Abstract
Hypothalamic expression of Kiss1 plays an essential role in the onset of puberty, gonadal development, and ovulation. Estrogens regulate the expression of Kiss1 in the hypothalamus through estrogen receptor-α. Kiss1 is also expressed in the ovary, where its expression correlates with the onset of puberty and progression of the estrous cycle. To date, estrogen regulation of Kiss1 expression in the ovary has not been investigated. We recently observed that gonadotropin-induced Kiss1 expression was absent in Esr2-null rat ovaries even though Esr1 was present. Wild-type granulosa cells abundantly expressed Kiss1 and oocytes expressed the Kiss1 receptor. We characterized estrogen receptor-β (ESR2) regulation of Kiss1 expression in granulosa cells by identifying granulosa cell–specific transcript variants and potential regulatory regions. The Kiss1 promoter, an upstream enhancer, and a downstream enhancer all possessed conserved estrogen response elements (EREs) and showed active histone marks in gonadotropin-stimulated granulosa cells. The transcriptionally active Kiss1 promoter, as well as the enhancers, also revealed enrichment for ESR2 binding. Furthermore, activity of a Kiss1 promoter construct was induced after overexpression of ESR2 and was blocked upon mutation of an ERE within the promoter. Finally, pregnant mare serum gonadotropin and human chorionic gonadotropin administration induced phosphorylation of ESR2 and upregulated the AP-1 proteins FOSL2 and JUNB in granulosa cells. Activated MAPK ERK2 was associated with the ESR2 phosphorylation in granulosa cells, and AP-1 factors could synergistically activate the Kiss1 promoter activity. These gonadotropin-induced changes paralleled Kiss1 expression in granulosa cells. We conclude that gonadotropin-stimulated Kiss1 expression in granulosa cells is dependent on both the activation of ESR2 and the upregulation of AP-1.
Kisspeptins are essential gatekeepers for the onset of puberty (1, 2) and critical regulators of ovulation (3–5). Kisspeptins bind to KISS1 receptor (KISS1R/GPR54) in the hypothalamus and within the hypothalamic-pituitary axis and confer the upstream signals to GnRH release that subsequently regulates gonadotropin secretion (1). The peripheral action of gonadotropins at the level of the gonads leads to estrogen production, and estrogens in turn regulate hypothalamic Kiss1 gene expression (6–8).
Kiss1 is expressed within the hypothalamic anteroventral periventricular nucleus (AVPV), arcuate nucleus (ARC), periventricular nucleus, and anterodorsal preoptic nucleus (9, 10). Studies have shown that Kiss1 expression is also widely detected throughout the central nervous system and other organs including ovary, placenta, testes, prostate, liver, lung, kidney, and pancreas (11–13). The potential roles of extrahypothalamic kisspeptins are diverse (14), but it remains unknown whether Kiss1 expression in extrahypothalamic tissues is also regulated by estrogen signaling.
In addition to the pivotal role of kisspeptins in hypothalamic control of reproductive function, a putative role of kisspeptin signaling in the direct control of ovarian function, including follicular development, oocyte maturation, steroidogenesis, and ovulation, has been suggested (4, 15–19). Expression of Kiss1 in the ovary shows changes with estrous cyclicity (15), and kisspeptin concentration in follicular fluid correlates with follicular as well as serum estrogen levels (18). It has been suggested that intrafollicular kisspeptins may play an important role in follicle maturation (4, 18). Recent studies have demonstrated that administration of exogenous kisspeptin-54 can induce egg maturation in women undergoing in vitro fertilization (20–22). Kisspeptins can also enhance the in vitro maturation of oocytes (23). Moreover, Kiss1r haploinsufficiency led to premature ovarian failure in mutant mice, which was not rescued by gonadotropin replacement, further suggesting an important role of kisspeptin signaling within the ovary (16). Kiss1r-mediated signaling has been reported as one of the factors contributing to activating PLC/IP3-dependent calcium mobilization, required for oocyte survival and maintenance of follicular integrity (17).
Several studies have investigated the molecular mechanisms regulating Kiss1 gene expression within hypothalamic nuclei (4, 24, 25). It has been reported that estrogen receptor-α (ESR1) but not estrogen receptor-β (ESR2) regulates the expression of Kiss1 in hypothalamic neurons (6, 8, 26). Previous studies have suggested that estrogens may play a role in regulating the expression of ovarian kisspeptins (15, 18). However, the potential regulatory mechanisms have not yet been investigated. We have recently observed that gonadotropin-induced Kiss1 gene expression in granulosa cells was absent in Esr2-null (Esr2−/−) rats (27), suggesting an essential regulatory role for ESR2. In the current study, we identified a granulosa cell–specific Kiss1 promoter and characterized the promoter regulation by ESR2 and the potential role of ESR2 in gonadotropin-induced Kiss1 expression in granulosa cells.
Materials and Methods
Animal models
All procedures were performed in accordance with the protocols approved by the University of Kansas Medical Center Animal Care and Use Committee. Wild-type and Esr2−/− Holtzman Sprague-Dawley rats were included in this study. Esr2−/− rats were generated by targeting exon 3 in the Esr2 gene, as described earlier (28). Deletion of exon 3 resulted in a frameshift and null mutation in the ESR2 coding sequence (28). All animals were screened for presence of the mutation by PCR using tail-tip DNA samples (RED Extract-N-Amp Tissue PCR Kit; Sigma-Aldrich, St. Louis, MO) and primers targeting the flanking intron sequences (28).
Collection of granulosa cells, oocytes, and thecal interstitial tissue
Four-week-old Esr2−/− and age-matched wild-type female rats were used to evaluate gonadotropin-induced follicular development and ovulation. Synchronized follicular growth was performed by intraperitoneal injection of 30 IU pregnant mare serum gonadotropin (PMSG; Lee Biosolutions, Maryland Heights, MO). Forty-eight hours after the PMSG injection, 30 IU of human chorionic gonadotropin (hCG; Lee Biosolutions) was injected intraperitoneally. Animals were euthanized before and after PMSG injections, as well as 4 and 10 hours after hCG injections, and ovaries were dissected and collected in M199 media (Hyclone; GE Healthcare Life Science, Pittsburgh, PA). After the oviducts, ovarian fat pad, and membranes were removed, ovaries were washed with M199 media and placed in a 35-mm noncoated sterile Petri dish containing the media with protease and phosphatase inhibitor cocktails (Sigma-Aldrich). Ovarian follicles were punctured under stereomicroscopic observation with 25-G needles fitted to 1 mL syringe and pressed to release the oocytes and the granulosa cells. Thecal interstitial tissues were separated, washed with M199 media in a microcentrifuge tube, and immediately frozen at −80°C until RNA or protein extraction. Cumulus granulosa cells were removed from the oocytes by repeated pipetting followed by repeated capillary suction under an inverted microscope. After the oocytes were removed in a microcentrifuge tube and stored at −80°C, remaining granulosa cells were passed through a 40-µm strainer (Fisher Scientific, Lenexa, KS) and collected by centrifugation. Washed granulosa cells were either frozen −80°C until RNA or protein extraction or processed for chromatin immunoprecipitation (ChIP) assays.
Detection of gene expression at mRNA level
Gene expression at the mRNA level was evaluated by conventional RT-PCR, quantitative RT-PCR (RT-qPCR), and RNA sequencing (RNA-seq) analyses. Total RNA was isolated from the granulosa cells with TRI Reagent (Sigma-Aldrich) and cDNAs were prepared from 2 µg of total RNA with oligo random primers and Superscript II reverse transcriptase (Thermo Fisher Scientific, Grand Island, NY). Conventional PCR amplification of cDNA was done in a 25 μL reaction volume by using DreamTaq Green DNA polymerase (Thermo Fisher Scientific). RT-qPCR amplification of cDNAs was carried out in a 20-μL reaction mixture containing Applied Biosystems Power SYBR Green PCR Master Mix (Thermo Fisher Scientific). Amplification and fluorescence detection of RT-qPCR were carried out on Applied Biosystems QuantStudio Flex 7 Real Time PCR System (Thermo Fisher Scientific). The ΔΔCT method was used for relative quantification of target mRNA normalized to 18S RNA. A list of quantitative PCR (qPCR) primer sequences is shown in Table 1.
Table 1.
Primers Used in qRT-PCR Studies
| Symbol | Reference mRNA | Forward Primer | Reverse Primer | Amplicon |
|---|---|---|---|---|
| Kiss1 | NM_181692.1 | 26F-TGCTGCTTCTCCTCTGTGTG | 174R-AGGCTTGCTCTCTGCATACC | 149 bp |
| Kiss1r | NM_001301151 | 502F-CTGGGAGACTTCATGTGCAA | 614R-GGGAACACAGTCACGTACCA | 113 bp |
| Esr1 | NM_012689.1 | 44F-TTCGAGCACATTCCTTCCTT | 235R-GCTTTGGTGTGAAGGGTCAT | 191 bp |
| Esr2 | NM_012754 | 1101F-GAAGCTGAACCACCCAATGT | 1310R-CAGTCCCACCATTAGCACCT | 209 bp |
| Erk2 | NM_053842 | 274F-GCCGCGCTACACTAATCTCT | 464R-TGCCGATGATGTTCTCATGT | 190 bp |
| Fosl2 | NM_001013146 | 107F-CTCCCGAAGAGGAGGAGAAG | 332R-GGACTGATTTTGCACACAGG | 226 bp |
| JunB | NM_021836 | 1148F-TGGAGGACAAGGTGAAGACA | 1436R-GCCAGAGTCCAGTGTGTGAG | 289 bp |
| Rn18s | NR_046237.1 | 1622F-GCAATTATTCCCCATGAACG | 1744R-GGCCTCACTAAACCATCCAA | 123 bp |
RNA-seq data were previously generated and analyzed (27, 29). Briefly, granulosa cells isolated 10 hours after hCG treatment of PMSG-primed immature rats were used for the RNA-seq analysis (27, 29). Granulosa cells were isolated as described above, and total RNA was purified with TRI Reagent (Sigma-Aldrich). We used 500 ng of total RNA for the RNA-seq library preparation. Libraries were prepared with the TruSeq standard mRNA kit (Illumina, San Diego, CA) according to the manufacturer’s instructions. The cDNA libraries were sequenced at the Molecular Biology Core Laboratory of Mayo Clinic (Rochester, MN). RNA-seq data were analyzed with the CLC Genomics Workbench (Qiagen Bioinformatics, Germantown, MD) to identify the differentially expressed genes. For this study, reads per kilobase million values were extracted from data deposited to Sequence Read Archive, under accession number SRX4060980-SRX4060980.
