Abstract
Point mutations in p21ras are associated with ∼30% of human tumors by disrupting its GTP hydrolysis cycle, which is critical to its molecular switch function in cellular signaling pathways. In this work, we investigate the impact of Gln 61 substitutions in the structure of the p21N-ras active site and particularly focus on water reorganization around GTP, which appears to be crucial to evaluate favorable and unfavorable hydration sites for hydrolysis. The NRas-GTP complex is analyzed using a hybrid quantum mechanics/molecular mechanics approach, treating for the first time to our knowledge transient water molecules at the ab initio level and leading to results that account for the electrostatic coupling between the protein complex and the solvent. We show that for the wild-type protein, water molecules are found around the GTP γ-phosphate group, forming an arch extended from residues 12 to 35. Two density peaks are observed, supporting previous results that suggest the presence of two water molecules in the active site, one in the vicinity of residue 35 and a second one stabilized by hydrogen bonds formed with nitrogen backbone atoms of residues 12 and 60. The structural changes observed in NRas Gln 61 mutants result in the drastic delocalization of water molecules that we discuss. In mutants Q61H and Q61K, for which water distribution is overlocalized next to residue 60, the second density peak supports the hypothesis of a second water molecule. We also conclude that Gly 60 indirectly participates in GTP hydrolysis by correctly positioning transient water molecules in the protein complex and that Gln 61 has an indirect steric effect in stabilizing the preorganized catalytic site.
Introduction
p21ras is a small GTPase protein known for its central role in cellular signal transduction (1). It functions as a molecular switch that, in the active GTP-bound conformation, enables the transmission of signals of cell proliferation that are subsequently switched off by the hydrolysis reaction, leading to the inactive guanosine diphosphate (GDP)-bound state. Although p21ras (referred to as Ras in this article) presents low GTPase activity and catalytic efficiency (2), the reaction rate is accelerated up to five orders of magnitude by GTPase-activating proteins (GAPs) (3, 4) that supply an arginine residue (Arg 789) to the protein complex active site.
Specific Ras mutations (5) found in ∼30% of human tumors have been shown to reduce its intrinsic GTPase capability (6, 7) and make it insensitive to GAPs (8, 9). Such mutations induce a hydrolysis rate drop that keeps the Ras molecular switch constitutively active, leading to abnormal cell proliferation.
These oncogenic mutations mainly consist in the substitution of highly conserved residues (namely Gly 12, Gly 13, and Gln 61) found in the three Ras isoforms p21H-ras, p21N-ras, and p21K-ras. In this study, we focus on the p21 N-ras isoform (referred to as NRas), which harbors activating mutations at position 61 in ∼60% of NRas tumors, mostly found in melanomas (10). According to the Catalogue of Somatic Mutations in Cancer (COSMIC) (11), Gln 61 can be replaced by arginine (∼45% Q61R), lysine (∼36% Q61K), leucine (∼10% Q61L), histidine (∼7% Q61H), proline (∼1% Q61P), and glutamic acid (<1% Q61E).
Because of Ras’s potential for the development of targeted therapeutic strategies, numerous experimental (12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24) and theoretical (15, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38) studies have been carried out during the last 40 years to understand Ras’s intrinsic GTPase mechanism and its enhancement by GAPs. Despite such efforts, the reaction pathway remains unclear, as results support both associative (12, 14, 15, 16, 17, 19, 21, 25, 26, 27, 28, 29, 34, 36, 38) and dissociative (20, 22, 23, 30, 32, 33) mechanisms for the GTP hydrolysis reaction within the protein complex.
Because Gln 61 substitutions, by any other residue, drastically decrease both the intrinsic and GAP-enhanced GTP hydrolysis rate, this residue is expected to have a crucial role in the enzymatic catalysis of the reaction. The most recurrent mechanisms proposed in previous studies notably involve 1) a direct role of Gln 61 in proton transfers (12, 13, 15, 32, 33, 38), 2) a stabilizing effect of the transition state by Gln 61 in a substrate-assisted catalysis in which GTP itself acts as the general base of the reaction (14, 15, 16, 17, 19, 21, 25, 34), 3) a role of accurate positioning of water molecules in the substrate assisted catalysis (27, 28, 30), and 4) an indirect steric effect of Gln 61 in stabilizing the preorganized catalytic configuration of the active site (26, 29).
In addition to this, although few studies focus on the role of water molecules present in the protein complex active site (31, 36, 37), it also remains controversial whether a single water molecule (12, 13, 14, 15, 16, 26, 27, 30, 31, 32) or two water molecules take part in the enzymatic catalysis of the GTP hydrolysis reaction (21, 25, 28, 33, 34, 38).
Concerning the theoretical approaches, molecular dynamics simulations have been carried out to study Ras (15, 28) and Ras-GAP complex (27) conformations and to determine their structural catalyzing effect. Hybrid quantum mechanics/molecular mechanics (QM/MM) approaches have also been used to treat the protein complex active site at the ab initio level and therefore elucidate the reaction pathway (30, 32, 33, 34, 35, 38). These simulation methods have also been used to study water molecules within the Ras active site and have brought insight into 1) water molecules that are proton-transfer competent (31), 2) energy barriers associated to one- and two-water-molecule mechanisms (38), and 3) the number of water molecules in the vicinity of the GTP γ-phosphate group (37). Nevertheless, both the role of these water molecules and their accurate positioning in the active site remain to be elucidated.
In this article, we present new insights, to our knowledge, into water molecule distribution within the active site of wild-type (WT) NRas and Gln 61 mutants reported in the COSMIC database. Hybrid QM/MM dynamics simulations are used to treat the complex active site, together with the water molecules included in it, at the ab initio level while accounting for the global dynamics of NRas and the bulk solvent. It is the first time to our knowledge that transient water molecules are treated quantum mechanically to account for the electrostatic coupling between the protein complex and the solvent. Although the initial sampling is short, we show that water molecules are precisely positioned within the WT protein, whereas this localization is lost upon Gln 61 substitutions because of the structural rearrangements undergone by the active site residues.
Methods
Molecular structures
WT NRas initial structure was extracted from the Brookhaven Protein Data Bank (PDB) entry 3CON (39) of p21N-ras in complex with GDP. This structure was then carefully aligned to HRas-GTP (PDB: 1QRA (21)) using PyMOL (40) to get the GTP coordinates together with two water molecules positioned in the vicinity of its γ-phosphate group. The magnesium cation from 1QRA, with two coordinated water molecules, was also included in the starting structure. Residues 61–71, which are missing in 3CON but conserved in NRas and HRas, were also extracted from 1QRA to complete the 3CON structure. To overcome a possible rearrangement of the protein due to the difference in GDP/GTP binding, the resulting system was energy minimized using the AMBER package (41) and a free molecular dynamic simulation was subsequently run in the Born implicit solvent model (42) for 1 ns, with a 1 fs time step, at a temperature of 300 K. This structure was subsequently aligned to the Ras-GAP complex (PDB: 1WQ1 (19)) to position GAP with respect to NRas. The alignment was done using PyMOL and the “align” command, which performs a sequence alignment and a structural superposition of the α-carbon atoms. 152 atoms were also used for the alignment (126 after refinement) with a final root mean square (RMS) equal to 0.527 Å. In our calculations, only residues 780–795 of GAP, corresponding to the so-called “arginine finger loop” that binds Ras, were included. This sequence contains the Arg 789 residue, the electrostatic effects of which have been reported to be crucial (19, 23, 26, 27, 28, 29, 35)—aside from its steric effects—for enhancing the rate of GTP hydrolysis reaction (3, 4) and that we consequently need to account for. Six oncogenic mutant structures were prepared from the WT structure, substituting Gln 61 with arginine (Q61R), lysine (Q61K), leucine (Q61L), histidine (Q61H), proline (Q61P), and glutamic acid (Q61E) using the PyMOL mutagenesis command (40). Indeed, a previous x-ray structure analysis (13) concluded that the overall structural changes between the WT and oncogenic mutant proteins are very small. The only consistent changes appear in loops L2 (residues 30–38) and L4 (residues 58–66), which have been reported to be very mobile by experimental studies (12, 43, 44). Each mutant structure was first minimized before adding the solvent, using the AMBER package (41).
