Abstract
Purpose
To determined KIAA1199 expression and investigate its correlation with the clinicopathologic data and prognosis of hepatocellular carcinoma (HCC), as well as markers of epithelial-mesenchymal transition (EMT); N-cadherin, E-cadherin and vimentin.
Materials and methods
Western blot, quantitative real-time PCR, and immunohistochemical staining were used to measure KIAA1199 expression in human HCC specimens. Subsequently, the correlation between KIAA1199 expression and the pathological characteristics of HCC patients was analyzed. Univariate and multivariate analyses were used to explore the risk factors associated with disease-free survival (DFS) and overall survival (OS).
Results
KIAA1199 expression was remarkably increased in hepatocellular carcinoma tissues compared to paracarcinomatous tissues. This phenomenon was accompanied by aberrant expression of EMT-associated markers. In addition, high KIAA1199 expression was associated with severe pathological symptoms, low DFS, and low OS. Results of the multivariate analysis showed that KIAA1199 expression may be an independent predictor of low disease-free survival and OS of HCC patients.
Conclusion
KIAA1199 overexpression in HCC patients is associated with aberrant expression of EMT-associated markers and severe clinicopathological symptoms, and thus may function as a marker of poor prognosis in HCC.
Keywords: KIAA1199, hepatocellular carcinoma, EMT, prognosis, biomarker
Introduction
Hepatocellular carcinoma (HCC) is a fatal cancer ranked as the leading cause of cancer related deaths among women, the fifth most commonly diagnosed tumor worldwide and the seventh most common type of cancer.1 The 5-year survival rate of HCC patients is <11% due to poor prognosis caused by the metastatic and rapid progression of the disease. Hepatectomy or orthotopic liver transplantation is the most effective treatment for HCC.2 However, most HCC patients are diagnosed at an advanced stage when surgical treatment is no longer effective or even possible due to local aggression and distant metastases. Therefore, there is a need to develop sensitive and specific biomarkers to enable early diagnosis of hepatocellular carcinoma and to predict unfavorable prognosis.
KIAA1199 (Hs.459088, chr.15q25.1, TMEM2 L) is located on the chromosome band 15q25.1 ~150 kb upstream of the KIAA gene cluster.3 Recent studies have shown that the KIAA1199 gene is closely associated with the development, as well as the spread, of tumors. Several studies have stated that KIAA1199 is overexpressed in different malignancies including; gastric cancer,6 breast cancer,7 oral squamous cell carcinoma,5 prostate cancer,4 and colorectal tumors.8,10 These findings suggest that KIAA1199 regulates proliferation, metastasis, and invasion of tumors.3 Currently, there is no report on KIAA1199 expression in hepatocellular carcinoma and its clinical value. Nevertheless, some studies have indicated that KIAA1199 is a potential biomarker of tumor growth, metastasis and invasiveness in some types of human malignancies.7
Here, we analyzed the expression of KIAA1199 in human HCC specimens and determined whether it is associated with epithelial-mesenchymal transition (EMT). We also explored the association between KIAA1199 expression and the growth, metastasis and invasion of HCC cells and its clinicopathological features.
Material and methods
Patient enrollment and follow-up
The study was carried out in the Qi Lu Hospital of Shandong University where 178 patients who had undergone curative resection for HCC were enrolled between 2010 and 2015. No anti-cancer treatment was given to the patients before surgery. A postoperative pathological examination was used to confirm the diagnosis of hepatocellular carcinoma. Patient clinical information such as age, lymph node metastasis, histological grade, tumor location, sex, tumor-node-metastasis and tumor size were recorded. To explore the prognostic value of KIAA1199 in patients after surgery, disease-free survival (DFS) and overall survival (OS) were examined. The average follow-up period was 28 months (ranging from 1 month to 5 years). We obtained written informed consent from all patients and received approval from the Human Research Ethics Committee of the hospital in accordance with the Helsinki Declaration guidelines before the study commenced.
