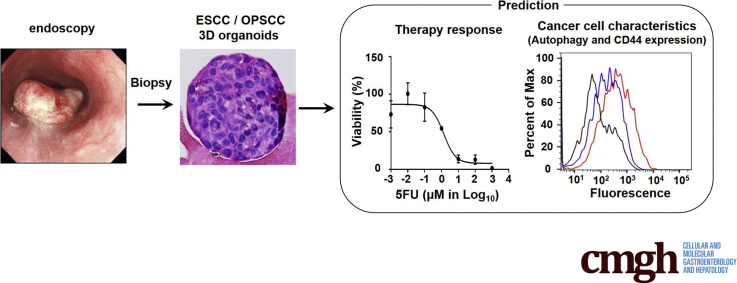
Abstract
Background & Aims
Oropharyngeal and esophageal squamous cell carcinomas, especially the latter, are a lethal disease, featuring intratumoral cancer cell heterogeneity and therapy resistance. To facilitate cancer therapy in personalized medicine, three-dimensional (3D) organoids may be useful for functional characterization of cancer cells ex vivo. We investigated the feasibility and the utility of patient-derived 3D organoids of esophageal and oropharyngeal squamous cell carcinomas.
Methods
We generated 3D organoids from paired biopsies representing tumors and adjacent normal mucosa from therapy-naïve patients and cell lines. We evaluated growth and structures of 3D organoids treated with 5-fluorouracil ex vivo.
Results
Tumor-derived 3D organoids were grown successfully from 15 out of 21 patients (71.4%) and passaged with recapitulation of the histopathology of the original tumors. Successful formation of tumor-derived 3D organoids was associated significantly with poor response to presurgical neoadjuvant chemotherapy or chemoradiation therapy in informative patients (P = 0.0357, progressive and stable diseases, n = 10 vs. partial response, n = 6). The 3D organoid formation capability and 5-fluorouracil resistance were accounted for by cancer cells with high CD44 expression and autophagy, respectively. Such cancer cells were found to be enriched in patient-derived 3D organoids surviving 5-fluorouracil treatment.
Conclusions
The single cell-based 3D organoid system may serve as a highly efficient platform to explore cancer therapeutics and therapy resistance mechanisms in conjunction with morphological and functional assays with implications for translation in personalized medicine.
Keywords: 3D Organoids, Autophagy, CD44, 5-Fluorouracil
Abbreviations used in this paper: AV, autophagy vesicle; CD44H, high expression of CD44; CQ, chloroquine; DMEM, Dulbecco’s modified Eagle medium; EMT, epithelial-mesenchymal transition; ESCC, esophageal squamous cell carcinoma; FBS, fetal bovine serum; 5FU, 5-fluorouracil; H&E, hematoxylin and eosin; IC50, half maximal inhibitory concentration; IHC, immunohistochemistry; LC3, light chain 3; OPSCC, oropharyngeal squamous cell carcinoma; PI, propidium iodide; SCCs, squamous cell carcinomas; TE11R, 5-fluorouracil–resistant derivative of TE11; 3D, 3-dimensional
Graphical abstract
Summary.
We have established 3 dimensional organoids from patients with esophageal and oropharyngeal squamous cell carcinomas. We show that 3-dimensional organoids reveal resistance mechanisms and provide a robust platform to predict therapy response in the setting of personalized medicine.
Squamous cell carcinomas (SCCs) of the oropharynx and the esophagus are anatomically and clinically distinct diseases, yet share a number of histopathological characteristics and genetic as well as environmental risk factors in their pathogenesis.1 In particular, esophageal SCC (ESCC) is the deadliest of all human SCCs, and is common worldwide.2 The poor clinical outcome of ESCC is associated with late diagnosis at advanced stages, therapy resistance, and early recurrence.3 To treat patients with ESCC and oropharyngeal SCC (OPSCC), combined chemotherapy with 5-fluorouracil (5FU), platinum drugs, or taxanes as well as radiation are performed along with or without endoscopic resection, ablation, or open surgery.4, 5 Therapy resistance can be attributed to multiple biological processes such as epithelial-mesenchymal transition (EMT)6 and autophagy.7, 8 Frequent genetic alterations in ESCC include tumor suppressor p53 mutations,1 which may negate therapy-induced apoptotic cell death while promoting EMT, invasive growth, and metastasis. Early recurrence may be accounted for by a subset of ESCC cells referred to as cancer stem-like cells with high tumor-initiating capability. Such cells are characterized by high CD44 expression (CD44H)9 and linked to cell invasion, metastasis, chemotherapy resistance, and poor prognosis.10, 11, 12 The generation and maintenance of intratumoral ESCC CD44H cells may involve EMT and autophagy, thereby contributing to poor prognosis in patients.13 It is currently difficult to evaluate functional properties of ESCC cells and predict therapeutic response in a clinical setting, hampering the development of effective personalized medicine.
The 3-dimensional (3D) organoid system is a cell culture–based physiologically relevant platform,14 featuring tissue-like architecture grown in a mount of basement membrane extract with media containing niche factors. The morphological and functional characteristics of a variety of tissue types have been recapitulated in 3D organoids generated from single-cell suspensions or cell aggregates isolated from murine and human tissues as well as stem cells propagated in culture.14, 15 The 3D organoid system has been utilized to study intestinal epithelial homeostatic mechanisms and malignant transformation.16, 17 3D organoids may serve as an excellent tool to explore genes and pathways altered during disease progression, gene-drug association, and personalized therapy design. Patient-derived tumor organoids have been generated and characterized for colorectal, pancreatic, and prostate cancers18, 19, 20; however, ESCC and OPSCC 3D organoids remain to be investigated. Herein, we have described for the first time tumor organoid formation from ESCC and OPSCC patients. Tumor-derived 3D organoids from therapy-naïve ESCC patients appear to predict therapy resistance. In tumor-derived 3D organoids, we find that chemotherapy may induce CD44H cells with high autophagic capability, thus revealing single cell–derived heterogeneous tumor cell populations with certain therapy resistance mechanisms.