QM/MM molecular dynamics simulations
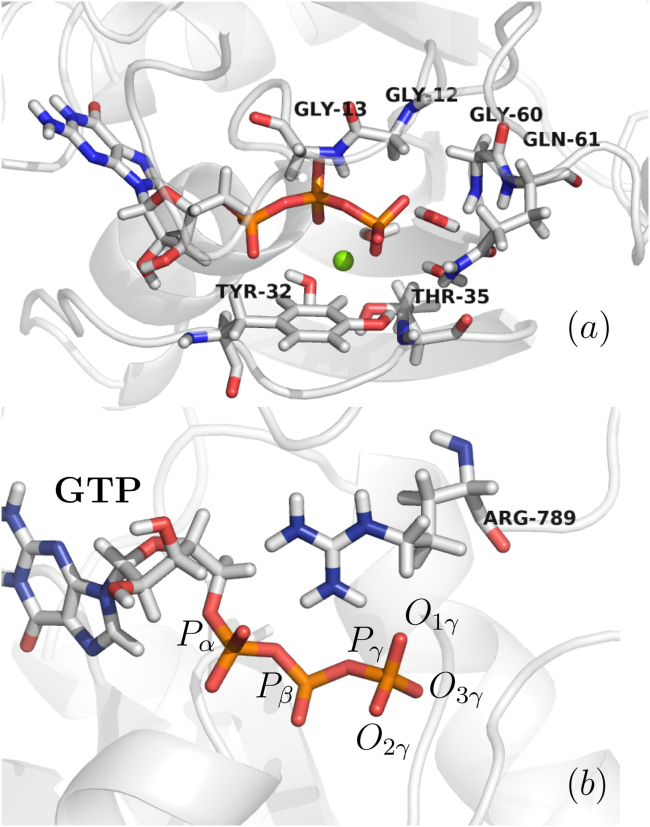
All QM/MM molecular dynamics simulations were carried out using the AMBER package (45). Each molecular system was partitioned into a QM and a MM region, the QM region consisting of critical residues 12–13, 32, 35, and 60–61 from Ras, Arg 789 from GAP, GTP, and a Mg2+ ion with two coordinated water molecules, found in the crystallographic structures of Ras (12, 21, 39, 46) (see Fig. 1). Water molecules found within 5 Å of the GTP Pγ atom (introduced from crystallographic data or from the explicit solvation model, as described below) were also treated quantum mechanically during the simulation. Because of diffusion, their MM or QM identity was updated when they exited the active site and when new water molecules reached the QM region.
Figure 1.
View of the quantum region consisting of (a) residues 12–13, 32, 35, and 60–61 from Ras, GTP, and Mg2+ ion with two coordinated water molecules and (b) GAP Arg 789 and GTP. To see this figure in color, go online.
The ab initio treatment of several residues from the active site leads to the introduction of 10 link atoms, one per QM-MM bond that was sectioned to define the QM region boundaries. These atoms were added following the recommended settings in the AMBER molecular dynamics package.
The semiempirical parameterized model number 3 Hamiltonian (47) and the classical ff99bsc0 force field (48) were used to treat the QM and the MM regions, respectively. This quantum potential is sufficient to observe the structural rearrangement of the protein complex active site upon Gln 61 substitution. Its approximation consists in using the core approximation and neglecting all integrals involving differential overlap (except for the one-electron core resonance and the one-center exchange integrals). The electron repulsion integrals are subsequently determined by empirical parameters so that the potential fits experimental heats of formation, dipole moments, ionization potentials, and geometries. Na+ ions were added to neutralize the total charge of the system, which was solvated in a periodic box of transferable intermolecular potential with 3 points (49) water extending up to a minimal cutoff of 8 Å from the protein boundary. To get rid of possible vacuum bubbles created during the solvation of the protein complex, 10,000 cycles of minimization were performed with a 500 kcal ⋅ mol−1 ⋅ Å−2 restraint applied to the solute. QM/MM minimization runs were subsequently carried out to remove bad contacts and relax the structure, applying a 5 kcal ⋅ mol−1 ⋅ Å−2 restraint on residues 780 and 795 from GAP only to avoid its undocking. As many runs were performed as needed to reach values of 10−2 Å for the RMS. The entire system was then gently heated from 0 to 300 K using the Langevin thermostat with a collision frequency of 2 ps, at constant volume, with weak restraints of 5 kcal ⋅ mol−1 ⋅ Å−2 applied to the entire protein-ligand complex.
Finally, the production setup was implemented in the constant number of particles, constant pressure, and constant temperature ensemble under periodic boundary conditions with 1 atm target pressure and using the Berendsen barostat with a pressure relaxation time of 1 ps. Long-range electrostatic interactions were treated using the particle mesh Ewald method (45). Covalent bonds involving hydrogen atoms from the MM region were constrained using the shake algorithm (50) so that a 1 fs step could be used. We consider that the first 100 ps of this production phase account for the equilibration phase, as the RMSD plots of the backbone atoms do not present a significant drift after this simulation time (each panel (f) of Figs. S1–S7). As no prior MM sampling was performed in explicit solvent, the simulation sampling can be considered as limited. However, the simulation includes the electrostatic coupling between the protein active site and water molecules, which is fundamental for addressing water distribution. The production was run for 1 ns by portions of 50 ps because of the lack of an automated procedure to quantum mechanically treat water molecules within 5 Å of the GTP Pγ atom. Indeed, water molecules initially present in the active site are free to move outside but can be replaced by water molecules from the bulk solvent. It is important to note that, for the first time to our knowledge, transient water molecules are treated quantum mechanically in the NRas active site in an adaptative way. We point out that, despite being completely unconstrained, both water molecules from the crystallographic structures coordinated to the Mg2+ ion remain in their initial positions during the entire simulation in accordance with previous theoretical studies (28).
WT NRas and its six oncogenic mutants at position 61 (Q61E, Q61P, Q61H, Q61L, Q61K, and Q61R) were all treated following this simulation protocol. For each molecule, system properties (energy, density, temperature, pressure, volume, and backbone RMSD) were monitored to check the quality of the obtained trajectories. The corresponding plots are presented in Figs. S1–S7.
Results
The trajectories obtained from the QM/MM dynamics simulations were analyzed to evaluate the conformational changes of key residues (Gly 12, Gly 13, Tyr 32, Thr 35, and Arg 789) of the active site impacted by the different Gln 61 substitutions. The water presence probability density function was also determined from these trajectories to compare water distribution within the NRas active site.
Active site stability
To evaluate how Gln 61 substitutions impact the active site conformation and stability, we plotted the root mean-square deviation (RMSD) (51) of each NRas residue (Gly 12, Gly 13, Tyr 32, Thr 35, Gly 60, and 61) treated quantum mechanically during the simulations as well as the lifetime of the strongest native contact (51) of GAP Arg 789 residue. The implementation of both analysis methods is further described in the Supporting Materials and Methods.
WT p21N-ras
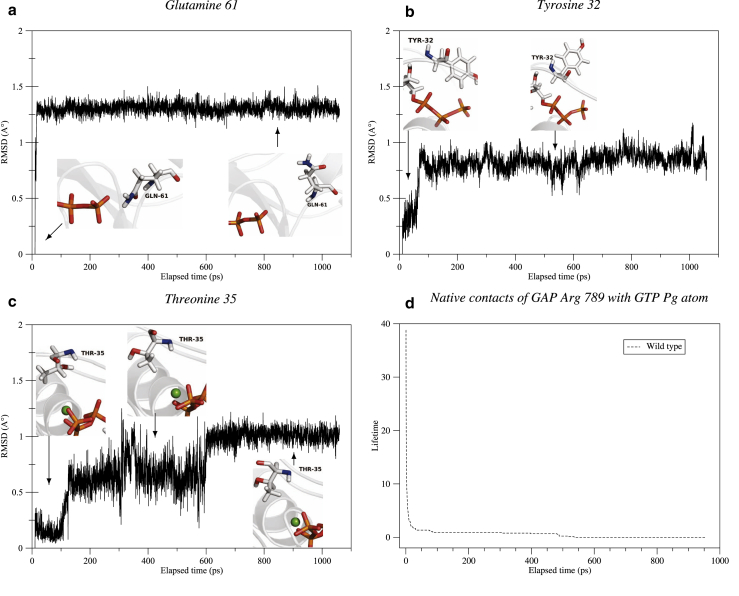
The RMSD plots of residues 12, 13, and 60 within the WT present very small fluctuations (∼0.1–0.2 Å (see Fig. S8)), indicating that these residues are stable in the active site of WT NRas. Residues 61, 32, and 35 from NRas adopt multiple conformations. Their RMSD plots are described in more detail in this section, as well as the behavior of the GAP arginine finger depicted by the lifetime curve of its stronger native contact.
Gln 61 fluctuations (Fig. 2 a) are also in the range ∼0.1–0.2 Å, although its RMSD presents an initial jump of ∼1.25 Å that occurs during the equilibration phase of the simulation. The Gln 61 side chain, initially bent toward the GTP accordingly to crystallographic structure 1WQ1 (19), rapidly unbends, getting outside of the active site, and remains in this state for the remaining simulation time as reported by previous dynamical studies (27, 28).
