Abstract
The highly reducing iterative polyketide synthases responsible for lovastatin biosynthesis contains a section homologous to condensation (CON) domain observed in nonribosomal peptide synthetases (NRPSs). In the present study, we expressed the isolated lovastatin CON domain and solved the crystal structure to 1.79 Å resolution. The overall structure shows similarity to canonical condensation domains of NRPSs, containing the N-terminal and C-terminal subdomains that resemble enzymes of chloramphenicol acetyltransferase family, whereas distinct structural features are observed at the active site. The acceptor entry of the substrate channel is blocked by a flexible loop, thereby preventing the loading of substrate for a new round of chain elongation. The mutation of conserved catalytic motif located at the midpoint of substrate channel agrees with the incapability of CON to catalyzed amide-bond formation. The structure helps to understand the function of CON in lovastatin biosynthesis.
1. Introduction
Polyketide synthases (PKSs) are responsible for assembly of structurally diverse polyketide natural products of which many are of therapeutic or commercial value. PKSs are structurally and functionally homologous to the fatty acid synthases (FASs). A minimal PKS consisting of a ketosynthase (KS), a malonyl-CoA:ACP acyltransferase (MAT), and an acyl carrier protein (ACP) is responsible for the elongation of a polyketide chain via decarboxylative Claisen condensation while the ketoreductase (KR), dehydratase (DH) and enoylreductase (ER) tailoring domains control the final oxidation state of the newly added extender units. Compared to the well-studied bacterial PKSs that operate in a modular fashion, fungal PKSs use iteratively a single set of catalytic domains in different combinations to generate natural products that vary in the length of polyketide chains and the state of reduction and dehydration [[1], [2], [3]]. How a single set of catalytic domains of a fungal iterative PKS discriminate between the polyketide intermediates and control the processing levels during the elongation of polyketide chains is still obscure.
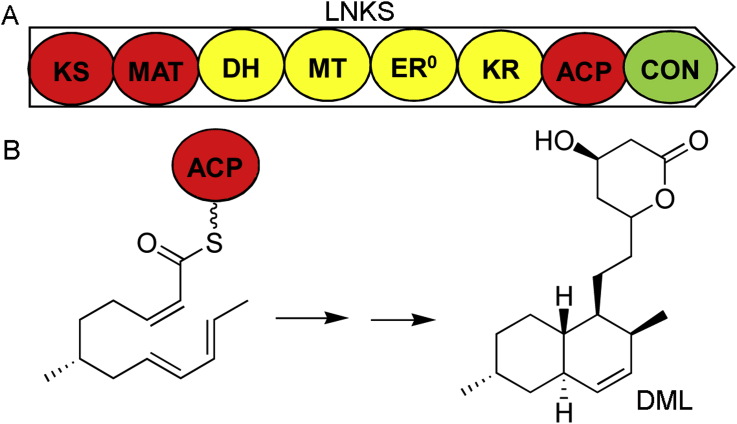
Lovastatin (also known as monacolin K, mevinolin, and mevastatin) is a polyketide produced by several fungal species including Aspergillus terreus [4], Monascus pilosus [5] and Paecilomyces viridis [6]. It has a similar structure to hydroxymethylglutaryl coenzyme A (HMG-CoA) and competitively inhibits the HMG-CoA reductase, the key enzyme in the cholesterol biosynthesis pathway. This activity confers on lovastatin clinical importance in treating hypercholesterolemia to reduce the risk of cardiovascular disease. The biosynthetic gene clusters for lovastatin in A. terreus and M. pilosus have been characterized and encode two highly reducing PKSs (HR-PKSs) that play central roles in the biosynthesis of lovastatin. The 277 kDa lovastatin diketide synthase (LDKS, LovF in A. terreus and MokB in M. pilosus) contains KS, AT, DH, ER, KR, and ACP domains that are arranged in the same order as in bacterial modular PKSs and an additional methyltransferase (MT) positioned between DH and ER domains. It behaves noniteratively, catalyzing a single decarboxylative condensation and the following methylation, ketoreduction, dehydration, and enoylreduction steps to produce the 2-methylbutyrate moiety. The 335 kDa lovastatin nonaketide synthase (LNKS, LovB in A. terreus and MokA in M. pilosus) in conjunction with a discrete trans-acting ER (LovC in A. terreus and MokE in M. pilosus) catalyze about 35 chemical reactions to generate the main skeleton of lovastatin, dihydromonacolin L (DML). LNKS is an iterative fungal HR-PKS, containing an inactive ER, and an additional C-terminal domain that resembles the condensation domain (CON) of nonribosomal peptide synthetases (NRPSs) compared to LDKS (Fig. 1).
