Abstract
Background
Cryptosporidium is a major cause of childhood diarrhea. Current modes of cryptosporidiosis diagnosis involve procedures that are costly and require both a well-equipped laboratory and technical expertise. Therefore, a cost-effective, user-friendly, and rapid method for point-of-care detection of Cryptosporidium is desirable.
Methods
A total of 832 diarrheal stool specimens collected from 200 children aged <2 years were tested by Giardia/Cryptosporidium QUIK CHEK, enzyme-linked immunosorbent assay (ELISA), and quantitative polymerase chain reaction (qPCR) to compare the performance of the individual techniques. We also tested for the presence of other diarrheal pathogens in qPCR-positive samples with a TaqMan Array Card (TAC) to assess whether Cryptosporidium was the sole causative agent for the diarrheal episodes.
Results
Of 832 samples, 4.4% were found positive for Cryptosporidium by QUIK CHEK, 3.6% by ELISA, and 8.8% by qPCR. Using TAC-attributed Cryptosporidium diarrhea as the gold standard, the sensitivities of QUIK CHEK, ELISA, and qPCR were 92.3%, 71.8%, and 100%, respectively; the specificities were 97.1%, 94.3%, and 0%, respectively. Analysis of the qPCR-positive and QUIK CHEK–negative samples by TAC identified other enteropathogens as more likely than Cryptosporidium to be the causative agents of diarrhea.
Conclusions
QUIK CHEK was more sensitive and specific than ELISA. While qPCR detected Cryptosporidium in more samples than QUIK CHEK, most of these were instances of qPCR detecting small quantities of Cryptosporidium DNA in a diarrheal episode caused by another enteropathogen. We concluded that QUIK CHEK was comparable in sensitivity and superior in specificity to qPCR for the diagnosis of Cryptosporidium diarrhea.
Keywords: cryptosporidiosis, childhood diarrhea, rapid diagnostic test, diagnostic performance, Bangladesh
The Giardia/Cryptosporidium QUIK CHEK device has a comparable sensitivity and higher specificity than quantitative polymerase chain reaction for the diagnosis of Cryptosporidium-attributed diarrhea. It also showed better sensitivity and specificity than enzyme-linked immunosorbent assay.
Diarrhea is one of the most widely recognized diseases and a major cause of childhood mortality [1], with intestinal protozoan infections being important causes of acute and persistent diarrhea [2]. Cryptosporidium species is the leading protozoal cause of diarrhea worldwide in immunocompetent and immunocompromised subjects [3–5], with 8%–19% of cases attributed to Cryptosporidium species in low-income nations, and it has a significant effect on mortality [6, 7]. Among the diarrheagenic protozoan pathogens, Cryptosporidium species results in the most deaths among children <5 years of age, while the other 2 enteric protozoan parasites, Giardia species and Entamoeba histolytica, also contribute, but to a lesser extent [8, 9]. Although several Cryptosporidium species have been identified in humans, Cryptosporidium hominis and Cryptosporidium parvum cause >90% of human cases of cryptosporidiosis, while other species that are less commonly associated with human infection include Cryptosporidium meleagridis, Cryptosporidium cuniculus, Cryptosporidium felis, and Cryptosporidium canis; this is, however, dependent on setting [10–12]. The parasite infects the microvillous region of epithelial cells in the digestive and respiratory tract of humans, ultimately causing infectious diarrhea [13, 14]. The Global Enteric Multicenter Study identified Cryptosporidium as one of the 4 major contributors to moderate-to-severe diarrheal diseases during the first 2 years of life and showed Cryptosporidium as a key pathogen in diarrheal disease, even among otherwise healthy children. In fact, Cryptosporidium is second only to rotavirus as an agent of moderate to extreme diarrhea in children <2 years of age. There is a 2–3 times higher risk of death among children aged 12–23 months with cryptosporidiosis than in subjects of a similar age group without diarrhea [8, 15]. Enteric infection caused by Cryptosporidium in children can have devastating consequences by affecting intestinal absorption of nutrients and perturb childhood development [16]. This parasite is transmitted through fecal–oral routes either by consumption of contaminated food or water, or by person-to-person (anthroponotic) or animal-to-human (zoonotic) transmission [17].
