Abstract
Background
Ethionamide is used to treat multidrug-resistant tuberculosis (MDR-TB). The antimicrobial pharmacokinetics/pharmacodynamics, the contribution of ethionamide to the multidrug regimen, and events that lead to acquired drug resistance (ADR) are unclear.
Methods
We performed a multidose hollow fiber system model of tuberculosis (HFS-TB) study to identify the 0–24 hour area under the concentration-time curve (AUC0-24) to minimum inhibitory concentration (MIC) ratios that achieved maximal kill and ADR suppression, defined as target exposures. Ethionamide-resistant isolates underwent whole-genome and targeted Sanger sequencing. We utilized Monte Carlo experiments (MCEs) to identify ethionamide doses that would achieve the target exposures in 10000 patients with pulmonary tuberculosis. We also identified predictors of time-to-sputum conversion in Tanzanian patients on ethionamide- and levofloxacin-based regimens using multivariate adaptive regression splines (MARS).
Results
An AUC0-24/MIC >56.2 was identified as the target exposure in the HFS-TB. Early efflux pump induction to ethionamide monotherapy led to simultaneous ethambutol and isoniazid ADR, which abrogated microbial kill of an isoniazid-ethambutol-ethionamide regimen. Genome sequencing of isolates that arose during ethionamide monotherapy revealed mutations in both ethA and embA. In MCEs, 20 mg/kg/day achieved the AUC0-24/MIC >56.2 in >95% of patients, provided the Sensititre assay MIC was <2.5 mg/L. In the clinic, MARS revealed that ethionamide Sensititre MIC had linear negative relationships with time-to-sputum conversion until an MIC of 2.5 mg/L, above which patients with MDR-TB failed combination therapy.
Conclusions
Ethionamide is an important contributor to MDR-TB treatment regimens, at Sensititre MIC <2.5 mg/L. Suboptimal ethionamide exposures led to efflux pump-mediated ADR.
Keywords: efflux pumps, hollow fiber system model, artificial intelligence, MIC vs clinical outcome, tuberculous meningitis
Ethionamide is used in both the long and shorter forms of multidrug-resistant (MDR) tuberculosis treatment, together with first- and second-line agents [1]. Ethionamide is administered at a dose of 15–20 mg/kg/day, usually as 2 to 3 divided doses; there are no pharmacokinetics/pharmacodynamics (PK/PD) studies to support this dose choice. Moreover, there is still considerable debate as to what exactly ethionamide adds to an MDR tuberculosis regimen with such highly active agents such as fluoroquinolones. There are 2 possible approaches to identify the contribution of ethionamide to the MDR tuberculosis regimens: preclinical PK/PD studies or a clinical study indicating that ethionamide resistance is associated with worse outcomes [2–4]. Here, we performed PK/PD studies for ethionamide in the hollow fiber system model of tuberculosis (HFS-TB), a model with good clinical predictive accuracy [5–8]. Results were used to identify a dose of ethionamide that could achieve the identified optimal exposures in patients. In parallel, we also used machine-learning algorithms to identify the most important predictors of outcome in Tanzanian patients on an ethionamide-containing World Health Organization (WHO)–based regimen.
Ethionamide is a prodrug that is activated by the Baeyer–Villiger monooxygenase EthA (encoded by Rv3854c), which is regulated by the transcriptional repressor EthR. EthA and EthR mutations are associated with ethionamide resistance. Once activated, both ethionamide and its structural analogue isoniazid kill Mtb by inhibiting the molecular target InhA; mutations lead to simultaneous resistance to either drug, so that each can generate resistance to the other [9]. Large proportions of isoniazid-resistant (approximately 30%) MDR tuberculosis (>62%) and extensively drug-resistant tuberculosis (>92%) isolates carry inhA promoter mutations, and most carry catalase peroxidase (katG) mutations so that simultaneous resistance is common. The hazard ratios for relapse were 4.3 with katG mutations and 8.7 with inhA promoter mutations [10–12]. The shorter-form WHO regimen is dependent on high-dose isoniazid and ethionamide to overcome “low-level” isoniazid resistance conferred by the inhA mutations. The benefit of high-dose isoniazid and ethionamide with higher ethionamide minimum inhibitory concentrations (MICs) is unclear; indeed, ethionamide MICs are often not determined in treatment programs. On the other hand, while efflux pump induction leads to simultaneous resistance between isoniazid and ethambutol, the role of efflux pumps in ethionamide resistance is unknown [13–15]. Therefore, we sought to determine if exposure to ethionamide confers cross-resistance, or at least cross-tolerance, to isoniazid and ethambutol. This would help determine if it is wise to use these drugs in companion with ethionamide in MDR-tuberculosis regimens.
METHODS
Materials, Organisms, and Reagents
Mtb H37Ra (American Type Culture Collection 25177) was utilized for the HFS-TB experiments as described elsewhere [16]. Ethionamide was purchased from Sigma Aldrich (St. Louis, MO). Ethionamide-d3 (internal standard) was purchased from Santa Cruz Biotechnology (Dallas, TX). Hollow fiber cartridges were purchased from FiberCell (Frederick, MD). We utilized the BACTEC MGIT 960 Mycobacterial Growth Indicator Tube (Mycobacterial Growth Indicator Tube; Franklin Lakes, NJ) system to monitor time to positivity (TTP).
MICs and Screening for Exposure Effect of Ethionamide Against Mtb
MICs for the laboratory strain were identified using the MGIT and microbroth assays as described elsewhere [17]. We also examined MICs using the MYCOTB Sensititre assay, per manufacturer’s instructions. Next, we coincubated Mtb with static concentrations of ethionamide (0, 1, 2, 4, 8, 16, 32, 64, and 128 mg/L) for 7 days in test tubes and 12-well plates as described elsewhere [17, 18].
