transporters (Lickteig et al., 2008; Maher et al., 2005). Regarding the enterotype specificity, lack of gut microbiota augmented the PCB-mediated increase in BAs in liver, SIP (low dose) and LIP (high dose) of many primary and conjugated BAs (Figure 5B). A likely explanation for these enterotype specific differences is that PCB congeners present in the Fox River mixture and/or metabolites without microbiota interaction have more potent effects on primary BA synthesis/conjugation in GF mice.
In addition, the present study has unveiled novel microbial biomarkers and that were altered following oral exposure to the PCB Fox River mixture in mice in a dose-dependent manner; most notably, the low PCB dose-mediated increase in A. muciniphila correlated with the increase in BAs in serum in a gut microbiota-dependent manner. The low PCB dose-mediated increase in A. muciniphila coincided with an increase in serum and SIP BAs of CV mice (Figs. 2D, 3, and 5). Although A. muciniphila has not been shown to directly participate in secondary BA synthesis, it positively correlated with higher levels of circulating primary BAs during high fat diet induced obesity (Pierre et al., 2016). This suggests that A. muciniphila communicates with host receptors involved in primary BA synthesis via its microbial metabolites. Indeed, in intestine of GF mice and intestinal organoids, genes affected by A. muciniphila colonization were involved in cell growth and survival, as well as metabolism of fatty acid, cholesterol, and BAs (Derrien et al., 2011; Lukovac et al., 2014). Consistent with the literature, our data showed that the PCB low dose-mediated increase in A. muciniphila positively correlated with an increase in serum- and small intestinal-primary BAs. In addition, the present study revealed that A. muciniphila also displayed a strong positive correlation with the total pool of most secondary BAs. This observation suggests that either A. muciniphila has genes encoding BA-metabolizing enzymes that are yet to be discovered, or it facilitates the growth of other BA-metabolizing bacteria via symbiosis mechanisms.
The present study also unveiled pathways that may contribute to the toxicities of PCBs. The high PCB dose resulted in an increase of the Prevotella genus. The Prevotella abundance at mucosal sites have linked to localized and systemic disease, including periodontitis, bacterial vaginosis, rheumatoid arthritis, metabolic disorder, and low-grade systemic inflammation in emerging studies in humans (Choi et al., 2013; Larsen, 2017). These findings suggest that Prevotella may be a critical diagnostic biomarker for human environmental pollutants induced inflammatory diseases. Presumably, studies in our mice support a causal role of PCB high dose-mediated Prevotella in promotion of clinical inflammatory bowel disease (IBD) and inflammatory related diseases. Considering that PCBs have been shown to initiate inflammatory response in the vasculature (Petriello et al., 2014), it is possible that PCBs may also contribute to increased risks of gastrointestinal inflammation through enriching this bacterial genus.
Similar to the host, bacterial P450s play important roles in the oxidative metabolism of structurally diverse organic chemicals from both exogenous and endogenous sources, including aromatic hydrocarbons and their derivatives (Lewis and Wiseman, 2005). The apparent up-regulation of microbial P450s at the low PCB dose in this study suggests that the intestinal bacteria attempt to metabolize and potentially detoxify the PCB parent compounds and other secondary metabolites. In addition to P450 enzymes, transporters such as the ABC efflux transporters also appeared to be increased by PCB exposure. In the host system, ABC transporters serve as an important cyto-protective mechanism by eliminating toxicants out of cells. Bacterial ABC transporters are also important exporters that may protect the microorganisms from xenobiotic insult (PMID: 8302219). The apparent up-regulation of ABC transporters as well as other transporters, preferably at the high PCB dose, together with the P450-mediated pathways (induced at the low dose) provides additional protections against toxic insults. Indeed, many xenobiotic metabolism pathways appeared to increase in a dose-dependent manner, including the degradation of aminobenzoate, naphthalene, polyaromatic hydrocarbon, bisphenol, benzoate, ethylbenzene, nitrotoluenen, chloroalkane, dioxin, and xylene (Figure 6B). Other microbial xenobiotic metabolizing pathways were only increased by PCB low dose, including the degradation of styrene, atrazine, chlorobenzene, and caprolactam (Figure 6B).