Immunohistochemistry
Immunohistochemistry was performed with a two-step protocol following the manufacturer’s instructions. Specimens were embedded in paraffin wax, fixed in 10% formaldehyde and cut into 4-µm serial sections. Paraffin was removed from the sections using a graded alcohol series of 100%, 95% in xylene and rehydrated in 75%, and finally washed with PBS. Slides were treated with citrate buffer in a microwave oven for 20 minutes at a pH level of 6.0 and then cooled to room temperature. Thereafter, 3% hydrogen peroxide was used to quench the endogenous peroxidase activity. Next, the specimens were treated with corresponding antibodies at 4°C overnight in a moist chamber: polyclonal rabbit anti-KIAA1199 antibody (21129–1-AP; Proteintech; Wuhan, China; diluted 1:100), anti-E-cadherin antibody (20874-1-AP; Proteintech; diluted 1:200), anti-N-cadherin antibody (22018-1-AP; Proteintech; diluted 1:200), and anti-vimentin antibody (10366-1-AP; Proteintech; diluted 1:200). Subsequently, they were washed with PBS and then incubated for 10 minutes at 37°C with the biotinylated secondary antibody (ZB-2301; Zhongshan Jin qiao Co., Beijing, China; diluted 1:300). Finally, the tissue sections were examined by applying 3,3-Diaminobenzidine (GK600505; Gen Tech co, Shanghai, China). The sections were dehydrated and mounted after counterstaining with hematoxylin. Sections treated with the same conditions without primary antibodies served as negative controls.
Immunohistochemical assessment
Semi-quantitative assessment of the immunohistochemical results of KIAA1199, N-cadherin, E-cadherin and vimentin was performed in terms of intensity of staining (range, 0–3; 0= absent, 1= weak, 2= moderate, 3= strong) and percentage of stained cells (range, 0–3; <10%=score 1, 10%–50%= score 2, >50%=score 3). A final total score ≥3 was considered low expression, and score >3 was recorded as high expression.18 All the immunostained slides were analyzed by two experienced and specialized pathologists working independently, and both were blinded to each other’s results and the patients’ characteristics. A joint analysis by the two pathologists was performed where discrepancies were observed to reach a consensus.
Western blot assay
Proteins were extracted from HCC tissues and paracarcinomatous tissues by a cell lysis buffer kit containing a protease inhibitor cocktail which was kept on ice for 30 minutes before use. Supernatants containing the proteins were collected after centrifugation of the samples at 14,000 rpm for 15 minutes. SDS-PAGE was used to separate the proteins, which were then transferred onto polyvinylidene difluoride membranes. The membranes were then incubated at 4°C overnight with primary antibodies after blocking with 5% skim milk for 1 hour at room temperature. The bands were incubated with secondary antibody for 1 hour and quantified using a chemiluminescence imaging system. The following primary antibodies were used: polyclonal rabbit anti-KIAA1199 antibody (21129-1-AP; Proteintech; diluted 1:1,000), anti-β-actin antibody (60008-1-Ig; Proteintech; diluted 1:3,000), horseradish peroxidase-conjugated secondary antibody (ZB-2301, rabbit IgG; Zhongshan Jinqiao Co.; diluted 1:10,000).
Quantitative real-time PCR
The total RNA isolated, according to manufacturer’s instructions, from tissues was used to synthesize cDNA. Thereafter, quantitative real-time PCR (qRT-PCR) was conducted using the SYBR Green reagent in the ABI Prism 7900HT™ sequence detection system (Thermo Fisher Scientific, Waltham, MA, USA). The following primers were used: KIAA1199, F primer 5′-AGGCGTGACACTGTCTCGGCTACAG-3′ and R primer 5′-CCACTCCACGTCTTGAACCCAC-3′. GAPDH, F primer 5′-TTGGTATCGTGGAAGGACTCA-3′ and R primer 5′-TGTCATCATATTTGGCAGGTT-3′.