Results
Generation of 3D Organoids From Endoscopic Biopsies From Patients
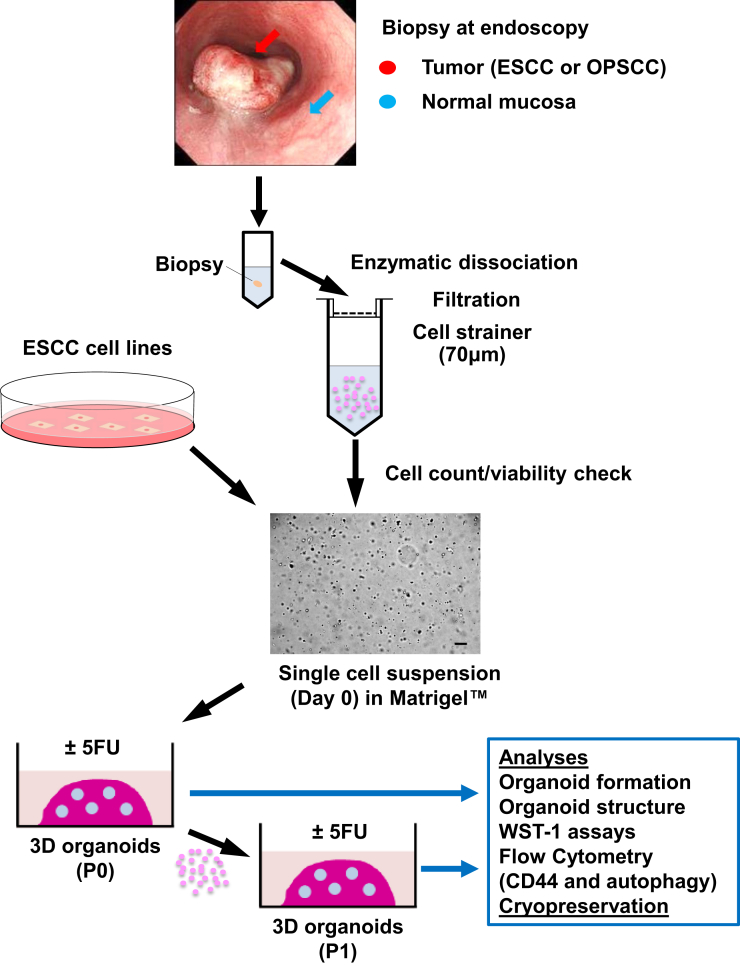
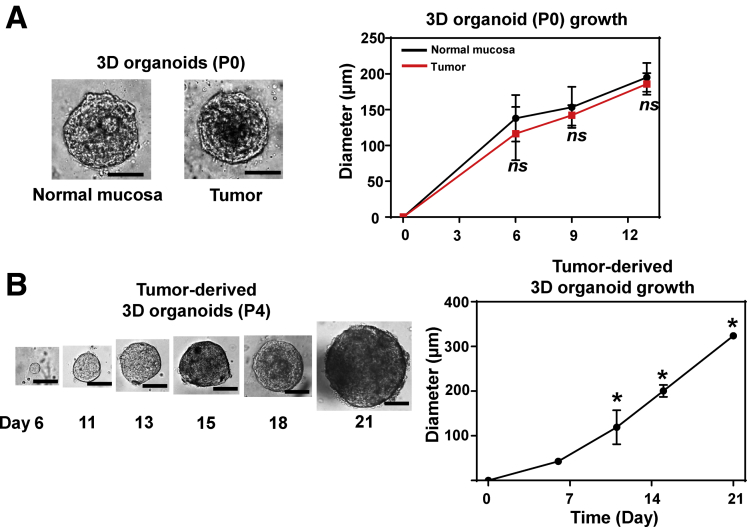
To generate 3D organoids from therapy-naïve patients with ESCC or OPSCC (Table 1), we prepared single cell suspensions from both tumors and adjacent normal mucosa biopsies (Figure 1). We fed the cells suspended in Matrigel with fully supplemented aDMEM/F12+ as utilized to generate 3D organoids for a variety of human gastrointestinal normal and neoplastic tissues21 and murine normal esophagus.22 We detected the growth of spherical structures typically by day 6 under phase-contrast microscopy. They continued to grow to 50–200 μm in diameter by day 14. We defined such structures as 3D organoids (Figure 2A). There was no significant difference in average size of primary 3D organoids from normal mucosa of 3 independent patients compared with their counterparts from tumors (Figure 2A). Excluding 4 tumor samples we had to discontinue culture due to fungal contamination, 3D structures were successfully generated from 15 of 21 tumors (71.4%) and 12 out of 18 adjacent normal mucosa (66.7%) at a 0.1%–1% organoid formation rate (Table 2). The normal mucosa-derived 3D structures reached a plateau in size by day 14 while a subset of tumor-derived 3D organoids continued to grow over the time period of 2–3 weeks. Six of 14 (42.9%) of tumor-derived, but not normal mucosa-derived, 3D organoids were successfully passaged at least once (P1-P7) to exhibit a continuous growth (Figure 2B).
Table 1.
Patients Subjected to Biopsy Before Surgery
| Patient | Type | Stage | Grade | 3D Organoids |
Neoadjuvant Therapya |
Surgical Resection | |||
|---|---|---|---|---|---|---|---|---|---|
| Normal | Tumor | Type | Clinical response | Histological response | |||||
| 1 | ESCC | III | mod | - | + | CT | PD | n/ab | - |
| 4 | ESCC | IV | poor | + | + | CT | PD | 1 | + |
| 5 | ESCC | III | well | + | + | CRT | SD | n/ab | - |
| 6 | ESCC | III | well | - | - | CRT | SD | 1 | + |
| 8 | ESCC | III | mod | + | + | CRT | SD | n/ab | - |
| 9 | ESCC | III | mod | + | - | CRT | PR | 3 | + |
| 10 | ESCC | III | poor | + | + | CT | PD | n/ab | - |
| 12 | ESCC | III | mod | + | + | CRT | SD | n/ab | - |
| 13 | ESCC | III | poor | - | - | CRT | PR | 3 | + |
| 14 | ESCC | III | mod | - | - | CRT | PR | 3 | + |
| 15 | ESCC | IV | well | + | + | CRT | SD | 1 | + |
| 16 | ESCC | III | mod | + | + | CT | SD | 1 | + |
| 17 | ESCC | III | poor | + | - | CRT | PR | n/ab | - |
| 18 | ESCC | III | mod | + | + | CRT | PR | 1 | + |
| 19 | ESCC | III | well | + | + | CRT | PR | 1 | + |
| 20 | ESCC | IV | poor | + | + | CRT | PD | n/ab | - |
| 23 | OPSCC | IV | mod | n.d. | + | n.d. | n/a | n/a | + |
| 24 | OPSCC | IV | mod | n.d. | + | n.d. | n/a | n/a | + |
| 25 | OPSCC | IV | poor | n.d. | + | n.d. | n/a | n/a | + |
| 26 | OPSCC | III | poor | n.d. | + | n.d. | n/a | n/a | + |
| 27 | OPSCC | III | mod | n.d. | - | n.d. | n/a | n/a | + |
NOTE. The formation of 3D organoids was evaluated every other day and determined at day 14. The failure of 3D organoid formation was confirmed at day 21 following an extended observation period. Four patients (2, 3, 7, 11) were excluded for analyses due to microbial (fungal) contamination of tumor organoid culture. Two patients (21, 22) were used for flow cytometry only (Figure 5). We excluded these 2 patients in this table. Two patients underwent surgery in stage IV. Patient 4 underwent esophagectomy following chemotherapy that decreased the size of metastatic liver tumor; however, clinical response was assessed as PD for multiple metastatic liver lesions newly identified during the esophagectomy. Patient 15 was eligible for surgery for distant lymph node metastasis based on a guideline by the Japanese classification of Esophageal Cancer, 11th edition.48
CRT, chemoradiation therapy; CT, chemotherapy; mod, moderately differentiated; n/a, not available; n.d., not determined (3D organoids) or not done (neoadjuvant therapy); PD, progressive disease; poor, poorly differentiated; PR, partial response; SD, stable disease; well, well-differentiated.
Neoadjuvant therapy was performed before surgery.
No surgery performed.
Figure 1.
3D organoids generation and analyses. Biopsies from tumors and adjacent normal mucosa were taken at the time of diagnostic endoscopy and enzymatically dissociated and filtrated to prepare single-cell suspensions in Matrigel to initiate 3D organoid culture. The resulting 3D products were subjected to morphological and functional assays at P0 and subsequent passages (P1 or later) with or without pharmacological treatments (eg, 5FU). Scale bar = 100 μm.
Figure 2.
Growth kinetics of patient-derived 3D organoids. 3D organoids grown from normal mucosa and tumors were photomicrographed under phase-contrast microscopy. (A) Representative images of primary 3D organoids at day 13 from paired normal mucosa and tumor biopsies. The average size (mean ± SD) of primary 3D organoids (P0) from 3 independent patients (biological replicates) were plotted at indicated time points as in the growth curve (right). ns, not significant vs normal mucosa. (B) The representative images of passaged tumor-derived 3D organoids (passage 4 [P4]) are shown (left) along with the corresponding growth curve (right). The growth curve demonstrates the average size (mean ± SD) of ESCC 3D organoids from 2 independent patients (biological replicates). *P < .05 vs all earlier indicated time points. Scale bar = 100 μm. All data were presented as mean ± SD. Student’s t test was used to examine statistical significance.
Table 2.