Although very little is known regarding the microbial metabolism of PCBs by intestinal bacteria, the predicted increase in these xenobiotic metabolism pathways indicates that the microbial enzymes that are responsible for PCB metabolism may also contribute to the biotransformation of the other xenobiotics. For example, as was reviewed recently, common microbial reactions shared among many persistent organic environmental toxicants include deconjugation of liver metabolites and reduction reactions (Claus et al., 2016; Lu et al., 2015). In addition, bacterial C-S-lyase plays an important role in the formation of methyl sulfone (MeSO2) metabolites of PCBs in vivo following dietary exposure to PCBs (Claus et al., 2016). In summary, the differential xenobiotic responses to low and high PCB doses by intestinal bacteria highlights the importance of the gut microbiome in contributing to the xenobiotic detoxification with the host system.
Taken together, the present study has unveiled for the first time that the human relevant PCB mixture, namely Fox River mixture, altered the gut microbiome composition and predicted functions, and modulate BA homeostasis jointly with the host BA processing genes in a dose- and bio-compartment-specific manner. Distinct microbial biomarkers that correlate with individual BA species provide lead taxa that may drive the functional changes in secondary BA synthesis. Future studies are needed to mechanistically characterize the individual taxa in modulating PCB-mediated metabolic changes using in vitro anaerobic culture and in vivo inoculation to GF mouse models.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Dr Zidong Donna Fu for technical assistance in discussion on BA analysis, and all members in the Cui Laboratory for proofreading the manuscript.
FUNDING
This work was supported by the National Institutes of Health (grant numbers P30 ES005605 [HJL] and P42 ES013661 [HJL]; R01 GM111381 [JYC]), the start-up funds [JYC] from University of Washington Center for Exposures, Diseases, Genomics, and Environment (P30 ES0007033 [TJK]), as well as the Murphy Endowment [JYC].
REFERENCES
- Aminov Z., Haase R. F., Pavuk M., Carpenter D. O.; Anniston Environmental Health Research Consortium (2013). Analysis of the effects of exposure to polychlorinated biphenyls and chlorinated pesticides on serum lipid levels in residents of Anniston, Alabama. Environ. Health 12, 108.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandara S. B., Sadowski R. N., Schantz S. L., Gilbert M. E. (2017). Developmental exposure to an environmental PCB mixture delays the propagation of electrical kindling from the amygdala. Neurotoxicology 58, 42–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell M. R. (2014). Endocrine-disrupting actions of PCBs on brain development and social and reproductive behaviors. Curr. Opin. Pharmacol. 19, 134–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byard J. L., Paulsen S. C., Tjeerdema R. S., Chiavelli D. (2015). DDT, chlordane, toxaphene and PCB residues in Newport Bay and Watershed: Assessment of hazard to wildlife and human health. Rev. Environ. Contam. Toxicol. 235, 49–168. [DOI] [PubMed] [Google Scholar]
- Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., Fierer N., Pena A. G., Goodrich J. K., Gordon J. I., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capra E. J., Laub M. T. (2012). Evolution of two-component signal transduction systems. Annu. Rev. Microbiol. 66, 325–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmody R. N., Turnbaugh P. J. (2014). Host-microbial interactions in the metabolism of therapeutic and diet-derived xenobiotics. J. Clin. Invest. 124, 4173–4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S. L., Bammler T. K., Cui J. Y. (2017). RNA sequencing reveals age and species differences of constitutive androstane receptor-targeted drug-processing genes in the liver. Drug Metab. Dispos. 45, 867–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang J. Y. (2013). Bile acid metabolism and signaling. Compr. Physiol. 3, 1191–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J. J., Eum S. Y., Rampersaud E., Daunert S., Abreu M. T., Toborek M. (2013). Exercise attenuates PCB-induced changes in the mouse gut microbiome. Environ. Health Perspect. 121, 725–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claus S. P., Guillou H., Ellero-Simatos S. (2016). The gut microbiota: A major player in the toxicity of environmental pollutants? NPJ Biofilms Microbiomes 2, 16003.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clore G. M., Venditti V. (2013). Structure, dynamics and biophysics of the cytoplasmic protein-protein complexes of the bacterial phosphoenolpyruvate: Sugar phosphotransferase system. Trends. Biochem. Sci. 38, 515–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien M., Van Baarlen P., Hooiveld G., Norin E., Muller M., de Vos W. M. (2011). Modulation of mucosal immune response, tolerance, and proliferation in mice colonized by the mucin-degrader Akkermansia muciniphila. Front Microbiol. 2, 166.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doull J. (2003). The “Red Book” and other risk assessment milestones. Hum. Ecol. Risk Assess. 9, 1229–1238. [Google Scholar]
- Duntas L. H., Stathatos N. (2015). Toxic chemicals and thyroid function: Hard facts and lateral thinking. Rev. Endocr. Metab. Disord. 16, 311–318. [DOI] [PubMed] [Google Scholar]
- EPA press release (1979). EPA bans PCB manufacture; phases out uses. https://archive.epa.gov.