Statistical analysis
Continuous statistics were presented as mean±SD. Chi-squared test was used to analyze the immunohistochemical data while the Fisher’s exact test and two-tailed chi-squared test were used to assess the relationship between the clinicopathologic characteristics and KIAA1199 expression status. The Spearman’s correlation test and two-tailed chi-squared test were applied to evaluate the relationships between KIAA1199 and vimentin, E-cadherin and N-cadherin. The association between the survival time of the patients and the level of KIAA1199 expression was assessed by the log-rank test and Kaplan–Meier survival analysis. Multivariate analysis was performed using the Cox proportional hazards regression model to determine the independent predictors that were significant in univariate analysis. SPSS 22 (IBM Corporation, Armonk, NY, USA) and GraphPad Prism 7 (GraphPad Software, San Diego, CA, USA) were used for statistical analyses. A P-value of ≥0.05 was regarded as statistically significant.
Results
KIAA1199 expression in paracarcinomatous and HCC tissues
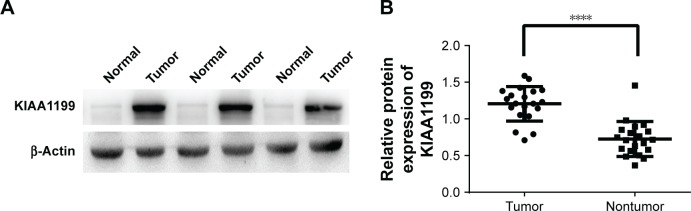
KIAA1199 expression in paracarcinomatous tissues and HCC tissues obtained from the 178 patients was measured by immunostaining. The results indicated that KIAA1199 expression was lower in paracarcinomatous tissues (66/178, 37.1%) (P=0.01) compared to carcinoma tissues (132/178, 74.2%) (Table 1). Figure 1 (A1 and A2) are typical photographs of the immunostaining. KIAA1199 mRNA expression was evaluated in 20 pairs of human specimens using qRT-PCR. As shown in Figure 2, KIAA1199 mRNA expression was considerably lower in adjacent normal tissues (3.58±0.38 vs 7.97±0.84, P=0.001) than in tumor tissues. The Western blot results on KIAA1199 protein levels were in agreement with those of qRT-PCR. Hence, KIAA1199 protein levels were significantly lower in adjacent non-cancerous liver tissues (0.725±0.05337 vs 1.20±0.05, P=0.0003; Figure 3A and B) than in HCC tissues.
Table 1.
Differential expression of KIAA1199 in HCC tissues and corresponding paracarcinoma tissues (n=178)
| Tissue | KIAA1199 expression
|
P-value | |
|---|---|---|---|
| Low (%) | High (%) | ||
|
| |||
| HCC tissues | 46 (25.8) | 132 (74.2) | 0.01 |
| Paracarcinoma tissues | 112 (62.9) | 66 (37.1) | |
Abbreviation: HCC, hepatocellular carcinoma.
Figure 1.
Immunohistochemical staining for KIAA1199, E-cadherin, N-cadherin and vimentin. Magnification 400×.
Notes: (A1) Negative KIAA1199 expression. (A2) Positive KIAA1199 expression. (B1) Positive E-cadherin expression. (B2) Negative E-cadherin expression. (C1) Negative N-cadherin expression. (C2) Positive N-cadherin expression. (D1) Negative vimentin expression. (D2) Positive vimentin expression. KIAA1199 is principally localized in cytoplasm of tumor cells; E-cadherin is mainly localized in the cytomembrane of cells; vimentin is mainly localized in the cytoplasm of tumor cells; N-cadherin is localized in the cytoplasm and on the membrane of the tumor cells.
Figure 2.