Figure 2.
RMSD plot alongside the associated conformational changes of Gln 61 (a), Tyr 32 (b), and Thr 35 (c) from WT p21N-ras and lifetime curve of native contacts between GTP Pγ atom and GAP Arg 789 residue (d) during QM/MM molecular dynamics. To see this figure in color, go online.
Tyr 32, which is part of the switch I region, is first closed with its side chain bent toward the ligand so that its hydroxy group engages hydrogen bonds with GTP Oγ atoms. Subsequently, it is found open as its side chain moves out of the active site. Both conformations are stable during the simulation and have already been identified in a previous NMR study (18). Indeed, Geyer et al. (18) showed that these two conformations of Tyr 32 induce a split resonance in the β-phosphate spectra resulting from at least two different chemical environments of this group due to the distant, i.e., open, conformation of Tyr 32 relative to the nucleotide and the closed one. During the simulated time and within the WT, the open one remains the longest (see Fig. 2 b), but it does not exclude that Tyr 32 side chain might get closed again, as another NMR spectroscopy study (44) showed that switch regions have internal motions in the nanosecond timescale.
The Thr 35 RMSD plot presents three main stages (Fig. 2 c). The transition between the first two steps is associated to a conformational change undergone by the side-chain hydroxy and methyl groups: the hydroxy group initially toward the inside of the active site moves outwards, and the methyl group initially outwards moves inwards.
When moving to the third stage, the Thr 35 side chain stretches further, its methyl group getting even closer to the Mg2+ ion. This jump in the RMSD plot also accounts for the backbone oxygen atom that was initially inwards and moves outwards the active site. The three configurations of this residue that also belongs to the switch I region are represented in the insets of Fig. 2 c.
Finally, because the conformational changes of the GAP Arg 789 side chain are poorly depicted by its RMSD plot, contrary to the previously described residues, we have represented instead the different interactions it can engage in. To this end, we have plotted the lifetime of its strongest native contact (51) inferred from the initial structure on a distance criterion (Fig. 2 d). This contact is made with the GTP Pγ atom, and although its lifetime decays during the simulation, at least one Arg 789 atom satisfies the selected criterion during the entire simulation. Moreover, other strong native contacts with GTP γ-phosphate-group atoms are present (O1γ and O2γ), and several non-native contacts develop during the simulation, notably with O3β and O3γ atoms. We point out that non-native contacts also arise with Gln 61 residue, in accordance with a time-resolved Fourier transform infrared spectroscopy study (24) that describes important interactions between the two, but these non-native contacts are not depicted here for the sake of clarity.
Q61 mutants
As for WT NRas, the RMSD plots of residues 12, 13, and 60 within Gln 61 mutants present small fluctuations (∼0.1–0.2 Å). They are not affected by the Gln 61 substitutions and remain stable in the active site of the oncogenic mutant proteins (Figs. S9–S14).
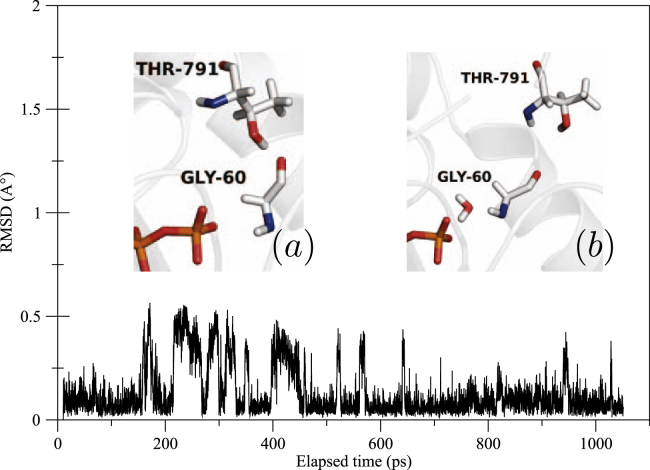
Q61R is an exception. Within this mutant, even though RMSD fluctuations of Gly 60 remain small (∼0.2 Å), two different stages can be identified (see Fig. 3). They correspond to two conformations adopted by the backbone oxygen atom. Initially pointing out of the active site, it turns inwards because of remarkably stable hydrogen bonds formed with residue Thr 791 of GAP as depicted in Fig. 3, inset (a). This hydrogen bonding cannot occur within the WT because of different orientations adopted by both residues that make impossible such interaction as shown in Fig. 3, inset (b).
Figure 3.
RMSD plot of Gly 60 within Q61R mutant during QM/MM molecular dynamics simulation. In the first inset, this residue is shown forming hydrogen bonds with GAP Thr 791 (a), compared to its conformation in WT NRas when forming hydrogen bonds with a transient water molecule (b). To see this figure in color, go online.
From a structural point of view in this study, which accounts for the arginine-binding loop of GAP only, Q61R mutation modifies the docking of this portion of GAP to NRas, which results not only in a difference of the network of interactions existing between both but also in a difference of interactions with water molecules in the active site of the NRas-GTP complex. Indeed, within WT NRas, Gly 60 mainly engages in hydrogen bonds through its backbone nitrogen atom with transient water molecules that get in the active site, whereas within Q61R, hydrogen bonds are formed with GTP Oγ atoms. Such differences are depicted in Fig. 3.
Q61E
Within the Q61E mutant, the RMSD plot of Glu 61 (Fig. S15 a) presents slightly greater fluctuations (∼0.2–0.3 Å) than those of Gln 61 within WT NRas (Fig. 2 a). Glu 61 is thus more mobile than Gln 61 within the WT. Furthermore, its side chain undergoes a conformation change that enables the carboxylate group to point toward the GTP γ-phosphate group. This state remains stable during our simulation, as shown in Fig. S15 a.
As described above for WT NRas, Tyr 32 is found in both closed and open conformations; however, within Q61E, the closed conformation is the one that remains the longest within the simulation time, as shown in Fig. S15 b.
In the Q61E NRas active site, Thr 35 is found in the two first conformations identified within the WT and mostly remains in the second one (see Fig. S15 c).
Arg 789 residue is initially engaged in native contacts with the GTP Pγ atom, as within the WT, because of the adopted procedure to obtain Q61E’s initial structure. However, these contacts are lost during the simulation, and none of them remain after 500 ps (Fig. S15 d). Moreover, from the beginning, fewer native contacts are made compared to the WT because several are already lost during the preceding QM/MM minimization cycles described in the Methods. Non-native contacts also develop during the simulation, mainly with Tyr 32 residue.
Q61P
In Q61P NRas, Pro 61 residue remains stable with small fluctuations around a unique conformation (∼0.1–0.2 Å) as shown in Fig. S16 a.
Tyr 32 residue switches from an initial closed configuration to an open configuration during the equilibration phase (see Fig. S16 b) and remains in this state during the simulation.
Thr 35 in Q61P NRas adopts the second and the third conformations identified within WT NRas, although in the third one, the backbone oxygen atom does not follow the conformational change as within the WT (Fig. S16 c).
Finally, the side chain of GAP Arg 789, initially engaged in native contacts with GTP Pγ atom, loses them all after 500 ps (Fig. S16 d). Non-native contacts develop mainly with Tyr 32 residue.
Q61H
Two main stages can be identified on the RMSD plot of the His 61 residue (Fig. S17 a), associated to two configurations of its side chain (bent and unbent toward the ligand), as depicted on the insets of Fig. S17 a. The corresponding RMSD fluctuations are very small (∼0.1–0.2 Å).
Tyr 32 remains in the open conformation during the simulation, although it was closed in the initial structure, as shown in Fig. S17 b.
The Thr 35 side chain adopts the second and third configurations previously identified within WT NRas although, in the third one, the backbone oxygen atom does not follow the conformational change (Fig. S17 c). It mainly remains in the second one (i.e., the side-chain hydroxy group pointing out of the protein active site and the methyl group pointing inwards).
Within Q61H mutant, the initial native contacts between Arg 789 and the GTP Pγ atom are rapidly lost: none of them remain after 150 ps (Fig. S17 d). As within Q61E and Q61P, non-native contacts appear with Tyr 32 residue.
Q61L
The RMSD plot alongside the associated conformational changes of Leu 61 residue (Fig. S18 a) presents small fluctuations (∼0.1–0.2 Å) around two main configurations adopted during our simulation. As shown in Fig. S18 a, the conformational change is achieved by a 90° rotation about the carbon 3-carbon 4 single bond.