3D Organoids Formation Frequency per Organoid Size From ESCC Patients
| Patient | Normal Mucosa-Derived 3D Organoids |
Tumor-Derived 3D Organoids |
||||
|---|---|---|---|---|---|---|
| 50–100 μm | 100–200 μm | ≥200 μm | 50–100 μm | 100–200 μm | ≥200 μm | |
| 1 | 0 | 0 | 0 | 22 ± 11 | 4 ± 1.3 | 0 |
| 4 | 43 ± 6.7 | 25 ± 5.0 | 8 ± 2.6 | 70 ± 12 | 26 ± 6.5 | 7 ± 3.2 |
| 5 | 9 ± 1.3 | 0.8 ± 0.4 | 0 | 78 ± 12 | 39 ± 7.5 | 0 |
| 8 | 56 ± 12 | 15 ± 4.0 | 0 | 22 ± 3.2 | 9 ± 2.4 | 0 |
| 9 | 52 ± 1.5 | 28 ± 1.7 | 0 | 0 | 0 | 0 |
| 10 | 27a | 11a | 0 | 31 ± 3.2 | 33 ± 5.4 | 2.1 ± 0.7 |
| 12 | 25 ± 6.1 | 13 ± 4.0 | 0 | 105 ± 7.8 | 58 ± 3.3 | 0.3 ± 0.2 |
| 15 | n.d. | n.d. | n.d. | 93 ± 20 | 4 ± 1.8 | 0 |
| 16 | n.d. | n.d. | n.d. | 113 ± 8.7 | 16 ± 2.8 | 1.1 ± 0.4 |
| 17 | >200 | 237 ± 56 | 4 ± 0.3 | 0 | 0 | 0 |
| 18 | 120 ± 5.3 | 74 ± 3.8 | 3 ± 0.7 | 83 ± 6.4 | 56 ± 4.5 | 21 ± 1.9 |
| 19 | >200 | 92 ± 4.3 | 8 ± 1.2 | 84 ± 17 | 55 ± 9.2 | 22 ± 4.5 |
| 20 | >200 | 107 ± 5.4 | 4 ± 0.8 | >200 | 27 ± 4.2 | 0.2 ± 0.1 |
NOTE. Values are mean ± SD. The 3D organoid formation rate from ESCC patients was determined at day 14 as the average number of organoids formed from 2 × 104 cells seeded per well. 3–10 independent wells were used except patient 10.
n.d., not determined (due to a small number of viable cells isolated from a biopsy).
Single well only for 3D organoids from normal mucosa.
Morphological Characterization of 3D Organoids
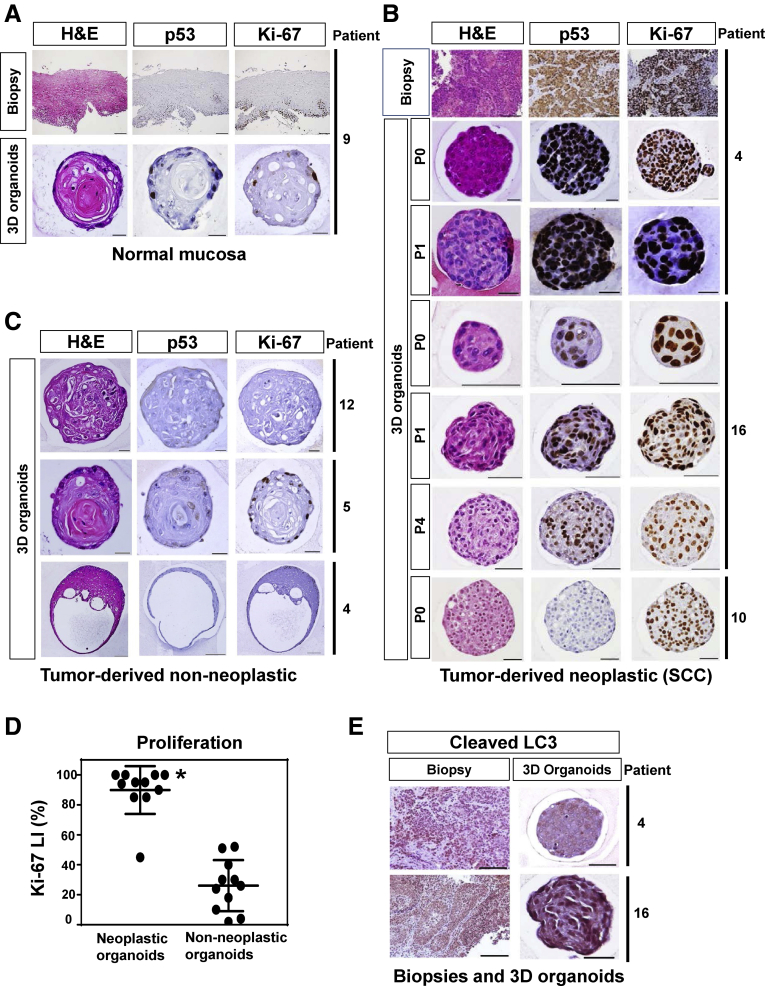
We examined next the paraffin-embedded organoid culture products morphologically. 3D organoids from normal mucosa displayed histologically non-neoplastic epithelial cell nests with normal nuclear features, limited proliferation and squamous cell differentiation as corroborated by keratinization (Figure 3A). The esophageal epithelium of human undergoes terminal differentiation, passive migration toward the luminal surface and ultimately desquamation into the lumen. Unlike the normal human esophageal epithelium, esophageal epithelium cells in the center of the 3D organoid cannot desquamate following terminal differentiation. They show keratinization (cornification) in the center of the 3D organoid after the differentiation gradient, representing an artefactual aspect of this model system.23, 24 By contrast, 3D organoids from tumors contained structures with neoplastic characteristics, hereafter referred to as SCC 3D organoids, which were compatible with poorly differentiated SCC, featuring atypical cells with high nuclear/cytoplasmic ratio (Figure 3B). IHC for Ki-67 revealed the highly proliferative nature of SCC cells in 3D organoids. A subset of patients displayed upregulation of the tumor suppressor p53 protein in SCC 3D organoids, indicating accumulation of dysfunctional mutant p53 protein,25, 26 and thus recapitulating this aspect of the original tumors. 3D organoids arising from the primary tumors contained the above described histologically non-neoplastic structures from normal mucosa (Table 3 and Figure 3C). The non-neoplastic nature of such structures was corroborated by their disappearance in passaged tumor-derived 3D organoids (data not shown), the lack of p53 stabilization and lower proliferation (Ki67 labeling index) (Figure 3C and D) compared with their SCC counterparts (eg, patient 4) (Figure 3B). Five of 12 (41.7%) of tumor biopsies gave rise to 3D organoids comprising >30% SCC-compatible structures (Table 3). In such patients, 3D organoids were readily passaged (P2-P7) with growth properties, proliferation and p53 expression that were comparable from passage to passage (Figure 3B); however, the majority of tumor biopsies produced predominantly non-neoplastic structures over SCC-compatible structures (Table 3), suggesting that the aDMEM/F12+ medium may be more permissive for normal cells to form 3D organoids than SCC cells.
Figure 3.
Morphological analysis of patient-derived 3D organoids. Biopsies and 3D organoids representing (A) normal mucosa and (B–E) tumors from indicated patients were morphologically analyzed by H&E staining, IHC for p53, Ki-67, and cleaved LC3. (A) Normal mucosa gave rise to non-neoplastic 3D organoids with a clear concentric stratification or stratified squamous cell differentiation gradient, approximately reproducing all layers of a stratified squamous epithelium with basaloid and spinous cells in the periphery and stratum corneum-like squames in the center. Tumor biopsies gave rise to both (B) neoplastic and (C) non-neoplastic 3D structures. (B) Tumor-derived neoplastic 3D structures (SCC 3D organoids) feature lack clear differentiation pattern although the eosinophilic cytoplasm of most cells point to a squamous differentiation, and thus, compatible with poorly differentiated squamous cell carcinoma. There is mild to moderate nuclear atypia noted. (C) Tumor-derived non-neoplastic 3D organoids contain cells with very little or no nuclear atypia (patient 12). The majority of non-neoplastic structures feature focal incomplete keratinization without clear stratification (patient 5). The minority of non-neoplastic structures display stratification (ie, a concentric complete keratinization [patient 4]), a rare cystic form with squamous differentiation with very little or no terminally differentiated squamous (horny layer-like) cells. Scale bar = 100 μm for tissue sections; (A, C) 20 μm for 3D organoids. Scale bar = 100 μm for tissue sections (patient 4); 20 μm for 3D organoids (patient 4); and (B) 50 μm for 3D organoids (patients 10 and 16). (D) Ki67 labeling index was determined in tumor-derived neoplastic (SCC compatible) and non-neoplastic 3D structures (10 per each group) from multiple patients as biological replicates (3 independent patients for neoplastic 3D organoids and 7 independent patients for non-neoplastic 3D organoids). Data were presented as mean ± SD. Student’s t test was used to examine statistical significance. *P < .05, vs non-neoplastic organoids. (E) Cleaved LC3 was stained in tumor biopsies and corresponding SCC 3D organoids. Scale bar = 100 μm for tissue sections; and 50 μm for 3D organoids.