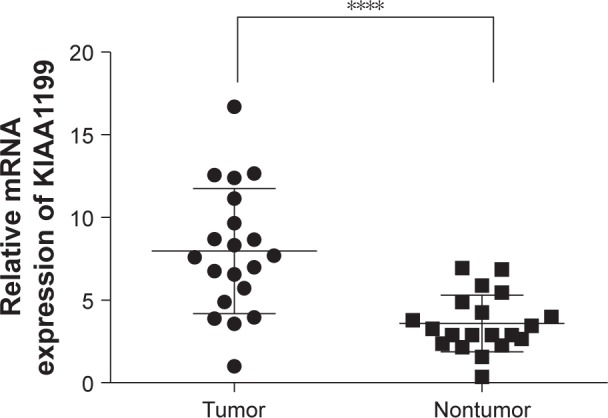
qPCR analysis of KIAA1199 expression in 20 pairs of HCC tissues and matched adjacent normal tissues.
Note: ****P<0.05.
Figure 3.
(A) Representative images of KIAA1199 protein expression in HCC tissues and paired normal tissues from three patients. (B) Western blot analysis of KIAA1199 expression in 20 pairs of HCC tissues and matched adjacent normal tissues.
Note: ****P<0.05.
Abbreviation: HCC, hepatocellular carcinoma.
Relationship between KIAA1199 expression and E-cadherin, N-cadherin, and vimentin expression in HCC tissues
We found high N-cadherin expression in 128 out of 178 HCC samples (71.9%; Figure 1: C1 and C2). Spearman’s rank correlation test showed a considerably positive correlation between KIAA1199 expression and N-cadherin expression in HCC specimen (r=0.402, P<0.05; Table 2). On the contrary, a profound negative relationship was noted between KIAA1199 expression and E-cadherin expression in HCC samples (r=−0.242, P=0.001; Table 2). Using the same test, we found high E-cadherin expression in 52 out of 178 HCC specimens (29.2%; Figure 1: B1 and B2). Additionally, the expression of vimentin was high in 145 out of 178 HCC tissues (81.5%; Figure 1: D1 and D2) and was similar to the expression level of N-cadherin. Moreover, a strong positive correlation was found between vimentin expression and KIAA1199 expression in HCC tissues as determined by the Spearman’s rank correlation test (r=0.313, P<0.001; Table 2).
Table 2.
Correlations of KIAA1199 expression with E-cadherin, N-cadherin and vimentin in HCC
| Immunoreactivity | KIAA1199 expression
|
R-value | P-value | |
|---|---|---|---|---|
| Low | High | |||
|
| ||||
| E-cadherin expression | ||||
| Low | 24 | 102 | −0.242 | 0.001 |
| High | 22 | 30 | ||
| N-cadherin expression | ||||
| Low | 27 | 23 | 0.402 | <0.001 |
| High | 19 | 109 | ||
| Vimentin expression | ||||
| Low | 18 | 15 | 0.313 | <0.001 |
| High | 28 | 117 | ||
Abbreviation: HCC, hepatocellular carcinoma.
Relationship between KIAA1199 expression and patient clinicopathological characteristics
We further assessed the correlation between the clinical features of patients with HCC and KIAA1199 expression in HCC tissues. As shown in Table 3, upregulated KIAA1199 expression was associated with some severe clinicopathological parameters, such as vascular invasion (P<0.05 for KIAA1199), Edmondson grade (P<0.05 for KIAA1199), TNM staging (P<0.05 for KIAA1199) and alpha-fetoprotein (P<0.05 for KIAA1199). However, KIAA1199 expression was not correlated with age, gender, tumor size, tumor nodule number, tumor capsula, cirrhosis, HBeAg status, and Child-Pugh grade.
Table 3.