HFS Tuberculosis Ethionamide Study, Whole-genome Sequencing, and PK-PD Modeling
HFS-TB growth conditions for log-phase growth Mtb for ethionamide, were as described in the introduction articles and elsewhere [19–23]. Ethionamide treatment was infused via computerized syringe pumps over a 2-hour period to mimic time to maximum concentration encountered in patients and 0–24 hour area under the concentration time curve (AUC0-24) to MIC (AUC0-24/MIC) exposures of 0, 2.1, 4.2, 9.2, 17.9, 32.4, and 74.4, for a total duration of 35 days [24]. Ethionamide concentrations in each system were programmed to decline at a half-life of 3 hours. The central compartment was sampled at 0, 2, 4, 6, 9, 11, and 23.5 hours after the day 28 dose for ethionamide concentrations, which was measured using assays described in the Supplemental Methods. The peripheral compartment was sampled on days 0, 1, 2, 7, 14, 21, and 28 for quantification of total bacterial burden using both TTP and colony-forming unit (CFU) counts on Middlebrook 7H10 agar plus 10% oleic acid-dextrose-catalase (OADC) [17, 20, 25]. Ethionamide-resistant CFUs, defined as growth on agar supplemented with 5 times the ethionamide MIC alone or in the presence of the efflux pump blocker verapamil, were also captured.
Beginning on day 28, each of the ethambutol-treated HFS-TB units was also treated with isoniazid and ethambutol at concentration-time profiles achieved by daily 10 mg/kg of isoniazid and 25 mg/kg of ethambutol for 7 days. HFS-TB units were sampled as described above for drug concentrations after the day 28 triple dose, while the peripheral compartment was sampled for bacterial cultures on days 28, 30, and 35 for estimation of Mtb burden, as well as for colonies resistant to ethambutol (3 × MIC), isoniazid (3 × MIC), and ethionamide. Ethionamide-resistant colonies underwent whole-genome sequencing as described elsewhere, and mutations were confirmed using Sanger sequencing [17, 26, 27]. The relationship between PK/PD exposures such as AUC0-24/ MIC and microbial kill and resistance was modeled as outlined in the accompanying introduction article, and the EC80 (exposure associated with 80% of maximal kill) was calculated as the target exposure [19].
Monte Carlo Experiments
The rationale and steps for Monte Carlo experiments (MCEs) are detailed in the introduction article [19]. We utilized the ethionamide population pharmacokinetic parameters and variance by Zhu et al (Table 1) [28]. We also utilized the published spreadsheets of Donald and Seifert [29] as well as those of Hughes and Smith [30] of ethionamide cerebrospinal fluid (CSF) concentration-time profiles to identify CSF population pharmacokinetic parameter estimates using ADAPT, which we then used for tuberculous meningitis MCEs. Conte et al have identified an epithelial lining fluid-to-serum ratio of 8.7, which we used for pulmonary tuberculosis MCEs [31]. We examined doses of ethionamide at 10, 15, 20, 25, and 30 mg/kg per day for the probability to achieve or exceed the EC80 (target exposure) in the lung or CSF of 10000 tuberculosis patients at each dose. For MIC distribution, we used the results of Huang et al [32]. We also examined the performance of each dose using the MIC distribution from Tanzanian patients based on the MYCOTB Sensititre assay (described below). For each MIC method, we calculated the PK/PD target exposure using the MIC identified with the particular MIC assay in our laboratory strain.
Table 1.
Ethionamide Pharmacokinetic Parameters and Variances
| Pharmacokinetic Parameter | Domain of Input (Mean ± SD) [2, 3] |
10000 Simulated Patients (Mean ± SD) |
US Food and Drug Administration Ethionamide Label Informationa (Mean ± SD) |
|---|---|---|---|
| Clearance, L/h/kg | 1.88 ± 2.26 | 1.92 ± 1.56 | … |
| Volume, L/kg | 3.95 ± 1.20 | 3.93 ± 2.12 | (93.5 ± 19.2)b |
| Absorption constant, hour-1 | 0.25 ± 0.10 | 0.25 ± 0.32 | … |
| 0–24 hour area under the concentration-time curve, mg*h/L for 250 mg | … | 7.14 ± 5.21 | 7.67 ± 1.69 |
| Peak, mg/L for 250 mg | … | 1.42 ± 1.46 | 2.16 ± 0.61 |
Abbreviation: SD, standard deviation.
bData not published in milligrams per kilogram, and patient weight not published.
Machine Learning Analyses of Tanzanian Patients With MDR Tuberculosis
Multivariate adaptive regression spline (MARS) models, which are artificial intelligence algorithms that we have used previously to identify high-order interaction and nonlinear relationships, were used to identify predictors of time-to-sputum conversion in 41 MDR tuberculosis patients [33–39]. The patients were treated at Kibong’oto Infectious Diseases Hospital in Tanzania between May 2013 and August 2015. Further description of the patient cohort and details on the MARS modeling are provided in the Supplemental Methods.
RESULTS
Ethionamide Microbial Kill of Extracellular Mtb
The ethionamide MIC of the laboratory strain used for HFS-TB experiments was 1 mg/L based on the MGIT assay and 2.5 mg/L based on the Sensititre assay. Examination of static ethionamide concentrations coincubated with log-phase growth Mtb revealed a maximal kill (Emax) of 1.94 ± 0.12 log10 CFU/mL and the concentration mediating 50% of Emax (EC50) of 2.64 ± 0.36 times MIC (r2 = 0.95). For intracellular Mtb, the same concentrations achieved an Emax of 2.88 ± 0.26 log10 CFU/mL and an EC50 of 1.01 ± 0.15 times MIC (r2 = 0.90), which is better than for extracellular effect.