Table 3.
Frequency of Organoid Types Within Patient Tumor-Derived 3D Organoids
| Patient | Tumor-Derived 3D Organoids Content |
|
|---|---|---|
| Non-Neoplastic (%) | Neoplastic (SCC-Compatible) (%) | |
| 1 | n.d. | n.d. |
| 4 | 28 | 72 |
| 5 | 100 | 0 |
| 8 | 95 | 5 |
| 10 | 86 | 14 |
| 12 | 100 | 0 |
| 15 | n.d. | n.d. |
| 16 | 67 | 33 |
| 18 | 100 | 0 |
| 19 | 92 | 8 |
| 20 | 87 | 13 |
| 23 | 25 | 75 |
| 24 | 67 | 33 |
| 26 | 11 | 89 |
NOTE. The primary 3D organoids (P0) generated directly from ESCC and OPSCC patient biopsies were evaluated by H&E staining for the frequency of 3D structures with non-neoplastic and neoplastic characteristics. Each organoid type was determined by evaluating 15–119 spherical structures (>50 μm) for each patient except patient 16 where only 3 3D organoid structures were detected throughout all H&E slides examined. None of the 3D organoids from normal mucosa contained neoplastic organoids.
n.d., not determined.
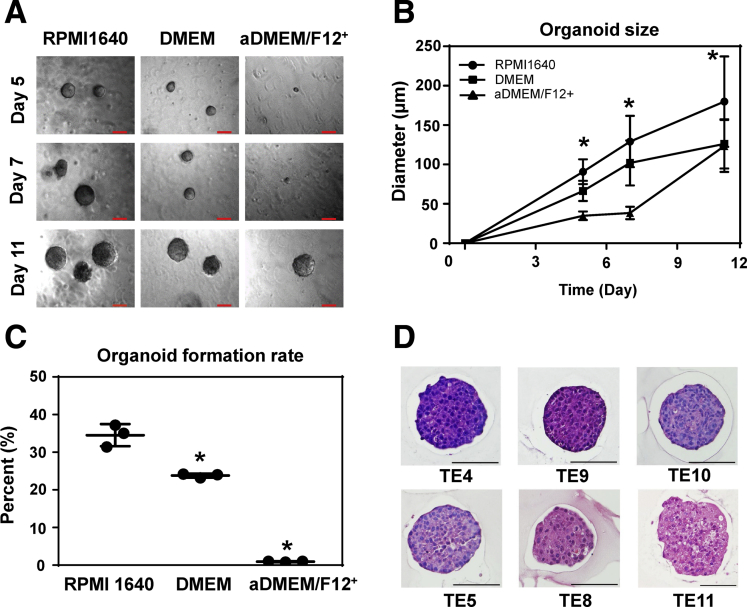
We next generated 3D organoids utilizing human ESCC cell lines (Figure 4A–D) to compare with biopsy-derived primary SCC 3D organoids (Figures 2 and 3). Their growth properties and morphological characteristics were comparable; however, ESCC cell lines formed 3D organoids less efficiently in the aDMEM/F12+ medium compared with conventional cell culture media (ie, RPMI1640 and DMEM supplemented with 10% FBS) used to establish and maintain ESCC cell lines (Figure 4; and data not shown). This showed that some reagents in aDMEM/F12+ medium would work as an inhibitor for growing SCC 3D organoids. Nevertheless, tumor biopsy-derived 3D organoids barely grew in RPMI1640 and DMEM containing 10% FBS (data not shown).
Figure 4.
Morphological characteristics and functional analysis of cell line–derived ESCC 3D organoids. ESCC 3D organoids were grown with TE11 cells in indicated cell culture media to acquire (A) phase contrast images and evaluate (B) organoid growth (size) at indicated time points. (C) Organoid formation rate was determined at day 11. RPMI1640 and DMEM are supplemented with 10% FBS. aDMEM/F12+ is fully supplemented as used to grow patient-derived 3D organoids. Scale bar =100 μm. (B) Data are presented as mean ± SD of at least 7 organoids. Data represent 3 independent experiments with similar results and 1-way analysis of variance with multiple comparisons (Tukey) was performed for each (B) time point in and (C) condition. *P < .05 vs RPMI1640, n = 3. (D) 3D organoids were grown with indicated ESCC cell lines in RPMI-10% FBS for morphological analyses by H&E staining. All tested cell lines give rise to solid 3D organoids comprising atypical cells with high nuclear/cytoplasmic ratio. Organoids from TE4, TE9 and TE10 show more basophilic cells with moderate to severe atypia. TE11 organoids show more eosinophilic cytoplasm and little nuclear atypia. TE8 and TE5 organoids represent intermediate variants with moderate atypia and eosinophilic cytoplasm. Scale bar = 100 μm.
Interestingly, the successful formation of patient-derived tumor organoids was significantly associated with poor therapy response in the patients from whom biopsies were taken before chemotherapy or chemoradiotherapy (Tables 4 and 5). Such a finding suggests that the current 3D organoid culture conditions may be utilized clinically to predict therapy response because they may be more permissive for the formation of 3D organoids by SCC cells with a capability of adaptation or survival under therapy-related stress.
Table 4.
Tumor Organoids Formation and Response to Chemotherapy or Chemoradiation Therapy in ESCC Patients
| Tumor-Derived 3D Organoids formation | Therapeutic Response |
||
|---|---|---|---|
| PD+SD (n = 10) | PR (n = 6) | P value | |
| Success (n = 11) | 9 | 2 | .0357 |
| Failure (n = 5) | 1 | 4 | |
NOTE. There was no patient with complete response. Although oncologists typically classify SD as a clinical response, stable disease (SD), either alone or combination with partial response (PR), was not associated with successful organoid formation. Fisher's exact test was used to examine statistical significance.
PD, progressive disease.
Table 5.
Tumor Organoids Formation and Histological Response to Chemotherapy or Chemoradiation Therapy in ESCC Patients
| Tumor-Derived 3D organoids formation | Histological Response |
||
|---|---|---|---|
| Grade 1 (n = 6) | Grade 2 or 3 (n = 3) | P Value | |
| Success (n = 5) | 5 | 0 | .0476 |
| Failure (n = 4) | 1 | 3 | |
NOTE. There was no patient with Grade 0 after chemotherapy or chemoradiation therapy. Fisher's exact test was used to examine statistical significance.