Current methods of diagnosis of Cryptosporidium species include identification of Cryptosporidium oocysts by microscopy, antigen detection by enzyme-linked immunosorbent assay (ELISA), and DNA detection by polymerase chain reaction (PCR). Stool microscopy has low sensitivity in detecting Cryptosporidium species [18], and acid-fast staining is a prerequisite to differentiate the Cryptosporidium oocysts from other parasites and also from the yeast cells that are frequently present in stool [19]. The diagnostic methods becoming more widely used therefore are based on either fecal antigen detection or parasite DNA, but both require considerable technical expertise. Quantitative PCR (qPCR) is considered the most sensitive method, but because of this sensitivity may detect clinically insignificant amounts of cryptosporidial DNA in a diarrheal episode due to another pathogen. qPCR is also expensive and requires skilled personnel, which limits its use [20, 21]. Rapid detection techniques of fecal antigen have the potential to provide easy and cost-effective diagnosis of this pathogen in resource-limited settings.
Rapid antigen device tests are now available for the detection of Cryptosporidium include ImmunoCard STAT! Cryptosporidium/Giardia (Meridian Bioscience), Xpect Giardia/ Cryptosporidium (Remel), and Giardia/Cryptosporidium QUIK CHEK (TechLab) [22–24]. Using ELISA as the reference standard, the QUIK CHEK had a sensitivity and specificity of 100% and 100%, respectively, for Cryptosporidium, which compared favorably to the result of ImmunoCard STAT! and Xpect test [24, 25].
Here, we report the performance of a rapid membrane immunoassay, QUIK CHEK, for the qualitative detection of Cryptosporidium species causing diarrheal episodes. In this study, the performance of this rapid antigen point-of-care test for detection of Cryptosporidium species was compared to ELISA and qPCR. The TaqMan Array Card (TAC), which provides quantitative results for a broad panel of enteropathogens, was used to attribute the cause of diarrhea and was considered the gold standard for diagnosis of Cryptosporidium diarrhea.
MATERIALS AND METHODS
Ethics Approval
The study was approved by the research and ethical review committees of the International Centre for Diarrhoeal Disease Research, Bangladesh (icddr,b). Informed written consent was obtained from parents/guardians for the participation of their child in the study.
Study Area, Population, and Sampling
The study was conducted using 832 diarrheal stool specimens from a cohort of 200 children (aged up to 2 years) from November 2014 to July 2016 in Mirpur, Dhaka, Bangladesh. All of the fresh fecal specimens were tested by QUIK CHEK (TechLab) at the field site (Mirpur, Dhaka) and then transported to the Emerging Infections and Parasitology Lab, icddr,b, maintaining a cold chain and thereafter stored at –80°C. Then, ELISA (Cryptosporidium II test, TechLab) and (after DNA extraction) parasite-specific qPCR assays were performed with all of the stool samples in batches. The 74 samples found to be positive for Cryptosporidium by qPCR (37 QUIK CHEK positive and 37 QUIK CHEK negative) were further tested by TAC assay, which can detect the other enteropathogens that are common in Bangladeshi infants.
Rapid Antigen Point-of-care Test (QUIK CHEK)
The rapid antigen point-of-care test or QUIK CHEK assay was used in accordance with the manufacturer’s instructions. All reagents and specimens were brought to room temperature prior to testing. Twenty-five microliters of fresh fecal specimens was used for testing. Then, 0.5 mL of diluent was added to the prelabeled test tube containing the fecal specimen. One drop of conjugate was then added. Tubes were then inverted and vortexed to ensure adequate suspension; 0.5 mL of the sample-conjugate mixture was then transferred into the sample well of the test device. Following the incubation of the test device for 15 minutes, 300 µL of wash buffer was added to the reaction window, followed by 2 drops of substrate, after which the device was left for 10 minutes at room temperature. Test results were read immediately after the final incubation [24].
Cryptosporidium Oocyst Antigen Test (ELISA)
The Cryptosporidium II kit (TechLab) was used for the Cryptosporidium antigen detection in samples according to the principles of ELISA. The kit was used in accordance with the manufacturer’s instructions [26]. Interpretation of the assay was based on optical density (OD) readings at a single wavelength of 450 nm with the OD values ≥0.15 being considered positive for Cryptosporidium. Positive and negative controls were run with each batch of test specimens.
Extraction of Nucleic Acid From Fecal Specimens
Total nucleic acid extraction was performed using the modified QIAamp fast stool DNA extraction protocol, which incorporates a 3-minute bead-beating step to lyse the Cryptosporidium oocysts [27]. Total DNA was purified with the QIAamp Fast DNA Stool Mini Kit (Qiagen, Hilden, Germany) from fresh or frozen stool samples. An internal control, PhHV (phocine herpesvirus) was used for inhibition control with each sample during extraction, which was then measured by qPCR reaction. In addition, 1 negative control (purified water instead of stool) was included in each batch (24 samples usually) as an extraction blank to control for potential carryover contamination.