Fig. 1.
Synthesis of dihydromonacolin L (DML) catalyzed by lovastatin nonaketide synthase (LNKS). (A) Domain organization of LNKS. A trans-acting enoylreductase is required for DML synthesis. (B) The Diels-Alder reaction involved in DML synthesis.
To initiate DML synthesis, LNKS is covalently attached to an acetyl starter unit derived from decarboxylation of malonyl-CoA. The KS domain then catalyzes repeatedly decarboxylative condensations to elongate the acyl chain on the ACP. Nine malonyl-CoA, eight NADPH and an S-adenosyl-l-methionine (SAM) are required for the synthesis of DML. Different combinations of tailoring domains are used to processing the nascent polyketide intermediate. The MT transfers a methyl group from SAM to the tetraketide; ketoreduction fellows each of the eight chain-extension steps; the DH is active in the first six elongations; enoylreduction occurs at the tetraketide, pentaketide, and heptaketide stages to produce fully reduced sections. Kinetic assays of the MT and KR domains using a panel of acyl-S-N-acetylcysteamine (SNAC) substrate mimics reveal that these two tailoring domains compete for the β-ketoacyl intermediates generate by the KS domain [7]. The MT domain is highly selective for the natural tetraketide and outcompetes the KR domain at this step only, whereas, the catalytic efficiency of KR domain is less substrate-dependent. Consequently, the relative efficiency of catalytic domains determines the tailoring outcome of the polyketide intermediate. A Diels-Alder cycloaddition occurs at the hexaketide stage to form the fused rings of DML decalin system. A triene-SNAC substrate mimics has been synthesized to test LNKS for Diels-Alderase activity. The product with the same stereochemistry as DML is only generated in the presence of undenatured LNKS, suggesting the Diels-Alder cycloaddition is enzymatically catalyzed [8].
The CON domain in the C terminus of LNKS has been proposed to catalyze the release of DML [9]. However, in vitro reconstitution experiments indicate that it does play a crucial role in the formation of DML but is not responsible for product release [10]. The truncated LNKS lacking CON domain in combination with the trans-acting ER does not produce detectable amounts of DML but generates a few shunt polyketide products of different length, suggesting that the functions of the minimal PKS and the tailoring domains is not affected. Here, we report the heterologous expression and structural characterization of the isolated CON domain. The CON structure reveals a similar overall fold but distinct substrate channel compared to the previously reported canonical NRPS condensation domains.
2. Materials and methods
2.1. Protein expression and purification
The DNA fragment encoding CON domain was amplified from Monascus sp. CPCC 400017 genomic DNA using primers 5′- ATCGTAATCCATATGCTCGTGGCAGCAAGCGAAGG-3′ and 5′- TGATTCGATGAATTCAAGCCAACTTCAACGCGGGATTC -3′ (restriction sites underlined). The PCR product was digested with NdeI and EcoRI and cloned into the same sites in pET28a (Novagen). The resulting plasmid was verified by sequence and transformed into Escherichia coli BL21(DE3). The His-tagged CON protein was overexpressed in LB medium containing 50 μg/mL of kanamycin at 37 °C, grown to OD600 = 0.4, and induced with 0.5 mM IPTG. After 12 h at 16 °C, cells were harvested by centrifugation and resuspended in lysis buffer (300 mM NaCl, 50 mM Tris, pH 7.5). Following sonication, cell debris was removed by centrifugation (20,000 g for 40 min). The supernatant was loaded onto a Ni-NTA column, which was then washed with 50 mL lysis buffer containing 15 mM imidazole and eluted with 5 mL of lysis buffer containing 300 mM imidazole. Proteins were further purified by using a Superdex 200 gel filtration column (GE Healthcare Life Sciences) equilibrated with buffer containing 25 mM NaCl, 10 mM Tris and 1 mM DTT (pH 7.5). The proteins were concentrated to 10 mg/mL and stored at −80 °C.