KIAA1199 expression status in relation to clinicopathologic features in 178 HCC patients
| Parameters | Case number | KIAA1199 expression
|
χ2 | P-value | |
|---|---|---|---|---|---|
| Low | High | ||||
|
| |||||
| Age (years) | |||||
| ≤60 | 130 | 30 | 100 | 1.924 | 0.165 |
| >60 | 48 | 16 | 32 | ||
| Gender | |||||
| Male | 108 | 27 | 81 | 0.141 | 0.707 |
| Female | 69 | 19 | 50 | ||
| Tumor size (cm) | |||||
| ≤5 | 63 | 14 | 49 | 0.667 | 0.414 |
| >5 | 115 | 32 | 83 | ||
| Tumor nodule number | |||||
| Single | 136 | 37 | 99 | 0.559 | 0.455 |
| Multiple | 42 | 9 | 33 | ||
| Vascular invasion | |||||
| Yes | 88 | 8 | 80 | 25.485 | <0.05 |
| No | 90 | 38 | 52 | ||
| Tumor capsula | |||||
| Absent | 72 | 20 | 52 | 0.236 | 0.627 |
| Present | 106 | 26 | 80 | ||
| Edmondson grade | |||||
| I–II | 87 | 35 | 52 | 18.381 | <0.05 |
| III–IV | 91 | 11 | 80 | ||
| Cirrhosis | |||||
| Absent | 72 | 17 | 55 | 0.314 | 0.575 |
| Present | 106 | 29 | 77 | ||
| lHBeAg status | |||||
| Positive | 126 | 29 | 97 | 1.798 | 0.180 |
| Negative | 52 | 17 | 35 | ||
| Child-Pugh grade | |||||
| A | 168 | 43 | 125 | 0.096 | 0.757 |
| B | 10 | 3 | 7 | ||
| TNM stage | |||||
| I–II | 109 | 39 | 70 | 14.489 | <0.05 |
| III–IV | 69 | 7 | 62 | ||
| AFP (ng/mL), U/mL | |||||
| ≤20 | 37 | 28 | 9 | 60.526 | <0.05 |
| >20 | 141 | 18 | 123 | ||
Abbreviations: AFP, alpha-fetoprotein; HCC, hepatocellular carcinoma.
Survival assessment
Survival curves were plotted using the Kaplan–Meier method and the survival time was assessed by the log-rank test (Figure 4). The results showed that a low KIAA1199 expression in HCC patients (P<0.001 for DFS and OS) was associated with considerably longer DFS and OS compared with the high KIAA1199 expression. The median DFS corresponding to high KIAA1199 expression was 9 months, and 17 months for low KIAA1199 expression. The median OS for high KIAA1199 expression was 17 months, and 20 months for low KIAA1199 expression.
Figure 4.
Kaplan-Meier analysis of DFS and OS in patients with HCC according to KIAA1199 staining.
Note: Patients with high KIAA1199 expression had a shorter OS and DFS than did those with low KIAA1199 expression (n=178; P<0.05; log rank test).
Abbreviations: DFS, disease-free survival; HCC, hepatocellular carcinoma; OS, overall survival.
It was observed from the univariate analysis results (Table 4), that high KIAA1199 expression, high vascular invasion, Edmondson grade, TNM and alpha-fetoprotein were associated with reduced OS and DFS. A further multivariate survival analysis demonstrated that KIAA1199 expression was an independent predictive factor for DFS (HR =4.223, 95% CI: 2.552–5.756, P<0.001) and OS (HR =4.835, 95% CI: 2.655–6.229; P<0.001).
Table 4.