Identification of Kiss1 regulatory sequences in granulosa cells
The Kiss1 transcription start site in rat granulosa cells was detected by a modified 5′ rapid amplification of cDNA ends (RACE) PCR and sequencing of the PCR products (30, 31). Briefly, 2 μg total RNA from granulosa cells was reverse transcribed with a gene-specific primer located on the last exon of Kiss1 gene. The 3′ end of the first strand of cDNA was polyadenylated, and a second strand of cDNA was synthesized by template switching (TS) PCR with a TS oligo with poly dT sequences. Then the double-stranded cDNA products were PCR amplified with TS oligo and gene-specific primers and cloned into a TA cloning vector (Promega, Madison, WI) and sequenced (Genewiz, South Plainfield, NJ). This step was complemented by conventional RT-PCR with variant specific forward primers and a common reverse primer. All the primers used in RT, RACE-PCR, and RT-PCR processes are shown in Tables 2 and 3. After we determined the transcription start site, Kiss1 proximal promoter and distant enhancer sequences were compared with known promoter and enhancer sequences (25), and bioinformatic analyses (rVista 2.0, MULTITF) (32) were done to identify the potential binding sites for ESR2 and other transcriptional regulators.
Table 2.
Primers for 5′ RACE PCR to Determine the Kiss1 Transcription Start Site
| Exon | Chromosome Location | Forward Primer | Reverse Primer/Gene-Specific Primer | Application |
|---|---|---|---|---|
| Exon 3 | Ch13: 50529847 | Not applicable | 3R-CTTTTGCCAGGCATTAACGA | First strand cDNA synthesis |
| TSS | Not applicable | QT.Fd-AGCAGTGGTATCAACGCAGAA-TTTTTTTTTTTTTTTTTT | Not applicable | Second strand/5′ cDNA end synthesis |
| TSS-Exon 3 | Not applicable; Ch13: 50529847 | QO.Fd-AGCAGTGGTATCAACGCAGAA | 3R-CTTTTGCCAGGCATTAACGA | 5′ cDNA end amplification |
Abbreviations: Ch, chromosome; TSS, transcription start site.
Table 3.
Primers for Conventional RT-PCR to Detect Kiss1 Transcript Variants
| Exon | Chromosome Location | Forward Primer | Reverse Primer | Amplicon |
|---|---|---|---|---|
| Exon 2-3 | Ch13: 50529847-50532103 | 2F-ATGATCTCGCTGGCTTCTTG | 3R-CTTTTGCCAGGCATTAACGA | 147 bp |
| Exon 1-3 | Ch13: 50529847-50535078 | 1F-CTCAGTGTGCTCCAACTACC | 3R-CTTTTGCCAGGCATTAACGA | 322 bp |
| 247 bp |
Abbreviation: Ch, chromosome.
Transcriptionally active histone marks within the Kiss1 locus
Kiss1 promoter and enhancer sites were screened for active histone marks of H3K27ac and H3K4Me3 enrichment by ChIP. ChIP assays were performed on granulosa cells isolated from wild-type rat ovaries 48 hours after PMSG followed by 4 hours after hCG administration. Briefly, granulosa cells were isolated from ovarian follicles as described above and fixed with 0.75% formaldehyde for 10 minutes at room temperature and quenched with 0.125 M glycine for 5 minutes. Fixed cells were washed twice with cold PBS containing 0.5% IGEPAL CA-630 and 0.5% Triton X-100 and resuspended in cold Lysis Buffer (5 mM PIPES pH 8.0, 85 mM KCl, 0.5% NP-40; catalog no. sc-45000; Santa Cruz Biotechnology, Dallas, TX) in the presence of phenylmethylsulfonyl fluoride and protease inhibitor cocktail (Sigma-Aldrich) and kept on ice for 15 minutes. Crude nuclear extracts were collected by microcentrifugation at 4°C (2000g) for 5 minutes and resuspended in Lysis Buffer High Salt (1XPBS; 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS; catalog no. sc-45001; Santa Cruz Biotechnology) in the presence of phenylmethylsulfonyl fluoride and protease inhibitor cocktail (Sigma-Aldrich) and sonicated for 60 cycles (20 seconds on, 59 seconds off) at 70% amplitude to produce an average fragment size range of 300 to 600 bp. Immunoprecipitation was performed with 5 µg antibody [anti-H3K27ac , catalog no. 8173, RRID: AB_10949503 (33); anti-H3K4me3, catalog no. 9751, RRID:AB_2616028; (34); and normal rabbit IgG, catalog no. 2729, RRID:AB_1031062 (35) all from Cell Signaling Technology, Danvers, MA] conjugated to 20 µL protein A/G magnetic beads (Dynabeads; Thermo Fisher Scientific) overnight. Bead-chromatin complexes were washed with Low Salt Buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.1, 150 mM NaCl), High Salt Buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.1, 500 mM NaCl), LiCl Buffer (0.25 M LiCl, 1% IGEPAL, 1% deoxycholic acid, 1 mM EDTA, 10 mM Tris-HCl, pH 8.1), and TE Buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0), with each wash performed twice for 5 minutes. Cell lysis, sonication, immunoprecipitation, and cleanup steps were all performed at 4°C. Finally, chromatin DNA was eluted from the magnetic beads with 200 µL of elution buffer (1% SDS, 0.1 M NaHCO3), treated with RNase and proteinase K, and protein-DNA crosslinks were reversed with the addition of 12 µL 5M NaCl and heating in a shaker incubator overnight and purified with Qiaquick columns (Qiagen). DNA was eluted in 100 µL of 10 mM Tris-HCl pH 8.0, and 2.5 µL aliquots were used in qPCR analyses. qPCR primers for the target sites are shown in Table 4.
Table 4.
Primers for ChIP-qPCR for 5′ Enhancer, Promoter, and 3′ Enhancer Sites in Rat Kiss1 Gene
| Target | Chromosome Location | Forward Primer | Reverse Primer | Amplicon |
|---|---|---|---|---|
| 5′ Enhancer | Ch13: 50552612-50552768 | GCGAAAGGCTCATACACACA | CAGTCACCTGCCCAGATTTC | 157 bp |
| Promoter site 1 | Ch13: 50535655-50536033 | CCTCCACCAGAATTGACTCC | CCCCCTTCCTAACAACTGCT | 172 bp |
| Promoter site 2 | Ch13: 50535275-50535440 | CCATCCAAGGCTGTTCTCAT | TTGGCAGAGAAAGGCAAGTC | 166 bp |
| 3′ Enhancer | Ch13: 50526910-50527070 | TCAGGTTGTGCTTGCTTACG | GTGCTTCCTCCTTTGCTGAC | 161 bp |
Abbreviation: Ch, chromosome.
ESR2 regulation of Kiss1 promoter activity
The regulatory role of ESR2 on Kiss1 promoter was determined by its binding to Kiss1 promoter and enhancer sequences and by activation of a Kiss1 promoter-reporter construct. ESR2 binding to Kiss1 promoter or enhancer sequences was detected by ChIP assays (as described above), whereas regulation of promoter activity by ESR2 was assessed by promoter-reporter assays. Reporter assays were performed in a spontaneously immortalized rat granulosa cell line (SIGC) obtained from Prof. Robert C. Burghardt (Texas A&M University, College Station, TX) (36). Wild type or DNA-binding domain deleted (DBD−/−) Esr2 coding sequences (28) subcloned into a lentiviral vector pLV-EF1a-IRESneo (37) were stably transduced into SIGC cells to assess transcriptional activation potential of Kiss1 promoter by ESR2. A 1.5-kbp or 0.4-kbp fragment of the rat Kiss1 promoter was cloned into the Kpn I and Xho I sites of pGL4.25 firefly luciferase vector (Promega). PCR primers for subcloning the Kiss1 promoter into pGL4.25 vector are listed in Table 5. Potential ESR2 binding sites [estrogen response element (ERE)] in the Kiss1 promoter were identified with rVista 2.0 MULTITF (32). A putative ERE site (AGGTCAgatTGGCCT) within the 1.5-kbp Kiss1 promoter was mutated (ATTAGTgatATGAAT) with Q5 Site-Directed Mutagenesis Kit (New England Biolabs, Ipswich, MA). Oligonucleotides used for mutagenesis of the promoter are listed in Table 6. For the promoter-reporter assays, 100 ng of the promoter-reporter vectors were transfected into SIGC cells expressing wild-type ESR2, mutant ESR2, or vector control with Lipofectamine 2000 (Thermo Fisher Scientific). A Renilla vector (pGL4.74[hRluc/TK]) was used as an internal control. Twenty-four hours after transfection, cells were stimulated with 10 nM 17β estradiol for an additional 24 hours. Standard dual-luciferase assays were performed on the cell lysates, with dual-luciferase reporter assay reagents (Promega).
Table 5.
PCR Primers for Cloning the Kiss1 Promotor Into pGL4.25 Reporter Vector
| Target | Chromosome Location | Forward Primer | Reverse Primer | Amplicon |
|---|---|---|---|---|
| −0.4 kbp Kiss1 promoter | Ch13: 50535090-50535438 | ggtaccGGCAGAGAAAGGCAAGTCAT | ctcgagGCTCCTGGGCTTCCTCTAT | 348 bp |
| −1.5 kbp Kiss1 promoter | Ch13: 50535090-50536569 | ggtaccAGCAACCTTGAGGGAAGAA | ctcgagGCTCCTGGGCTTCCTCTAT | 1479 bp |
Sequences in lowercase letters indicate restriction enzyme sites for cloning.
Abbreviation: Ch, chromosome.
Table 6.
PCR Primers for Mutating the ERE Site in Kiss1 Promoter
| Target | Chromosome Location | Forward Primer | Reverse Primer |
|---|---|---|---|
| Kiss1 promoter ERE | Ch13: 50535090-50536569 | ATatgaatCCACCAGAATTGACTCCTTG | CactaaTGGCTTCTTTCTCTCAGAGAGG |
Sequences in lowercase letters indicate mutant ERE sites.
Abbreviation: Ch, chromosome.