Selenomethionine-labeled CON domain protein was obtained by the pathway inhibition method. E. coli BL21 (DE3) transformed with the expression plasmid was grown to an OD600 of 0.4 in M9 medium containing 0.4% glucose as carbon source at 37 °C. Then, amino acids lysine, phenylalanine, and threonine were added to a final concentration of 100 mg/L each; isoleucine, leucine, and valine were added to a final concentration of 50 mg/L each; and SeMet was added to a final concentration of 60 mg/L. The culture was incubated for another 15 min at 37 °C and then moved to 20 °C and induced by 1 mM IPTG for 12 h. The purification of the SeMet-labeled protein followed the same protocol as described above.
2.2. Size estimation
Samples (0.1 mL) were injected onto a Superdex 200 gel filtration column equilibrated with 150 mM NaCl and 10 mM Tris (pH 7.5). The molecular weight of the CON domain was determined through comparisons to known standards (Gel Filtration Standard; Biorad Laboratories).
2.3. Crystallization and structure determination
Crystals of the CON domain were grown by sitting drop vapor diffusion at 20 °C by mixing 2 μL of protein solution (10 mg/mL in 25 mM NaCl, 1 mM DTT, 10 mM Tris, pH 7.5) with 1.0 μL of crystallization buffer (18% (w/v) polyethylene glycol 3350, 0.2 M potassium acetate, pH 7.5). Crystals of SeMet-labeled protein were obtained at 8 °C with precipitant solution containing 20% (w/v) polyethylene glycol 3350, 0.2 M potassium acetate, pH 8.2. Crystals were flash frozen in well solution containing 20% glycerol prior to data collection.
Data were collected at Shanghai Synchrotron Radiation Facility Beamline BL18U and BL19U, and processed with HKL2000. The structure was solved to 2.5 Å resolution by single-wavelength anomalous dispersion (SAD) phasing using the program Phenix [11]. A 1.79 Å-resolution data set collected from a crystal of native CON protein was used to iteratively build and refine the model through the programs Coot [12] and Refmac5 [13]. Atomic coordinate and structure factor of the CON domain have been deposited in the Protein Data Bank with accession code 6AD3 (Table 1).
Table 1.
Data collection and refinement statistics.
| CON (6AD3) | SeMet CON | |
|---|---|---|
| Data collection | ||
| Wavelength (Å) | 0.9792 | 0.9792 |
| Space group | P21221 | P21221 |
| a, b, c (Å) | 51.1, 64.0, 173.2 | 50.9, 64.0, 173.0 |
| Resolution (Å) | 50–1.79 (1.82–1.99) | 50–2.50 (2.59-2.50) |
| Rmerge | 0.076 (0.422) | 0.130 (0.459) |
| I/σI | 15.4 (3.2) | 20.3 (10.2) |
| Completeness (%) | 99.9 (100) | 99.9 (100) |
| Redundancy | 6.6 (6.6) | 7.1 (7.6) |
| Refinement | ||
| Resolution (Å) | 50–1.79 | |
| No. reflections | 51687 | |
| Rwork/Rfree | 0.176/0.206 | |
| No. atoms | ||
| Protein | 3528 | |
| Water | 260 | |
| B-factors (Å2) | ||
| Protein | 25.5 | |
| Water | 25.8 | |
| rmsd | ||
| Bond lengths (Å) | 0.013 | |
| Bond angles (°) | 1.203 | |
2.4. Computational modeling
Simulated docking of the cyclization product (1R,2R,4aS,6R,8aR)–1,2,4a,5,6,7,8,8a-Octahydro-2,6-dimethylnaphthalene-1-carboxylic acid, N-acetylcysteamine thioester to the CON domain was performed using Autodock Vina 31. The ligand was docked to the CON structure in a grid box with a size of 10 Å on each dimension centered on the catalytic histidine. Nine different possible binding modes were obtained from the docking simulation with a maximum energy difference of −7.8 kcaL/mol and maximum upper bound RMSD difference of 8.42 Å. The binding modes that have the dimethylnaphthalene oriented towards the interior of the cavity were chosen for further analysis.