HFS-TB Pharmacokinetics and Pharmacodynamics
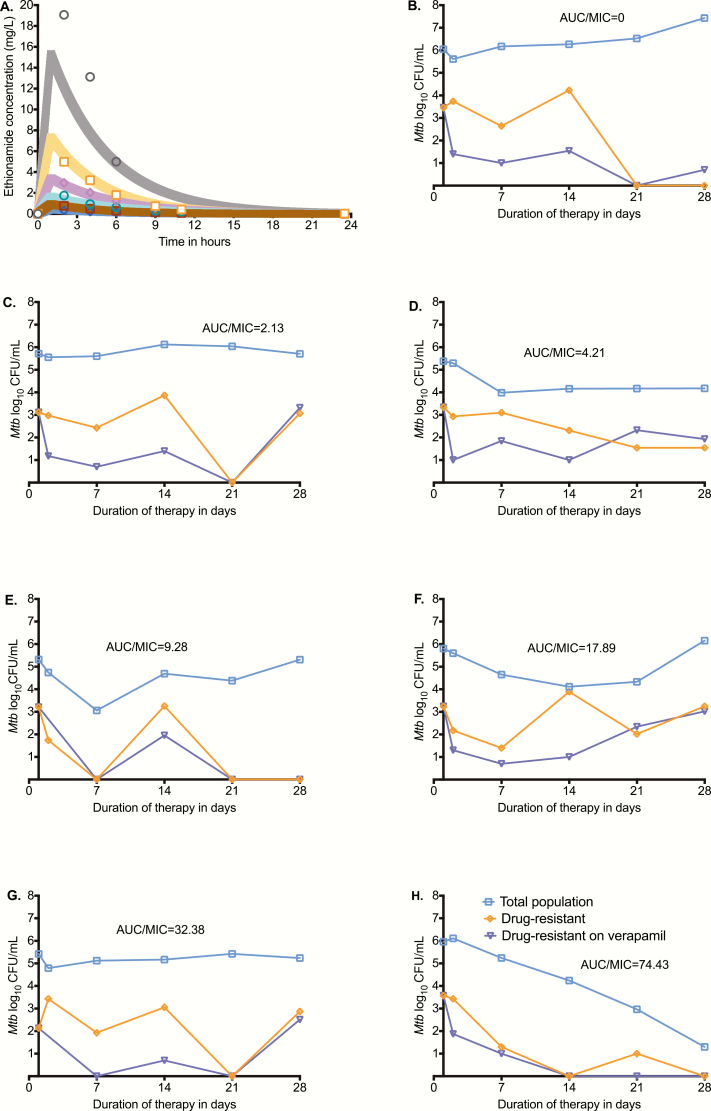
Ethionamide pharmacokinetic parameters were a clearance of 0.06 ± 0.00 L/h, volume of 0.24 ± 0.02 L, and a half-life of 3.04 ± 0.39 hours. Thus, the variability between HFS-TB units was low, indicating minimum technical variability. Figure 1A shows the model predicted vs observed concentrations in the HFS-TB units. Figure 1B–1H show the CFU/mL vs time curves of the [i] total bacterial population, [ii] ethionamide-resistant subpopulation, and [iii] ethionamide-resistant subpopulation in the presence of ethionamide plus verapamil (efflux pump inhibitor) for each AUC/MIC. In nontreated controls the percentage of the ethionamide-resistant subpopulation was unchanged and stayed below 1% throughout the 28 days. In Figure 1C–1G the intermediate exposures amplified the proportion of the ethionamide-resistant subpopulation, which was mostly efflux-pump driven in the beginning but was explained by another mechanism by day 28. Sequencing of ethionamide-resistant isolates picked from ethionamide-treated HFS-TB units on day 28 revealed an ethA 394G>T (Glu132*) mutation. In addition, 1 resistant isolate also had an Ala15Pro embA mutation (captured from ethambutol-resistance plates), even though the systems had not been exposed to ethambutol as of yet. There were no katG or inhA or ethR promoter or embB mutations. In Figure 1H, the highest ethionamide exposure killed drug-susceptible and drug-resistant subpopulations in parallel and thus totally suppressed emergence of the drug-resistant subpopulation.
Figure 1.
Ethionamide pharmacokinetics and time-kill curves in the hollow fiber system model of tuberculosis (HFS-TB). (A) Observed extracellular concentration time-profile in HFS-TB (symbols) against model-derived curves (shaded lines) show that intended concentrations were achieved. The slope of observed vs model-predicted concentrations was 1.03 ± 0.02 (r2 = 0.99), which means there was minimal bias. (B) On day 0, the total bacterial population was 5.42 log10 colony-forming units/mL and Mtb grew in the absence of ethionamide treatment. Without any ethionamide exposure, the % of the ethionamide-resistant to the total Mtb population was <1% throughout the study. (C) The lowest exposure examined held the bacterial burden constant throughout the duration of the experiment. The size of the ethionamide-resistant subpopulation was smaller in the presence of verapamil for the first 3 weeks, which means that most of the drug-resistant subpopulation was accounted for by efflux pumps. However, on day 28, the entire subpopulation was accounted for by a mechanism other than efflux pumps. (D–G) Across the intermediate exposures, the microbial kill plateaued within the first 2 weeks; the efflux pump–driven resistance pattern was similar to that of the lowest exposure. (H) In the highest ethionamide exposure, all ethionamide-resistant and susceptible subpopulations were killed in parallel, including efflux pump–mediated resistance, demonstrating that ethambutol efflux pump induction is a saturable process. Abbreviations: AUC, area under the concentration time curve; MIC, minimum inhibitory concentration.
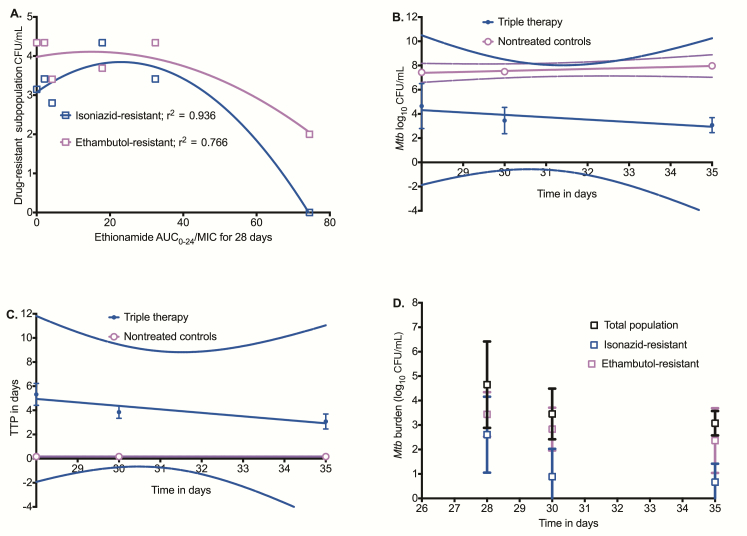
On day 28, there were sizeable ethambutol- and isoniazid-resistant subpopulations in HFS-TB not yet exposed to ethambutol and isoniazid, with the typical inverted “U” shape vs ethionamide AUC/MIC shown in Figure 2A [22]. The mean kill rates (ie, slopes of log10 CFU/mL vs time) on treatment with the triple drug combination of ethionamide, isoniazid, and ethambutol on days 28–35 are shown in Figure 2B and demonstrate that there was no effective kill. If slopes were examined for each HFS-TB unit, none of the slopes in triple drug–treated systems differed from the nontreated HFS-TB slope, confirming earlier results. Similar results were obtained when TTP was used as the bacterial burden readout (Figure 2C). Thus, the prior treatment with ethionamide had made the entire Mtb population refractory or tolerant to the isoniazid-ethambutol therapy, even though the cultures had no katG or inhA mutations (Figure 2D). Thus, ethionamide monotherapy led to phenotypic resistance to both isoniazid and ethambutol, mostly via efflux pump induction.