Assessment of hCG/LH/choriogonadotropin receptor signaling in Kiss1 regulation
The link between hCG/LH/choriogonadotropin receptor (LHCGR) stimulation and ESR2-signaling with transcriptional regulation of Kiss1 was studied by assessing changes in MAPK phosphorylation and expression of AP-1 factors. We also investigated the phosphorylation of ESR2 after hCG administration. Expression of mRNAs was detected by RT-qPCR (as described above), whereas proteins were evaluated by Western blotting. For Western blotting, primary granulosa cell lysates were prepared in 1× SDS sample buffer (62.5 mM Tris-HCl pH 6.8, 2% SDS, 42 mM dithiothreitol, 10% glycerol, and 0.01% bromophenol blue; Cell Signaling Technology), sonicated to shear DNA and reduce viscosity and then heat denatured. Proteins were separated on 4% to 20% SDS-PAGE and transferred to polyvinylidene difluoride membranes. Membranes were blocked with 5% milk and incubated with primary antibodies for 1 hour at room temperature. Membranes were subsequently incubated with the following primary antibodies at the appropriate dilution in blocking buffer: anti-pESR2 (catalog no. ab109122; Assay Biotechnology, Little Rock, AR; RRID: AB_10681401; 1:2000) (38), anti-ESR2 (catalog no. 05-824; EMD Millipore, Burlington, MA; RRID: AB_11212759; 1:5000) (39), anti-pERK2 (catalog no. 4370; Cell Signaling Technology; RRID: AB_2315112; 1:2000) (40), anti-ERK2 (catalog no. 4695; Cell Signaling Technology; RRID: AB_390779; 1:2000) (41), anti-JUNB (catalog no. sc-8051; Santa Cruz Biotechnology; RRID: AB_2130023; 1:1000) (42), anti-FOSL2 (catalog no. TA809661; Origene Technologies Inc., Rockville, MD; RRID: AB_2744660; 1:1000) (43), and anti-ACTB (catalog no. A5441; Sigma-Aldrich; RRID: AB_476692; 1:30,000) (44). ACTB was detected as the loading control. Membranes were washed, blocked, and incubated with peroxidase-conjugated anti-mouse, or anti-rabbit secondary antibodies (Jackson Immunoresearch, West Grove, PA) at a dilution of 1:25,000 to 50,000, and immunoreactivity signals were visualized with Luminata Crescendo Western HRP substrate (Millipore Sigma, Burlington, MA). Western blot signal intensities were quantified by ImageJ analyses (National Institutes of Health, Bethesda, MD; https://imagej.nih.gov/ij/). Phosphorylated estrogen receptor-β (pESR2) signals were normalized with ESR2, and phosphorylated ERK2 (pERK2) signals were normalized with ERK2. ACTB signals were used to normalize the signals of FOSL2 and JUNB. The signal intensities detected in the PMSG-primed hCG 4-hour group were presented as relative fold over the PMSG group.
Activated ERK2 interaction with ESR2
We further investigated whether activated ERK2 targets ESR2 in granulosa cells isolated from PMSG-primed hCG-treated rats. Activated ERK2 interaction with ESR2 was detected by coimmunoprecipitation of pERK1/2 in granulosa cell proteins. Granulosa cells collected from PMSG-hCG treated wild-type rat ovaries in the presence of protease and phosphatase inhibitor cocktails (Sigma-Aldrich) were lysed in nondenaturing buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4, and 1 µg/mL leupeptin) (cell lysis buffer; catalog no. 9803; Cell Signaling Technology) for preparation of protein samples. Cell lysates were briefly sonicated to shear genomic DNA. After sonication, the cell lysates were centrifuged (10,000g) at 4°C for 10 minutes, and the supernatants were collected for immunoprecipitation assays. Protein samples were precleared with normal rabbit IgG (catalog no. 2729; Cell Signaling Technology) and protein A magnetic beads (Dynabeads; Thermo Fisher Scientific) to prevent nonspecific binding. An aliquot of the lysates was preserved to use as input, and for each immunoprecipitation reaction, lysates prepared from granulosa cells collected from one animal equivalent in 1 mL of lysis solution was used. Immunoprecipitation was performed overnight with a pERK antibody (catalog no. 4370; Cell Signaling Technologies; 1:50 dilution) or equivalent amount of normal rabbit IgG at 4°C on a rotating device. Then 20 µL of protein A magnetic beads (Dynabeads; Thermo Fisher Scientific) was added to each reaction tube and incubated at 4°C on a rotating device for another 4 hours. The magnetic beads were pelleted out by placing in a magnetic separation rack, and the pellet was washed eight times with cell lysis buffer (catalog no. 9803; Cell Signaling Technologies). After the final wash, the pellets were suspended in 150 µL of 1XSDS sample buffer (catalog no. 7722; Cell Signaling Technology). Immunoprecipitation samples were boiled for 10 minutes and separated from magnetic beads, and Western blot analyses were performed with pERK, ESR2, and pESR2 antibodies as described above.
AP-1 activation of Kiss1 promoter
Potential AP-1 binding sites (AP-1 response element) in the Kiss1 promoter were identified with rVista 2.0 MULTITF (32). Enrichment of JUNB binding within the Kiss1 promoter locus was investigated by ChIP assays with a rabbit monoclonal antibody (catalog no. 3753; Cell Signaling Technology; RRID: AB_2130002; 1:50 dilution) (45). ChIP assays were performed in granulosa cells isolated from wild-type rats 4 hours after hCG treatment as described above. qPCR primers for the target sites are shown in Table 4.
Regulation of Kiss1 promoter activity by AP-1 factors (JUNB and FOSL2) was assessed by promoter-reporter assays. An empty pGL4.25 firefly luciferase vector (Promega) or the pGL4.25 vector containing a 1.5-kbp fragment of the rat Kiss1 promoter was used in the reporter assays. Reporter assays were performed in SIGC cells as described above. Briefly, 100 ng of the empty pGL4.25 firefly luciferase vector or the promoter-reporter vector was cotransfected into SIGC cells with 100 ng of empty vector, JUNB, FOSL2, or both JUNB and FOSL2 expression vectors with Lipofectamine 2000 (Thermo Fisher Scientific). A Renilla vector (pGL4.74[hRluc/TK]) was used as an internal control. Twenty-four hours after transfection, transfection media was replaced with fresh cell culture media and incubated for an additional 24 hours. Standard dual-luciferase assays were performed on the cell lysates, with dual-luciferase reporter assay reagents (Promega).
Statistical analysis
Each experimental group consisted of a minimum of six rats, and the procedures were repeated at least two times for reproducibility of the results. The experimental results are expressed as mean ± SE. The results were analyzed for one-way ANOVA, and the significance of mean differences was determined by Duncan post hoc test, with P ≤ 0.05 considered a significant level of difference. All statistical calculations were done with SPSS 22 (IBM, Armonk, NY).
Results
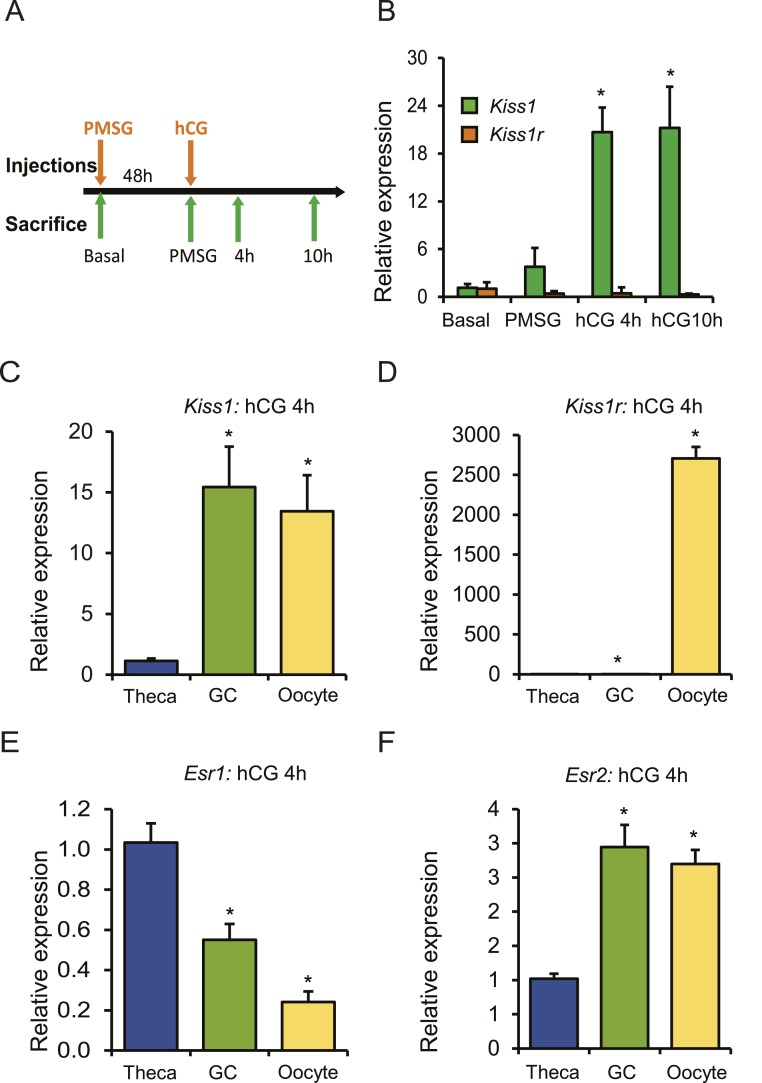
Gonadotropins stimulate Kiss1 expression in the rat ovary
Kiss1 gene expression was analyzed in ovary samples collected from prepubertal rats treated with exogenous gonadotropins (Fig. 1A). Gonadotropin treatment induced expression of Kiss1 in rat ovaries; however, expression Kiss1r remained unchanged (Fig. 1B). Kiss1 expression increased remarkably 4 hours after hCG injection and remained elevated at 10 hours after hCG (Fig. 1B). Expression of Kiss1 and Kss1r was further examined in theca and granulosa cells, as well as oocytes isolated from gonadotropin treated ovaries. Compared with theca cells, granulosa cells and oocytes expressed higher levels of Kiss1 mRNA (Fig. 1C). In contrast, expression of Kiss1r was detected predominantly in the oocytes (Fig. 1D). Interestingly, expression of Esr2 but not Esr1 correlated with Kiss1 mRNA levels in granulosa cells and oocytes (Fig. 1E and 1F).
Figure 1.
Expression of Kiss1 is induced by gonadotropin administration. (A) Four-week-old wild-type female rats were treated with gonadotropins, and the ovaries were collected at the indicated time points for RNA isolation and qRT-PCR analyses. (B) Kiss1 expression increased significantly 4 h after hCG injection and remained elevated at 10 h after hCG. However, the expression of Kiss1r did not show any changes after gonadotropin stimulation. (C) Ovaries were collected 4 h after hCG administration for the isolation of oocytes, granulosa cells, and thecal-interstitial tissues. qRT-PCR analyses showed that Kiss1 expression was significantly higher in granulosa cells and oocytes compared with theca cells. (D) But expression of Kiss1r was detected exclusively in the oocytes. (E) Expression of Esr1 was significantly higher in theca cells compared with granulosa cells or oocytes. (F) In contrast, expression of Esr2 was significantly higher in granulosa cells and oocytes coinciding with Kiss1 expression. qRT-PCR data are represented as mean ± SEM. n = 6. *P ≤ 0.05. GC, granulosa cell.