3. Results and discussions
3.1. Crystallization of isolated CON
The genomic DNA of Monascus Sp. CPCC 400017, a lovastatin producer, was used as the template to amplify the CON domain with the primers designed according to the mokA gene of Monascus pilosus BCRC38072. L2578, which is 2 residues downstream of the ACP domain, was selected as the N terminus, while A3075, the last residue of MokA, was selected as the C terminus [5,14]. The resulting DNA fragment encoding a polypeptide consisting of 498 amino acid residues was ligated into the NdeI and EcoRI restriction sites of pET28b. The N-terminally His6-tagged CON was expressed in E. coli BL21 (DE3) and purified to homogeneity by affinity and size-exclusion chromatography. The purified CON migrates at ∼57 kDa on a size exclusion column as estimated by comparison to molecular weight standards, consistent with the expected monomer mass of 55 kDa.
CON domain purified to homogeneity was subjected to high-throughput crystallization trials. Diffraction-quality crystals were obtained by sitting drop vapor diffusion method using polyethylene glycol 3350 as a precipitant. The protein crystallizes in space group P21221 with one monomer per asymmetric unit. Phasing trials using molecular replacement with different search models derived from known condensation domains failed to identify a solution because of low sequence identities (<24%). Selenomethionine-derivatized protein was then purified and crystallized by the same protocols. The structure was solved via SAD phasing and refine to 1.79 Å resolution, with Rworking and Rfree values of 0.18 and 0.21, respectively. The electron density for the N-terminal 48 residues and an internal disordered region (263–266) was not observed. The final refined model contains 446 out of 498 residues of the monomer (Table 1).
3.2. Overall structure
CON domain closely resembles the free-standing condensation domain VibH (PDB code 1L5A, 2.9 Å Cα rmsd, sequence identity, 14%) [15] and the canonical condensation domain from the first module of the calcium-dependent antibiotic synthetase (CDA-C1, PDB code 5DU9, 2.6 Å Cα rmsd, sequence identity, 17%) [16], the second module of bimodular DhbF (PDB code 5U89, 2.6 Å Cα rmsd, sequence identity 22%) [17], the AB3403 from the human pathogen Acinetobacter baumannii (PDB code 4ZXI, 3.0 Å Cα rmsd, sequence identity, 15%) [18], the enterobactin synthase from Escherichia coli (PDB code 5T3D, 3.0 Å Cα rmsd, sequence identity, 18%) [18], the sixth module of tyrocidine synthetase (PDB code 2JGP, 3.2 Å Cα rmsd, sequence identity 23%) [19], and the terminal module SrfA-C of surfactin biosynthesis (PDB code 2VSQ, 3.8 Å Cα rmsd, sequence identity, 18%) [20]. An automatic search of CON domain for structurally homologous proteins using the Dali server revealed similarity to the heterocyclization (Cy) domain of bacillamide synthetase (PDB code 5T3E, 2.8 Å Cα rmsd, sequence identity, 12%) [21] and epothilone EpoB protein (PDB code 5T81, 3.2 Å Cα rmsd, sequence identity, 17%) [22], the terminal condensation-like (CT) domain catalyzing macrocyclization (PDB code 5DIJ, 3.6 Å Cα rmsd, sequence identity, 15%) [23], the epimerization domain from the first module of the cyclic peptide antibiotic gramicidin synthetase (PDB code 5ISX, 2.8 Å Cα rmsd, sequence identity, 13%) [24] and tyrocidine synthetase A (PDB code 2XHG, 2.8 Å Cα rmsd, sequence identity, 14%) [25], and the X-domain from the final NRPS module of teicoplanin biosynthesis (PDB code 4TX2, 3.1 Å Cα rmsd, sequence identity, 21%) [26].