The Tyr 32 side chain mainly remains, during the simulation time, in the open configuration as within WT NRas.
Thr 35 adopts the second conformation encountered within WT NRas (i.e., the side-chain hydroxy group pointing out of the protein active site and the methyl group pointing inwards), as well as a new orientation such that both hydroxy and methyl groups point out of the active site (see Fig. S18 c). This last conformation is the longer lasting within the simulation time.
Finally, the side chain of GAP Arg 789 rapidly loses the few initial native contacts with the GTP Pγ atom, and non-native contacts appear with Tyr 32 residue (see Fig. S18 d).
Q61K
The RMSD plot of Lys 61 presents larger fluctuations (Fig. S19 a) than in the case of Gln 61 within WT NRas (see Fig. 2 a). Q61K consequently appears to be more mobile. Indeed, its side chain undergoes a conformational change, as shown in Fig. S19 a, even though its amine group forms stable hydrogen bonds with GTP Oγ atoms during most of the simulation time.
During the same time, Tyr 32 remains in the closed configuration (Fig. S19 b).
Two main stages can be identified from the Thr 35 RMSD plot (see Fig. S19 c). The first one corresponds to the configuration encountered within Q61L mutant in which both the hydroxy and the methyl group of the side chain point out of the active site. The second one corresponds to the second configuration observed within the WT. Both are depicted in Fig. S19 c.
GAP Arg 789 maintains the initial native contacts made with the GTP Pγ atom almost as long as within the WT (Fig. S19 d). Non-native contacts are made with the Tyr 32 residue.
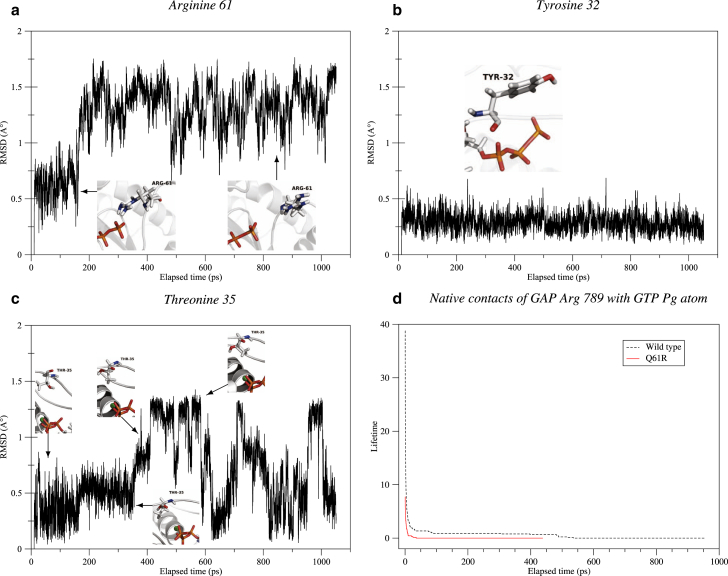
Q61R
The RMSD plot of Arg 61 within Q61R NRas (Fig. 4 a) presents significantly greater fluctuations (∼0.5–0.6 Å) than for WT Gln 61. This residue is actually found in two main configurations. In the first one, the side-chain amine groups engage hydrogen bonds with GTP Oγ atoms. In the second conformation, the Arg 61 side chain moves out from the active site. The open conformation lasts for longer during the simulation time. Both can be seen on Fig. 4 a.
Figure 4.
RMSD plots alongside the associated conformational changes of Arg 61 (a), Tyr 32 (b), and Thr 35 (c) from Q61R NRas and lifetime curve of native contacts between GTP Pγ atom and GAP Arg 789 (d) during QM/MM molecular dynamics. To see this figure in color, go online.
From the closed configuration in the initial structure, Tyr 32 switches to the open one during the first steps of our simulation, although the amplitude of the side chain motion is limited compared to the WT case (see Fig. 4 b).
Q61R appears to be the mutant in which Thr 35 is the least stable, as it continuously switches between the three main conformations identified within WT NRas and a fourth already identified within the Q61L mutant (see Fig. 4 c).
As with other Gln 61 mutants, Arg 789, initially involved in native contacts with the GTP Pγ atom, loses them totally after 500 ps (Fig. 4 d). Non-native contacts are subsequently identified with the Glu 31 residue.
From the previous comparisons between WT NRas and Gln 61 mutant proteins, it appears that Gln 61 substitution has a structural impact mainly on residues 32 and 35 from NRas, on residue 60 in the case of Q61R only, and residue 789 from the arginine-binding loop of GAP, although to assess the impact of Gln 61 mutants on the NRas-GAP complex, further calculations are needed. Depending on the mutant, the open or the closed conformation is mainly adopted by the Tyr 32 residue during the simulation time. Thr 35 adopts multiple conformations observed within both WT and mutant proteins but with different relative stabilities. For Q61R only, the interaction network of Gly 60 residue is modified. The arginine finger supplied by the portion of GAP considered in this study rapidly loses the initially present native contacts with the GTP Pγ atom within all Gln 61 substitutions except Q61K and forms non-native contacts with residue 61 in the WT case only.
Water occupancy within the active site
The structural changes described in the previous section brought into question the presence of solvent in the active site. Indeed, trajectory analysis revealed that the accessibility and remaining time of water molecules in the protein active site are different within WT NRas and Gln 61 mutants. To evaluate these differences, we computed the radial distribution function (RDF) of water molecules around GTP phosphorus atoms and the water distribution in the protein active site projected on a plane containing GTP Pβ and Pγ atoms.
RDF of water molecules
In this study, to evaluate the RDF of water molecules within the NRas active site, we implemented an algorithm allowing us to overcome certain limitations of available RDF codes. This custom-designed tool is consequently adapted to our study case and makes it possible to define optimal parameters like the accuracy but also 1) to account for molecules treated with different energy models according to the hybrid QM/MM approach used here, 2) to discard specific molecules (such as the two water molecules coordinated to the Mg2+ ion), and 3) to process all the trajectories in a single run. To validate this algorithm, it was tested on a trajectory accounting for the same molecules within the same threshold distance but without its specific commands and led to the same results as given by the RDF analysis tool provided by the Visual Molecular Dynamics package (52).
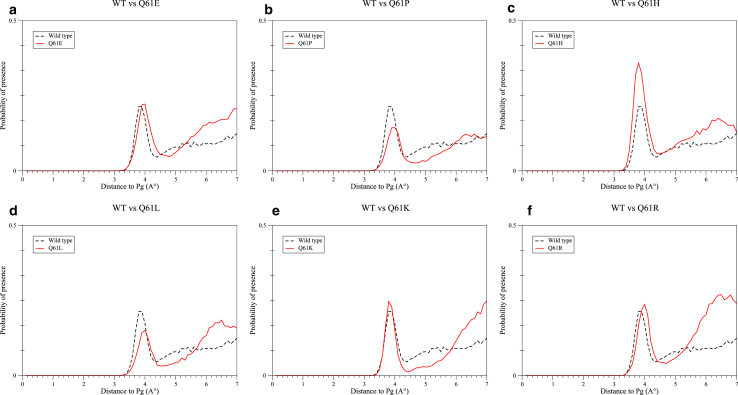
Fig. 5 represents the RDF of water molecules around the GTP Pγ atom for WT NRas and each Gln 61 mutant. For WT NRas, a first peak is found centered at ∼3.8 Å from Pγ, as reported in a previous molecular dynamics study of the Hras-GTP complex (37). The same peak is observed for the six studied mutants. It is shifted to ∼4 Å for Q61E, Q61P, Q61L, and Q61R and presents noticeable amplitude variations for Q61P, Q61H, and Q61L (0.14, 0.36, and 0.15, respectively, vs. 0.21 for WT NRas).
Figure 5.
RDF of water molecules around GTP Pγ atom within WT NRas versus (a) Q61E, (b) Q61P, (c) Q61H, (d) Q61L, (e) Q61K, and (f) Q61R mutants. To see this figure in color, go online.
At ∼6–7 Å from the GTP Pγ atom, the difference between the RDF of WT NRas and Gln 61 mutants is noticeable (Fig. 5). The probability of finding a water molecule in this area is lower within WT NRas than within any of the mutated forms. Indeed, for WT NRas, the RDF curve fluctuates around a density value of 0.08, whereas it reaches values of 0.20, 0.11, 0.17, 0.16, 0.24, and 0.25 for Q61E, Q61P, Q61H, Q61L, Q61K, and Q61R, respectively. Furthermore, the integral of the RDF curve in this region reveals the presence of 1.12 water molecules for WT NRas compared with 1.92, 1.24, 1.71, 1.80, 2.01, and 2.74 water molecules for Q61E, Q61P, Q61H, Q61L, Q61K, and Q61R, respectively.