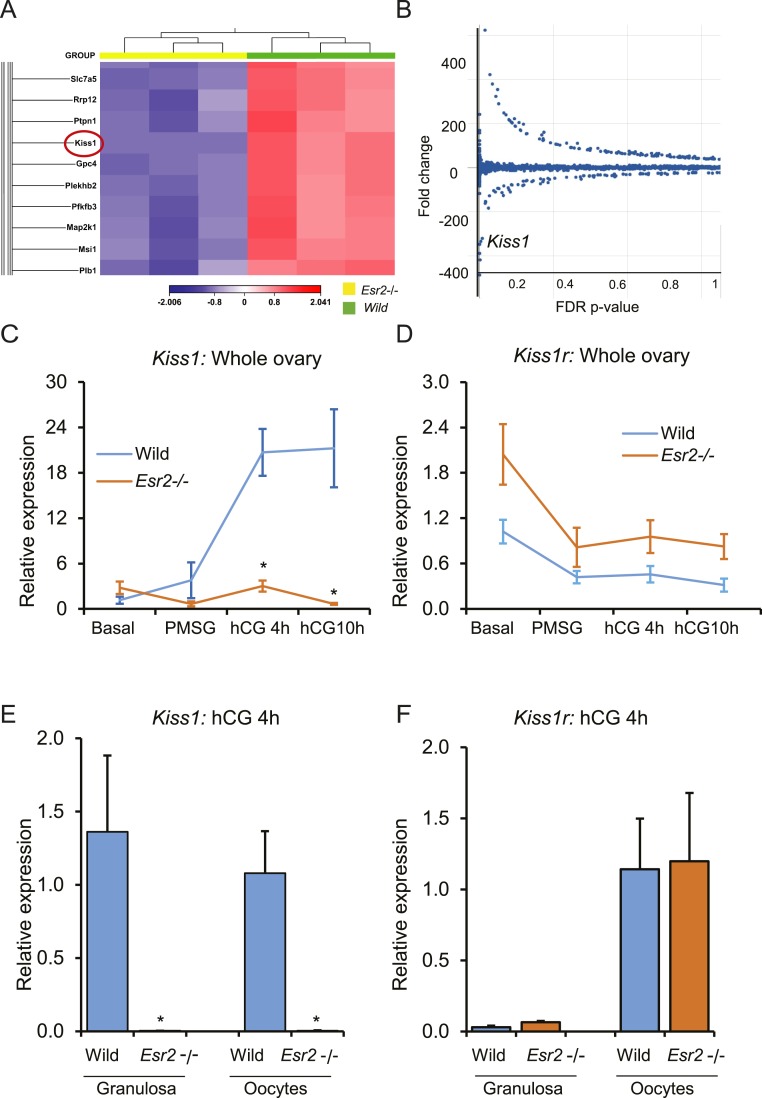
Gonadotropins failed to induce Kiss1 expression in the Esr2−/− rat ovary
RNA-seq results showed that expression of Kiss1 was highly downregulated (−260 fold, false discovery rate P = 0.0002) in gonadotropin-treated Esr2−/− granulosa cells from PMSG-hCG primed immature animals 10 hours after hCG (Fig. 2A and 2B). Unlike wild-type ovaries, gonadotropin treatment failed to induce Kiss1 expression in Esr2−/− ovaries (Fig. 2C). Conversely, expression of Kiss1r was slightly higher in the Esr2−/− ovaries before and after gonadotropin treatment; however, it was not statistically significant (Fig. 2D). Kiss1 expression was absent in Esr2−/− granulosa cells and oocytes (Fig. 2E) isolated 4 hours after hCG injection. However, expression of Kiss1r in granulosa cells or oocytes was comparable between the wild-type and Esr2−/− rats (Fig. 2F).
Figure 2.
ESR2 is essential for gonadotropin-induced Kiss1 expression in rat granulosa cells. (A, B) RNA-seq analyses identified Kiss1 as one of the highly downregulated genes [−260 fold, false discovery rate (FDR) P = 0.0002] in gonadotropin-induced rat granulosa cells. (C) Gonadotropin-induced upregulation of Kiss1 expression was absent in Esr2−/− rat ovaries. (D) Although Kiss1r expression in Esr2−/− ovaries appeared elevated, it was not statistically significant. (E) Induction of Kiss1 expression was absent in Esr2−/− granulosa cells and oocytes, but (F) Kiss1r expression was not affected in Esr2−/− granulosa cells or oocytes after gonadotropin stimulation. qRT-PCR data represent the mean ± SEM. n = 6. *P ≤ 0.05.
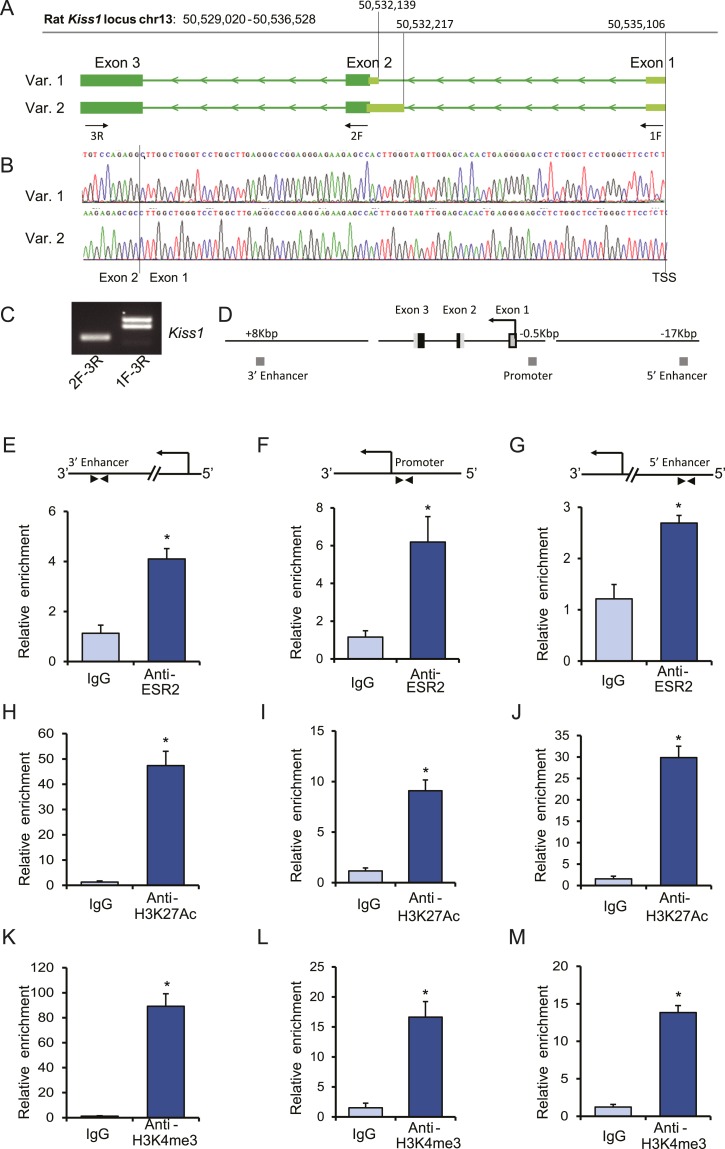
Identification of granulosa cell–specific Kiss1 promoter and enhancers
A granulosa cell–specific Kiss1 transcription start site was identified with a modified 5′ RACE PCR. Sequencing of the RACE PCR products showed that the transcription start site is identical to the previously reported Kiss1 transcription start site in kisspeptin neurons from the mouse (25) (Fig. 3A and 3B). Like murine kisspeptin neurons, rat granulosa cells expressed two transcript variants due to an alternative splicing of the second exon (Fig. 3B). RT-PCR using exon-specific primers further verified that granulosa cells express two transcript variants (Fig. 3C). It has previously been reported that the proximal promoter region and the distant 5′ and 3′ enhancers of the murine Kiss1 gene possess conserved ERE sites (25). To evaluate this within the rat gene, ChIP assays were performed on gonadotropin-stimulated (4 hours after hCG) rat granulosa cells with anti-ESR2, anti-H3K27ac, and anti-H2K4me3 rabbit monoclonal antibodies and normal rabbit IgG as a negative control. Both the proximal promoter and the enhancers showed marked enrichment for ESR2 binding (Fig. 3E–3G), which was associated with enriched histone marks of H3K27ac (Fig. 3H–3J) and H2K4me3 (Fig. 3K–3M). These findings suggest that ESR2 can bind to and regulate Kiss1 in rat granulosa cells.
Figure 3.
Assessment of chromatin activation marks and ESR2 binding to Kiss1 regulatory sequences in rat granulosa cells. (A) Schematic presentation of transcription start sites and transcript variants of Kiss1 expressed in rat granulosa cells, which are based on the (B) cDNA sequences obtained by a modified 5′ RACE PCR. (C) Conventional RT-PCR results also confirmed the expression of two transcript variants of Kiss1 in rat granulosa cells. ChIP assays were performed on gonadotropin-stimulated rat granulosa cells with anti-ESR2, anti-H3K27ac, and anti-H2K4me3 rabbit monoclonal antibodies. Normal rabbit IgG was used as a negative control. (D) Schematic representation of Kiss1 gene locus showing a 3′ enhancer region (~8 kbp downstream of the start site), promoter with an ERE site (~0.5 kbp upstream of the start site), and a 5′ enhancer region (~17 kbp upstream of Kiss1 start-site). ChIP-qPCR results demonstrated relative enrichment of (E–G) ESR2, (H–J) H3K27ac, and (K–M) H3K4me3 at the 3′ enhancer, promoter with ERE, and 5′ enhancer sites. ChIP-qPCR data are presented as mean ± SEM. n = 3. *P ≤ 0.05. chr, chromosome; TSS, transcription start site; Var, variant.