Univariate and multivariate analysis of factors associated with OS and DFS
| Parameter | OS
|
DFS
|
||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value | |
|
| ||||||
| Univariate analysis | ||||||
| Age (≤60 vs >60 years) | 0.956 | 0.832–1.029 | 0.866 | 0.865 | 0.726–1.328 | 0.863 |
| Gender (male vs female) | 0.865 | 0.647–1.329 | 0.876 | 0.843 | 0.756–1.312 | 0.722 |
| Tumor size (≤5 vs >5 cm) | 0.952 | 0.683–1.464 | 0.812 | 0.733 | 0.697–1.527 | 0.912 |
| Tumor nodule number (single vs multiple) | 1.982 | 0.926–2.007 | 0.125 | 1.731 | 0.936–1.982 | 0.163 |
| Vascular invasion (no vs yes) | 4.357 | 3.874–8.543 | <0.001 | 5.374 | 4.387–9.614 | <0.001 |
| Tumor capsula (absent vs present) | 0.613 | 0.432–0.926 | 0.079 | 0.567 | 0.432–0.946 | 0.081 |
| Edmondson grade (I–II vs III–IV) | 2.957 | 1.852–4.237 | <0.001 | 3.327 | 1.928–3.658 | <0.001 |
| Cirrhosis (absent vs present) | 1.169 | 0.829–2.211 | 0.521 | 1.087 | 0.810–1.921 | 0.832 |
| HBeAg status (positive vs negative) | 0.936 | 0.689–1.431 | 0.897 | 0.811 | 0.481–1.235 | 0.336 |
| Child-Pugh grade (A vs B) | 1.231 | 0.753–1.564 | 0.877 | 1.012 | 0.789–1.515 | 0.802 |
| TNM stage (I–II vs III–IV) | 2.569 | 1.556–3.283 | <0.001 | 2.693 | 1.894–3.324 | <0.001 |
| AFP (≤20 vs >20 U/mL) | 2.322 | 1.274–3.285 | 0.018 | 2.153 | 1.201–2.894 | 0.043 |
| KIAA1199 expression (high vs low) | 2.342 | 1.923–2.888 | <0.001 | 2.796 | 1.568–3.368 | <0.001 |
| Multivariate analysis | ||||||
| Vascular invasion (no vs yes) | 7.151 | 5.534–9.427 | <0.001 | 15.635 | 8.007–27.832 | <0.001 |
| Edmondson grade (I–II vs III–IV) | 4.447 | 2.957–5.232 | <0.001 | 3.987 | 2.988–4.958 | <0.001 |
| TNM stage (I–II vs III–IV) | 3.596 | 2.589–4.129 | <0.001 | 5.351 | 3.110–7.654 | <0.001 |
| AFP (≤20 vs >20 U/mL) | 2.754 | 1.524–3.153 | 0.014 | 1.654 | 1.740–2.845 | 0.023 |
| KIAA1199 expression (low vs high) | 4.835 | 2.655–6.229 | <0.001 | 4.223 | 2.552–5.756 | <0.001 |
Abbreviations: AFP, alpha-fetoprotein; DFS, disease-free survival; HCC, hepatocellular carcinoma; OS, overall survival.
Discussion
Here, we explored the expression of KIAA1199 in HCC tissues and its association with the clinical outlook of HCC patients. The results showed that KIAA1199 was upregulated in tissues isolated from HCC patients compared with non-cancerous tissues. The results revealed that KIAA1199 expression was significantly increased in tumors characterized with advanced TNM stage, perineural invasion, distant metastasis and poor histological grade. Importantly, there was a positive correlation between the KIAA1199 expression and that of vimentin and N-cadherin; and a negative correlation between E-cadherin and KIAA1199 expression. Furthermore, the high expression of KIAA1199 in HCC tissues was associated with poor prognosis, suggesting that it is an independent predictor of unfavorable outcomes in patients with HCC.
Previous studies demonstrate that upregulation of KIAA1199 promotes tumorigenesis, infiltration, invasion, migration, and apoptosis. Zhang et al3 reported the involvement of KIAA1199 in metastasis, invasion, and cell motility across various cell types. Additionally, Chanthammachat et al5 stated that KIAA1199 may regulate the processes involved in cancer cell growth, survival, and metastasis. A study by Matsuzaki et al6 found that KIAA1199 expression was elevated in gastric cancer tissues. Collectively, these findings suggest that upregulation of KIAA1199 mRNA may influence the peritoneal dissemination, distant metastasis, lymph node metastasis, tumor differentiation and poor prognosis of cancer. Birkenkamp-Demtroder et al9 demonstrated that, in colorectal cancer tissues, KIAA1199 mRNA and protein levels are increased, and that KIAA1199 signals through the Wnt-pathway to influence cell adhesion, motility and proliferation. Kuscu et al11 also reported that the expression of KIAA1199 was markedly elevated in invasive MAD-MB-231 breast cancer cells and invasion breast cancer specimens. In addition, they found that KIAA1199 was highly expressed in invasive DU145 prostate cancer cells. Collectively, Kuscu’s findings indicate that KIAA1199 is associated with the invasiveness and progression of cancer cells. Other studies found that KIAA1199 has dual effects depending on the tumor type.2 Compared to normal tissues, our study showed that the expression of KIAA1199 was markedly increased in hepatocellular cancer tissues, particularly in patients with severe clinicopathological symptoms. These datasets indicate that increased KIAA1199 expression contributes to the development of HCC.