3D Organoids Recapitulate Intratumoral Functional Cancer Cell Heterogeneity
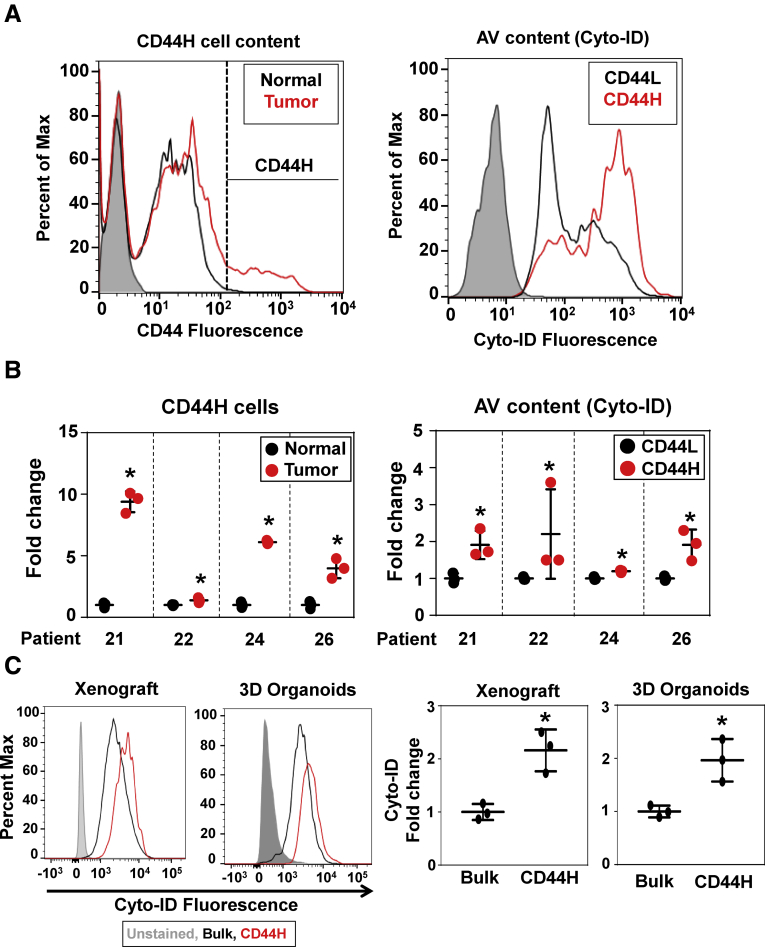
High autophagic activity in tumors correlates with poor clinical outcome in ESCC patients.13 SCC tissues comprise heterogeneous cancer cells and other cell types. Tumor cell heterogeneity was documented in patient biopsies, representing both ESCC and OPSCC, via flow cytometry that revealed a broader range of cell surface CD44 expression within tumors compared with adjacent normal mucosa (Figure 5A and B) where >99% of CD44-positive cells were negative for CD45, a marker for immune cells. Moreover, tumor cells with high CD44 expression (CD44H) contained more autophagic vesicles (AVs) (Figure 5A and B), suggesting that CD44H cells may have a higher autophagic capacity to cope with a variety of stressors present in the tumor microenvironment. This is in line with the role of autophagy-mediated redox regulation in ESCC CD44H cell homeostasis.13 To our knowledge this is the first report documenting high autophagic activities in live CD44H SCC cells within patient tumor biopsies. Nonepithelial cell types such as inflammatory cells and fibroblasts barely grew in aDMEM/F12+ medium, indicating that the cell culture medium is more permissive for epithelial cells to grow in 3D. We next evaluated the extent to which the organoid culture conditions may recapitulate the intratumoral SCC cell heterogeneity in single cell-derived SCC 3D organoids. To that end, we utilized TE11, an extensively characterized ESCC cell line13, 27, 28 to compare cell surface expression of CD44 in 3D organoids and xenograft tumors grown in immunodeficient mice. Flow cytometry revealed a similar cell surface CD44 expression pattern in both 3D organoids and xenograft tumors (data not shown). Moreover, AV content was increased within CD44H cells and comparable between 3D organoids and xenograft tumors (Figure 5C). Thus, the 3D organoid culture conditions may maintain the functional heterogeneity of intratumoral SCC cells.
Figure 5.
Tumor biopsies and TE11 3D organoids contain CD44H SCC cells with high autophagic activities. Intratumoral heterogeneity was evaluated by flow cytometry for cell surface markers and autophagic activities. Biopsies from tumors and adjacent normal mucosa from 2 ESCC and 2 OPSCC patients were dissociated for flow cytometry to detect CD44H cells and AV content (Cyto-ID). (A) The representative histograms of patient 21. (B) All 4 patients (biological replicates) showed an increased CD44H cell content in tumors compared with normal mucosa (*P < .05, vs normal, n = 3 by Student’s t test) and an increased AV content in CD44H cells as compared with CD44L cells (*P < .05, vs CD44L, n = 3 by Student’s t test) within tumor biopsies. (C) The cell surface expression pattern of CD44 of TE11 ESCC cells was analyzed in xenograft tumors and 3D organoids. The AV content (Cyto-ID) in CD44H cells was compared with the bulk population of TE11 cells in xenograft tumors and 3D organoids. Representative histogram plots (left) are shown with the corresponding quantification (right) from 3 independent experiments with similar results (*P < .05 vs bulk, n = 3 by Student’s t test). All data are presented as mean ± SD.
3D Organoids Reveal Potentially Therapy Resistant SCC Cell Populations Characterized by High CD44 Expression and Autophagy
Given the observed relationship between increased tumor organoid formation and therapy resistance in patients (Tables 4 and 5), we hypothesized that therapy-refractory CD44H cells are more capable of SCC 3D organoid formation.
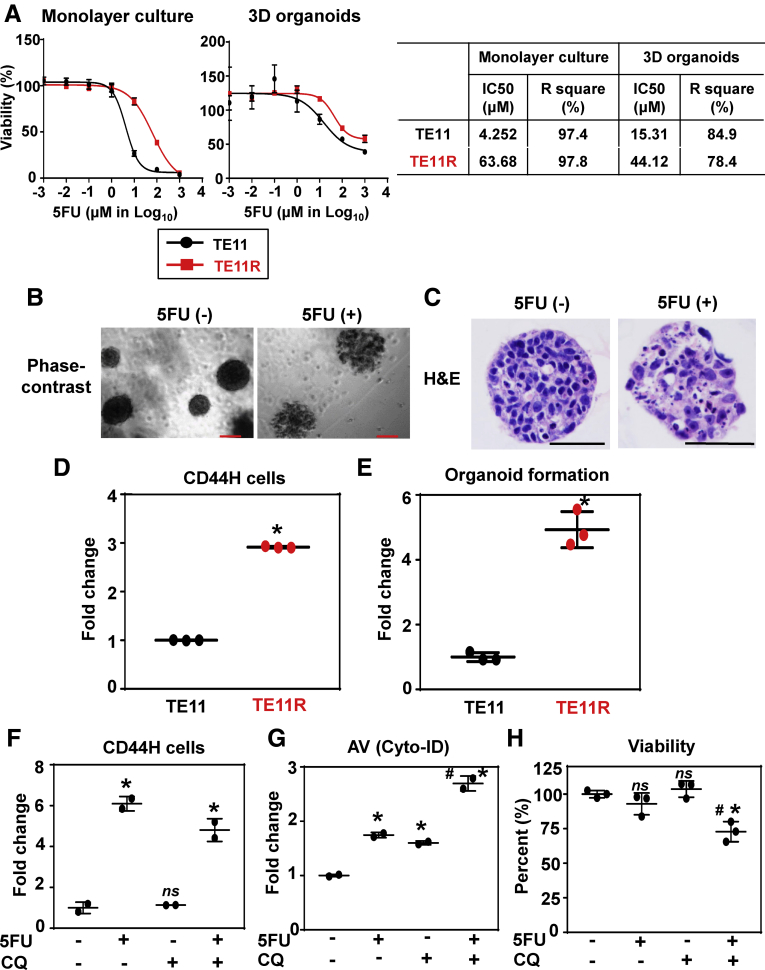
We asked first how 3D organoid culture conditions may influence SCC cell response to 5FU, a standard SCC chemotherapy agent. To that end, TE11 and 5FU-resistant derivative of TE11 (TE11R) cells were exposed to a variety of 5FU concentrations in monolayer culture as well as established 3D organoid structures for 72 hours in 96-well plates. Following 5FU treatment, we have determined the half maximal inhibitory concentration (IC50) via WST1 assays. The IC50 confirmed higher 5FU resistance of TE11R cells as compared with parental TE11 cells in both monolayer and 3D organoid culture conditions. Additionally, the IC50 revealed increased 5FU resistance of parental TE11 cells in 3D organoids compared with monolayer culture conditions (Figure 6A). Phase-contrast imaging documented disintegrated 3D structures in response to 5FU treatment (Figure 6B). H&E staining confirmed apoptotic cells with increased vacuolization and nuclear fragmentation as well as decreased cell adhesiveness and increased intercellular spaces (Figure 6C).
Figure 6.