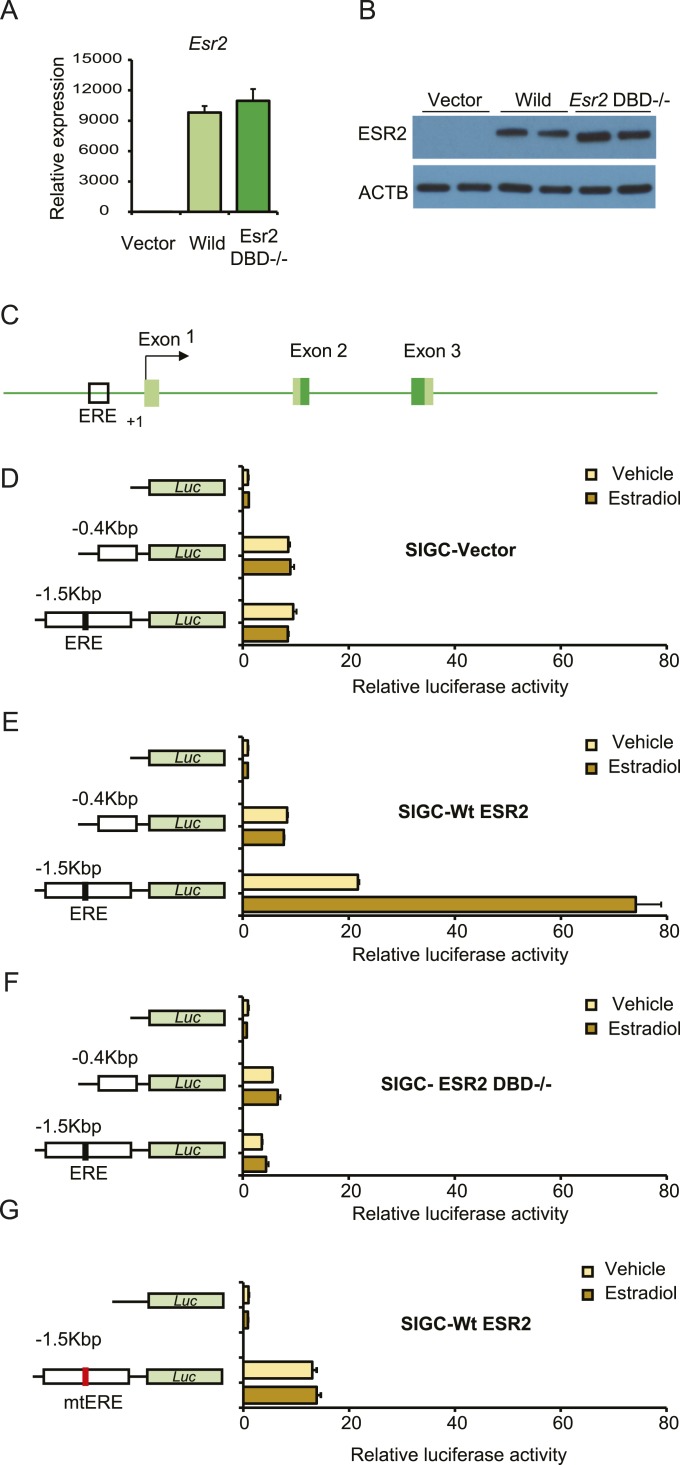
Kiss1 promoter regulation by ESR2 in granulosa cells
We further investigated the mechanisms that led to failure of Kiss1 expression in granulosa cells in absence of ESR2 expression. We identified that the Kiss1 proximal promoter possesses an ERE ~0.5 kbp upstream of the transcription start site and subsequently investigated its functional significance. An immortalized rat granulosa cell line (SIGC) was used for Kiss1 promoter-reporter assays. SIGC cells were stably transduced with lentiviral vectors expressing wild-type or DBD−/− rat ESR2 or a vector control (Fig. 4A and 4B). Kiss1 promoter activity was assessed after transfecting promoter-reporter constructs carrying different lengths of Kiss1 promoters (Fig. 4D–4G) and in the absence or presence of estradiol. Promoter constructs with (−1.5 kb) or without (−0.4 kb) ERE sites exhibited similar reporter activity in SIGC transfected with an empty pLV-EF1a-IRES-Neo vector (Fig. 4D) and were unresponsive to estradiol. Activity of the −1.5 Kb Kiss1 promoter harboring an ERE site was markedly upregulated in SIGC cells expressing wild-type ESR2 and treated with estradiol (Fig. 4E) but not in cells expressing a DBD−/− ESR2 (Fig. 4F). Importantly, a promoter construct with a mutant ERE sequence failed to exhibit any such promoter induction in SIGC cells expressing the wild-type ESR2 (Fig. 4G).
Figure 4.
ESR2 regulation of Kiss1 promoter activity in rat granulosa cells. An SIGC was used for Kiss1 promoter-reporter assays. SIGC cells were stably transduced with lentiviral vectors expressing wild-type or DBD−/− rat ESR2. (A) qRT-PCR and (B) Western blot analyses confirmed the ectopic expression of Esr2 in rat SIGC cells. (C) Schematic diagram of Kiss1 promoter indicating presence of a potential ESR2 binding site (ERE). (D, E) Luciferase assays were performed after transfection of the empty pGL4.12 vector or pGL4.25 reporter constructs carrying different lengths of Kiss1 promoters, as shown in the left panels. Promoter constructs with or without ERE sites exhibited similar reporter activity in (D) SIGC transfected with the empty pGL4.12 vector, whereas it was markedly upregulated in (E) SIGC cells expressing wild-type ESR2 but not in (F) cells expressing a DBD−/− ESR2. However, promoter constructs harboring a mutant ERE site did not exhibit such upregulation in (G) SIGC cells expressing wild-type ESR2. Luciferase activity data are presented as fold induction relative to that of empty pGL4.12 vector. Light and dark yellow bars indicate the absence and presence of estradiol in culture media. Each bar represents mean ± SEM of four independent experiments. Luc, luciferase; Wt, wild-type.
Gonadotropin-induced Kiss1 expression in granulosa cells
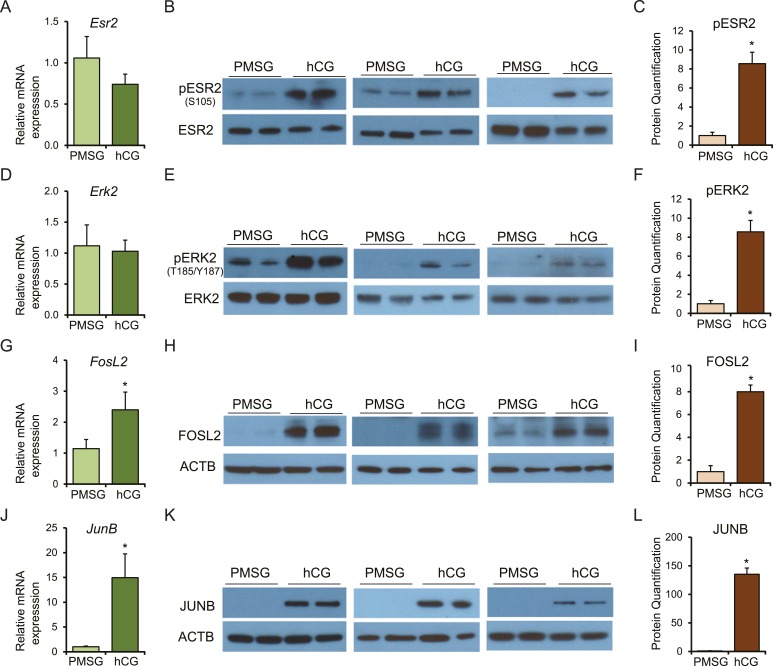
Finally, we explored the link between hCG/LHCGR signaling and ESR2 activation that induces Kiss1 expression in rat granulosa cells. We observed that although the expression of ESR2 (Fig. 5A and 5B) or ERK2 (Fig. 5D and 5E) did not change after hCG administration, gonadotropin stimulation resulted in phosphorylation of ESR2 (Fig. 5B and 5C) and ERK2 (Fig. 5E and 5F) in granulosa cells. hCG also markedly induced the expression of the AP-1 transcription factors FOSL2 (Fig. 5G–5I) and JUNB (Fig. 5J–5L).
Figure 5.
Establishing a link between hCG/LHCGR signaling and ESR2 activation that induces Kiss1 expression in rat granulosa cells. Four-week-old wild-type female rats were injected with 30 IU PMSG, and 48 h after PMSG administration, 30 IU of hCG was injected intraperitoneally. Granulosa cells were isolated 48 h after PMSG treatment or 4 h after hCG treatment in PMSG-primed rats. (A, D, G, and J) Total RNAs purified from granulosa cells were used for RT-qPCR analyses, and (B, E, H and K) protein extracts were subjected to Western blot analyses. Western blot data from three different Western blots performed on six independent samples were quantified by (C, F, I and L) ImageJ analyses. (A, B) Esr2 mRNA and protein levels in wild-type rat granulosa cells 48 h after PMSG injection did not show any significant difference from that of 4 h after hCG injection in PMSG-primed rats. (B, C) However, pESR2 (S105) protein levels were significantly higher 4 h after hCG injection. (D, E) Erk2 mRNA and protein levels in wild-type rat granulosa cells 48 h after PMSG injection were also not different from those of 4 h after hCG injection in PMSG-primed rats. (E, F) However, pERK2 (T185/Y187) protein levels were significantly higher after hCG injection. In contrast, expression of Fosl2 and JunB was markedly upregulated in granulosa cells after hCG injection at both (G, J) mRNA and (H, I, K, L) protein levels. RT-qPCR and Western blot signal quantification data are presented as mean ± SEM. n = 6. *P ≤ 0.05.
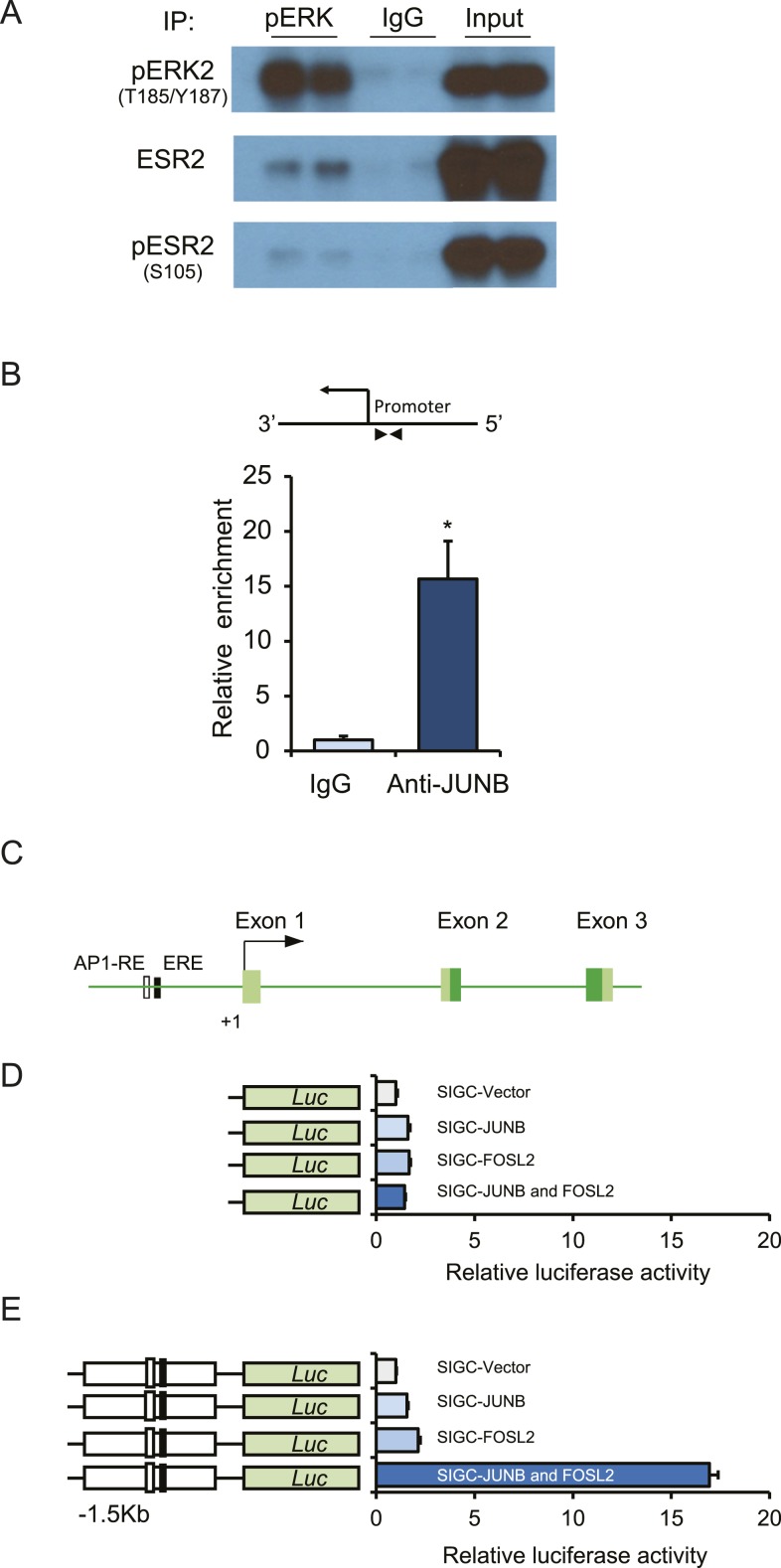
We observed that pERK2 interacted with ESR2 in granulosa cells isolated from PMSG-hCG–treated rats (Fig. 6A). Presence of pESR2 was also detected in the pERK2 interactome complex (Fig. 6A). Direct binding of the AP-1 transcription factor JUNB was detected in the Kiss1 promoter locus of granulosa cells isolated from PMSG-hCG–treated rats (Fig. 6B). Furthermore, activity of the −1.5 Kb Kiss1 promoter harboring an AP-1 RE site was markedly upregulated in SIGC cells expressing either JUNB or FOSL2 (Fig. 6E). Remarkably, expression of both JUNB and FOSL2 together resulted in a synergistic upregulation of Kiss1 promoter activity (Fig. 6E).