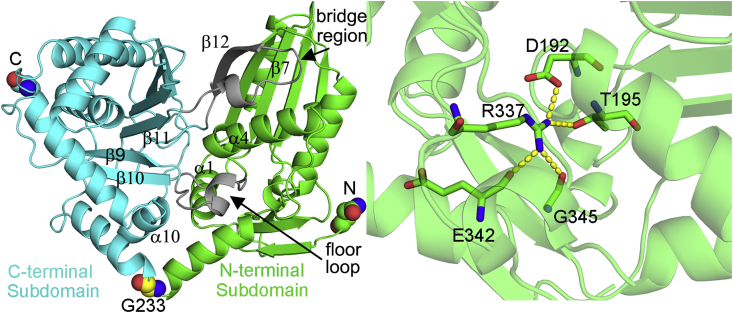
Like all previously reported structures of condensation domain superfamily, CON is a pseudodimer consisting of two subdomains. The overall structure is V-shaped, with the N- (Asn50-Gly233) and C-terminal (Gln234-Ala499) subdomains comprising approximately one half of the V-shape (Fig. 2). Both subdomains adopt αβα sandwich folds resembling chloramphenicol acetyltransferases (CATs). It has been noticed that the angle of the V-shape can vary slightly with the swiveling motion centered on the residue corresponding to G233 in CON (Fig. 2A). The two subdomains of CON adopt the closed orientation observed in CDA-C1. There are only two major bridge regions between these two CAT-like subdomains. The first region (residues 420–440) includes β12 and its upstream loop (Fig. 2A). The strand β12 extends the N-terminal four-stranded β sheet that is positioned almost perpendicular to the six-stranded mixed β-sheet of the C-terminal subdomain. The second region is formed by a loop connecting β9 and β10 (residues 335–350) which is at the floor of the active site canyon and referred as “floor loop” (Fig. 2A). H-bond interactions between the side chain of a conserved R337 and the peptide carbonyls of E342 and G345 stabilize the floor loop (Fig. 2B). The conserved R337 is also involved in the interactions between two subdomains by the hydrogen-bond to the T195 side chain and the salt bridge with D192. These two bridge regions appear to be rather flexible since they display slightly higher B-factors than average.
Fig. 2.
Structure of CON domain. (A) The V-shaped fold of CON. Two bridge regions are colored grey. The N-terminal and C-terminal residues and the G233 related to swiveling motion are shown as spheres. (B) Interactions stabilizing the floor loop plugged into the N-terminal subdomain.
3.3. Substrate channel
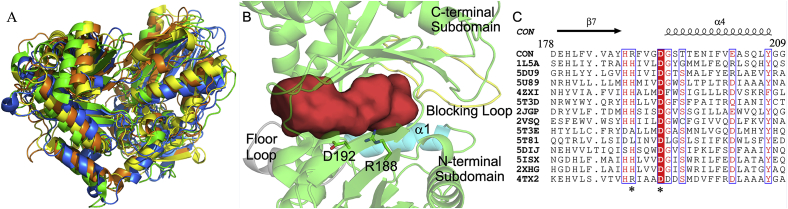
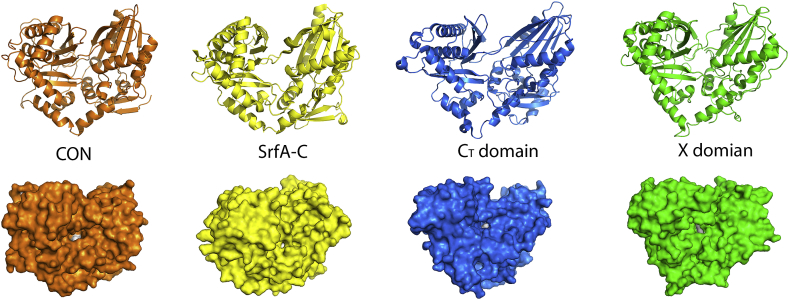
A condensation domain elongates the peptide chain by catalyzing amide bond formation in the assembly of amino acid build blocks. Thus, it is accessible to substrates attached to the phosphopantetheine arms of both an upstream donor peptide carrier protein (PCP) and a downstream acceptor PCP. A channel running through the protein is formed at the interface of the two subdomains, starting near the floor loop of the C-terminal subdomain and ending near the helix α1 of the N-terminal subdomains with the conserved HHxxxD motif located at the midpoint of the channel. The PCP-C didomain structures reveal that the donor PCP is positioned on the C domain at the entrance near the floor loop [19,23,24], whereas, the structure of AB3403 C-A-PCP-TE termination module suggests that the acceptor PCP delivers the substrate into the substrate channel form the entrance near the helix α1 of the N-terminal subdomain [18]. The substrate channel of CON is similarly organized with the exception that the acceptor entry side is blocked by the loop between helix α10 and strand β11 (Fig. 3A and B). This structural feature is also observed in the X and CT domains that do not require downstream acceptor substrates [23,26]. The second histidine of the conserved HHxxxD motif is proposed to play an important role in positioning the α-amino group for nucleophilic attack [16]. However, this histidine is replaced by arginine in CON (Fig. 3C). The X-domain (PDB code 4TX2) recruiting cytochrome P450 oxygenases in glycopeptide biosynthesis contains the same residue replacement [26]. Structure comparison between CON, the Srf A-C condensation domain, the CT domain, and the X-domain has shown that they all present a similar V shape fold with a substrate channel running through the protein. The conserved resides of HHxxxD motif locate at the midpoint of the substrate channel (Fig. 4).