Evaluation of 5FU treatment and the relationship between CD44H cells and autophagy in TE11 3D organoids. TE11 and TE11R cells were analyzed for 5FU sensitivity. (A) WST1 assays were performed to determine 5FU IC50. 3D organoids were grown with TE11 and TE11R cells for 8 days. The established 3D organoids and subconfluent monolayer culture (control) were treated with indicated concentrations of 5FU for 72 hours. Representative dose response curves for TE11 and TE11R under indicated conditions are shown (left) along with the IC50 and R2 values in the table (right). Cell morphology was assessed (B) under a phase-contrast microscopy and (C) by H&E staining. (B) Note that TE11 organoids treated with 5FU displayed disorganized structure (right). Scale bar = (B) 100 μm, (C) 50 μm. (A–C) Data represent 2 independent experiments with similar results with 3 samples per condition in each experiment. TE11 and TE11R were analyzed for (D) CD44H cell content and (E) organoid formation capability. (D) Flow cytometry was used to evaluate cell surface CD44 expression in monolayer culture. Representative histogram plots (top) and quantification (bottom) of CD44 expression are shown. (E) Organoid formation rate was compared by seeding an equal number of TE11 and TE11R cells. (D, E) Data represent 2 independent experiments with similar results with 3 samples per condition in each experiment. (D, E) *P < .05 vs TE11; n = 3 by Student’s t test. (F) CD44H cell content, (G) autophagy activity (Cyto-ID), and (H) cell viability (4',6-diamidino-2-phenylindole) were evaluated by flow cytometry in established 3D organoids following treatment for 72 hours with or without 5FU at a sublethal concentration (100 μM) along with or without 1 μg/mL CQ, a pharmacological inhibitor of autophagic flux. (G) Representative histogram plots (left) are shown with the corresponding quantification (right). All data are presented as mean ± SD. *P < .05 vs 5FU (-) CQ (-); #P < .05 vs 5FU (+) CQ (-). ns, not significant; n = 2 to 3 by 1-way analysis of variance with multiple comparisons (Tukey). (F–H) Data represent 2 independent experiments with similar results.
We next carried out flow cytometry analysis of TE11 and TE11R cells to determine cell surface CD44 expression. TE11R cells showed higher CD44 expression than parental TE11 cells (Figure 6D) and that TE11R cells were more capable of forming 3D organoids than parental TE11 cells (Figure 6E), suggesting the link between organoid formation capacity to therapy resistant CD44H subset of SCC cells.
We investigated further the utility of 3D organoids as a platform to characterize therapy resistant SCC cells. To that end, we analyzed SCC cells surviving 5FU exposure in 3D organoids. When 3D organoids were analyzed by flow cytometry following 5FU treatment, dissociated 3D organoids demonstrated an enrichment of CD44H cell populations (Figure 6F). Since CD44H cells display a higher capacity of autophagy,13 we determined autophagic flux in TE11 3D organoids. The presence of basal autophagic flux was documented by flow cytometry as increased AV content in established 3D organoid structures in the presence of Chloroquine (CQ), a pharmacological inhibitor of autophagic flux. 5FU increased AV content that was augmented further by concurrent CQ treatment (Figure 6G), indicating the presence of the autophagic flux in cells treated with 5FU. Moreover, CQ increased 5FU-mediated cytotoxicity in TE11 3D organoids (Figure 6H), suggesting that autophagy may facilitate the survival of TE11 cells upon 5FU treatment.
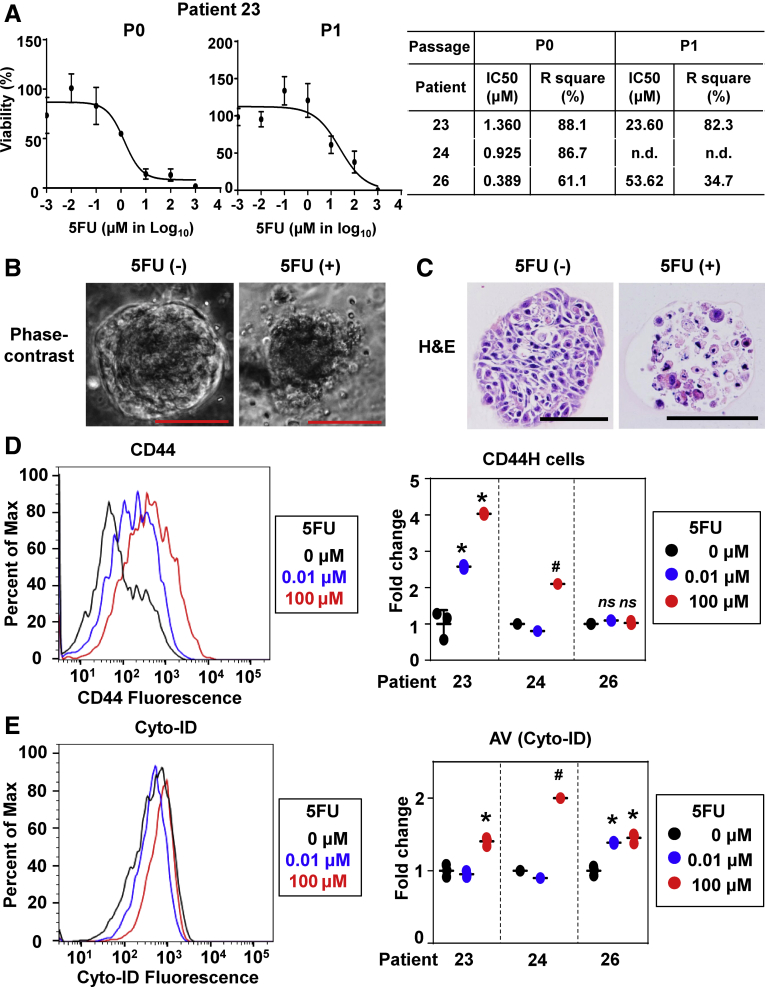
Finally, we tested patient tumor-derived 3D organoids for 5FU treatment. Tumor-derived primary 3D organoids (passage 0) from 3 independent OPSCC patients responded to 5FU to a variable degree, reflecting upon the IC50 values (Figure 7A). As observed in TE11 3D organoids treated with 5FU (Figure 6), phase-contrast imaging and H&E staining showed that 5FU treatment decreased cell adhesiveness to increase the intracellular spaces, thereby disintegrating the 3D structures with concurrent nuclear fragmentation in SCC cells (Figure 7B and C), suggesting 5FU-induced apoptotic cell death in primary tumor-derived 3D organoids. We suspected that coexisting non-neoplastic 3D structures within primary tumor-derived 3D organoids (Figure 3C, Table 3) may increase the overall 5FU sensitivity, leading to an underestimation of the IC50 values for SCC cells present in 3D organoids. Therefore, we exposed passaged tumor-derived 3D organoids where non-neoplastic structures were eliminated. There was a significant increase in IC50 for the passaged 3D organoids in 2 independent patients tested (patients 23 and 26) (Figure 7A), suggesting that 5FU resistance may have increased in secondary 3D organoids (P1) compared with primary 3D organoids (P0). The increased IC50 in the passaged 3D organoids may be accounted for by an enrichment of SCC cells. However, tumor-derived primary 3D organoids from the above patients contained 25% or a smaller fraction of non-neoplastic 3D structures (Table 3). Thus, the increased IC50 may be explained by an increase in therapy-resistant CD44H cells or activation of cytoprotective mechanisms such as autophagy. We analyzed the 5FU-surviving SCC cells in patient tumor-derived 3D organoids by flow cytometry, which revealed an enrichment of CD44H cells in 5FU-treated 3D organoids from patient 23, but not patient 26 (Figure 7D). However, 5FU increased the overall AV contents in 3D organoids from both patients 23 and 26 (Figure 7E). These observations suggest that 5FU-treated patient tumor-derived 3D organoids may reveal a patient-to-patient variability in mechanisms for 5FU-resistance.
Figure 7.
Evaluation of 5FU-induced cytotoxicity, CD44H cells and autophagy in patient tumor-derived 3D organoids. OPSCC patient tumor-derived 3D organoids were treated with indicated concentrations of 5FU for 72 hours. WST1 assays were performed to determine 5FU IC50. (A) Representative dose response curves for primary (P0) and secondary (P1) 3D organoids from patient 23 (left) are shown along with the IC50 and R square values in the table for 3 independent patients as biological replicates (right). Cell morphology was assessed (B) under phase-contrast microscopy and (C) by H&E staining. Scale bar = 100 μm. (D, E) Tumor-derived 3D organoids from 3 independent patients were grown for 7 days followed by treatment with indicated concentrations of 5FU for 72 hours. (D) CD44H cell content and (E) AV content (Cyto-ID) were evaluated by flow cytometry. The representative histograms of patient 23 (left) are shown with the corresponding quantification (right); data of patients 23, 24, and 26 (3 biological replicates) are presented as mean ± SD. (D, E) One-way analysis of variance with multiple comparisons (Tukey) was used to examine statistical significance. *P < .05 vs 0 μM (n = 3) in each patient. not significant (ns) vs 0 μM (n = 3) in each patient. #No technical replicate was available due to the small number of viable cells that were recovered from 3D organoids in patient 24.