Figure 2.
Ethionamide exposure and cross-resistance to isoniazid and ethambutol. (A) The isoniazid- and ethambutol-resistant subpopulations vs ethionamide area under the concentration time curve/minimum inhibitory concentration (AUC/MIC) best fit the quadratic equation model for acquired resistance on day 28 with an r2 similar to that of the ethionamide-resistant subpopulation, even though the hollow fiber system model of tuberculosis (HFS-TB) units had not yet been treated with isoniazid and ethambutol. (B) The exposures achieved in all HFS-tuberculosis were isoniazid AUC0-24/MIC of 480 and an ethambutol peak/MIC of 3.5, in addition to ethionamide. The kill slopes for the triple drug-treated HFS-TB were −0.196 ± 0.116 log10 colony-forming units (CFU)/mL/day (r2 = 0.742) and did not significantly deviate from zero (P = .339) compared to the slope of 0.081 ± 0.015 (r2 = 0.968) in nontreated HFS-tuberculosis, which also did not deviate from zero (P = .115). (C) A similar pattern is observed using time to positivity (TTP) as a measure of Mtb burden. The slopes of nontreated vs triple drug treated were not significantly different from each other by TTP (P = .154), and the slopes did not differ significantly from zero. (D) The overlap of the total population with the isoniazid- and/or ethambutol-resistant log10 CFU/mL values that was seen means that the total bacterial burden on day 28 to day 35 was mostly constituted of drug-resistant subpopulations. Abbreviations: AUC, area under the concentration time curve; MIC, minimum inhibitory concentration; TTP, time to positivity.
HFS-TB–derived Optimal Ethionamide Exposures
The day 28 inhibitory sigmoid Emax model for total bacterial burden vs AUC0-24/MIC was:
| (1) |
where the EC50 is an AUC0-24/MIC of 10.78 and the EC80 is an AUC0-24/MIC of 56.2. On the other hand, a 1.0 log10 CFU/mL kill compared to day 0, defined as cidal effect in PK/PD models with other bacteria, was an AUC0-24/MIC of 10. Similarly, based on TTP, the EC50 was an AUC0-24/MIC of 6.34 (r2 = 0.997), not significantly different from the CFU/mL readout.
The quadratic function model for AUC0-24/MIC vs ethionamide-resistant CFU/mL was:
| (2) |
Based on equation 2, total suppression of the resistant subpopulation was at an AUC0-24/MIC of 42.
Monte Carlo Experiments
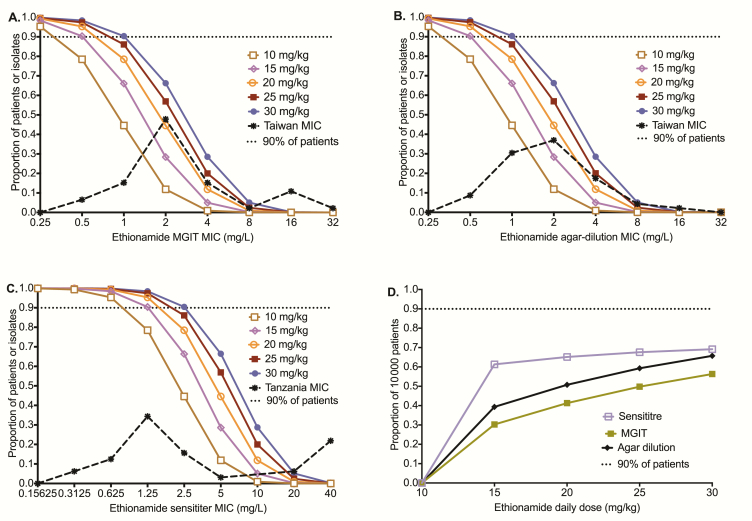
Table 1 shows that MCEs accurately recapitulated ethionamide pharmacokinetics encountered in patients, including the AUC0-24 and peak concentrations achieved with the dose of 20 mg/kg. Figure 3A shows the target attainment probability (TAP) in pulmonary tuberculosis at each MIC, based on the MGIT assay MICs, while Figure 3B was based on agar dilution method MICs; they show virtually the same TAP vs MIC. As would be expected, the MIC at which TAP falls below 90% was the same for each dose by either MIC method. However, the Sensititre assay had different TAP vs MIC relationships (Figure 3C), with higher proportions of patients achieving TAP >90% until an MIC of 2.5 mg/L with the currently used doses and until an MIC of 5.0 mg/L for the dose of 30 mg/kg. Figure 3D is a summation of how well each dose performed over the entire MIC distribution; none of the doses tested achieved cumulative fraction of response (CFR) in >90% of patients. If on the other hand we used a proposed Sensititre MIC susceptibility breakpoint <2.5 mg/L to triage patients who should be treated with ethionamide, then the doses of 20 mg/kg/day and 30 mg/kg/day would achieve optimal exposures in >95% of patients.
Figure 3.
Monte Carlo simulation results for 10000 patients with pulmonary tuberculosis. Target attainment probability (TAP) is the proportion of 10000 patients who achieve target exposure at a minimum inhibitory concentration (MIC). The TAP fell as the MIC rose. Since the type of MIC assay affects the MIC distribution, we examined the TAP using 3 MIC assays. (A) For MGIT-based MICs from Taiwan isolates, the TAP for the standard dose of 15–20 mg/kg/day falls below 90% at an MIC of 1.0 mg/L, which should be the clinical breakpoint for this dose. (B) TAP using agar dilution–based MICs, which are the gold standard, was similar to that for MGIT based. (C) When we used the Sensititre assay-based MIC for our laboratory strain to calculate target area under the concentration time curve (AUC)/MIC and the Sensititre-based MIC of clinical isolates, the ethionamide 15–20 mg/kg/day dose achieved a TAP which fell below 90% at the MIC of 2.5 mg/L, which should be the proposed MIC clinical breakpoint at these doses. At a dose of 30 mg/kg/day, the proposed clinical breakpoint MIC is 5 mg/L. (D) The cumulative fraction of response for different doses examined is a summation over all MICs. Abbreviation: MIC, minimum inhibitory concentration.