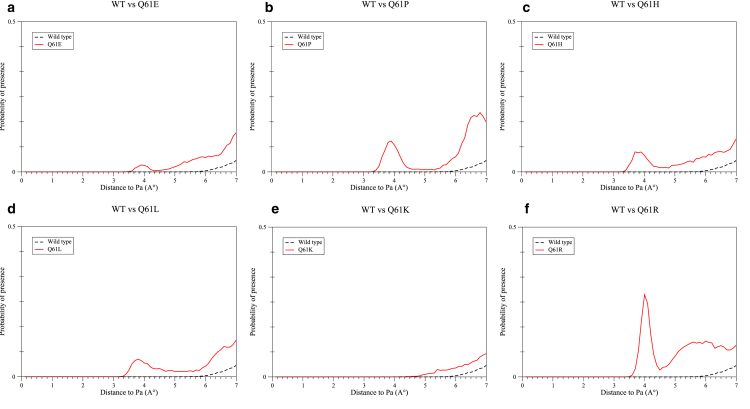
The RDF of water molecules around the GTP Pα atom was also calculated, showing a significant difference associated with a large amount of water found at ∼4 Å from this atom in the case of the Q61R mutant (see Fig. 6 f).
Figure 6.
RDF of water molecules around GTP Pα atom within the WT protein versus (a) Q61E, (b) Q61P, (c) Q61H, (d) Q61L, (e) Q61K, and (f) Q61R mutants. To see this figure in color, go online.
A similar peak is observed on the Pα RDF representations of Q61E, Q61P, Q61H, and Q61L (Fig. 6, a–d). This peak is shifted to a distance of ∼3.9 Å from the GTP Pα atom for Q61E and Q61P, ∼3.7 Å for Q61H, and ∼3.8 Å for Q61L, with significantly smaller amplitudes for these four mutants (i.e., 0.02, 0.10, 0.07, and 0.06 for Q61E, Q61P, Q61H, and Q61L, respectively, compared to 0.27 for Q61R). As for WT NRas, this peak does not appear on the RDF plot of Q61K. The integration of this curve leads to 0.76 water molecules in a 7 Å radius of the GTP Pα atom for Q61K and to 0.20 water molecules for WT NRas. Although both RDF curves have similar shapes, there are more water molecules in the vicinity of the GTP α-phosphorus atom in the active site of Q61K than within WT NRas.
Although the amount of water molecules is expected to be a crucial prerequisite for hydrolysis, these results suggest that their accurate positioning is even more important. To get more insight into the distribution of water molecules, we propose to precisely localize them in the NRas active site. To this end, we plotted the water probability density in two dimensions for WT NRas and Gln 61 mutants.
2D RDF of water molecules
To map and visualize the regions of the active site where water molecules tend to remain during the simulation, we implemented an algorithm that counts the number of water molecules within a distance of a given atom and projects them on a specific plane. More details are provided in the Supporting Materials and Methods.
In this study, a plane containing GTP Pβ and Pγ atoms was chosen as the projection plane (see Fig. S20). All water molecules found within 7 Å of the GTP Pγ atom were projected on this plane. The MG2+ ion, GTP phosphorus atoms, and nitrogen backbone atoms of several residues were also projected to see how water molecules are positioned in the active site with respect to these atoms. Nitrogen backbone atoms were selected because of their hydrogen bond donor/acceptor competence, which makes them critical in the establishment of a hydrogen-bond network within the protein active site. Results for WT NRas and Gln 61 mutants are presented in Figs. 7 and 8, respectively.
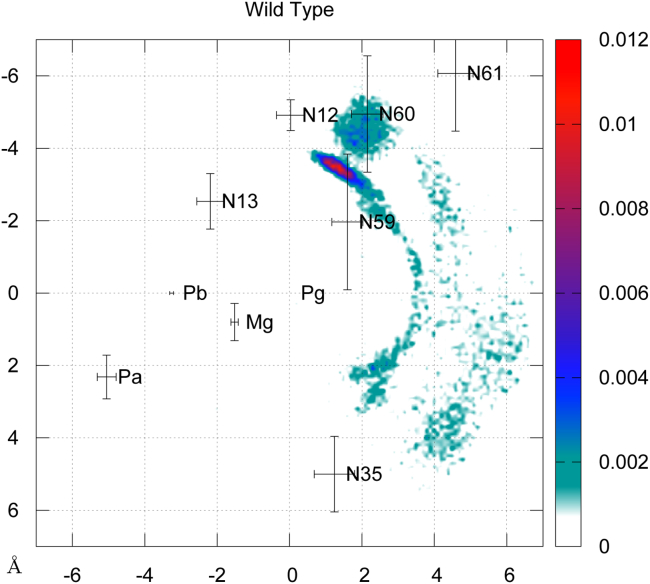
Figure 7.
2D RDF of water molecules within the active site of WT p21N-ras. The average coordinates of nitrogen backbone atoms from residues 12, 13, 35, 59, 60, and 61 have also been projected as well as the GTP phosphorus atoms and Mg2+ ion. The corresponding SD values are represented with error bars. To see this figure in color, go online.
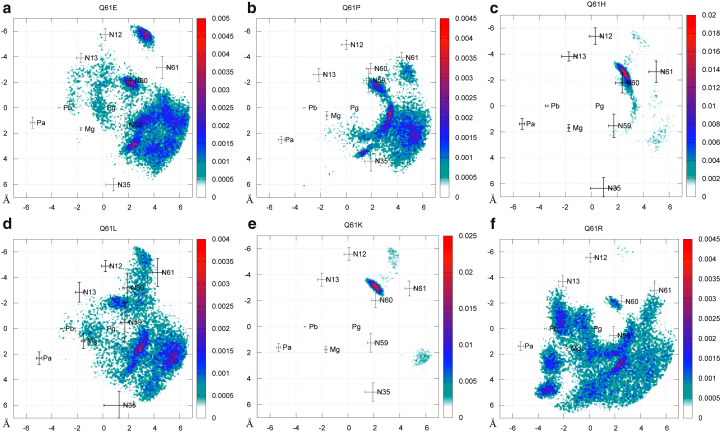
Figure 8.
2D RDF of water molecules within the active site of (a) Q61E, (b) Q61P, (c) Q61H, (d) Q61L, (e) Q61K, and (f) Q61R mutants. The average coordinates of nitrogen backbone atoms from residues 12, 13, 35, 59, 60, and 61 have also been projected as well as the GTP phosphorus atoms and Mg2+ ion. The corresponding SD values are represented with error bars. To see this figure in color, go online.
WT p21N-ras
For WT NRas, water molecules stay close to the GTP Pγ atom, forming an arch of density around the γ-phosphate group extended from residues 12 to 35 (see Fig. 7). The highest water density is localized in a small region between residues 12, 59, and 60. Indeed, although water molecules are unconstrained during the whole QM/MM simulation, we observed that they naturally tend to stay in this area because of stable hydrogen bonds formed with the nitrogen backbone atoms of Gly12 and Gly60 residues:
(donor) - - - N atom of Gly12 (acceptor)
N atom of Gly60 (donor) - - - (acceptor)
A second region of high water density is found in the vicinity of residue Thr 35 (only its nitrogen backbone atom N35 is represented), although the associated amplitude is significantly smaller. This region corresponds to the place in the protein active site where earlier studies of WT Ras (12, 16, 19, 21, 28) identified a presumed lytic water molecule. Finally, no water molecule is found in the vicinity of GTP Pα or Pβ atoms.
Gln 61 mutations
RDF around the GTP Pγ atom of WT NRas and Q61E is similar, with an excess of solvent found at ∼6–7 Å from this atom for Q61E (Fig. 5 a). However, the two-dimensional (2D) RDF plotted for this mutant (Fig. 8 a) shows that water molecules are delocalized within its active site compared to WT NRas. Three water density peaks arise between residues 35 and 59, next to residue 60, and between residues 12 and 61. The amplitude of each of these three peaks is one order of magnitude smaller than for the highest peak found within WT NRas. Water density is higher in other regions of the active site, in particular in the vicinity of the GTP Pβ atom.
Previously in this article, RDF plots have shown that Q61P and Q61L mutations lead to a lower probability of finding water molecules at ∼4 Å from the GTP Pγ atom than in WT NRas (Fig. 5, b and d), whereas a peak of density arises at the same distance from the Pα atom (Fig. 6, b and d). In the 2D RDF of Q61P, the arch of water molecules observed for WT NRas can also be observed, although it is scattered compared to the WT (see Fig. 8 b). The peak found between residues 12 and 60 is now shifted toward residue 59. Another peak of water density arises close to GTP Pγ atom, following the direction. Both present similar amplitudes, one order of magnitude smaller than the peak held by residues 12 and 60 within WT NRas, and again, water density is higher in other parts of the active site, in particular in the vicinity of the MG2+ ion.