- Eskenazi B., Rauch S. A., Tenerelli R., Huen K., Holland N. T., Lustig R. H., Kogut K., Bradman A., Sjodin A., Harley K. G. (2017). In utero and childhood DDT, DDE, PBDE and PCBs exposure and sex hormones in adolescent boys: The CHAMACOS study. Int. J. Hyg. Environ. Health 220, 364–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman M. D., Kohles S. S. (2012). Plasma levels of polychlorinated biphenyls, non-Hodgkin lymphoma, and causation. J. Environ. Public Health 2012, 258981.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z. D., Cui J. Y. (2017). Remote sensing between liver and intestine: Importance of microbial metabolites. Curr. Pharmacol. Rep. 3, 101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z. D., Selwyn F. P., Cui J. Y., Klaassen C. D. (2017). RNA-Seq profiling of intestinal expression of xenobiotic processing genes in germ-free mice. Drug Metab. Dispos. 45, 1225–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S., Murinova L., Trnovec T., Loffredo C. A., Washington K., Mitra P. S., Dutta S. K. (2014). Biomarkers linking PCB exposure and obesity. Curr. Pharm. Biotechnol. 15, 1058–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandjean P., Grønlund C., Kjær I. M., Jensen T. K., Sørensen N., Andersson A.-M., Juul A., Skakkebæk N. E., Budtz-Jørgensen E., Weihe P. (2012). Reproductive hormone profile and pubertal development in 14-year-old boys prenatally exposed to polychlorinated biphenyls. Reprod. Toxicol. 34, 498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm F.A., Hu D., Kania-Korwel I., Lehmler H.J., Ludewig G., Hornbuckle K.C., Duffel M.W., Bergman A., Robertson L.W. (2015) Metabolism and metabolites of polychlorinated biphenyls. Crit. Rev. Toxicol. 45, 245–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guengerich F. P. (2001) Analysis and characterization of enzymes and nucleic acids In Principles and Methods of Toxicology (Hayes A. W., Ed.), pp. 1625–1688. Taylor & Francis, Boca Raton. [Google Scholar]
- Gupta P., Thompson B. L., Wahlang B., Jordan C. T., Zach Hilt J., Hennig B., Dziubla T. (2017). The environmental pollutant, polychlorinated biphenyls, and cardiovascular disease: A potential target for antioxidant nanotherapeutics. Drug. Deliv. Transl. Res. 8, 740–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiser H. J., Turnbaugh P. J. (2013). Developing a metagenomic view of xenobiotic metabolism. Pharmacol. Res. 69, 21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Adamcakova-Dodd A., Lehmler H. J., Gibson-Corley K., Thorne P. S. (2015). Toxicity evaluation of exposure to an atmospheric mixture of polychlorinated biphenyls by nose-only and whole-body inhalation regimens. Environ. Sci. Technol. 49, 11875–11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamshidi A., Hunter S., Hazrati S., Harrad S. (2007). Concentrations and chiral signatures of polychlorinated biphenyls in outdoor and indoor air and soil in a major U.K. conurbation. Environ. Sci. Technol. 41, 2153–2158. [DOI] [PubMed] [Google Scholar]
- Jia W., Xie G., Jia W. (2017). Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat. Rev. Gastroenterol. Hepatol. 15, 111–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D. J., Ridlon J. M., Moore D. R. 2nd, Barnes S., Hylemon P. B. (2008). Clostridium scindens baiCD and baiH genes encode stereo-specific 7alpha/7beta-hydroxy-3-oxo-delta4-cholenoic acid oxidoreductases. Biochim. Biophys. Acta 1781, 16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kania-Korwel I., Barnhart C. D., Stamou M., Truong K. M., El-Komy M. H., Lein P. J., Veng-Pedersen P., Lehmler H. J. (2012). 2, 2′, 3, 5′, 6-Pentachlorobiphenyl (PCB 95) and its hydroxylated metabolites are enantiomerically enriched in female mice. Environ. Sci. Technol. 46, 11393–11401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kania-Korwel I., El-Komy M. H., Veng-Pedersen P., Lehmler H. J. (2010). Clearance of polychlorinated biphenyl atropisomers is enantioselective in female C57Bl/6 mice. Environ. Sci. Technol. 44, 2828–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kania-Korwel I., Hornbuckle K. C., Robertson L. W., Lehmler H. J. (2008). Dose-dependent enantiomeric enrichment of 2, 2′, 3, 3′, 6, 6′-hexachlorobiphenyl in female mice. Environ. Toxicol. Chem. 27, 299–305. [DOI] [PubMed] [Google Scholar]