Multiplex qPCR
The Cryptosporidium species real-time PCR assay was performed as part of a multiplex assay including Giardia intestinalis and E. histolytica. The multiplex qPCR also included an internal control (PhHV) to determine efficiency of the qPCR and detect inhibition in the sample. Positive and negative controls were used in each run of qPCR. Amplification consisted of 15 minutes at 95°C followed by 40 cycles of 15 seconds at 95°C, 30 seconds at 60°C, and 30 seconds at 72°C [28]. The 16S small-subunit ribosomal RNA gene primers and TaqMan probes for Cryptosporidium species, G. intestinalis, E. histolytica, and PhHV were used [28, 29]. Amplification, detection, and data analysis were performed with the CFX96 real-time detection system (Bio-Rad). Fluorescence was measured during the annealing step of each cycle.
TAC Assay
Seventy-four samples found to be positive for Cryptosporidium by qPCR were tested by TAC assay according to the protocol as described by Liu et al [30] for the identification of other enteric pathogens in the diarrheal episodes. We attributed pathogens as the cause of diarrhea if they were diarrhea-associated pathogens present at the diarrhea-associated quantities described by Liu et al [27].
Data Analysis
Descriptive statistics including mean and percentage were explored. To assess our experimental diagnostic test (QUIK CHEK) in comparison with reference standards (ELISA and qPCR), we used accuracy measurements which included sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and kappa (κ) coefficient. The diagti and kap commands in Stata 13.0 were used to calculate these accuracy measurements, and the 95% confidence interval (CI) for those accuracy measurements was calculated using the exact binomial distribution approach. A 2-tailed P value ≤.05 was considered significant.
RESULTS
A total of 832 diarrheal stool samples were tested by QUIK CHEK, Cryptosporidium II ELISA, and qPCR. Of these 832 diarrheal stool samples, 4.4% (n = 37) were found positive for Cryptosporidium by QUIK CHEK assay, 3.6% (n = 30) by ELISA assay, and 8.9% (n = 74) by qPCR testing (Table 1). Thirty-seven of the 832 samples had been found to be both QUIK CHEK and qPCR positive, in which the average qPCR quantification cycle (Cq) value was 23. Among these 37 samples, 29 were found to be Cryptosporidium positive by all the 3 techniques with an average qPCR Cq value of 22 and ELISA OD value of 1.34 (range, 0.16–2.87). On the other hand, the mean Cq value of qPCR-positive but QUIK CHEK–negative samples was 31, suggesting that these samples contained substantially less parasite DNA.
Table 1.
Sample Testing Profile for the Evaluation of Different Diagnostic Techniques for Cryptosporidium Detection
| qPCR Testing (n = 832) | QUIK CHEK Assay | ELISA | TAC Assay | |||
|---|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | Positive | Negative | |
| Positive: n = 74 (8.8%) | 37 (50) | 37 (50) | 30 (40.5) | 44 (59.5) | 74 (100) | 0 (0) |
| Negative: n = 758 (91.1%) | 0 (0) | 758 (100) | 0 (0) | 758 (100) | Not tested | Not tested |
All 832 samples were tested by QUIK CHEK, ELISA, and qPCR. Only the qPCR-positive samples were tested by TAC.
Abbreviations: ELISA, enzyme-linked immunosorbent assay; qPCR, quantitative polymerase chain reaction; TAC, TaqMan Array Card.
Agreement between QUIK CHEK and ELISA was 86.0% (κ = 0.860 and P < .0001), with sensitivity of 96.7% and specificity of 99.0% and PPV and NPV of 78.4% and 99.9%, respectively (ELISA as the gold standard). Likewise, agreement between QUIK CHEK and qPCR was 64.6% (κ = 0.646 and P < .0001), with sensitivity of 50% and specificity of 100%; the PPV and NPV had been 100% and 95.3%, respectively (qPCR as the gold standard). When using qPCR as the reference, the measured agreement, sensitivity, specificity, PPV, and NPV of ELISA were 55.4%, 40.5%, 100%, 100%, and 94.5%, respectively (Table 2).
Table 2.
Performance Comparison of the Techniques (QUIK CHEK®, ELISA and qPCR) in Detecting Cryptosporidium spp.
| Methods Comparison (n = 832) |
Sensitivity | Specificity | PPV | NPV | Agreement Analysis |
|---|---|---|---|---|---|
| QUIK CHEK vs ELISAa | 96.7% | 99.0% | 78.4% | 99.9% | κ = 0.860 (P < .0001) |
| QUIK CHEK vs qPCRa | 50.0% | 100.0% | 100% | 95.3% | κ = 0.646 (P < .0001) |
| ELISA vs qPCRa | 40.5% | 100.0% | 100% | 94.5% | κ = 0.554 (P < .0001) |
Abbreviations: ELISA, enzyme-linked immunosorbent assay; NPV, negative predictive value; PPV, positive predictive value; qPCR, quantitative polymerase chain reaction.
aGold standard.