Epithelial-mesenchymal transition (EMT) involves modifications of gene expression leading to the repression of epithelial phenotype and activation of mesenchymal phenotype. EMT is an early event in the process of distant metastasis of tumor cells.12,13 EMT also promotes rapid growth of tumor cells14 and it is a key process that regulates tumor invasion and metastasis. E-cadherin, N-cadherin and vimentin are important protein markers of EMT. E-cadherin is a cell adhesion molecule found in the epithelium which is formed by intracellular catenin and peptide proteins linked to the cytoskeletal actin. E-cadherin participates in cell adhesion, growth and proliferation, and maintains epithelial cell morphology and structural integrity.15 Vimentin, a major intermediate silk filament protein present in stromal cells, participates in cancer cells adhesion, invasion, migration and signaling by regulating the interaction between cytoskeletal proteins and cell adhesion molecules.16 During EMT, changes to the N-cadherin expression influence cell invasion, migration and survival by regulating intracellular signaling pathways.17 As the biomarkers and regulators of EMT, E-cadherin, N-cadherin, and vimentin have been reported to be expressed in tumor tissues and are thought to promote poor prognosis.14,16 In this retrospective study, E-cadherin expression was negatively associated with the expression of KIAA1199 in HCC. In contrast, KIAA1199 expression was positively correlated with vimentin and N-cadherin expression in HCC. Therefore, upregulated KIAA1199 expression in HCC is closely associated with abnormal levels of proteins associated with EMT; which implies that KIAA1199 modulates the progression of hepatocellular cancer by influencing the E-cadherin-, N-cadherin- and vimentin-mediated EMT process.
Further, our results show that high KIAA1199 expression in HCC patients led to shorter survival time, while low KIAA1199 expression produced opposite effects. Interestingly, KIAA1199 overexpression was found to be an independent predictor of poor prognosis as revealed by multivariate survival analysis. These findings suggest that high KIAA1199 expression leads to poor prognosis in HCC patients and may be used as a prognostic biomarker.
Limitations
The sample size used in this study was small. Selection bias cannot be ignored as this is a retrospective study design. The mechanism by which KIAA1199 promoted the invasion and metastasis of HCC cells was not explored. Hence, we propose that future studies should apply advanced tools such as cell invasion assays, gene knock-out platforms, gene transfection and cell migration assays to validate our findings.
Conclusion
HCC, a malignant tumor with strong invasive and metastatic properties, is usually diagnosed late which contributes to poor prognosis. Our investigation suggests that KIAA1199 overexpression promotes the progression of HCC. Moreover, our study shows that KIAA1199 is associated with the EMT process, and may be an independent prognostic biomarker for poor prognosis in hepatocellular carcinoma. Further studies are warranted to study the mechanism of KIAA1199 in HCC. Finally, KIAA1199 could be a potential biomarker that can be exploited for drug discovery and to facilitate early diagnosis of HCC.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (No 81571367).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Blechacz B, Mishra L. Hepatocellular carcinoma biology. Recent Results Cancer Res. 2013;190:1–20. doi: 10.1007/978-3-642-16037-0_1. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y, Jia S, Jiang WG. KIAA1199 and its biological role in human cancer and cancer cells (review) Oncol Rep. 2014;31(4):1503–1508. doi: 10.3892/or.2014.3038. [DOI] [PubMed] [Google Scholar]