- Kania-Korwel I., Hornbuckle K. C., Robertson L. W., Lehmler H. J. (2008). Influence of dietary fat on the enantioselective disposition of 2, 2′, 3, 3′, 6, 6′-hexachlorobiphenyl (PCB 136) in female mice. Food Chem. Toxicol. 46, 637–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kania-Korwel I., Lukasiewicz T., Barnhart C. D., Stamou M., Chung H., Kelly K. M., Bandiera S., Lein P. J., Lehmler H. J. (2017). Editor’s highlight: Congener-specific disposition of chiral polychlorinated biphenyls in lactating mice and their offspring: Implications for PCB developmental neurotoxicity. Toxicol. Sci. 158, 101–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kania-Korwel I., Wu X., Wang K., Lehmler H. J. (2017). Identification of lipidomic markers of chronic 3, 3′, 4, 4′, 5-pentachlorobiphenyl (PCB 126) exposure in the male rat liver. Toxicology 390, 124–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kania-Korwel I., Xie W., Hornbuckle K. C., Robertson L. W., Lehmler H. J. (2008). Enantiomeric enrichment of 2, 2′, 3, 3′, 6, 6′-hexachlorobiphenyl (PCB 136) in mice after induction of CYP enzymes. Arch. Environ. Contam. Toxicol. 55, 510–517. [DOI] [PubMed] [Google Scholar]
- Kim H. M., Youn C. H., Ko H. J., Lee S. H., Lee Y. M. (2017). The relationship between the blood level of persistent organic pollutants and common gastrointestinal symptoms. Korean J. Fam. Med. 38, 233–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaassen C. D., Cui J. Y. (2015). Review: Mechanisms of how the intestinal microbiota alters the effects of drugs and bile acids. Drug Metab. Dispos. 43, 1505–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl K. D., Cary T. L., Karasov W. H., Dearing M. D. (2015). Larval exposure to polychlorinated biphenyl 126 (PCB-126) causes persistent alteration of the amphibian gut microbiota. Environ. Toxicol. Chem. 34, 1113–1118. [DOI] [PubMed] [Google Scholar]
- Kostyniak P. J., Hansen L. G., Widholm J. J., Fitzpatrick R. D., Olson J. R., Helferich J. L., Kim K. H., Sable H. J., Seegal R. F., Pessah I. N., et al. (2005). Formulation and characterization of an experimental PCB mixture designed to mimic human exposure from contaminated fish. Toxicol. Sci. 88, 400–411. [DOI] [PubMed] [Google Scholar]
- Kostyniak P. J., Hansen L. G., Widholm J. J., Fitzpatrick R. D., Olson J. R., Helferich J. L., Kim K. H., Sable H. J. K., Seegal R. F., Pessah I. N., et al. (2005). Formulation and characterization of an experimental PCB mixture designed to mimic human exposure from contaminated fish. Toxicol. Sci. 88, 400–411. [DOI] [PubMed] [Google Scholar]
- Kuczynski J., Stombaugh J., Walters W. A., Gonzalez A., Caporaso J. G., Knight R. (2012). Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr. Protoc. Microbiol. Chapter 1: Unit 1E 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille M. G., Zaneveld J., Caporaso J. G., McDonald D., Knights D., Reyes J. A., Clemente J. C., Burkepile D. E., Vega Thurber R. L., Knight R., et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen J. M. (2017). The immune response to Prevotella bacteria in chronic inflammatory disease. Immunology 151, 363–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis D. F. V., Wiseman A. (2005). A selective review of bacterial forms of cytochrome P450 enzymes. Enzyme Microb. Technol. 36(2005), 377–384. [Google Scholar]