Fig. 3.
Active site of CON. (A) Secondary structure alignment of CON and its homologues shows their similarity. Orange: CON. Yellow: Srf A-C condensation domain (2VSQ). Marine: the CT domain (5DIJ). Green: the X domain (4TX2). Conserved residues of the HHxxxD motif are shown as sticks in color magenta. The first histidine is replaced by serine in 5DIJ, and the second histidine is replaced by arginine in CON and 4TX2. (B) The substrate channel between two subdomains. The floor loop at the donor entry and the helix α1 at the acceptor entry are colored grey and cyan respectively. The loop blocking the acceptor entry is colored yellow. Residues R188 and D192 corresponding to conserved catalytic motif of condensation domain are shown as sticks. (C) The sequence alignment showing the conserved HHxxxD motif of condensation domains. The second histidine is replaced by arginine in CON and X domain (4TX2). 1L5A, 5DU9, 5U89, 4ZXI, 5T3D, 2JGP and 2VSQ are condensation domains. 5T3E and 5T81 are heterocyclization domain. 5DIJ is a condensation-like domain (CT) for macrocyclization. 5ISX and 2XHG are epimerization domains. 4TX2 is an X-domain for recruiting another enzyme.
Fig. 4.
Structural comparison of CON, the Srf A-C condensation domain, the CT domain, and the X domain. They all show a similar V shape fold with a substrate channel running through the protein. The surface structural comparison of CON (orange), the Srf A-C condensation domain (2VSQ, yellow), the CT domain (5DIJ, marine), and the X domain (4TX2, green) shows that conserved resides of HHxxxD motif (white) locate at the midpoint of the substrate channel.
3.4. Function of CON
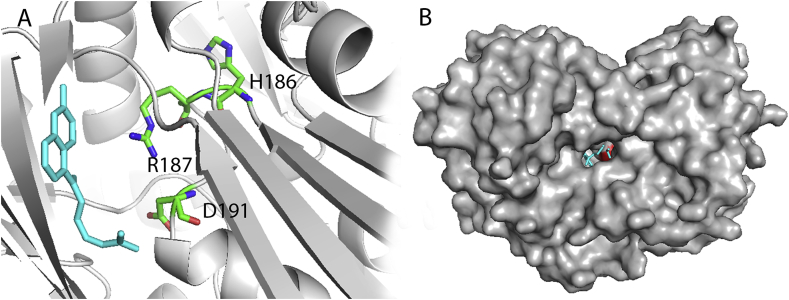
The LNKS LovB and the trans-acting ER LovC contain all catalytic activities required for DML synthesis, including the enzymatically catalyzed Diels-Alder reaction to yield the decalin ring [8,27]. Catalytic domains related to assembly of polyketide chains have been well-characterized. The CON with unknown function is therefore a reasonable candidate domain that is responsible for the Diels−Alderase activity [28]. Both in vitro and in vivo assays have been carried out to inspect the CON function. The heterologously expressed LovB variant without the CON domain produces only truncated pyrones. Supplementation of the CON domain in trans can restore the generation of expected DML intermediate [10]. In A. terreus, addition of the adenylation, thiolation, and reductase domains of cytochalasin synthase CheA onto the full lovB gene shows no obvious impact, whereas, replacing the CON by the CheA condensation domain completely abolishes lovastatin production [28]. Encouragingly, a pre-Diels−Alder product is identified from fermentations of the Fusarium heterosporum expressing truncated LovB without the CON domain, providing strong evidence for the Diels−Alderase hypothesis referred previously. The structure of CON shows no similarity to the previously reported SpnF [29], PyrE 3 [30], and PyrI4 [31] Diels−Alderase, suggesting that they use different catalytic mechanism. The cyclized product of the Diels-Alder reaction could be docked into active reasonably (Fig. 5). It enters the CON active site channel from the entrance that has been observed in the CT-ACP complex (PDB code: 5JED) [23]. The CON active site has enough room to accommodate the cyclized product. However, it is difficult to propose the exact roles of each active site residues. The CON active site may provide suitable shape and environment for the substrate to form the cyclized product with proper stereochemistry. Further biochemical analyses of CON domain are necessary to decode its function in lovastatin biosynthesis.