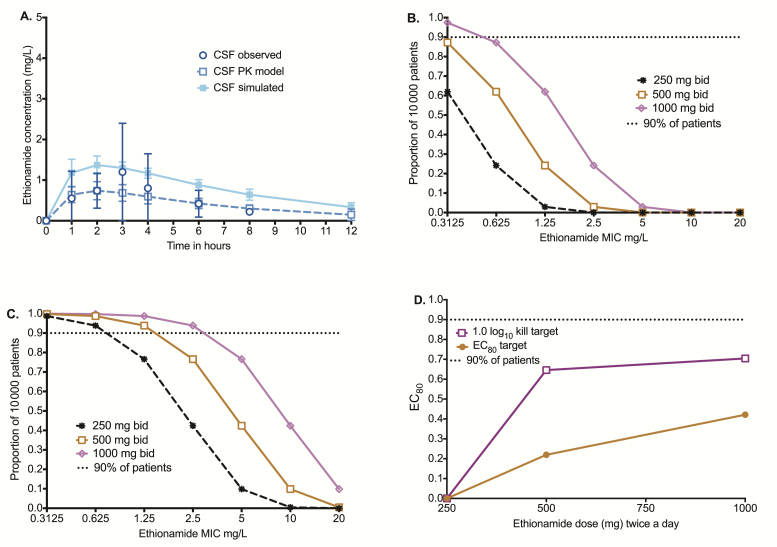
As regards to tuberculous meningitis, our pharmacokinetic modeling of data reported in the literature [31] revealed a serum half-life of 1.06 ± 1.23 hours, which was considerably shorter than the CSF half-life of 3.96 hours in the patients. Figure 4A shows the 10000-patient simulated concentration-time profiles superimposed on data observed in patients treated with 250 mg every 12 hours. The MCEs were performed to examine TAP for doses of 250 mg every 12 hours, 500 mg every 12 hours, and 1000 mg every 12 hours for the EC80 target, with results shown in Figure 4B. We also examined the probability of these doses achieving the CSF AUC0-24/MIC of 10, which mediates 1.0 log10 CFU/mL, with results shown in Figure 4C. The CFRs for those doses were as shown in Figure 4D using the Sensititre-based MICs for both the EC80 and cidal effect target. None of the doses achieved a CFR >90%.
Figure 4.
Target attainment in 10000 patients with tuberculous meningitis. (A) Population pharmacokinetics-based simulation of concentration-time profiles in the cerebral spinal fluid vs that observed in patients shows that simulation-derived concentrations were similar to those seen in patients. (B) The target attainment probability (TAP) of doses of 250–1000 mg twice a day for the exposure associated with 80% of maximal kill (EC80 target) was poor regardless of minimum inhibitory concentration (MIC). (C) For the target of 1.0 log10 colony-forming units/mL kill (cidal effect), the TAP for 1000 mg twice a day was >90% up to an MIC of 2.5 mg/L. (D) For the 250 mg twice a day dose, the cumulative fraction of response (CFR) was 0% for both the cidal effect and EC80 targets. As regards to the 500-mg twice a day dose, the CFRs were 64% and 21% for the cidal effect and EC80 targets, respectively. The CFRs for the 1.0 log10 kill did not improve much with the 1000-mg BID dose but went up substantially for the EC80 target. Abbreviations: bid, twice daily; CSF, cerebrospinal fluid; EC80, exposure associated with 80% of maximal kill; MIC, minimum inhibitory concentration; PK, pharmacokinetic.
MARS Output for Pulmonary Sputum Time-to-Culture Conversion in Tanzanian Patients
Among the 41 patients treated for MDR tuberculosis in Tanzania, the MYCOTB Sensititre MIC distributions for 4 drugs were as shown in Supplemental Figure S1. MARS identified 4 factors predictive of time-to-sputum conversion, shown as basis functions (BFs) in Table 2, which explained more than one third of the variance in time-to-cure. A history of prior tuberculosis (BF1) had the highest variable importance score of 100%. Ethionamide MIC (BF2 and BF3) had an importance score of 78%, followed by cavities on chest X-ray (BF4 and BF5) and patient weight. BF1 is a simple hinge function “max (0, Prior tuberculosis episodes – 0),” which means that the value of the expression is zero unless there is a history of prior tuberculosis, which history when present leads to a worse outcome (negative coefficient of −2.154). BF3 is a composite BF, conditional on BF2, described by the hinge function “max (0, 2.5 – ethionamide MIC),” which means that there is a linear relationship between time-to-sputum conversion and ethionamide MIC. The negative coefficient of −1.49 in Table 2 means that as ethionamide MIC increases, time-to-sputum conversion is longer until the hinge at an MIC of 2.5 mg/L, above which the negative effect remains unchanged at the maximum value. The receiver operating characteristics (ROCs) for the predictions of time-to-sputum conversion in learn set was 84%, and the cross-validated ROC (used to assess the reliability of the parameter estimates and robustness of the final model in a future dataset) was 81%. The final linearized model for time-to-sputum conversion (Y) in months was:
| (3) |
Table 2.
Multivariate Adaptive Regression Spline Model Output for Predictors Time-to-Sputum Conversion in 41 Patients
| BF | Number of Patients Affected, n (%) | Coefficient |
|---|---|---|
| BF1 = max (0, prior tuberculosis episodes – 0) | 41 (100) | –2.154 |
| BF2 = (data on ethionamide MIC present) | 32 (78) | 3.545 |
| BF3 = max (0, 2.5 – ethionamide MIC)* BF2 | 15 (37) | –1.490 |
| BF4 = (data on chest X-ray cavities present) | 29 (71) | 0.676 |
| BF5 = max (0, chest X-ray cavities present – 1)* BF4 | 14 (34) | –3.023 |
| BF6 = (data on weight present) | 39 (95) | 6.115 |
Abbreviations: BF, basis function; MIC, minimum inhibitory concentration.
These results mean that ethionamide MIC (BF2 and BF3) is a major determinant of time-to-sputum conversion and shows a stepwise increase in worse outcome with each increase in value of MIC in the classic PK/PD fashion [40]. The result also means that the worse outcomes as MIC increases are encountered even below the threshold MIC value of 2.5 mg/L. Conversely, kanamycin and cycloserine MICs were not identified as important predictors for time-to-sputum conversion, which suggests less contribution of these 2 drugs to the regimen in the MIC range investigated and is a major surprise.