Figure 6.
Activated ERK2 interacts with ESR2, and AP-1 factors can activate Kiss1 promoter. Protein complex formation was determined by a coimmunoprecipitation assay using protein extracts prepared from granulosa cells isolated from PMSG-primed, hCG-treated immature rats. Immunoprecipitation was performed with a rabbit monoclonal antibody to pERK1/2 or normal rabbit IgG. The immunoprecipitates were then subjected to Western blot analyses with pERK1/2, ESR2, and pESR2 antibodies. (A) We observed that pERK2 was coimmunoprecipitated with ESR2. (B) ChIP assays were performed on gonadotropin-stimulated rat granulosa cells with an anti-JUNB rabbit monoclonal antibody. Normal rabbit IgG was used as a negative control. ChIP-qPCR results demonstrated a relative enrichment of JUNB at the Kiss1 promoter. (C) Schematic diagram of Kiss1 promoter indicating presence of a potential ESR2 binding (ERE) and AP-1 binding sites. Luciferase assays were performed after cotransfecting (D) the empty pGL4.12 luciferase vector or (E) pGL4.25 vector carrying 1.5 kb of the Kiss1 promoter along with AP-1 factors JUNB or FOSL2 into SIGC cells. (D, E) Empty pGL4.12 vector exhibited a minimal induction of luciferase activity after either JUNB or FOSL2 overexpression, whereas (E) the relative luciferase activity of the reporter constructs with 1.5 kb of Kiss1 promoter harboring an AP-1 RE site showed a marked upregulation when both JUNB and FOSL2 were cotransfected. Relative luciferase data are presented as mean ± SEM of three independent experiments in duplicate wells. n = 6. *P ≤ 0.05. Luc, luciferase.
Discussion
Estrogen signaling can regulate hypothalamic Kiss1 by either activating or repressing gene expression (6–8). In the AVPV nucleus, estrogens activate Kiss1 expression, whereas in the ARC, estrogens repress it (24, 25, 46, 47). Both negative and positive regulation of hypothalamic kisspeptin neurons is mediated by ESR1 (6, 46, 48, 49). In contrast to what is occurring in the hypothalamus, we discovered that gonadotropin-induced Kiss1 expression in granulosa cells is dependent on and induced by ESR2.
Administration of gonadotropins, particularly hCG, stimulates Kiss1 gene expression in the ovary; however, the expression of Kiss1r remains unchanged. Previous reports have shown that Kiss1 and Kiss1r receptor are expressed in all the cellular compartments in the ovary (19, 50). In contrast, we detected Kiss1 expression in rat granulosa cells and oocytes, and Kiss1r only in the oocytes, suggesting that intraovarian kisspeptins may act on oocytes during follicle maturation but only during and after the LH surge. This possibility is supported by previous studies reporting that kisspeptins produced in the ovaries may regulate oocyte growth and maturation (16, 17). We identified that expression of Kiss1 correlates with the expression of Esr2 in granulosa cells and oocytes but not with Esr1 expression. This finding is highly suggestive of an important regulatory role for ESR2 in gonadotropin-induced Kiss1 expression in the ovary that cannot be achieved by ESR1. Our RNA-seq data also demonstrated that Esr2−/− granulosa cells did not induce Kiss1 expression, even though estrogen is being produced by the follicles (27). This finding was corroborated in the current study in that expression of Kiss1 could not be induced in rat granulosa cells or oocytes in the absence of ESR2 expression. In this regard, regulation of ESR1-dependent Kiss1 expression in AVPV or ARC nuclei (6, 46, 48, 49) differs from that of granulosa cells or oocytes.
The Kiss1 gene has been reported to use alternative transcription start sites and express multiple transcript variants through alternative splicing (25, 51, 52). In this study, we identified the granulosa cell–specific transcription start site of Kiss1. Interestingly, this transcription start site was identical to the Kiss1 start site reported in kisspeptin neurons of the mouse (25). We detected two different transcript variants of Kiss1 in rat granulosa cells, which are in line with the observations in mouse kisspeptins neurons (25). These transcripts have differing 5′ untranslated sequence but encode the same primary amino acid sequence. The rat Kiss1 promoter also showed a high degree of homology with the mouse promoter, including a conserved ERE site in the proximal promoter, an upstream 5′ enhancer, and a downstream 3′ enhancer sequence (25). Enrichment of active histone marks including H3K4me3 and H3K27Ac was detected in the Kiss1 promoter, as well as both enhancer sites in gonadotropin-stimulated rat granulosa cells. These regions were also associated with enriched binding of ESR2 (Kiss1 promoter and enhancer sites) and indicate that the Kiss1 gene locus is a direct target of ESR2. Binding to these cognate ERE sites, ESR2 plays an essential regulatory role in gonadotropin-induced Kiss1 expression in granulosa cells. Because of the similarity in the conserved ERE sites, we speculate that the molecular mechanisms used by ESR2 may have similarities to the mechanisms that have been shown for ESR1-mediated positive or negative regulation of Kiss1 expression in AVPV or ARC nuclei (24–26, 47, 53, 54). Nonetheless, it is highly likely that ESR1 and ESR2 recruit and interact with different cohorts of nuclear proteins in the hypothalamus and ovary.
To further confirm the role of ESR2 in Kiss1 promoter regulation, luciferase reporter assays were performed in an immortalized rat granulosa cell line (SIGC). Although a 0.4-kbp proximal promoter exhibited a basal level of promoter activity (greater than that of a promoterless control), it was independent of ESR2 expression and induction by estradiol. In contrast, the 1.5-kbp fragment of the Kiss1 promoter harboring an ERE site showed marked upregulation of reporter activity upon ESR2 overexpression and exposure to estradiol. Our results are in line with the human Kiss1 promoter, where maximum activity was detected with reporter construct containing 1.0 kbp of upstream promoter sequence (55). Loss of estradiol induced promoter activity in SIGC cells expressing DBD-mutant ESR2, and after mutating the ERE site within the promoter highlights the critical role played by ESR2 in regulating the Kiss1 promoter in granulosa cells.
Although the expression of Kiss1 in granulosa cells or oocytes is ESR2 dependent, our data indicate that it is induced only after the hCG administration. To explore the link between hCG/LHCGR signaling and ESR2 regulation of Kiss1 expression, we evaluated changes in gene expression, protein levels, and phosphorylation before and after hCG administration. pESR2 was observed after hCG and correlated with pERK2). We also detected interaction of pERK2 with ESR2 in granulosa cells. This interactome complex also demonstrated the presence of pESR2. These findings suggest a potential role for MAPK-dependent activation of ESR2 in granulosa cells. It has been reported that gonadotropin stimulation can activate the MAPK pathway and specifically MAPK1/3 (ERK1/2) (56). Activated ERK1/2 can phosphorylate ESR2 (57–59), and this phosphorylation of ESR2 increases its transcriptional activation potential (60).
AP-1 binding sites are reported to be present near the ERE sites within the Kiss1 promoter (55, 61). It has also been shown that estrogen receptors can activate gene expression by interacting with AP-1 factors and increasing the transcriptional potential (62–64). We observed that expression of AP-1 factors FOSL2 and JunB was remarkably upregulated in granulosa cells after hCG administration. JUNB was enriched within the Kiss1 promoter locus of granulosa cells isolated from gonadotropin-treated rats. Moreover, AP-1 factors FOSL2 and JunB exhibited a synergistic activation of the Kiss1 promoter activity. AP-1 factors are ubiquitously expressed and activated by Ras-MAP kinase signaling, and in cooperation with other transcriptional regulators they can activate cell type–specific enhancers (65).
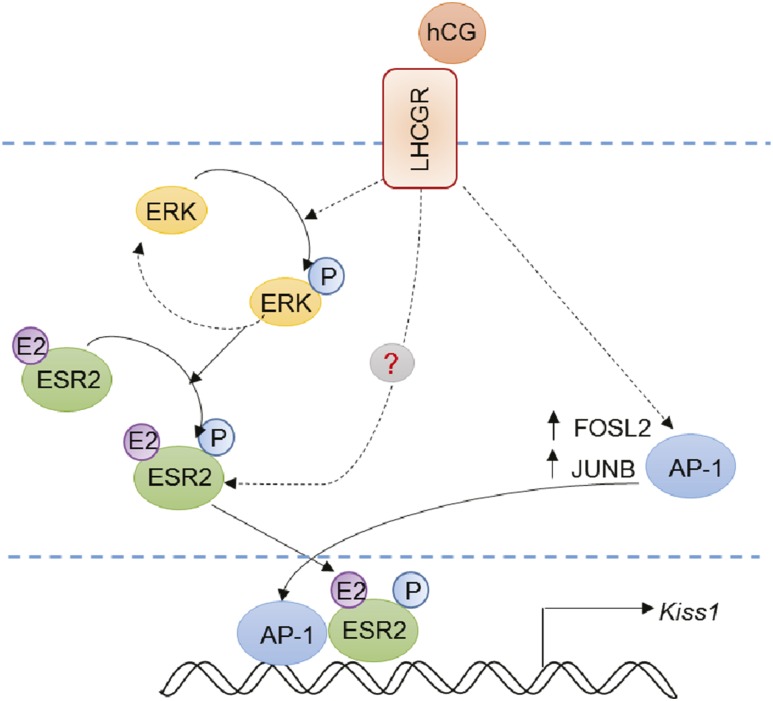
We hypothesize that hCG/LHCGR signaling leads to phosphorylation of ERK2, which in turn phosphorylates and activates ESR2 (Fig. 7). Gonadotropin stimulation also leads to marked upregulation of AP-1 factors, which along with activated pESR2 can induce Kiss1 expression in granulosa cells (Fig. 7). However, this effect does not exclude roles for other serine threonine kinases downstream of LHCGR activation. It is important to note that disruption of ESR2 reduces the expression of Lhcgr on granulosa cells (27, 28). Thus, a reduced level of Lhcgr expression in Esr2−/− granulosa cells may also contribute to the failure of Kiss1 induction.