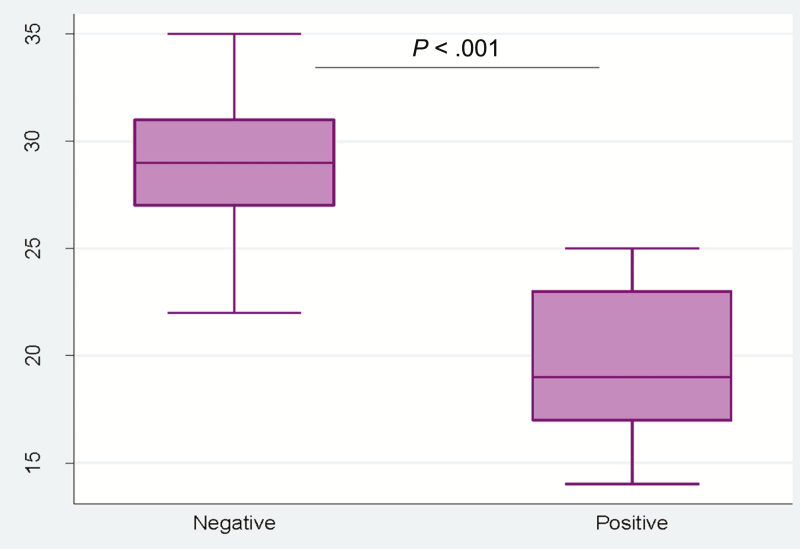
We further assessed the burden of pathogens in all 74 qPCR-defined Cryptosporidium-positive samples by TAC assay. The QUIK CHEK–positive samples had lower TAC Cq (mean Cq value = 19) values (ie, more parasite DNA) than QUIK CHEK–negative samples (mean Cq value = 29) (Figure 1).
Figure 1.
Distribution of QUIK CHEK–positive and –negative samples (quantification cycle) among the samples tested by TaqMan Array Card assay. Abbreviations: Ct, cycle threshold; TAC, TaqMan Array Card.
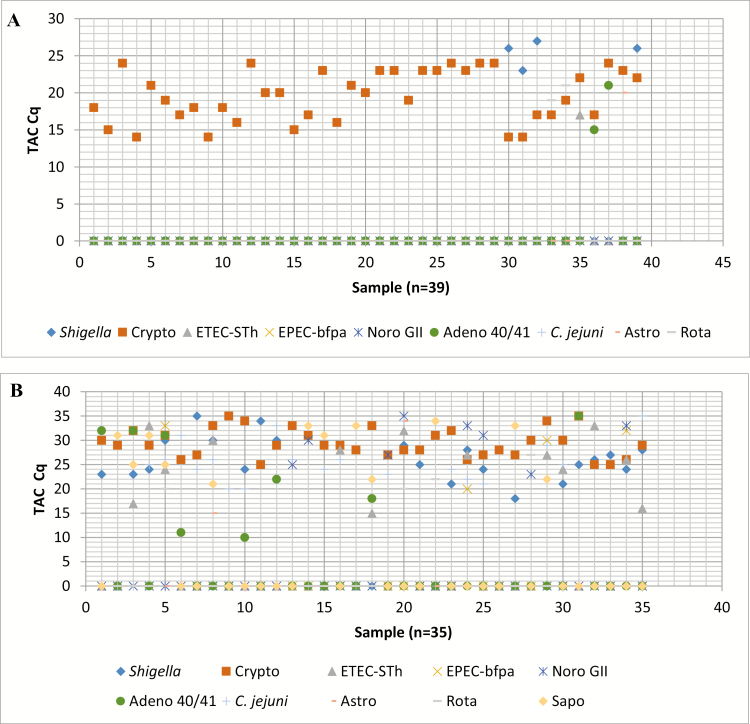
The TAC assay was used on all cryptosporidia qPCR-positive samples to test for the presence of other potential causes of diarrhea. The TAC assay was designed with a total of 76 enteropathogen targets including the positive controls and different strains. The major pathogens identified in the diarrheal stool samples are given in Table 3. We found that 39 of the 74 samples had a Cryptosporidium quantity that was highly diarrhea associated per a large multisite study (Cq ≤ 24) [27]. Thus, the TAC assay demonstrated that those 39 diarrheal episodes were caused by this high Cryptosporidium burden as well as categorized to “TAC-attributed Cryptosporidium diarrhea” (Figure 2A). For the rest (35/74) of the samples, the Cryptosporidium quantity was lower and not highly diarrhea associated (Cq > 24) and there were other diarrhea-associated pathogens present; those were categorized as “mixed infections” (Figure 2B). Interestingly, 92.3% (36 of 39) of the samples categorized by TAC as “Cryptosporidium diarrhea” were revealed positive for Cryptosporidium by the QUIK CHEK assay. For the diagnosis of Cryptosporidium causing diarrhea, the sensitivity and specificity of the QUIK CHEK assay had been calculated to be 92.3% and 97.1%, respectively (Table 4); sensitivity and specificity of ELISA were found to be 71.8% and 94.3%, respectively (Table 4); and for qPCR those were 100% and 0%, respectively (considering “TAC-attributed Cryptosporidium diarrhea” as the gold standard); see Table 4.