For the Q61L mutant (Fig. 8 d), water positioning in the active site is delocalized compared to WT NRas. The peak of water density previously found, held by residues 12 and 60 within WT NRas, does not appear anymore, but two others are observed between residues 59 and 35. Their amplitude is one order of magnitude smaller than the highest density peak within WT NRas. We also note that water density is higher in other regions of the active site, in particular in the vicinity of residue 13 and the GTP Pβ atom.
For mutations Q61H and Q61K (Fig. 8, c and e), water distribution is more localized than within the active site of WT NRas. Indeed, within Q61H (Fig. 8 c), the peak of density held by residues 12 and 60 presents a higher amplitude than within WT NRas, like the RDF plot around the Pγ atom (Fig. 5 c). We also observe that the arch of water molecules together with the second highest peak of density placed in the vicinity of residue Thr 35 disappear.
Mutation Q61K also results in a higher amplitude for the density peak held by residues 12 and 60 and in the disappearance of the peak previously found next to residue 35 (Fig. 8 e).
Q61R is the mutation that leads to the most delocalized water distribution in the active site (Fig. 8 f). The peak held by residues 12 and 60 is not observed anymore, but according to the RDF plots, the probability of finding water molecules at 4 Å from Pγ atom is similar to that of WT NRas (Fig. 5 f). This is due to water molecules placed all around the γ-phosphate group (Fig. 8 f). The second peak of water density, found close to Thr 35, has a higher amplitude within Q61R than within WT NRas. High densities of water molecules are observed around α- and β-phosphate groups.
From the previous descriptions of water distribution in NRas active site, we conclude that the water molecule found next to the GTP Pγ atom, held by hydrogen bonds formed with residues 12 and 60, has an important role in the enzymatic catalysis of the GTP hydrolysis reaction. Indeed, water density is noticeably high in this region within the WT NRas active site but negligible within four of the six studied oncogenic mutants (Q61E, Q61P, Q61L, and Q61R). Nevertheless, further calculations are needed to elucidate its role and determine if it 1) reacts with the ligand, 2) participates in concerted proton transfers, or 3) has a structuring role without being chemically involved in hydrolysis. Our calculations show that residues 12 and 60 ensure its positioning by hydrogen bonds.
It also appears that although the density peak observed next to residue 35 presents an amplitude negligible compared to that of the peak held by residues 12 and 60 (Fig. 7), water presence in both regions of the active site is crucial for catalyzing the hydrolysis reaction. Indeed, this peak is absent from two of the six studied oncogenic mutants (Q61H and Q61K), showing that a single water density peak is not sufficient despite having a higher amplitude within these mutant proteins than within WT NRas. The presence of a second water molecule in the active site is thus necessary for an effective catalysis, in accordance with previous theoretical studies of Ras (21, 25, 28, 33, 34, 38). It does not imply that both water molecules participate chemically in hydrolysis; further calculations are needed to determine the role of the second water molecule in the catalysis of the reaction by the protein complex.
Concerning the presence of quantumly treated solvent in the vicinity of α- and β-phosphate groups within most of the oncogenic mutants, we suppose that water molecules in this region of the active site disturb the electronic density of the ligand, making the Oβ-Pγ bond breakage more difficult to achieve as reported in a study of the role of the Mg2+ ion (35), for which a change in its coordination sphere induced changes in the charge distribution of the GTP. Nevertheless, this assumption is beyond the scope of our study; further calculations are needed to understand the impact of water distribution on the electronic density of the GTP in the active site of the protein complex.
Discussion
In this section, we examine how the position of water molecules is related to the structural rearrangements of the active site under Gln 61 substitution. For more clarity, RMSD, RDF, and 2D RDF results are summarized in Table S1.
A non-negligible difference between WT NRas and Gln 61 mutants pertains to GAP Arg 789 native/non-native contacts determined from the initial structures based on a distance criterion. Within the WT, initial native contacts with the GTP Pγ atom remain during the entire simulation, whereas they are completely lost after a certain time within all the studied Gln 61 mutants except for Q61K, in which a native contact remains almost as long as for the WT. Moreover, WT NRas is the only system in which non-native contacts appear between GAP Arg 789 and Gln 61, whereas for all the studied mutant proteins, non-native contacts are formed with the Tyr 32 residue. Q61R, which develops non-native contacts with Glu 31, is an exception. It is therefore of interest to understand why in this study (which only accounts for the binding loop of GAP), Arg 789 native/non-native contacts are remarkably different for the WT and the mutated NRas, especially because previous experimental studies (8, 9) showed that such mutations render Ras insensitive to its activation by GAPs. Nevertheless, further calculations are needed, taking into account the entire GAP in the MM part, to assess how Gln 61 substitutions impact the interaction network of the so-called arginine finger.
In this study, which only considers a small portion of GAP, we presume that for Q61E, Q61P, Q61L, and Q61R, Arg 789 native contacts with the GTP Pγ atom are lost because of the delocalized positioning of water molecules, described in the 2D water distribution (Fig. 8, a, b, d, and f). The displaced quantumly treated solvent prevents the Arg 789 side chain from remaining in the vicinity of this atom because of steric hindrance. The electrostatic interactions between the arginine binding loop of GAP, NRas, and/or the ligand might also be damped by these water molecules.
For Q61H, which presents a more localized 2D water distribution than the WT, we presume that the higher water density near the GTP Pγ atom, depicted by the higher amplitude peak in both its RDF curve (Fig. 5 c) and its 2 RDF mapping (Fig. 8 c), is responsible for the vanishing of native contacts between Arg 789 and the GTP Pγ atom.
Concerning Q61K, which presents more localized water distribution than WT NRas (Fig. 8 e) and has the most similar RDF plots to those of the WT (Figs. 5 c and 6 c), native contacts between the arginine finger and the GTP Pγ atom can persist almost as long as within the WT.
The GAP Arg 789 non-native contact results for WT NRas and Gln 61 mutants shed light on possible explanations for why these NRas mutants hinder inactivation by GAPs. The chemical nature of the replacing residue might itself prevent the formation of non-native contacts between it and the GAP Arg residue. The active site structural reorganization induced by Gln 61 substitutions might well 1) prevent Arg 789 from adopting the position taken when interacting with residue 61 within the WT or 2) change the water molecules’ positioning so that they also prevent Arg 789 from adopting the position taken within WT NRas by steric hindrance and/or by damping the interactions between GAP and NRas. Nevertheless, further calculations, including the entire GAP protein, are needed to assess the impact of Gln 61 substitutions on NRas-GTP-GAP interactions.
Table S1 shows that the Q61R mutation destabilizes the most the active site. Indeed, although Thr 35 is able to adopt the conformations encountered within the WT, it is unstable within the active site of Q61R, as it constantly switches between these conformations. Such behavior might prevent this residue from interacting in a stable manner with a presumed lytic water molecule reported by earlier studies of WT Ras (12, 16, 19, 21, 28).
Q61R also results in a change of interactions between NRas and the arginine-binding loop of GAP, as Gly 60 interacts with Thr 791 from the considered portion of GAP through hydrogen bonds that cannot exist within the WT (inset of Fig. 3).
The 2D RDF plot of Q61R revealed the absence of the peak of water density placed next to the GTP Pγ atom held by residues 12 and 60 (Fig. 8 f). We infer that the lack of water molecules in this part of the active site is due to the previously described changes of the interactions in which Gly 60 is involved in the active site of the mutant. We deduce that within WT NRas, through hydrogen bonds formed with water molecules that get in the active site, Gly 60—a highly conserved residue—is indirectly involved in the hydrolysis reaction by maintaining water molecules in a specific region.
This inference is consistent with previous studies (26, 29) that concluded that Gln 61 within WT Ras has an indirect steric effect in the catalysis of the GTP hydrolysis reaction. Because this residue is coupled to residues from regions of NRas that play a major role in its function, its mutation destroys the preorganized catalytic configuration adopted by Ras upon binding with GAP. Thus, in our study, we conclude that Gln 61 takes action in an indirect manner in the hydrolysis reaction by enabling Gly 60 to correctly position water molecules.