Figure 7.
Schematic model summarizing our observation. Our findings indicate that hCG/LHCGR signaling leads to phosphorylation of ERK2, which in turn interacts with ESR2. pERK2 can phosphorylate ESR2, but it does not exclude involvement of other serine threonine kinases downstream of LHCGR activation. hCG/LHCGR signaling also increases the expression of AP-1 factors, including FOSL2 and JUNB, which can play a synergistic role in Kiss1 promoter activation. Thus, activated pESR2 along with AP-1 factors induces the expression of Kiss1 in granulosa cells after gonadotropin stimulation. E2, estradiol; P, phosphorylation.
Collectively, our data reveal the essential role played by ESR2 in regulating expression of Kiss1 within granulosa cells. In the absence of ESR2, the Kiss1 gene is unable to respond to the signals from pituitary gonadotropes (FSH and LH) that are responsible for regulating follicular maturation and ovulation. This lack of response occurs even though granulosa cells express ESR1. Thus, ESR1 is incapable of compensating for the loss of ESR2 and maintaining Kiss1 expression in granulosa cells of Esr2−/− rats. It is also important to highlight the fact that ESR2 alone is insufficient for robust Kiss1 expression in granulosa cells. Expression remains low after PMSG stimulation and at a time when ample ligand (estrogen) for ESR2 is present. Our data indicate that stimulation of LHCGR is necessary to induce Kiss1. We hypothesize that hCG/LH induces ESR2 phosphorylation and the induction of other transcription factors (e.g., AP-1 binding proteins) to elicit an increase in Kiss1 expression. ESR2 is essential in this response and may serve a key role in organizing and promoting the gonadotropin-induced expression of Kiss1 in granulosa cells.
Acknowledgments
We acknowledge Professor Robert C. Burghardt of Texas A&M University for providing the spontaneously immortalized rat granulosa cell line.
Financial Support: This study was supported in part by the National Institute of General Medical Sciences of the National Institutes of Health (NIH) (Grant P20 GM103418 to V.P.C.) awarded to Kansas INBRE and an NIH Clinical and Translational Science Award (Grant UL1TR002366 to M.A.K.R.) awarded to University of Kansas Medical Center (KUMC), and the Lied Basic Science Grant Program of the KUMC Research Institute.
Disclosure Summary: The authors have nothing to disclose.
Glossary
Abbreviations:
- ARC
arcuate nucleus
- AVPV
anteroventral periventricular nucleus
- ChIP
chromatin immunoprecipitation, DBD−/−, DNA-binding domain deleted
- ERE
estrogen response element
- ESR1
estrogen receptor-α
- ESR2
estrogen receptor-β
- hCG
human chorionic gonadotropin
- KISS1R
KISS1 receptor
- LHCGR
LH/choriogonadotropin receptor
- pERK2
phosphorylated ERK2
- pESR2
phosphorylated estrogen receptor-β
- PMSG
pregnant mare serum gonadotropin
- qPCR
quantitative PCR
- RNA-seq
RNA sequencing
- RT-qPCR
quantitative RT-PCR
- SIGC
spontaneously immortalized rat granulosa cell line
- TS
template switching
References
- 1. de Roux N, Genin E, Carel JC, Matsuda F, Chaussain JL, Milgrom E. Hypogonadotropic hypogonadism due to loss of function of the KiSS1-derived peptide receptor GPR54. Proc Natl Acad Sci USA. 2003;100(19):10972–10976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Seminara SB, Messager S, Chatzidaki EE, Thresher RR, Acierno JS Jr, Shagoury JK, Bo-Abbas Y, Kuohung W, Schwinof KM, Hendrick AG, Zahn D, Dixon J, Kaiser UB, Slaugenhaupt SA, Gusella JF, O’Rahilly S, Carlton MB, Crowley WF Jr, Aparicio SA, Colledge WH. The GPR54 gene as a regulator of puberty. N Engl J Med. 2003;349(17):1614–1627. [DOI] [PubMed] [Google Scholar]
- 3. Caraty A, Smith JT, Lomet D, Ben Saïd S, Morrissey A, Cognie J, Doughton B, Baril G, Briant C, Clarke IJ. Kisspeptin synchronizes preovulatory surges in cyclical ewes and causes ovulation in seasonally acyclic ewes. Endocrinology. 2007;148(11):5258–5267. [DOI] [PubMed] [Google Scholar]
- 4. Castellano JM, Gaytan M, Roa J, Vigo E, Navarro VM, Bellido C, Dieguez C, Aguilar E, Sánchez-Criado JE, Pellicer A, Pinilla L, Gaytan F, Tena-Sempere M. Expression of KiSS-1 in rat ovary: putative local regulator of ovulation? Endocrinology. 2006;147(10):4852–4862. [DOI] [PubMed] [Google Scholar]
- 5. Matsui H, Takatsu Y, Kumano S, Matsumoto H, Ohtaki T. Peripheral administration of metastin induces marked gonadotropin release and ovulation in the rat. Biochem Biophys Res Commun. 2004;320(2):383–388. [DOI] [PubMed] [Google Scholar]
- 6. Smith JT, Cunningham MJ, Rissman EF, Clifton DK, Steiner RA. Regulation of Kiss1 gene expression in the brain of the female mouse. Endocrinology. 2005;146(9):3686–3692. [DOI] [PubMed] [Google Scholar]
- 7. Kinoshita M, Tsukamura H, Adachi S, Matsui H, Uenoyama Y, Iwata K, Yamada S, Inoue K, Ohtaki T, Matsumoto H, Maeda K. Involvement of central metastin in the regulation of preovulatory luteinizing hormone surge and estrous cyclicity in female rats. Endocrinology. 2005;146(10):4431–4436. [DOI] [PubMed] [Google Scholar]
- 8. Adachi S, Yamada S, Takatsu Y, Matsui H, Kinoshita M, Takase K, Sugiura H, Ohtaki T, Matsumoto H, Uenoyama Y, Tsukamura H, Inoue K, Maeda K. Involvement of anteroventral periventricular metastin/kisspeptin neurons in estrogen positive feedback action on luteinizing hormone release in female rats. J Reprod Dev. 2007;53(2):367–378. [DOI] [PubMed] [Google Scholar]
- 9. d’Anglemont de Tassigny X, Colledge WH. The role of kisspeptin signaling in reproduction. Physiology (Bethesda). 2010;25(4):207–217. [DOI] [PubMed] [Google Scholar]
- 10. Revel FG, Saboureau M, Masson-Pévet M, Pévet P, Mikkelsen JD, Simonneaux V. Kisspeptin mediates the photoperiodic control of reproduction in hamsters. Curr Biol. 2006;16(17):1730–1735. [DOI] [PubMed] [Google Scholar]
- 11. Muir AI, Chamberlain L, Elshourbagy NA, Michalovich D, Moore DJ, Calamari A, Szekeres PG, Sarau HM, Chambers JK, Murdock P, Steplewski K, Shabon U, Miller JE, Middleton SE, Darker JG, Larminie CG, Wilson S, Bergsma DJ, Emson P, Faull R, Philpott KL, Harrison DC. AXOR12, a novel human G protein-coupled receptor, activated by the peptide KiSS-1. J Biol Chem. 2001;276(31):28969–28975. [DOI] [PubMed] [Google Scholar]
- 12. Ohtaki T, Shintani Y, Honda S, Matsumoto H, Hori A, Kanehashi K, Terao Y, Kumano S, Takatsu Y, Masuda Y, Ishibashi Y, Watanabe T, Asada M, Yamada T, Suenaga M, Kitada C, Usuki S, Kurokawa T, Onda H, Nishimura O, Fujino M. Metastasis suppressor gene KiSS-1 encodes peptide ligand of a G-protein-coupled receptor. Nature. 2001;411(6837):613–617. [DOI] [PubMed] [Google Scholar]
- 13. Hauge-Evans AC, Richardson CC, Milne HM, Christie MR, Persaud SJ, Jones PM. A role for kisspeptin in islet function. Diabetologia. 2006;49(9):2131–2135. [DOI] [PubMed] [Google Scholar]
- 14. Oakley AE, Clifton DK, Steiner RA. Kisspeptin signaling in the brain. Endocr Rev. 2009;30(6):713–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ricu MA, Ramirez VD, Paredes AH, Lara HE. Evidence for a celiac ganglion-ovarian kisspeptin neural network in the rat: intraovarian anti-kisspeptin delays vaginal opening and alters estrous cyclicity. Endocrinology. 2012;153(10):4966–4977. [DOI] [PubMed] [Google Scholar]
- 16. Gaytan F, Garcia-Galiano D, Dorfman MD, Manfredi-Lozano M, Castellano JM, Dissen GA, Ojeda SR, Tena-Sempere M. Kisspeptin receptor haplo-insufficiency causes premature ovarian failure despite preserved gonadotropin secretion. Endocrinology. 2014;155(8):3088–3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dorfman MD, Garcia-Rudaz C, Alderman Z, Kerr B, Lomniczi A, Dissen GA, Castellano JM, Garcia-Galiano D, Gaytan F, Xu B, Tena-Sempere M, Ojeda SR. Loss of Ntrk2/Kiss1r signaling in oocytes causes premature ovarian failure. Endocrinology. 2014;155(8):3098–3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Taniguchi Y, Kuwahara A, Tachibana A, Yano Y, Yano K, Yamamoto Y, Yamasaki M, Iwasa T, Hinokio K, Matsuzaki T, Irahara M. Intra-follicular kisspeptin levels are related to oocyte maturation and gonadal hormones in patients who are undergoing assisted reproductive technology. Reprod Med Biol. 2017;16(4):380–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hu KL, Zhao H, Chang HM, Yu Y, Qiao J. Kisspeptin/kisspeptin receptor system in the ovary. Front Endocrinol (Lausanne). 2018;8:365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jayasena CN, Abbara A, Comninos AN, Nijher GM, Christopoulos G, Narayanaswamy S, Izzi-Engbeaya C, Sridharan M, Mason AJ, Warwick J, Ashby D, Ghatei MA, Bloom SR, Carby A, Trew GH, Dhillo WS. Kisspeptin-54 triggers egg maturation in women undergoing in vitro fertilization. J Clin Invest. 2014;124(8):3667–3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jayasena CN, Comninos AN, Nijher GM, Abbara A, De Silva A, Veldhuis JD, Ratnasabapathy R, Izzi-Engbeaya C, Lim A, Patel DA, Ghatei MA, Bloom SR, Dhillo WS. Twice-daily subcutaneous injection of kisspeptin-54 does not abolish menstrual cyclicity in healthy female volunteers. J Clin Endocrinol Metab. 