Fig. 5.
Possible binding model for the cyclization product to the CON domain obtained from AutoDock Vina. (A) The CON active channel has enough room to accommodate the cyclized product (cyan), residues of the conserved motif is displayed as green sticks. (B) Surface diagram depicting the simulated docking of cyclization product in the CON structure. The cyclization product are shown as cyan sticks. HRxxxD motif is colored in red.
Competing financial interests
The authors declare no competing financial interests.
Acknowledgments
We thank the staff at beamline BL18U1 and BL19U1 of the Shanghai Synchrotron Radiation Facility (China) and National Center for Protein Science Shanghai (China) for access and help with data collection. This work was supported by National Natural Science Foundation of China (31570056, 31770068) and Fundamental Research Funds for the Central Universities.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
References
- 1.Simpson T.J. Fungal polyketide biosynthesis - a personal perspective. Nat Prod Rep. 2014;31(10):1247–1252. doi: 10.1039/c4np00065j. [DOI] [PubMed] [Google Scholar]
- 2.Townsend C.A. Aflatoxin and deconstruction of type I, iterative polyketide synthase function. Nat Prod Rep. 2014;31(10):1260–1265. doi: 10.1039/c4np00092g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vederas J.C. Explorations of fungal biosynthesis of reduced polyketides - a personal viewpoint. Nat Prod Rep. 2014;31(10):1253–1259. doi: 10.1039/c4np00091a. [DOI] [PubMed] [Google Scholar]
- 4.Kennedy J. Modulation of polyketide synthase activity by accessory proteins during lovastatin biosynthesis. Science. 1999;284(5418):1368–1372. doi: 10.1126/science.284.5418.1368. [DOI] [PubMed] [Google Scholar]
- 5.Chen Y.P. Cloning and characterization of monacolin K biosynthetic gene cluster from Monascus pilosus. J Agric Food Chem. 2008;56(14):5639–5646. doi: 10.1021/jf800595k. [DOI] [PubMed] [Google Scholar]
- 6.Kimura K. Biosynthesis of monacolins: conversion of monacolin J to monacolin K (mevinolin) J Antibiot (Tokyo) 1990;43(12):1621–1622. doi: 10.7164/antibiotics.43.1621. [DOI] [PubMed] [Google Scholar]
- 7.Cacho R.A. Understanding programming of fungal iterative polyketide synthases: the biochemical basis for regioselectivity by the methyltransferase domain in the lovastatin megasynthase. J Am Chem Soc. 2015;137(50):15688–15691. doi: 10.1021/jacs.5b11814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Auclair K. Lovastatin nonaketide synthase catalyzes an intramolecular Diels-Alder reaction of a substrate analogue. J Am Chem Soc. 2000;122(46):11519–11520. [Google Scholar]
- 9.Cox R.J. Polyketides, proteins and genes in fungi: programmed nano-machines begin to reveal their secrets. Org Biomol Chem. 2007;5(13):2010–2026. doi: 10.1039/b704420h. [DOI] [PubMed] [Google Scholar]
- 10.Ma S.M. Complete reconstitution of a highly reducing iterative polyketide synthase. Science. 2009;326(5952):589–592. doi: 10.1126/science.1175602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adams P.D. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emsley P. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vagin A.A. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 14.Ma S.M., Tang Y. Biochemical characterization of the minimal polyketide synthase domains in the lovastatin nonaketide synthase LovB. FEBS J. 2007;274(11):2854–2864. doi: 10.1111/j.1742-4658.2007.05818.x. [DOI] [PubMed] [Google Scholar]
- 15.Keating T.A. The structure of VibH represents nonribosomal peptide synthetase condensation, cyclization and epimerization domains. Nat Struct Biol. 2002;9(7):522–526. doi: 10.1038/nsb810. [DOI] [PubMed] [Google Scholar]