Indeed, as shown in the active site stability results of WT NRas, the Gln 61 side chain gets out of the active site from the beginning of the simulation. Its carbonyl and amide groups do not interact with any of the ligand atoms and appear to be too far from the reactants to stabilize the transition state. Although Gln 61 interacts with transient water molecules, this interaction is taken over by the Gly 60 residue, which holds water molecules along with the Gly 12 residue, by hydrogen bonds formed through its nitrogen backbone atom. Moreover, in the 2D RDF plots, the projection of nitrogen backbone atoms clearly shows the rearrangement of the protein active site residues upon Gln 61 substitution. In the case of the Q61R mutation, the nitrogen backbone atom of Thr 35 does not appear, as it has moved further than the threshold we selected to study the active site. The corresponding SDs show the previously reported flexibility loss (13) of the protein backbone due to oncogenic mutations. The Q61R mutation, besides changing the solvent behavior in the protein complex active site because of its different chemical nature, disturbs the correct water molecule positioning in the active site by steric effects that displace Gly 60 such that it can no longer participate in maintaining water molecules in a specific side of the GTP γ-phosphate group.
Although the substitution of Gln 61 by a histidine (Q61H) or by a lysine (Q61K) results in a localized positioning of water molecules, which induces a peak of density maintained by residues 12 and 60 as within WT NRas (Fig. 8, c and e), the absence of the peak of water density in the vicinity of Thr 35 residue appears to impact the protein enzymatic action. This supports previous studies (21, 25, 28, 33, 34, 38) that considered the presence of a second water molecule in the active site for the catalysis of the hydrolysis reaction. Indeed, the double integral of the WT 2D RDF plot leads to a total amount of 1.5 and 3.4 water molecules present in the active site of the protein complex when taking into account water molecules within 5 and 7 Å of the GTP Pγ atom, respectively (discarding the two water molecules coordinated to the Mg2+ ion). As observed in Fig. 7, these water molecules do not accumulate in one region of the active site of WT NRas but are precisely positioned, forming an arch of water density between GTP γ-phosphate group extending from residues 12 to 35 of the active site. This observation does not imply that all the water molecules present in the active site of WT NRas participate chemically in the hydrolysis reaction. To elucidate their role, energy barriers have to be calculated for all the possible mechanisms of GTP hydrolysis within the NRas active site, taking the water molecules’ presence into account.
Conclusion
This study sheds light on the distribution of water molecules in the active site of WT NRas and Gln 61 mutants. These results can bring new perspectives on the role of water in the possible mechanisms involved in catalyzing the GTP hydrolysis within WT NRas, as well as explanations for the reaction rate drop within oncogenic mutant proteins.
We found that within the WT NRas active site, water molecules stay close to the GTP Pγ atom, forming an arch of density around the γ-phosphate group extended from residues 12 to 35 of the protein. The highest water density is localized between residues 12, 59, and 60, as water molecules are held in this region by hydrogen bonds formed with the nitrogen backbone atoms of Gly 12 and Gly 60 residues. A second peak of density is observed in the vicinity of the Thr 35 residue. The presence of two water density peaks supports the presence of an additional water molecule besides the lytic water molecule in the protein complex active site for an effective catalysis of the hydrolysis reaction.
Within NRas structures, all Gln 61 substitutions induce a structural rearrangement of the active site and modify the distribution of water molecules.
Residues Tyr 32, Thr 35, and Gly 60’s mobilities, stabilities, and interaction networks are particularly affected depending on the substitution. This is particularly true for Q61R, as Tyr 32 residue flexibility decreases, Thr 35 becomes unstable, and Gly 60 interacts with the GAP Thr 791 residue instead of forming hydrogen bonds with transient water molecules from the active site.
We also show that within Q61E, Q61P, Q61L, and Q61R, water molecule distribution is delocalized compared to the WT. This delocalization might prevent the GAP arginine finger from keeping its native contacts, inferred from the initial structures based on a distance criterion, with the GTP Pγ atom as well as forming non-native contacts with residue 61 by steric hindrance. It might also disturb GAP Arg 789’s crucial electrostatic effect.
The water distribution observed for Q61H and Q61K, much more localized than in the previous cases, results in the absence of the second peak of water density found in the vicinity of Thr 35 within WT NRas. This reinforces the need for the presence of a second water molecule for an effective enzymatic catalysis of the GTP hydrolysis reaction. It remains to be elucidated whether both water molecules participate in the chemical reaction or just one and hence to determine the role of the second water molecule.
Because Gly 60 residue participates in holding the highest water density peak by hydrogen bonds, we propose that this residue is involved in the catalysis of the GTP hydrolysis reaction by maintaining water molecules on a specific side of the GTP γ-phosphate group. The role of Gln 61 then appears to be consistent with an indirect steric effect in stabilizing the preorganized catalytic configuration of the active site. Indeed, it enables Gly 60 residue to correctly position water molecules.
Such insights about how Gln 61 substitutions impact the structure of the p21N-ras active site and on water reorganization around GTP appear to be crucial to evaluate favorable and unfavorable hydration sites for hydrolysis. These results are expected to shed light on dysfunctional NRas mutated forms. However, further calculations are needed to elucidate the catalytic mechanisms of GTP hydrolysis in the protein complex and correlate our predictions.
Acknowledgments
The authors thank CALMIP Supercomputer Center for central processing unit resources and support on project P1237.
Editor: Alan Grossfield.
Footnotes
Supporting Materials and Methods, twenty figures, and one table are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(18)30977-9.
Supporting Material
References
- 1.Bourne H.R., Sanders D.A., McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 2.Barbacid M. ras Genes. Annu. Rev. Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- 3.McCormick F. ras GTPase activating protein: signal transmitter and signal terminator. Cell. 1989;56:5–8. doi: 10.1016/0092-8674(89)90976-8. [DOI] [PubMed] [Google Scholar]
- 4.McCormick F. The world according to GAP. Oncogene. 1990;5:1281–1283. [PubMed] [Google Scholar]
- 5.Bos J.L. ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- 6.McGrath J.P., Capon D.J., Levinson A.D. Comparative biochemical properties of normal and activated human ras p21 protein. Nature. 1984;310:644–649. doi: 10.1038/310644a0. [DOI] [PubMed] [Google Scholar]
- 7.Sweet R.W., Yokoyama S., Gross M. The product of ras is a GTPase and the T24 oncogenic mutant is deficient in this activity. Nature. 1984;311:273–275. doi: 10.1038/311273a0. [DOI] [PubMed] [Google Scholar]
- 8.Trahey M., McCormick F. A cytoplasmic protein stimulates normal N-ras p21 GTPase, but does not affect oncogenic mutants. Science. 1987;238:542–545. doi: 10.1126/science.2821624. [DOI] [PubMed] [Google Scholar]
- 9.Adari H., Lowy D.R., McCormick F. Guanosine triphosphatase activating protein (GAP) interacts with the p21 ras effector binding domain. Science. 1988;240:518–521. doi: 10.1126/science.2833817. [DOI] [PubMed] [Google Scholar]
- 10.Pylayeva-Gupta Y., Grabocka E., Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat. Rev. Cancer. 2011;11:761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forbes S.A., Bhamra G., Stratton M.R. The catalogue of somatic mutations in cancer (COSMIC) Curr. Protoc. Hum. Genet. 2008;Chapter 10:Unit 10.11. doi: 10.1002/0471142905.hg1011s57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pai E.F., Krengel U., Wittinghofer A. Refined crystal structure of the triphosphate conformation of H-ras p21 at 1.35 A resolution: implications for the mechanism of GTP hydrolysis. EMBO J. 1990;9:2351–2359. doi: 10.1002/j.1460-2075.1990.tb07409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krengel U., Schlichting I., Wittinghofer A. Three-dimensional structures of H-ras p21 mutants: molecular basis for their inability to function as signal switch molecules. Cell. 1990;62:539–548. doi: 10.1016/0092-8674(90)90018-a. [DOI] [PubMed] [Google Scholar]