DISCUSSION
First, our PK/PD study demonstrates that ethionamide had a maximal kill rate or slope of 0.21 log10 CFU/mL/day in the HFS-TB, which is equivalent to isoniazid and ethambutol in the same model [14, 16, 22, 34, 40]. Thus, ethionamide clearly has a reasonable microbial kill rate. A major finding in the Tanzanian clinical study was the identification of MIC as a major determinant of time-to-sputum conversion by MARS. That finding has 3 major implications. First, the MARS ranking of ethionamide MIC as a predictor of time-to-sputum conversion is prima facie evidence that ethionamide plays a major role in the MDR tuberculosis combination regimen that included levofloxacin.
A second implication central to PK/PD theory, was the linear relationship between time-to-sputum conversion and MIC up to 2.5 mg/L, beyond which it had the same maximal deleterious effect [40]. This is reminiscent of a foundational rat model of Pseudomonas aeruginosa PK/PD study that demonstrated the central role of MIC in PK/PD exposure determination, discussed in detail in the introduction paper [19, 41]. Our study shows that the role of MICs in determining outcomes is not just a preclinical PK/PD phenomenon but is clearly of clinical import [42]. A final important implication is the following corollary: increases in MIC are a sine qua non of increasing resistance and the resistance breakpoints represent a concentration above which there is uniform failure at that dose of drug used [43, 44]. Conversely, cycloserine MICs were not important predictors of outcome consistent with HFS-TB and MCEs elsewhere [45].
Second, emergence of acquired drug resistance (ADR) terminated the microbial kill of ethionamide. The sequence of events comprising early efflux pump induction initiated by subtherapeutic ethionamide therapy, with eventual emergence of chromosomal mutations, conforms to our “antibiotic resistance arrow of time” model [13, 14, 46, 47]. Given both the receptor (efflux pump) and ligand (drug) promiscuity, efflux pumps induced by one drug can transport out unrelated pharmacophores, while each drug can induce several efflux pumps [13, 14, 47–52]. This “tolerance” or “phenotypic resistance” buys time to allow multiple rounds of replication by Mtb until chromosomal mutations arise in genes encoding the specific drug’s target protein. Here, we show for the first time with ethionamide that this also promotes mutations in genes such as embA, encoding resistance to structurally unrelated drugs such as ethambutol. This is likely because the mutations arise in a stochastic fashion throughout the genome. This phenomenon could explain the relative discordance of phenotypic ethambutol resistance and mutation in the resistance determining region of embB, the region encompassed on commercially available line probe assays, in comparison to other phenotypic/genotypic concordance for other anti-tuberculosis drugs [53].
Third, we utilized MCEs to identify ethionamide doses for both pulmonary and meningeal tuberculosis. We propose that the drug be administered on condition of MIC <2.5 mg/L by Sensititre assay and 1.0 mg/L based on MGIT and agar dilution methods. Thus, MIC knowledge is likely of clinical importance not only in use of ethionamide but also accompanying drugs such as isoniazid, ethambutol, gatifloxacin, and levofloxacin [16, 19, 34–36, 43]. Under these conditions, an ethionamide dose of 20 mg/kg/day achieved target exposures in >90% of patients with pulmonary tuberculosis. We acknowledge that these recommendations have significant clinical implications when in many MDR tuberculosis endemic locations ethionamide susceptibility is not performed or is performed with a single “critical concentration” based on the 1% proportion method of susceptibility testing. We therefore anticipate the importance of further field-oriented studies of the MYCOTB version of the Sensititre plate or other MIC assays. Also, we look forward to study of sequence-specific mutation in ethA and ethR associated with quantitative change in ethionamide MIC that may inform the best use of ethionamide in shorter-course regimens for MDR tuberculosis that include isoniazid and ethambutol.
In the case of tuberculous meningitis, it was difficult to achieve the EC80 target, even with a dose of 1000 mg twice a day. However, a cidal effect target was achievable. The idea is that the bacterial burden in CSF is low, and thus it may not be necessary to achieve the exposure target of ADR suppression. On the other hand, long-term outcomes in tuberculous meningitis are still poor, as is the rate of severe neurological sequelae [54]. Thus, it could be argued that high kill rates are a premium.
There are some limitations and a notable strength to our study. First, we used a single strain in our HFS-TB study. Use of multiple strains is preferable. However, MCEs take into account isolates with a wide range of MICs in calculating optimal dose and thus partially offset this limitation. Second, the optimal dose identified in MCEs will need to be validated in patients. Last, the dataset utilized to predict microbiological outcomes was small; however, we leveraged the versatility of machine learning methods to identify meaningful patterns in the data. In summary, ethionamide has an effect on Mtb that is comparable to first-line agents such as isoniazid and ethambutol. Resistance emerges via induction of efflux pumps at suboptimal doses.
Supplemental Data
Supplemental materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Author contributions. T. G. and D. D designed the study; T. G., D. D., and P. B. performed the hollow fiber studies; D. D. wrote the first draft of the manuscript; T. K. performed DNA extraction; P. S. L. performed drug concentration assays; T. G. and S. S. performed the whole-genome sequencing analysis; T. G. performed PK/PD modeling and MCE; J. P. and T. G. performed MARS analysis of clinical data; S. G. M., S. K. H., and H. M. designed the clinical studies and performed the clinical studies and susceptibility testing of clinical isolates; D. D., J. P., and T. G. wrote the initial draft manuscript; all authors edited the manuscript and contributed to the final version of the manuscript.
Financial support. This work was supported by the Baylor Research Institute, Dallas, Texas, and portions from Tanzania were funded by the National Institutes of Health and the National Institute of Allergy and Infectious Diseases (U01AI119954; to S. K. H.). Supplement sponsored by Baylor Research Institute, Dallas, Texas.