Table 3.
Major Enteropathogens Other Than Cryptosporidium Detected by TaqMan Array Card Assay in the Diarrheal Samples
| Serial | Pathogen | Prevalence Rate, % |
|---|---|---|
| 1 | Campylobacter jejuni | 59.4 |
| 2 | EPEC-bfpa and ETEC-STh | 45.9 |
| 3 | Shigella | 44.6 |
| 4 | Sapovirus | 37.8 |
| 5 | Adenovirus 40/41 | 36.5 |
| 6 | Giardia | 31.1 |
| 7 | Norovirus GII | 27.0 |
| 8 | Rotavirus | 10.8 |
| 9 | Astrovirus | 8.1 |
| 10 | Vibrio cholerae | 2.7 |
Abbreviations: bfpa, bundle-forming pilus A; EPEC, enteropathogenic Escherichia coli; ETEC, enterotoxigenic Escherichia coli; STh, heat-stable toxin.
Figure 2.
A, Diarrheal episodes (n = 39) with highly diarrhea-associated Cryptosporidium (quantification cycle [Cq] ≤24) revealed by TaqMan Array Card (TAC) assay and categorized as diarrhea attributed to Cryptosporidium. The y-axis indicates the highly diarrhea-associated Cq of different pathogens including Shigella, Cryptosporidium species (Crypto), enterotoxigenic Escherichia coli (ETEC-STh), enteropathogenic E. coli (EPEC-bfpa), norovirus GII (Noro), adenovirus 40/41 (Adeno), Campylobacter jejuni, astrovirus (Astro), and rotavirus (Rota). The x-axis indicates 39 highly diarrhea-associated Cryptosporidium samples (1–39). For most of the cases, “0” in the TAC Cq line indicates “not detected” by the TAC assay, although in some cases, “0” defines the pathogen’s Cq value that exceeds the highly diarrhea-associated quantity (Cq) denoted by Liu et al [27]. B, Diarrheal episodes (n = 35) caused by mixed infections with various enteropathogens including lower Cryptosporidium burden (Cq >24). The y-axis indicates the diarrhea-associated Cq of different pathogens including Shigella, Cryptosporidium species, ETEC-STh, EPEC-bfpa, norovirus GII, adenovirus 40/41, C. jejuni, astrovirus, rotavirus, and sapovirus (Sapo). The x-axis indicates 35 samples with lower Cryptosporidium burden (1–35). The symbol “0” in the TAC Cq line indicates “not detected” by the TAC assay.
Table 4.
Performance of QUIK CHEK, Enzyme-linked Immunosorbent Assay, and Quantitative Polymerase Chain Reaction in Detecting Diarrhea Attributed to Cryptosporidium
| Diagnostic Technique | TAC-attributed Cryptosporidium Diarrhea | Diarrhea Attributed to Other Pathogens | Total |
|---|---|---|---|
| QUIK CHEK (n = 74) | |||
| Positive | 36 | 1 | 37 |
| Negative | 3 | 34 | 37 |
| Total | 39 | 35 | 74 |
| ELISA (n = 74) | |||
| Positive | 28 | 2 | 30 |
| Negative | 11 | 33 | 44 |
| Total | 39 | 35 | 74 |
| qPCR (n = 74) | |||
| Positive | 39 | 35 | 74 |
| Negative | 0 | 0 | 0 |
| Total | 39 | 35 | 74 |
Abbreviations: ELISA, enzyme-linked immunosorbent assay; qPCR, quantitative polymerase chain reaction; TAC, TaqMan Array Card.