- 14.Privé G.G., Milburn M.V., Kim S.H. X-ray crystal structures of transforming p21 ras mutants suggest a transition-state stabilization mechanism for GTP hydrolysis. Proc. Natl. Acad. Sci. USA. 1992;89:3649–3653. doi: 10.1073/pnas.89.8.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frech M., Darden T.A., Wittinghofer A. Role of glutamine-61 in the hydrolysis of GTP by p21H-ras: an experimental and theoretical study. Biochemistry. 1994;33:3237–3244. doi: 10.1021/bi00177a014. [DOI] [PubMed] [Google Scholar]
- 16.Schweins T., Geyer M., Wittinghofer A. Substrate-assisted catalysis as a mechanism for GTP hydrolysis of p21ras and other GTP-binding proteins. Nat. Struct. Biol. 1995;2:36–44. doi: 10.1038/nsb0195-36. [DOI] [PubMed] [Google Scholar]
- 17.Schweins T., Warshel A. Mechanistic analysis of the observed linear free energy relationships in p21ras and related systems. Biochemistry. 1996;35:14232–14243. doi: 10.1021/bi961119g. [DOI] [PubMed] [Google Scholar]
- 18.Geyer M., Schweins T., Kalbitzer H.R. Conformational transitions in p21ras and in its complexes with the effector protein Raf-RBD and the GTPase activating protein GAP. Biochemistry. 1996;35:10308–10320. doi: 10.1021/bi952858k. [DOI] [PubMed] [Google Scholar]
- 19.Scheffzek K., Ahmadian M.R., Wittinghofer A. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277:333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- 20.Cepus V., Scheidig A.J., Gerwert K. Time-resolved FTIR studies of the GTPase reaction of H-ras p21 reveal a key role for the β-phosphate. Biochemistry. 1998;37:10263–10271. doi: 10.1021/bi973183j. [DOI] [PubMed] [Google Scholar]
- 21.Scheidig A.J., Burmester C., Goody R.S. The pre-hydrolysis state of p21(ras) in complex with GTP: new insights into the role of water molecules in the GTP hydrolysis reaction of ras-like proteins. Structure. 1999;7:1311–1324. doi: 10.1016/s0969-2126(00)80021-0. [DOI] [PubMed] [Google Scholar]
- 22.Du X., Frei H., Kim S.H. The mechanism of GTP hydrolysis by Ras probed by Fourier transform infrared spectroscopy. J. Biol. Chem. 2000;275:8492–8500. doi: 10.1074/jbc.275.12.8492. [DOI] [PubMed] [Google Scholar]
- 23.Allin C., Ahmadian M.R., Gerwert K. Monitoring the GAP catalyzed H-Ras GTPase reaction at atomic resolution in real time. Proc. Natl. Acad. Sci. USA. 2001;98:7754–7759. doi: 10.1073/pnas.131549798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kötting C., Kallenbach A., Gerwert K. The GAP arginine finger movement into the catalytic site of Ras increases the activation entropy. Proc. Natl. Acad. Sci. USA. 2008;105:6260–6265. doi: 10.1073/pnas.0712095105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langen R., Schweins T., Warshel A. On the mechanism of guanosine triphosphate hydrolysis in ras p21 proteins. Biochemistry. 1992;31:8691–8696. doi: 10.1021/bi00152a002. [DOI] [PubMed] [Google Scholar]
- 26.Glennon T.M., Villà J., Warshel A. How does GAP catalyze the GTPase reaction of Ras? A computer simulation study. Biochemistry. 2000;39:9641–9651. doi: 10.1021/bi000640e. [DOI] [PubMed] [Google Scholar]
- 27.Resat H., Straatsma T.P., Miller J.H. The arginine finger of RasGAP helps Gln-61 align the nucleophilic water in GAP-stimulated hydrolysis of GTP. Proc. Natl. Acad. Sci. USA. 2001;98:6033–6038. doi: 10.1073/pnas.091506998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soares T.A., Miller J.H., Straatsma T.P. Revisiting the structural flexibility of the complex p21(ras)-GTP: the catalytic conformation of the molecular switch II. Proteins. 2001;45:297–312. doi: 10.1002/prot.1150. [DOI] [PubMed] [Google Scholar]
- 29.Shurki A., Warshel A. Why does the ras switch “break” by oncogenic mutations? Proteins. 2004;55:1–10. doi: 10.1002/prot.20004. [DOI] [PubMed] [Google Scholar]
- 30.Topol I.A., Cachau R.E., Burt S.K. Quantum chemical modeling of the GTP hydrolysis by the RAS-GAP protein complex. Biochim. Biophys. Acta. 2004;1700:125–136. doi: 10.1016/j.bbapap.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 31.Friedman Z.Y., Devary Y. Dissection of the GTPase mechanism of Ras protein by MD analysis of Ras mutants. Proteins. 2005;59:528–533. doi: 10.1002/prot.20423. [DOI] [PubMed] [Google Scholar]
- 32.Grigorenko B.L., Nemukhin A.V., Burt S.K. QM/MM modeling the Ras-GAP catalyzed hydrolysis of guanosine triphosphate. Proteins. 2005;60:495–503. doi: 10.1002/prot.20472. [DOI] [PubMed] [Google Scholar]
- 33.Grigorenko B.L., Nemukhin A.V., Burt S.K. Mechanisms of guanosine triphosphate hydrolysis by Ras and Ras-GAP proteins as rationalized by ab initio QM/MM simulations. Proteins. 2007;66:456–466. doi: 10.1002/prot.21228. [DOI] [PubMed] [Google Scholar]
- 34.Martín-García F., Mendieta-Moreno J.I., Mendieta J. The role of Gln61 in HRas GTP hydrolysis: a quantum mechanics/molecular mechanics study. Biophys. J. 2012;102:152–157. doi: 10.1016/j.bpj.2011.11.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rudack T., Xia F., Gerwert K. The role of magnesium for geometry and charge in GTP hydrolysis, revealed by quantum mechanics/molecular mechanics simulations. Biophys. J. 2012;103:293–302. doi: 10.1016/j.bpj.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prasad B.R., Plotnikov N.V., Warshel A. Addressing open questions about phosphate hydrolysis pathways by careful free energy mapping. J. Phys. Chem. B. 2013;117:153–163. doi: 10.1021/jp309778n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miyakawa T., Morikawa R., Nagao H. Analysis of water molecules around the Hras-GTP complex and GDP complex by molecular dynamics simulations. Mol. Physics. 2013;112:526–532. [Google Scholar]
- 38.B R.P., Plotnikov N.V., Warshel A. Quantitative exploration of the molecular origin of the activation of GTPase. Proc. Natl. Acad. Sci. USA. 2013;110:20509–20514. doi: 10.1073/pnas.1319854110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nedyalkova, L., Y. Tong, …, H. Park. 2008. Crystal structure of the human NRas GTPase bound with GDP. Brookhaven Protein Data Bank entry 3CON. https://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?uid=3CON.
- 40.Schrödinger L.L.C. Schrödinger, LLC; New York: 2015. The PyMOL Molecular Graphics System, Version 1.8. [Google Scholar]
- 41.Case D.A., Cerutti D.S., Kollman P. University of California; San Francisco, CA: 2017. AMBER 2017. [Google Scholar]
- 42.Hawkins G.D., Cramer C.J., Truhlar D.G. Parametrized models of aqueous free energies of solvation based on pairwise descreening of solute atomic charges from a dielectric medium. J. Phys. Chem. 1996;100:19824–19839. [Google Scholar]
- 43.Milburn M.V., Tong L., Kim S.H. Molecular switch for signal transduction: structural differences between active and inactive forms of protooncogenic ras proteins. Science. 1990;247:939–945. doi: 10.1126/science.2406906. [DOI] [PubMed] [Google Scholar]
- 44.Kraulis P.J., Domaille P.J., Laue E.D. Solution structure and dynamics of ras p21.GDP determined by heteronuclear three- and four-dimensional NMR spectroscopy. Biochemistry. 1994;33:3515–3531. doi: 10.1021/bi00178a008. [DOI] [PubMed] [Google Scholar]
- 45.Walker R.C., Crowley M.F., Case D.A. The implementation of a fast and accurate QM/MM potential method in Amber. J. Comput. Chem. 2008;29:1019–1031. doi: 10.1002/jcc.20857. [DOI] [PubMed] [Google Scholar]
- 46.Johnson C.W., Reid D., Mattos C. The small GTPases K-Ras, N-Ras, and H-Ras have distinct biochemical properties determined by allosteric effects. J. Biol. Chem. 2017;292:12981–12993. doi: 10.1074/jbc.M117.778886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stewart J.J. Optimization of parameters for semiempirical methods I. Method. J. Comput. Chem. 1989;10:209–220. [Google Scholar]
- 48.Pérez A., Marchán I., Orozco M. Refinement of the AMBER force field for nucleic acids: improving the description of alpha/gamma conformers. Biophys. J. 2007;92:3817–3829. doi: 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Price D.J., Brooks 3rd C.L. A modified TIP3P water potential for simulation with Ewald summation. J. Chem. Phys. 2004;121:10096–10103. doi: 10.1063/1.1808117. [DOI] [PubMed] [Google Scholar]
- 50.Ryckaert J.-P., Ciccotti G., Berendsen H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Chem. Phys. 1977;23:327–341. [Google Scholar]
- 51.Roe D.R., Cheatham T.E., III PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 52.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.