- Li T., Chiang J. Y. (2015). Bile acids as metabolic regulators. Curr. Opin. Gastroenterol. 31, 159–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lickteig A. J., Cheng X., Augustine L. M., Klaassen C. D., Cherrington N. J. (2008). Tissue distribution, ontogeny and induction of the transporters multidrug and toxin extrusion (MATE) 1 and MATE2 mRNA expression levels in mice. Life Sci. 83, 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K., Mahbub R., Fox J. G. (2015). Xenobiotics: Interaction with the intestinal microflora. ILAR J. 56, 218–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludewig G., Robertson L. W. (2013). Polychlorinated biphenyls (PCBs) as initiating agents in hepatocellular carcinoma. Cancer Lett. 334, 46–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukovac S., Belzer C., Pellis L., Keijser B. J., de Vos W. M., Montijn R. C., Roeselers G. (2014). Differential modulation by Akkermansia muciniphila and Faecalibacterium prausnitzii of host peripheral lipid metabolism and histone acetylation in mouse gut organoids. MBio 5, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher J. M., Cheng X., Slitt A. L., Dieter M. Z., Klaassen C. D. (2005). Induction of the multidrug resistance-associated protein family of transporters by chemical activators of receptor-mediated pathways in mouse liver. Drug Metab. Dispos. 33, 956–962. [DOI] [PubMed] [Google Scholar]
- Meeker J. D., Maity A., Missmer S. A., Williams P. L., Mahalingaiah S., Ehrlich S., Berry K. F., Altshul L., Perry M. J., Cramer D. W., et al. (2011). Serum concentrations of polychlorinated biphenyls in relation to in vitro fertilization outcomes. Environ. Health Perspect. 119, 1010–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaoka S., Miyazaki H., Aoyama Y., Yoshida A. (1990). Effects of dietary polychlorinated biphenyls on cholesterol catabolism in rats. Br. J. Nutr. 64, 161–169. [DOI] [PubMed] [Google Scholar]
- Patterson D. G. Jr, Wong L. Y., Turner W. E., Caudill S. P., Dipietro E. S., McClure P. C., Cash T. P., Osterloh J. D., Pirkle J. L., Sampson E. J., et al. (2009). Levels in the U.S. population of those persistent organic pollutants (2003–2004) included in the Stockholm Convention or in other long range transboundary air pollution agreements. Environ. Sci. Technol. 43, 1211–1218. [DOI] [PubMed] [Google Scholar]
- Pesatori A. C., Grillo P., Consonni D., Caironi M., Sampietro G., Olivari L., Ghisleni S., Bertazzi P. A. (2013). Update of the mortality study of workers exposed to polychlorinated biphenyls (Pcbs) in two Italian capacitor manufacturing plants. Med. Lav. 104, 107–114. [PubMed] [Google Scholar]
- Petriello M. C., Newsome B., Hennig B. (2014). Influence of nutrition in PCB-induced vascular inflammation. Environ. Sci. Pollut. Res. Int. 21, 6410–6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre J. F., Martinez K. B., Ye H., Nadimpalli A., Morton T. C., Yang J., Wang Q., Patno N., Chang E. B., Yin D. P. (2016). Activation of bile acid signaling improves metabolic phenotypes in high-fat diet-induced obese mice. Am. J. Physiol. Gastrointest. Liver Physiol. 311, G286–G304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon E., Monaikul S., Kostyniak P. J., Chi L. H., Schantz S. L., Sable H. J. (2013). Developmental exposure to polychlorinated biphenyls reduces amphetamine behavioral sensitization in Long-Evans rats. Neurotoxicol. Teratol. 38, 6–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon E., Powers B. E., McAlonan R. M., Ferguson D. C., Schantz S. L. (2011). Effects of developmental exposure to polychlorinated biphenyls and/or polybrominated diphenyl ethers on cochlear function. Toxicol. Sci. 124, 161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potera C. (2013). Running interference? Exercise and PCB-induced changes in the gut microbiome. Environ. Health Perspect. 121, A199.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quazi S., Yokogoshi H., Yoshida A. (1983). Effect of dietary fiber on hypercholesterolemia induced by dietary PCB or cholesterol in rats. J. Nutr. 113, 1109–1118. [DOI] [PubMed] [Google Scholar]
- Quinete N., Schettgen T., Bertram J., Kraus T. (2014). Occurrence and distribution of PCB metabolites in blood and their potential health effects in humans: A review. Environ. Sci. Pollut. Res. Int. 21, 11951–11972. [DOI] [PubMed] [Google Scholar]
- Ramirez-Perez O., Cruz-Ramon V., Chinchilla-Lopez P., Mendez-Sanchez N. (2017). The role of the gut microbiota in bile acid metabolism. Ann. Hepatol. 16, 15–s20. [DOI] [PubMed] [Google Scholar]