We have previously demonstrated that poor prognosis in ESCC patients is associated with high ESCC cell expression of cleaved LC3, a marker of active autophagic flux.11 When we evaluated cleaved LC3 by IHC in patient biopsies and tumor-derived 3D organoids, 44% of tumor biopsies (n = 16) displayed moderate to intense expression of cleaved LC3 as expected11 and that was maintained in 3D organoids (Figure 3E); however, there was no correlation between expression of cleaved LC3 in tumor biopsies and successful formation of tumor-derived 3D organoids (data not shown), albeit a small sample size. Thus, basal autophagy level was not predictive of the organoid formation.
Discussion
We describe for the first time the successful generation and characterization of tumor-derived 3D organoids from patients with SCCs and provide proof of principle about their utility as a robust platform to analyze cancer cell heterogeneity, evaluate therapy response and explore therapy resistance mechanisms with the potential of translation in the setting of personalized medicine for ESCC and OPSCC. To date, current therapies for these cancers require ongoing attention for improvement in survival rates. Unlike other esophageal 3D culture systems such as multicellular spheroid culture under free-floating conditions (see Whelan et al24 for an extensive review), our 3D organoids are defined as single cell-derived spherical structures grown in Matrigel with advantages (Table 6) including the capacity of determining stemness (self-renewal) and fate of cells. Importantly, analyses of SCC 3D organoids for proliferation, p53 expression, cell surface CD44 expression, and autophagy of SCC organoids revealed faithful recapitulation of key genetic, morphological and functional characteristics of the original tumors.
Table 6.
Characteristics of the Patient Tumor Biopsy–Derived SCC 3D Organoids in this Study
| Unique Features/Advantages | Technical Limitations/Unknown Factors |
|---|---|
|
|
Conventional SCC cell lines are established in monolayer culture over 1–3 months at a low success rate (<40%).29 Neoadjuvant therapy in current standard ESCC care makes isolation of viable SCC cells difficult from surgically resected tumors. Circumventing these problems, our protocol permits 3D organoid growth in a relatively short time period (typically within 2 weeks) with a small number of cells (2 × 104 cells/well and ∼4–5 × 105 live cells/biopsy) available via diagnostic biopsies from patients before therapeutic intervention. Moreover, 96-well plates were used to determine 3D organoid formation rate and cell viability under a phase-contrast microscopy and WST1 assays, respectively, indicating that tumor-derived 3D organoids could be evaluated in a moderate-to-high throughput manner.
Limitations (Table 6) in our protocol include sporadic fungal contamination despite prophylactic Amphotericin B (Fungizone) treatment, owing to likely a unique microbiota in the stenotic tumor-bearing oropharyngeal and esophageal tract. Excluding the patients with fungal contamination, the success rate of 3D organoid formation from tumor samples was approximately 70%, potentially due to impaired cell viability during cell isolation from biopsies or less epithelial cell contents (ie, more stroma cell contents) in biopsies. Moreover, a subset of tumor-derived 3D organoids contained histologically non-neoplastic cells and that 9 of 12 (75%) tumor-derived 3D organoids contained more non-neoplastic than neoplastic structures under our cell culture conditions (Table 3). Additionally, none of our SCC 3D organoids, unlike normal mucosa-derived organoids, displayed apparent keratinization or cornification, a hallmark of well-differentiated squamous cell carcinomas, despite the histological grade of the originating tumors. These observations indicated that the aDMEM/F12+-based medium may not be permissive for propagation of well-differentiated SCC cells or ex vivo differentiation of SCC cells. The higher success rate of ESCC 3D organoid formation from patients whose tumors display therapeutic resistance (Tables 4 and 5), albeit the small sample size, suggests also that our organoid culture conditions may be permissive for SCC cells with an increased capability of coping with stress. Providing cytoprotection under oxidative stress, autophagy is activated in ESCC patients who experience early recurrence and poor prognosis and that autophagy-mediated redox homeostasis is essential for CD44H cell generation and maintenance in ESCC13; however, the success of patient tumor-derived 3D organoid formation was not linked to the autophagic activity in biopsies (data not shown). Although CD44H cells in patient biopsies are likely to display a high organoid formation capability as suggested by TE11 CD44H cells (Figure 5C), we could analyze CD44H cell content in 4 patients only (Figure 5B). Extensive studies with larger sample sizes are warranted as a future direction to compare patient-derived 3D organoids from therapy-responders and non-responders, and establish the relationship between autophagy activity or CD44H cell content in tumor biopsies and the organoid formation. While CD44 is a well-established marker of stemness and malignant attributes in ESCC and OPSCC,13, 28, 30 other markers may influence 3D organoid formation, and can be explored.
The fully supplemented aDMEM/F12+ medium contains factors and agents that facilitate self-renewal of normal murine esophageal stem cells and has been successfully utilized for murine esophageal 3D organoids22, 31, 32; however, our normal mucosa-derived 3D organoids failed to be passaged, suggesting a potential species difference in the requirement of medium components. In this regard, we have optimized a condition that permits multiple passages for both murine and human normal esophageal 3D organoids,23 although it is currently unknown whether this condition is permissive for SCC 3D organoid growth. In addition, agents such as SB202190, a p38 MAPK inhibitor used in the aDMEM/F12+-based medium may be inhibitory for tumor growth.33 The aDMEM/F12+-based medium without SB2022190 and other supplements has been utilized to grow organoids from chemical carcinogen-induced murine tongue squamous cell carcinoma.34 Thus, further optimization may be needed for 3D organoid formation from human ESCC, OPSCC, and potentially other squamous cell carcinomas.
Experiments using clinically relevant concentrations of 5FU35 revealed lower 5FU sensitivity (ie, higher IC50) in 3D organoids compared with monolayer cultures (Figures 6A and 7A). drug resistance may be accounted for by cell intrinsic properties such as CD44 expression, autophagy and EMT. Although we have not explored EMT in this study, EMT has been documented in murine ESCC 3D organoids.28 Additionally, Matrigel and the 3D structures may limit drug diffusion; however, the cytotoxic effect of 5FU was not limited to the periphery of the 3D structures (Figures 6C and 7C). Patient-derived 3D organoids with evidence of 5FU resistance contained CD44H cells with increased autophagy capacity (Figure 7D and E). Thus, 3D organoids can be used as a tool to identify therapy resistance mechanisms while predicting therapy response. SCC cell lines, including unique drug-resistant derivatives,36 provide references of IC50 for therapeutic agents. Additionally, they will provide quality control to calibrate the influence of variables such as different lots of Matrigel and medium components to grow multiple biopsy-derived 3D organoids in expanded studies. The successful use of 96-well plates to evaluate cell proliferation and drug response has established the feasibility for SCC 3D organoids to serve a platform in moderate-to-high throughput drug screening. In addition to chemotherapeutics, radiation therapy and molecularly targeted therapeutics could be tested either alone or in combination. Identification of molecular determinants in therapy resistance will be facilitated by unbiased sequencing studies of patient-derived 3D organoids for genomic and epigenetic landscape1 and gene expression profiling of a library of well-annotated patient tumor-derived 3D organoids. Such a study is currently underway.
Given the importance of the tumor microenvironment, investigation of SCC-relevant niche factors and optimization of conditions for co-culturing with immune cells, endothelial cells, and cancer-associated fibroblasts is meritorious,24 especially given the advent and expansion for indications of immune checkpoint inhibitors in cancer therapy. Integration of the immune system in 3D organoid culture may open up new avenues of investigation. Additional future applications of 3D organoids in the clinical setting include analysis of precancerous conditions (ie, dysplasia) and metastatic lesions.