The average TAC Cq value of Cryptosporidium target in diarrhea attributed to Cryptosporidium parasite was 20 ± 3.43 and average TAC Cq of Cryptosporidium for diarrhea caused by other pathogens was 29 ± 2.92. Notably, a total of 37 samples, including 36 Cryptosporidium diarrheal episodes, were detected positive by the QUIK CHEK assay; those were also positive by qPCR. Moreover, there were another 37 samples found to be positive by qPCR but designated as negative by QUIK CHEK. All of the 37 diarrheal samples those were QUIK CHEK negative but qPCR positive for cryptosporidia were also evaluated by TAC to determine the causative agents according to Liu et al [27]. Analysis of qPCR-positive but QUIK CHEK–negative samples revealed, 92% (34/37) had lower quantities of Cryptosporidium DNA and other enteropathogens identified as the probable cause of diarrhea (mixed infections) by TAC assay.
DISCUSSION
This study shows the potential of the QUIK CHEK assay for rapid point-of-care diagnosis of highly diarrhea-associated Cryptosporidium. Using the TAC as the gold standard to identify Cryptosporidium-attributable diarrhea, the QUIK CHEK was more specific than qPCR, which is the most important finding of this present study.
For detection of Cryptosporidium in diarrheal samples, TAC assay and qPCR showed similar results, whereas only 50% (n = 37) of those positive samples were detected by the QUIK CHEK assay. Thirty-six of these 37 QUIK CHEK–positive samples were, however, cryptosporidia causing diarrheal episodes, demonstrated by TAC assay, whereas TAC assay revealed 39 of the 74 qPCR-positive samples as highly diarrhea-associated Cryptosporidium. Among 39 highly diarrhea-associated Cryptosporidium diarrheal samples detected by TAC assay, in 24 samples Cryptosporidium was the only infection detected. In addition, 28 of the 30 ELISA-positive samples were revealed as Cryptosporidium causing diarrheal episodes by TAC analysis. Therefore the present study demonstrated that the QUIK CHEK is more specific than qPCR and ELISA for the detection of Cryptosporidium as a cause of diarrhea.
In this study, the relation between QUIK CHEK–positive results and higher pathogen load, as expressed by lower Cq values, is particularly noteworthy. To our knowledge, this is the only report thus far where TAC assay has been performed to specifically differentiate the Cryptosporidium causing diarrheal infections from other causes. Ninety-two percent of the Cryptosporidium causing diarrheal episodes was successfully detected by QUIK CHEK in this study, which is close to qPCR and higher than ELISA.
The performance (sensitivity and specificity) of diagnostic tests in different epidemiological and pathogenic scenarios has practical implications for the design of surveillance and/or control programs for Cryptosporidium. In previous studies, sensitivity rates reported for various commercially available immunochromatographic and enzyme immunoassays had a detection range from 63% to 100% [31–34].
Sensitive and specific detection of Cryptosporidium infection is required to ensure that patients receive appropriate treatment. The clinical symptoms seen in the cases of mixed infections are due to complex interactions involving some or all of the other enteric pathogens present. If the clinicians start to use newer molecular methods such as TAC or qPCR for detection of enteropathogens in diarrheal stool samples, they will be able to identify the burden of enteropathogens responsible for the diarrhea. Based on the results of new molecular assays and the signs and symptoms of the patients, clinician will be able to figure out the causative agent of any diarrheal episode. Because molecular diagnosis such as qPCR/TAC and ELISA remain expensive and require skilled technicians, the clinician can use the QUIK CHEK test for rapid diagnosis of Cryptosporidium diarrhea as an alternative to molecular techniques. In our study, the QUIK CHEK’s specificity revealed results that were better than that of ELISA and qPCR, whereas the sensitivity is close to qPCR and higher than ELISA in detecting Cryptosporidium diarrhea. In comparison with ELISA and qPCR, the rapid point-of-care test QUIK CHEK is more feasible due to its enhanced convenience for use in field sites in remote areas, its cost-effectiveness, and its practicality, as it does not require highly skilled technicians or well-equipped laboratory settings.
In conclusion, the Giardia/Cryptosporidium QUIK CHEK has a comparable sensitivity and higher specificity than that of qPCR for the diagnosis of Cryptosporidium-attributable diarrhea. Conversely, qPCR was more sensitive yet less specific. Therefore, rapid point-of-care antigen testing provides a robust means with which to diagnose Cryptosporidium-attributable diarrhea.