Supplement sponsorship. This supplement is sponsored by the Baylor Institute of Immunology Research of the Baylor Research Institute.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Falzon D, Schunemann HJ, Harausz E, et al. . World Health Organization treatment guidelines for drug-resistant tuberculosis, 2016 update. Eur Respir J 2017; 49: pii: 1602308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pietersen E, Peter J, Streicher E, et al. . High frequency of resistance, lack of clinical benefit, and poor outcomes in capreomycin treated South African patients with extensively drug-resistant tuberculosis. PLoS One 2015; 10:e0123655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Modongo C, Pasipanodya JG, Magazi BT, et al. . Artificial intelligence and amikacin exposures predictive of outcomes in multidrug-resistant tuberculosis patients. Antimicrob Agents Chemother 2016; 60:5928–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Srivastava S, Modongo C, Siyambalapitiyage Dona CW, Pasipanodya JG, Deshpande D, Gumbo T. Amikacin optimal exposure targets in the hollow-fiber system model of tuberculosis. Antimicrob Agents Chemother 2016; 60:5922–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pasipanodya JG, Nuermberger E, Romero K, Hanna D, Gumbo T. Systematic analysis of hollow fiber model of tuberculosis experiments. Clin Infect Dis 2015; 61(Suppl 1):S10–7. [DOI] [PubMed] [Google Scholar]
- 6. Cavaleri M, Manolis E. Hollow fiber system model for tuberculosis: the European Medicines Agency experience. Clin Infect Dis 2015; 61(Suppl 1):S1–4. [DOI] [PubMed] [Google Scholar]
- 7. Chilukuri D, McMaster O, Bergman K, Colangelo P, Snow K, Toerner JG. The hollow fiber system model in the nonclinical evaluation of antituberculosis drug regimens. Clin Infect Dis 2015; 61(Suppl 1):S32–3. [DOI] [PubMed] [Google Scholar]
- 8. EMA. Qualification opinion. In-vitro hollow fiber system model of tuberculosis (HSF-TB). In: Agency EM. EMEA/H/SAB/049/1/QO/2014/SME Vol. London, UK: EMA, 2015. Available at: http://www.ema.europa.eu/docs/en_GB/document_library/Regulatory_and_procedural_guideline/2015/02/WC500181899.pdf. [Google Scholar]
- 9. Banerjee A, Dubnau E, Quemard A, et al. . inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science 1994; 263:227–30. [DOI] [PubMed] [Google Scholar]
- 10. Huyen MN, Cobelens FG, Buu TN, et al. . Epidemiology of isoniazid resistance mutations and their effect on tuberculosis treatment outcomes. Antimicrob Agents Chemother 2013; 57:3620–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Müller B, Streicher EM, Hoek KG, et al. . inhA promoter mutations: a gateway to extensively drug-resistant tuberculosis in South Africa?Int J Tuberc Lung Dis 2011; 15:344–51. [PubMed] [Google Scholar]
- 12. Niehaus AJ, Mlisana K, Gandhi NR, Mathema B, Brust JC. High prevalence of inhA promoter mutations among patients with drug-resistant tuberculosis in KwaZulu-Natal, South Africa. PLoS One 2015; 10:e0135003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schmalstieg AM, Srivastava S, Belkaya S, et al. . The antibiotic resistance arrow of time: efflux pump induction is a general first step in the evolution of mycobacterial drug resistance. Antimicrob Agents Chemother 2012; 56:4806–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Srivastava S, Musuka S, Sherman C, Meek C, Leff R, Gumbo T. Efflux-pump-derived multiple drug resistance to ethambutol monotherapy in Mycobacterium tuberculosis and the pharmacokinetics and pharmacodynamics of ethambutol. J Infect Dis 2010; 201:1225–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Colangeli R, Helb D, Sridharan S, et al. . The Mycobacterium tuberculosis iniA gene is essential for activity of an efflux pump that confers drug tolerance to both isoniazid and ethambutol. Mol Microbiol 2005; 55:1829–40. [DOI] [PubMed] [Google Scholar]
- 16. Deshpande D, Pasipanodya JG, Srivastava S, et al. . Gatifloxacin pharmacokinetics/pharmacodynamics-based optimal dosing for pulmonary and meningeal multidrug-resistant tuberculosis. Clin Infect Dis 2018; 67(Suppl 3):S293–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Deshpande D, Srivastava S, Chapagain M, et al. . Ceftazidime-avibactam has potent sterilizing activity against highly drug-resistant tuberculosis. Sci Adv 2017; 3:e1701102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Deshpande D, Srivastava S, Bendet P, et al. . On the antibacterial and sterilizing effect of benzylpenicillin in tuberculosis. Antimicrob Agents Chemother 2018; 62:e02232–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gumbo T, Alffenaar JWC. An introduction to pharmacokinetics/pharmacodynamics methods and scientific evidence base for dosing of second line tuberculosis drugs. Clin Infect Dis 2018; 67(Suppl 3):S267–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gumbo T, Louie A, Deziel MR, Parsons LM, Salfinger M, Drusano GL. Selection of a moxifloxacin dose that suppresses drug resistance in Mycobacterium tuberculosis, by use of an in vitro pharmacodynamic infection model and mathematical modeling. J Infect Dis 2004; 190:1642–51. [DOI] [PubMed] [Google Scholar]
- 21. Gumbo T, Lenaerts AJ, Hanna D, Romero K, Nuermberger E. Nonclinical models for antituberculosis drug development: a landscape analysis. J Infect Dis 2015; 211(Suppl 3):S83–95. [DOI] [PubMed] [Google Scholar]
- 22. Gumbo T, Louie A, Liu W, et al. . Isoniazid bactericidal activity and resistance emergence: integrating pharmacodynamics and pharmacogenomics to predict efficacy in different ethnic populations. Antimicrob Agents Chemother 2007; 51:2329–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Srivastava S, Pasipanodya JG, Meek C, Leff R, Gumbo T. Multidrug-resistant tuberculosis not due to noncompliance but to between-patient pharmacokinetic variability. J Infect Dis 2011; 204:1951–9.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Auclair B, Nix DE, Adam RD, James GT, Peloquin CA. Pharmacokinetics of ethionamide administered under fasting conditions or with orange juice, food, or antacids. Antimicrob Agents Chemother 2001; 45:810–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Deshpande D, Srivastava S, Nuermberger E, Pasipanodya JG, Swaminathan S, Gumbo T. A faropenem, linezolid, and moxifloxacin regimen for both drug-susceptible and multidrug-resistant tuberculosis in children: FLAME path on the milky way. Clin Infect Dis 2016; 63:S95–S101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Srivastava S, Garg A, Ayyagari A, Nyati KK, Dhole TN, Dwivedi SK. Nucleotide polymorphism associated with ethambutol resistance in clinical isolates of Mycobacterium tuberculosis. Curr Microbiol 2006; 53:401–5. [DOI] [PubMed] [Google Scholar]