- Ridlon J. M., Harris S. C., Bhowmik S., Kang D. J., Hylemon P. B. (2016). Consequences of bile salt biotransformations by intestinal bacteria. Gut. Microbes 7, 22–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridlon J. M., Hylemon P. B. (2012). Identification and characterization of two bile acid coenzyme A transferases from Clostridium scindens, a bile acid 7alpha-dehydroxylating intestinal bacterium. J. Lipid. Res. 53, 66–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridlon J. M., Kang D. J., Hylemon P. B. (2006). Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 47, 241–259. [DOI] [PubMed] [Google Scholar]
- Ridlon J. M., Kang D. J., Hylemon P. B., Bajaj J. S. (2014). Bile acids and the gut microbiome. Curr. Opin. Gastroenterol. 30, 332–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sable H. J., Monaikul S., Poon E., Eubig P. A., Schantz S. L. (2011). Discriminative stimulus effects of cocaine and amphetamine in rats following developmental exposure to polychlorinated biphenyls (PCBs). Neurotoxicol. Teratol. 33, 255–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sable H. J., Powers B. E., Wang V. C., Widholm J. J., Schantz S. L. (2006). Alterations in DRH and DRL performance in rats developmentally exposed to an environmental PCB mixture. Neurotoxicol. Teratol. 28, 548–556. [DOI] [PubMed] [Google Scholar]
- Schecter A., Colacino J., Haffner D., Patel K., Opel M., Papke O., Birnbaum L. (2010). Perfluorinated compounds, polychlorinated biphenyls, and organochlorine pesticide contamination in composite food samples from Dallas, Texas, USA. Environ. Health Perspect. 118, 796–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W. S., Huttenhower C. (2011). Metagenomic biomarker discovery and explanation. Genome. Biol. 12, R60.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selwyn F. P., Cheng S. L., Bammler T. K., Prasad B., Vrana M., Klaassen C., Cui J. Y. (2015). Developmental regulation of drug-processing genes in livers of germ-free mice. Toxicol. Sci. 147, 84–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selwyn F. P., Cheng S. L., Klaassen C. D., Cui J. Y. (2016). Regulation of hepatic drug-metabolizing enzymes in germ-free mice by conventionalization and probiotics. Drug Metab Dispos. 44, 262–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selwyn F. P., Csanaky I. L., Zhang Y., Klaassen C. D. (2015). Importance of large intestine in regulating bile acids and glucagon-like Peptide-1 in germ-free mice. Drug Metab. Dispos. 43, 1544–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selwyn F. P., Cui J. Y., Klaassen C. D. (2015). RNA-seq quantification of hepatic drug processing genes in germ-free mice. Drug Metab. Dispos. 43, 1572–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin E. S., Nguyen K. H., Kim J., Kim C. I., Chang Y.S. (2015). Progressive risk assessment of polychlorinated biphenyls through a Total Diet Study in the Korean population. Environ. Pollut. 207, 403-412. [DOI] [PubMed] [Google Scholar]
- Spanogiannopoulos P., Bess E. N., Carmody R. N., Turnbaugh P. J. (2016). The microbial pharmacists within us: A metagenomic view of xenobiotic metabolism. Nat. Rev. Microbiol. 14, 273–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudinger J. L., Goodwin B., Jones S. A., Hawkins-Brown D., MacKenzie K. I., LaTour A., Liu Y., Klaassen C. D., Brown K. K., Reinhard J., et al. (2001). The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc. Natl. Acad. Sci. U.S.A. 98, 3369–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S., Fukami T., Masuo Y., Brocker C. N., Xie C., Krausz K. W., Wolf C. R., Henderson C. J., Gonzalez F. J. (2016). Cyp2c70 is responsible for the species difference in bile acid metabolism between mice and humans. J. Lipid Res. 57, 2130–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M., Chen K., Yang F., Liu W. (2014). Exposure to organochlorine pollutants and type 2 diabetes: A systematic review and meta-analysis. PLoS One 9, e85556.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toft G. (2014). Persistent organochlorine pollutants and human reproductive health. Dan. Med. J. 61, B4967.. [PubMed] [Google Scholar]