Notes
Author contributions. R. H., W. A. P., C. A. G., and A. S. G. F. conceived and designed the study. Field work, data gathering, and laboratory experiments at icddr,b were performed by E. A., S. A., M. K., B. H. and M. A., with supervision of R. H. and A. S. G. F. M. K. and E. A. performed statistical analyses and also wrote the manuscript with input from R. H., E. R. H., M. T., and W. A. P. All authors contributed to revisions and approved the final version.
Acknowledgments. The icddr,b acknowledges with gratitude the governments of Bangladesh, Canada, Sweden, and the United Kingdom for providing core/unrestricted support. We thank the families of the Mirpur field area who participated in this study, and we also thank the work of the field and laboratory staff of the Emerging Infections and Parasitology Laboratory of icddr,b who worked on this project, and without whom we could not have completed this research.
Disclaimer. The sponsors had no role in study design, interpretation, or manuscript preparation.
Financial support. This work was supported by the National Institutes of Health (NIH) (grant number AI-043596) and the Bill & Melinda Gates Foundation (award number OPP1100514). We thank TechLab for the donation of Cryptosporidium diagnostics.
Potential conflicts of interest. W. A. P. is a consultant for TechLab and reports grants to his institution from Perrogo Nutritionals and payments for expert testimony for lectures from the University of California, Los Angeles and the University of Florida; patents and royalties from the University of Virginia; and travel expenses from the University of Virginia and the Bill & Melinda Gates Foundation. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed. S. A., M. A., E. A., R. H., B. H., A. F. and M. K. are employed by icddr,b and received grants from NIH. M. T., C. G., and E. H. employed by University of Virginia and received grants from NIH.
References
- 1. Boschi-Pinto C, Velebit L, Shibuya K. Estimating child mortality due to diarrhoea in developing countries. Bull World Health Organ 2008; 86:710–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Turkeltaub JA, McCarty TR 3rd, Hotez PJ. The intestinal protozoa: emerging impact on global health and development. Curr Opin Gastroenterol 2015; 31:38–44. [DOI] [PubMed] [Google Scholar]
- 3. Platts-Mills JA, Babji S, Bodhidatta L, et al. . MAL-ED Network Investigators Pathogen-specific burdens of community diarrhoea in developing countries: a multisite birth cohort study (MAL-ED). Lancet Glob Health 2015; 3:e564–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Checkley W, White AC Jr, Jaganath D, et al. . A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for Cryptosporidium. Lancet Infect Dis 2015; 15:85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Snelling WJ, Xiao L, Ortega-Pierres G, et al. . Cryptosporidiosis in developing countries. J Infect Dev Ctries 2007; 1:242–56. [PubMed] [Google Scholar]
- 6. Tzipori S, Ward H. Cryptosporidiosis: biology, pathogenesis and disease. Microbes Infect 2002; 4:1047–58. [DOI] [PubMed] [Google Scholar]
- 7. Mølbak K, Højlyng N, Gottschau A, et al. . Cryptosporidiosis in infancy and childhood mortality in Guinea Bissau, West Africa. BMJ 1993; 307:417–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kotloff KL, Nataro JP, Blackwelder WC, et al. . Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 2013; 382:209–22. [DOI] [PubMed] [Google Scholar]
- 9. Lozano R, Naghavi M, Foreman K, et al. . Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012; 380:2095–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bouzid M, Tyler KM, Christen R, Chalmers RM, Elwin K, Hunter PR. Multi-locus analysis of human infective Cryptosporidium species and subtypes using ten novel genetic loci. BMC Microbiol 2010; 10:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cacciò SM. Molecular epidemiology of human cryptosporidiosis. Parassitologia 2005; 47:185–92. [PubMed] [Google Scholar]
- 12. Chalmers R, Smith R, Elwin K, Clifton-Hadley F, Giles M. Epidemiology of anthroponotic and zoonotic human cryptosporidiosis in England and Wales, 2004–2006. Epidemiol Infect 2011; 139:700–12. [DOI] [PubMed] [Google Scholar]
- 13. Casemore DP, Armstrong M, Sands RL. Laboratory diagnosis of cryptosporidiosis. J Clin Pathol 1985; 38:1337–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Leitch GJ, He Q. Cryptosporidiosis—an overview. J Biomed Res 2012; 25:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sow SO, Muhsen K, Nasrin D, et al. . The burden of cryptosporidium diarrheal disease among children <24 months of age in moderate/high mortality regions of sub-Saharan Africa and South Asia, utilizing data from the Global Enteric Multicenter Study (GEMS). PLoS Negl Trop Dis 2016; 10:e0004729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Checkley W, Gilman RH, Epstein LD, et al. . Asymptomatic and symptomatic cryptosporidiosis: their acute effect on weight gain in Peruvian children. Am J Epidemiol 1997; 145:156–63. [DOI] [PubMed] [Google Scholar]