2013;98(11):4464–4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Narayanaswamy S, Prague JK, Jayasena CN, Papadopoulou DA, Mizamtsidi M, Shah AJ, Bassett P, Comninos AN, Abbara A, Bloom SR, Veldhuis JD, Dhillo WS. Investigating the KNDy hypothesis in humans by coadministration of kisspeptin, neurokinin B, and naltrexone in men. J Clin Endocrinol Metab. 2016;101(9):3429–3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Byri P, Gangineni A, Reddy KR, Raghavender KBP. Effect of kisspeptin on in vitro maturation of sheep oocytes. Vet World. 2017;10(3):276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Goto T, Tomikawa J, Ikegami K, Minabe S, Abe H, Fukanuma T, Imamura T, Takase K, Sanbo M, Tomita K, Hirabayashi M, Maeda K, Tsukamura H, Uenoyama Y. Identification of hypothalamic arcuate nucleus-specific enhancer region of Kiss1 gene in mice. Mol Endocrinol. 2015;29(1):121–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tomikawa J, Uenoyama Y, Ozawa M, Fukanuma T, Takase K, Goto T, Abe H, Ieda N, Minabe S, Deura C, Inoue N, Sanbo M, Tomita K, Hirabayashi M, Tanaka S, Imamura T, Okamura H, Maeda K, Tsukamura H. Epigenetic regulation of Kiss1 gene expression mediating estrogen-positive feedback action in the mouse brain. Proc Natl Acad Sci USA. 2012;109(20):E1294–E1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gottsch ML, Navarro VM, Zhao Z, Glidewell-Kenney C, Weiss J, Jameson JL, Clifton DK, Levine JE, Steiner RA. Regulation of Kiss1 and dynorphin gene expression in the murine brain by classical and nonclassical estrogen receptor pathways. J Neurosci. 2009;29(29):9390–9395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Khristi V, Chakravarthi VP, Singh P, Ghosh S, Pramanik A, Ratri A, Borosha S, Roby KF, Wolfe MW, Rumi MAK. ESR2 regulates granulosa cell genes essential for follicle maturation and ovulation. Mol Cell Endocrinol. 2018;474:214–226. [DOI] [PubMed] [Google Scholar]
- 28. Rumi MAK, Singh P, Roby KF, Zhao X, Iqbal K, Ratri A, Lei T, Cui W, Borosha S, Dhakal P, Kubota K, Chakraborty D, Vivian JL, Wolfe MW, Soares MJ. Defining the role of estrogen receptor β in the regulation of female fertility. Endocrinology. 2017;158(7):2330–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Khristi V, Chakravarthi VP, Singh P, Ghosh S, Pramanik A, Ratri A, Borosha S, Roby KF, Wolfe MW, Rumi MAK. Differentially regulated genes in Esr2-mutant rat granulosa cells. Data Brief. 2018;19:1008–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Frohman MA. 5′-End cDNA amplification using classic RACE. CSH Protocols. 2006;2006(1). [DOI] [PubMed] [Google Scholar]
- 31. Scotto-Lavino E, Du G, Frohman MA. Amplification of 5′ end cDNA with “new RACE.” Nat Protoc. 2006;1(6):3056–3061. [DOI] [PubMed] [Google Scholar]
- 32. Loots GG, Ovcharenko I. rVISTA 2.0: evolutionary analysis of transcription factor binding sites. Nucleic Acids Res. 2004;32(Web Server issue):W217–W221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. RRID: AB_10949503.
- 34. RRID: AB_2616028.
- 35. RRID: AB_1031062.
- 36. Stein LS, Stoica G, Tilley R, Burghardt RC. Rat ovarian granulosa cell culture: a model system for the study of cell-cell communication during multistep transformation. Cancer Res. 1991;51(2):696–706. [PubMed] [Google Scholar]
- 37. Hayer A, Shao L, Chung M, Joubert LM, Yang HW, Tsai FC, Bisaria A, Betzig E, Meyer T. Engulfed cadherin fingers are polarized junctional structures between collectively migrating endothelial cells. Nat Cell Biol. 2016;18(12):1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. RRID: AB_10681401.
- 39. RRID: AB_11212759.
- 40. RRID: AB_2315112.
- 41. RRID: AB_390779.
- 42. RRID: AB_2130023.
- 43. RRID: AB_2744660.
- 44. RRID: AB_476692.
- 45. RRID: AB_2130002.
- 46. Clarkson J, d’Anglemont de Tassigny X, Moreno AS, Colledge WH, Herbison AE. Kisspeptin-GPR54 signaling is essential for preovulatory gonadotropin-releasing hormone neuron activation and the luteinizing hormone surge. J Neurosci. 2008;28(35):8691–8697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Uenoyama Y, Tomikawa J, Inoue N, Goto T, Minabe S, Ieda N, Nakamura S, Watanabe Y, Ikegami K, Matsuda F, Ohkura S, Maeda K, Tsukamura H. Molecular and epigenetic mechanism regulating hypothalamic Kiss1 gene expression in mammals. Neuroendocrinology. 2016;103(6):640–649. [DOI] [PubMed] [Google Scholar]
- 48. Wintermantel TM, Campbell RE, Porteous R, Bock D, Gröne HJ, Todman MG, Korach KS, Greiner E, Pérez CA, Schütz G, Herbison AE. Definition of estrogen receptor pathway critical for estrogen positive feedback to gonadotropin-releasing hormone neurons and fertility. Neuron. 2006;52(2):271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Glidewell-Kenney C, Hurley LA, Pfaff L, Weiss J, Levine JE, Jameson JL. Nonclassical estrogen receptor alpha signaling mediates negative feedback in the female mouse reproductive axis. Proc Natl Acad Sci USA. 2007;104(19):8173–8177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cielesh ME, McGrath BM, Scott CJ, Norman ST, Stephen CP. The localization of kisspeptin and kisspeptin receptor in the canine ovary during different stages of the reproductive cycle. Reprod Domest Anim. 2017;52(suppl 2):24–28. [DOI] [PubMed] [Google Scholar]
- 51. Terao Y, Kumano S, Takatsu Y, Hattori M, Nishimura A, Ohtaki T, Shintani Y. Expression of KiSS-1, a metastasis suppressor gene, in trophoblast giant cells of the rat placenta. Biochim Biophys Acta. 2004;1678(2–3):102–110. [DOI] [PubMed] [Google Scholar]
- 52. Chan YM, Broder-Fingert S, Paraschos S, Lapatto R, Au M, Hughes V, Bianco SD, Min L, Plummer L, Cerrato F, De Guillebon A, Wu IH, Wahab F, Dwyer A, Kirsch S, Quinton R, Cheetham T, Ozata M, Ten S, Chanoine JP, Pitteloud N, Martin KA, Schiffmann R, Van der Kamp HJ, Nader S, Hall JE, Kaiser UB, Seminara SB. GnRH-deficient phenotypes in humans and mice with heterozygous variants in KISS1/Kiss1. J Clin Endocrinol Metab. 2011;96(11):E1771–E1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sun JM, Chen HY, Davie JR. Effect of estradiol on histone acetylation dynamics in human breast cancer cells. J Biol Chem. 2001;276(52):49435–49442. [DOI] [PubMed] [Google Scholar]
- 54. Fortress AM, Kim J, Poole RL, Gould TJ, Frick KM. 17β-Estradiol regulates histone alterations associated with memory consolidation and increases Bdnf promoter acetylation in middle-aged female mice. Learn Mem. 2014;21(9):457–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Li D, Mitchell D, Luo J, Yi Z, Cho SG, Guo J, Li X, Ning G, Wu X, Liu M. Estrogen regulates KiSS1 gene expression through estrogen receptor alpha and SP protein complexes. Endocrinology. 2007;148(10):4821–4828. [DOI] [PubMed] [Google Scholar]
- 56. Cameron MR, Foster JS, Bukovsky A, Wimalasena J. Activation of mitogen-activated protein kinases by gonadotropins and cyclic adenosine 5′-monophosphates in porcine granulosa cells. Biol Reprod. 1996;55(1):111–119. [DOI] [PubMed] [Google Scholar]
- 57. Tremblay A, Tremblay GB, Labrie F, Giguère V. Ligand-independent recruitment of SRC-1 to estrogen receptor beta through phosphorylation of activation function AF-1. Mol Cell. 1999;3(4):513–519. [DOI] [PubMed] [Google Scholar]
- 58. Lucas TF, Siu ER, Esteves CA, Monteiro HP, Oliveira CA, Porto CS, Lazari MF. 17beta-estradiol induces the translocation of the estrogen receptors ESR1 and ESR2 to the cell membrane, MAPK3/1 phosphorylation and proliferation of cultured immature rat Sertoli cells. Biol Reprod. 2008;78(1):101–114. [DOI] [PubMed] [Google Scholar]
- 59. Sanchez M, Picard N, Sauvé K, Tremblay A. Challenging estrogen receptor beta with phosphorylation. Trends Endocrinol Metab. 2010;21(2):104–110. [DOI] [PubMed] [Google Scholar]
- 60. Hamilton-Burke W, Coleman L, Cummings M, Green CA, Holliday DL, Horgan K, Maraqa L, Peter MB, Pollock S, Shaaban AM, Smith L, Speirs V. Phosphorylation of estrogen receptor beta at serine 105 is associated with good prognosis in breast cancer. Am J Pathol. 2010;177(3):1079–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Treen AK, Luo V, Chalmers JA, Dalvi PS, Tran D, Ye W, Kim GL, Friedman Z, Belsham DD. Divergent regulation of ER and Kiss genes by 17β-estradiol in hypothalamic ARC versus AVPV models. Mol Endocrinol. 2016;30(2):217–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gaub MP, Bellard M, Scheuer I, Chambon P, Sassone-Corsi P. Activation of the ovalbumin gene by the estrogen receptor involves the fos-jun complex. Cell. 1990;63(6):1267–1276. [DOI] [PubMed] [Google Scholar]
- 63. Tora L, Gaub MP, Mader S, Dierich A, Bellard M, Chambon P. Cell-specific activity of a GGTCA half-palindromic oestrogen-responsive element in the chicken ovalbumin gene promoter. EMBO J. 1988;7(12):3771–3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kushner PJ, Agard DA, Greene GL, Scanlan TS, Shiau AK, Uht RM, Webb P. Estrogen receptor pathways to AP-1. J Steroid Biochem Mol Biol. 2000;74(5):311–317. [DOI] [PubMed] [Google Scholar]
- 65. Madrigal P, Alasoo K. AP-1 takes centre stage in enhancer chromatin dynamics. Trends Cell Biol. 2018;28(7):509–511. [DOI] [PubMed] [Google Scholar]