- van den Berg M., Kypke K., Kotz A., Tritscher A., Lee S. Y., Magulova K., Fiedler H., Malisch R. (2017). WHO/UNEP global surveys of PCDDs, PCDFs, PCBs and DDTs in human milk and benefit-risk evaluation of breastfeeding. Arch. Toxicol. 91, 83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlang B., Falkner K. C., Clair H. B., Al-Eryani L., Prough R. A., States J. C., Coslo D. M., Omiecinski C. J., Cave M. C. (2014). Human receptor activation by aroclor 1260, a polychlorinated biphenyl mixture. Toxicol. Sci. 140, 283–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlang B., Song M., Beier J. I., Cameron Falkner K., Al-Eryani L., Clair H. B., Prough R. A., Osborne T. S., Malarkey D. E., Christopher States J., et al. (2014). Evaluation of Aroclor 1260 exposure in a mouse model of diet-induced obesity and non-alcoholic fatty liver disease. Toxicol. Appl. Pharmacol. 279, 380–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M., Houten S. M., Mataki C., Christoffolete M. A., Kim B. W., Sato H., Messaddeq N., Harney J. W., Ezaki O., Kodama T., et al. (2006). Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature 439, 484–489. [DOI] [PubMed] [Google Scholar]
- Wells J. E., Hylemon P. B. (2000) Identification and characterization of a bile acid 7alphadehydroxylation operon in Clostridium sp. strain TO-931, a highly active 7alphadehydroxylating strain isolated from human feces. Appl. Environ. Microbiol. 66, 1107–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Barnhart C., Lein P. J., Lehmler H. J. (2015). Hepatic metabolism affects the atropselective disposition of 2, 2′, 3, 3′, 6, 6′-hexachlorobiphenyl (PCB 136) in mice. Environ. Sci. Technol. 49, 616–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Duffel M., Lehmler H. J. (2013). Oxidation of polychlorinated biphenyls by liver tissue slices from phenobarbital-pretreated mice is congener-specific and atropselective. Chem. Res. Toxicol. 26, 1642–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Kim K. H., Phimister A., Bachstetter A. D., Ward T. R., Stackman R. W., Mervis R. F., Wisniewski A. B., Klein S. L., Kodavanti P. R., et al. (2009). Developmental exposure to polychlorinated biphenyls interferes with experience-dependent dendritic plasticity and ryanodine receptor expression in weanling rats. Environ. Health Perspect. 117, 426–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M., Hu T., Wang Y., Du Y., Hu C., Wu R. (2017). Polychlorinated biphenyls and its potential role in endometriosis. Environ. Pollut. 229, 837–845. [DOI] [PubMed] [Google Scholar]
- Zaneveld J. R., McMinds R., Vega Thurber R. (2017). Stress and stability: Applying the Anna Karenina principle to animal microbiomes. Nat. Microbiol. 2, 17121.. [DOI] [PubMed] [Google Scholar]
- Zani C., Toninelli G., Filisetti B., Donato F. (2013). Polychlorinated biphenyls and cancer: An epidemiological assessment. J. Environ. Sci. Health C. Environ. Carcinog. Ecotoxicol. Rev. 31, 99–144. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Klaassen C. D. (2010). Effects of feeding bile acids and a bile acid sequestrant on hepatic bile acid composition in mice. J. Lipid Res. 51, 3230–3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Nichols R. G., Correll J., Murray I. A., Tanaka N., Smith P. B., Hubbard T. D., Sebastian A., Albert I., Hatzakis E., et al. (2015). Persistent organic pollutants modify gut microbiota-host metabolic homeostasis in mice through aryl hydrocarbon receptor activation. Environ. Health Perspect 123, 679–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H. X., Adamcakova-Dodd A., Hu D., Hornbuckle K. C., Just C. L., Robertson L. W., Thorne P. S., Lehmler H. J. (2010). Development of a synthetic PCB mixture resembling the average polychlorinated biphenyl profile in Chicago air. Environ. Int. 36, 819–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.