- 4.Michishita E, Garcés G, Barrett JC, Horikawa I. Upregulation of the KIAA1199 gene is associated with cellular mortality. Cancer Lett. 2006;239(1):71–77. doi: 10.1016/j.canlet.2005.07.028. [DOI] [PubMed] [Google Scholar]
- 5.Chanthammachat P, Promwikorn W, Pruegsanusak K, et al. Comparative proteomic analysis of oral squamous cell carcinoma and adjacent non-tumour tissue from Thailand. Arch Oral Biol. 2013;58(11):1677–1685. doi: 10.1016/j.archoralbio.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 6.Matsuzaki S, Tanaka F, Mimori K, Tahara K, Inoue H, Mori M. Clinicopathologic significance of KIAA1199 overexpression in human gastric cancer. Ann Surg Oncol. 2009;16(7):2042–2051. doi: 10.1245/s10434-009-0469-6. [DOI] [PubMed] [Google Scholar]
- 7.Evensen NA, Kuscu C, Nguyen HL, et al. Unraveling the role of KIAA1199, a novel endoplasmic reticulum protein, in cancer cell migration. J Natl Cancer Inst. 2013;105(18):1402–1416. doi: 10.1093/jnci/djt224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tiwari A, Schneider M, Fiorino A, et al. Early insights into the function of KIAA1199, a markedly overexpressed protein in human colorectal tumors. PLoS One. 2013;8(7):e69473. doi: 10.1371/journal.pone.0069473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Birkenkamp-Demtroder K, Maghnouj A, Mansilla F, et al. Repression of KIAA1199 attenuates Wnt-signalling and decreases the proliferation of colon cancer cells. Br J Cancer. 2011;105(4):552–561. doi: 10.1038/bjc.2011.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lapointe LC, Pedersen SK, Dunne R, et al. Discovery and validation of molecular biomarkers for colorectal adenomas and cancer with application to blood testing. PLoS One. 2012;7(1):e29059. doi: 10.1371/journal.pone.0029059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuscu C, Evensen N, Kim D, Hu YJ, Zucker S, Cao J. Transcriptional and epigenetic regulation of KIAA1199 gene expression in human breast cancer. PLoS One. 2012;7(9):e44661. doi: 10.1371/journal.pone.0044661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baulida J. Epithelial-to-mesenchymal transition transcription factors in cancer-associated fibroblasts. Mol Oncol. 2017;11(7):847–859. doi: 10.1002/1878-0261.12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Puisieux A, Brabletz T, Caramel J. Oncogenic roles of EMT-inducing transcription factors. Nat Cell Biol. 2014;16(6):488–494. doi: 10.1038/ncb2976. [DOI] [PubMed] [Google Scholar]
- 14.Maier HJ, Wirth T, Beug H. Epithelial-mesenchymal transition in pancreatic carcinoma. Cancers. 2010;2(4):2058–2083. doi: 10.3390/cancers2042058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Solanas G, Porta-de-la-Riva M, Agustí C, et al. E-cadherin controls beta-catenin and NF-kappaB transcriptional activity in mesenchymal gene expression. J Cell Sci. 2008;121(Pt 13):2224–2234. doi: 10.1242/jcs.021667. [DOI] [PubMed] [Google Scholar]
- 16.Handra-Luca A, Hong SM, Walter K, Wolfgang C, Hruban R, Goggins M. Tumour epithelial vimentin expression and outcome of pancreatic ductal adenocarcinomas. Br J Cancer. 2011;104(8):1296–1302. doi: 10.1038/bjc.2011.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen J, Zhao J, Ma R, Lin H, Liang X, Cai X. Prognostic significance of E-cadherin expression in hepatocellular carcinoma: a meta-analysis. PLoS One. 2014;9(8):e103952. doi: 10.1371/journal.pone.0103952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhai LL, Wu Y, Cai CY, Tang ZG. Upregulated matrix metalloproteinase-2 and downregulated tissue factor pathway inhibitor-2 are risk factors for lymph node metastasis and perineural invasion in pancreatic carcinoma. Onco Targets Ther. 2015;8:2827–2834. doi: 10.2147/OTT.S90599. [DOI] [PMC free article] [PubMed] [Google Scholar]