- 16.Bloudoff K., Alonzo D.A., Schmeing T.M. Chemical probes allow structural insight into the condensation reaction of nonribosomal peptide synthetases. Cell Chem Biol. 2016;23(3):331–339. doi: 10.1016/j.chembiol.2016.02.012. [DOI] [PubMed] [Google Scholar]
- 17.Tarry M.J. X-ray crystallography and electron microscopy of cross- and multi-module nonribosomal peptide synthetase proteins reveal a flexible architecture. Structure. 2017;25(5):783–793 e4. doi: 10.1016/j.str.2017.03.014. [DOI] [PubMed] [Google Scholar]
- 18.Drake E.J. Structures of two distinct conformations of holo-non-ribosomal peptide synthetases. Nature. 2016;529(7585):235–238. doi: 10.1038/nature16163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Samel S.A. Structural and functional insights into a peptide bond-forming bidomain from a nonribosomal peptide synthetase. Structure. 2007;15(7):781–792. doi: 10.1016/j.str.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 20.Tanovic A. Crystal structure of the termination module of a nonribosomal peptide synthetase. Science. 2008;321(5889):659–663. doi: 10.1126/science.1159850. [DOI] [PubMed] [Google Scholar]
- 21.Bloudoff K. Structural and mutational analysis of the nonribosomal peptide synthetase heterocyclization domain provides insight into catalysis. Proc Natl Acad Sci U S A. 2017;114(1):95–100. doi: 10.1073/pnas.1614191114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dowling D.P. Structural elements of an NRPS cyclization domain and its intermodule docking domain. Proc Natl Acad Sci U S A. 2016;113(44):12432–12437. doi: 10.1073/pnas.1608615113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J. Structural basis of nonribosomal peptide macrocyclization in fungi. Nat Chem Biol. 2016;12(12):1001–1003. doi: 10.1038/nchembio.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen W.H. Interdomain and intermodule organization in epimerization domain containing nonribosomal peptide synthetases. ACS Chem Biol. 2016;11(8):2293–2303. doi: 10.1021/acschembio.6b00332. [DOI] [PubMed] [Google Scholar]
- 25.Samel S.A., Czodrowski P., Essen L.O. Structure of the epimerization domain of tyrocidine synthetase A. Acta Crystallogr D Biol Crystallogr. 2014;70(Pt 5):1442–1452. doi: 10.1107/S1399004714004398. [DOI] [PubMed] [Google Scholar]
- 26.Haslinger K. X-domain of peptide synthetases recruits oxygenases crucial for glycopeptide biosynthesis. Nature. 2015;521(7550):105–109. doi: 10.1038/nature14141. [DOI] [PubMed] [Google Scholar]
- 27.Witter D.J., Vederas J.C. Putative Diels-Alder-catalyzed cyclization during the biosynthesis of lovastatin. J Org Chem. 1996;61(8):2613–2623. doi: 10.1021/jo952117p. [DOI] [PubMed] [Google Scholar]
- 28.Boettger D. Evolutionary imprint of catalytic domains in fungal PKS-NRPS hybrids. Chembiochem. 2012;13(16):2363–2373. doi: 10.1002/cbic.201200449. [DOI] [PubMed] [Google Scholar]
- 29.Fage C.D. The structure of SpnF, a standalone enzyme that catalyzes [4 + 2] cycloaddition. Nat Chem Biol. 2015;11(4):256–258. doi: 10.1038/nchembio.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng Q. Structural insights into a flavin-dependent [4 + 2] cyclase that catalyzes trans-decalin formation in pyrroindomycin biosynthesis. Cell Chem Biol. 2018;25(6):718–727 e3. doi: 10.1016/j.chembiol.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 31.Zheng Q. Enzyme-dependent [4 + 2] cycloaddition depends on lid-like interaction of the N-terminal sequence with the catalytic core in PyrI4. Cell Chem Biol. 2016;23(3):352–360. doi: 10.1016/j.chembiol.2016.01.005. [DOI] [PubMed] [Google Scholar]