- 27. Srivastava S, Magombedze G, Koeuth T, et al. . Linezolid dose that maximizes sterilizing effect while minimizing toxicity and resistance emergence for tuberculosis. Antimicrob Agents Chemother 2017; 61:e00751–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhu M, Namdar R, Stambaugh JJ, et al. . Population pharmacokinetics of ethionamide in patients with tuberculosis. Tuberculosis 2002; 82:91–6. [DOI] [PubMed] [Google Scholar]
- 29. Donald PR, Seifart HI. Cerebrospinal fluid concentrations of ethionamide in children with tuberculous meningitis. J Pediatr 1989; 115:483–6. [DOI] [PubMed] [Google Scholar]
- 30. Hughes IE, Smith H. Ethionamide: its passage into the cerebrospinal fluid in man. Lancet 1962; 1:616–7. [DOI] [PubMed] [Google Scholar]
- 31. Conte JE Jr, Golden JA, McQuitty M, Kipps J, Lin ET, Zurlinden E. Effects of AIDS and gender on steady-state plasma and intrapulmonary ethionamide concentrations. Antimicrob Agents Chemother 2000; 44:1337–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huang TS, Lee SS, Tu HZ, et al. . Use of MGIT 960 for rapid quantitative measurement of the susceptibility of Mycobacterium tuberculosis complex to ciprofloxacin and ethionamide. J Antimicrob Chemother 2004; 53:600–3. [DOI] [PubMed] [Google Scholar]
- 33. Ebers A, Stroup S, Mpagama S, et al. . Determination of plasma concentrations of levofloxacin by high performance liquid chromatography for use at a multidrug-resistant tuberculosis hospital in Tanzania. PLoS One 2017; 12:e0170663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Deshpande D, Pasipanodya JG, Mpagama SG, et al. . Levofloxacin pharmacokinetics-pharmacodynamics, dosing, susceptibility breakpoints, and artificial intelligence in the treatment of multidrug-resistant tuberculosis. Clin Infect Dis 2018; 67(Suppl 3):S312–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chigutsa E, Pasipanodya JG, Visser ME, et al. . Impact of nonlinear interactions of pharmacokinetics and MICs on sputum bacillary kill rates as a marker of sterilizing effect in tuberculosis. Antimicrob Agents Chemother 2015; 59:38–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rockwood N, Pasipanodya JG, Denti P, et al. . Concentration-dependent antagonism and culture conversion in pulmonary tuberculosis. Clin Infect Dis 2017; 64:1350–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Friedman JH. Multivariate adaptive regression splines. Ann Statist 1991; 19:1–68. [DOI] [PubMed] [Google Scholar]
- 38. Friedman JH, Roosen CB. An introduction to multivariate adaptive regression splines. Stat Methods Med Res 1995; 4:197–217. [DOI] [PubMed] [Google Scholar]
- 39. Rogers Z, Hiruy H, Pasipanodya JG, et al. . The non-linear child: ontogeny, isoniazid concentration, and NAT2 genotype modulate enzyme reaction kinetics and metabolism. EBioMedicine 2016; 11:118–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gumbo T, Angulo-Barturen I, Ferrer-Bazaga S. Pharmacokinetic-pharmacodynamic and dose-response relationships of antituberculosis drugs: recommendations and standards for industry and academia. J Infect Dis 2015; 211(Suppl 3):S96–S106. [DOI] [PubMed] [Google Scholar]
- 41. Drusano GL, Johnson DE, Rosen M, Standiford HC. Pharmacodynamics of a fluoroquinolone antimicrobial agent in a neutropenic rat model of Pseudomonas sepsis. Antimicrob Agents Chemother 1993; 37:483–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Heysell SK, Pholwat S, Mpagama SG, et al. . Sensititre MYCOTB MIC plate compared to Bactec MGIT 960 for first- and second-line antituberculosis drug susceptibility testing in Tanzania: a call to operationalize MICs. Antimicrob Agents Chemother 2015; 59:7104–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gumbo T. New susceptibility breakpoints for first-line antituberculosis drugs based on antimicrobial pharmacokinetic/pharmacodynamic science and population pharmacokinetic variability. Antimicrob Agents Chemother 2010; 54:1484–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Drusano GL, Preston SL, Hardalo C, et al. . Use of preclinical data for selection of a phase II/III dose for evernimicin and identification of a preclinical MIC breakpoint. Antimicrob Agents Chemother 2001; 45:13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Deshpande D, Alffenaar JWC, Köser CU, et al. . D-cycloserine pharmacokinetics-pharmacodynamics, susceptibility, and dosing implications in multidrug-resistant tuberculosis: a Faustian deal. Clin Infect Dis 2018; 67(Suppl 3):S274–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gumbo T. Biological variability and the emergence of multidrug-resistant tuberculosis. Nat Genet 2013; 45:720–1. [DOI] [PubMed] [Google Scholar]
- 47. Pasipanodya JG, Gumbo T. A new evolutionary and pharmacokinetic-pharmacodynamic scenario for rapid emergence of resistance to single and multiple anti-tuberculosis drugs. Curr Opin Pharmacol 2011; 11:457–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kenakin T. Agonist-receptor efficacy. I: mechanisms of efficacy and receptor promiscuity. Trends Pharmacol Sci 1995; 16:188–92. [DOI] [PubMed] [Google Scholar]
- 49. Hopkins AL. Drug discovery: predicting promiscuity. Nature 2009; 462:167–8. [DOI] [PubMed] [Google Scholar]
- 50. Sturm N, Desaphy J, Quinn RJ, Rognan D, Kellenberger E. Structural insights into the molecular basis of the ligand promiscuity. J Chem Inf Model 2012; 52:2410–21. [DOI] [PubMed] [Google Scholar]
- 51. Brown JB, Okuno Y. Systems biology and systems chemistry: new directions for drug discovery. Chem Biol 2012; 19:23–8. [DOI] [PubMed] [Google Scholar]
- 52. Wong K, Ma J, Rothnie A, Biggin PC, Kerr ID. Towards understanding promiscuity in multidrug efflux pumps. Trends Biochem Sci 2014; 39:8–16. [DOI] [PubMed] [Google Scholar]
- 53. Cui Z, Li Y, Cheng S, et al. . Mutations in the embC-embA intergenic region contribute to Mycobacterium tuberculosis resistance to ethambutol. Antimicrob Agents Chemother 2014; 58:6837–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shaw JE, Pasipanodya JG, Gumbo T. Meningeal tuberculosis: high long-term mortality despite standard therapy. Medicine 2010; 89:189–95. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.