- 17. Fayer R, Morgan U, Upton SJ. Epidemiology of Cryptosporidium: transmission, detection and identification. Int J Parasitol 2000; 30:1305–22. [DOI] [PubMed] [Google Scholar]
- 18. Nazeer JT, El Sayed Khalifa K, von Thien H, et al. . Use of multiplex real-time PCR for detection of common diarrhea causing protozoan parasites in Egypt. Parasitol Res 2013; 112:595–601. [DOI] [PubMed] [Google Scholar]
- 19. Van den Bossche D, Cnops L, Verschueren J, Van Esbroeck M. Comparison of four rapid diagnostic tests, ELISA, microscopy and PCR for the detection of Giardia lamblia, Cryptosporidium spp. and Entamoeba histolytica in feces. J Microbiol Methods 2015; 110:78–84. [DOI] [PubMed] [Google Scholar]
- 20. Morgan UM, Pallant L, Dwyer BW, Forbes DA, Rich G, Thompson RC. Comparison of PCR and microscopy for detection of Cryptosporidium parvum in human fecal specimens: clinical trial. J Clin Microbiol 1998; 36:995–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stark D, Al-Qassab SE, Barratt JL, et al. . Evaluation of multiplex tandem real-time PCR for detection of Cryptosporidium spp., Dientamoeba fragilis, Entamoeba histolytica, and Giardia intestinalis in clinical stool samples. J Clin Microbiol 2011; 49:257–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Garcia LS, Shimizu RY, Novak S, Carroll M, Chan F. Commercial assay for detection of Giardia lamblia and Cryptosporidium parvum antigens in human fecal specimens by rapid solid-phase qualitative immunochromatography. J Clin Microbiol 2003; 41:209–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Leo M, Haque R, Kabir M, et al. . Evaluation of Entamoeba histolytica antigen and antibody point-of-care tests for the rapid diagnosis of amebiasis. J Clin Microbiol 2006; 44:4569–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Minak J, Kabir M, Mahmud I, et al. . Evaluation of rapid antigen point-of-care tests for detection of Giardia and Cryptosporidium species in human fecal specimens. J Clin Microbiol 2012; 50:154–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Youn S, Kabir M, Haque R, Petri WA Jr. Evaluation of a screening test for detection of Giardia and Cryptosporidium parasites. J Clin Microbiol 2009; 47:451–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hencke JD, Garcia LS, Herbein JF. Detection of Cryptosporidium spp. antigen in human fecal specimens using the Cryptosporidium II ELISA test. Am J Trop Med Hyg 2006; 75:220–1. [Google Scholar]
- 27. Liu J, Platts-Mills JA, Juma J, et al. . Use of quantitative molecular diagnostic methods to identify causes of diarrhoea in children: a reanalysis of the GEMS case-control study. Lancet 2016; 388:1291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Verweij JJ, Blangé RA, Templeton K, et al. . Simultaneous detection of Entamoeba histolytica, Giardia lamblia, and Cryptosporidium parvum in fecal samples by using multiplex real-time PCR. J Clin Microbiol 2004; 42:1220–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stroup SE, Roy S, Mchele J, et al. . Real-time PCR detection and speciation of Cryptosporidium infection using Scorpion probes. J Med Microbiol 2006; 55:1217–22. [DOI] [PubMed] [Google Scholar]
- 30. Liu J, Gratz J, Amour C, et al. . A laboratory-developed TaqMan Array Card for simultaneous detection of 19 enteropathogens. J Clin Microbiol 2013; 51:472–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Aldeen WE, Carroll K, Robison A, Morrison M, Hale D. Comparison of nine commercially available enzyme-linked immunosorbent assays for detection of Giardia lamblia in fecal specimens. J Clin Microbiol 1998; 36:1338–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Johnston SP, Ballard MM, Beach MJ, Causer L, Wilkins PP. Evaluation of three commercial assays for detection of Giardia and Cryptosporidium organisms in fecal specimens. J Clin Microbiol 2003; 41:623–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maraha B, Buiting AG. Evaluation of four enzyme immunoassays for the detection of Giardia lamblia antigen in stool specimens. Eur J Clin Microbiol Infect Dis 2000; 19:485–7. [DOI] [PubMed] [Google Scholar]
- 34. Weitzel T, Dittrich S, Möhl I, Adusu E, Jelinek T. Evaluation of seven commercial antigen detection tests for Giardia and Cryptosporidium in stool samples. Clin Microbiol Infect 2006; 12:656–9. [DOI] [PubMed] [Google Scholar]