Materials and Methods
Patients and Treatment
In accordance with Institutional Review Board–approved protocols and guidelines at each participating institution, biopsies were taken from tumors and adjacent normal mucosa upon diagnostic endoscopy from therapy-naïve ESCC and OPSCC patients between October 2015 and May 2018. Twenty ESCC patients were enrolled at Kagoshima University. One ESCC and 5 OPSCC patients were enrolled at the Hospital of the University of Pennsylvania. One ESCC patient was enrolled at Osaka University. Staging was done as described (Table 1).37 At Kagoshima University, 15 ESCC patients received chemoradiation therapy and 5 patients received chemotherapy alone according to published protocols.38 Therapy response was classified clinically into complete response, partial response, stable disease, and progressive disease as described.39 When ESCC patients underwent surgery 4 weeks after chemotherapy or chemoradiation therapy, histology of resected tumors was evaluated for therapeutic efficacy as described40, 41 where grade 0 or 1 was ineffective and grade 2 or 3 was effective.
Cell Lines, Monolayer, and 3D Organoid Culture
ESCC cell lines (TE series)42, 43 were grown in RPMI1640 (11875093, Thermo Fisher Scientific, Waltham, MA) containing 10% fetal bovine serum (FBS) (SH3007103, Thermo Fisher Scientific) and 100-U/mL penicillin and 100-μg/mL streptomycin (15140122, Thermo Fisher Scientific), at 37°C in a humidified atmosphere of 5% CO2. A 5FU-resistant derivative of TE11 (TE11R) was previously described.44, 45 Cell authentication was externally done by short-tandem repeat DNA profiling at the American Type Culture Collection (Manassas, VA).
To generate 3D organoids, each piece of the biopsies was rinsed in saline (0.9% NaCl) (S7653, Sigma-Aldrich, St. Louis, MO), and minced immediately with scissors and incubated for 45 minutes at 37°C in a 1-mL cocktail comprising 6.25-mg/mL dispase I (354235, BD Biosciences, San Jose, CA), 5-mg/mL collagenase (17018029, Thermo Fisher Scientific), 10-mM ROCK inhibitor Y27632 (1254, R&D Systems, Minneapolis, MN), and 0.5-mg/mL Fungizone (15290018, Thermo Fisher Scientific) with a periodical mixing (∼once every 10 minutes) via Vortex-Genie 2 (Scientific Industries, Bohemia, NY). The solution was replaced with - mL 0.25% trypsin/EDTA (R001100, Thermo Fisher Scientific) and incubated for 10 minutes at 37°C. The trypsin activity was then neutralized with 6-mL 250 mg/L soybean trypsin inhibitor (T9128, Sigma-Aldrich) in Dulbecco's phosphate-buffered saline (DPBS) (10010023, Thermo Fisher Scientific). The enzymatically dissociated tissue was forced through a 70-μm cell strainer (22-363-548, Thermo Fisher Scientific) and cells were pelleted via centrifugation at 2000 rpm for 5 minutes at room temperature. Cells were then resuspended in 1-mL advanced Dulbecco’s modified Eagle medium (DMEM)/F12 (12491015, Thermo Fisher Scientific) containing 100-U/mL penicillin and 100-μg/mL streptomycin (15140122, Thermo Fisher Scientific), 1×GlutaMAX (35050061, Thermo Fisher Scientific), and 1×HEPES (15630080, Thermo Fisher Scientific) (aDMEM/F12+). The Countess II FL Automated Cell Counter (Thermo Fisher Scientific) was used to count cells where Trypan blue exclusion test was done to determine cell viability. The cell suspension (4 × 105/mL) was mixed further with 10× volume of ice-cold Matrigel (354234, Corning, Corning, NY) and 2 × 104 cells in 50 μL of Matrigel were seeded into each well of the flat bottom 24-well plates. After solidification, 500 μL of aDMEM/F12+ supplemented with 1×N2 (17502048, Thermo Fisher Scientific), 1× B27 supplements (17504044, Thermo Fisher Scientific), 0.1-mM N-acetyl-L-cysteine (A7250, Sigma-Aldrich), 50-ng/mL recombinant human EGF (236-EG, R&D Systems), 2% Noggin/R-Spondin–conditioned media, 100-ng/mL recombinant human Wnt3A (5036-WN, R&D Systems), 500-nM A83-01 (2939, R&D Systems), 10-nM SB202190 (1264, R&D Systems), 10-nM gastrin (3006, R&D Systems), 10-nM nicotinamide (N0636, Sigma-Aldrich), and 10-μM Y27632, was added, a composition described for murine esophageal 3D organoids22 and replenished every other day. 3D organoids were grown for 10–14 days at 37°C in a humidified atmosphere of 5% CO2. When ESCC cell lines were used to generate 3D organoids, monolayer cultures were trypsinized to prepare cell suspensions. Under an inverted phase-contrast microscope (Apotome, Carl Zeiss, Jena, Germany), growing organoids were observed and photomicrographed to determine their number and size. Organoid formation rate was defined as the average number of ≥50-μm spherical structures at day 14 that was divided by the total number of cells seeded in each well at day 0. When indicated, 3D organoids were grown in the presence or absence of 5FU (F6627, Sigma-Aldrich) or chloroquine (CQ) (C6628, Sigma-Aldrich) at indicated concentrations after organoid structure was established at day 8. Dimethyl sulfoxide (BP231-1, Thermo Fisher Scientific) was used as vehicle for 5FU. CQ was reconstituted in water as described.13 3D organoids were recovered by digesting Matrigel with Dispase I (354235, BD Biosciences, 1U/mL), embedded in iPGell (GSPG20-1, GenoStaff, Tokyo, Japan) and fixed overnight in 4.0% paraformaldehyde for histological analyses. Alternatively, isolated organoids were dissociated by incubation with 0.25% Trypsin-EDTA, Y27632 and DNase I (1010415901, Sigma-Aldrich) into single cell suspensions and passaged.
WST1 Assays and Half Maximal Inhibitory Concentration Determination
Cell survival and the half maximal inhibitory concentration (IC50) for 5FU were determined as described.36 TE11 cells were seeded at the density of 200 cells in 5 μL Matrigel with 100-μL medium per well in 96-well plates. To treat patient-derived 3D organoids with 5FU, cells dissociated from biopsies were seeded at the density of 4,000 cells in 5 μL Matrigel with 100μL medium per well in 96-well plates. Established 3D organoids were untreated or treated with 5FU at a range of final concentrations (10–3–103 μM) for 72 hours, and further incubated with WST-1 reagent (11644807001, Sigma-Aldrich) (10 μL per well) for 2 hours before colorimetric cell proliferation assays, according to the manufacturer's instructions. Dose response curves were generated in GraphPad Prism 7.0 using the least squares fit (ordinary) with variable slope (4 parameters). Outliers were eliminated using the built-in outlier detection method. All experiments were performed in triplicate.
Histology and Immunohistochemistry
Paraffin-embedded biopsies and 3D organoid products were serially sectioned and subjected to hematoxylin and eosin (H&E) staining and immunohistochemistry (IHC) as described previously.13, 46 For IHC, sections were incubated with anti-p53 polyclonal antibody (M7001, 1:200; Dako, Copenhagen, Denmark) or anti-Ki67 monoclonal antibody (M7240, 1:800; Dako), or anticleaved light chain 3 (LC3) polyclonal antibody (AP1805a, 1:250; Abgent, San Diego, CA) overnight at 4°C. H&E stained slides were evaluated by 2 pathologists (SN and AKS) blind to clinical data and conditions. Ki-67 labeling index was determined by counting at least 10 organoids per group.
Xenograft Tumors
Xenograft tumors of TE11 cells were formed in immunodeficient mice (NU/J) for flow cytometry under an Institutional Animal Care and Use Committee-approved protocol. In brief, cells were suspended in 50% Matrigel and implanted subcutaneously into the dorsal flanks of 4 week old female athymic nu/nu mice (Taconic Biosciences, Hudson, NY, USA). Tumor growth was monitored using digital calipers. Tumors were harvested and minced into 1 mm3 pieces and incubated in Dulbecco's Modified Eagle Medium (DMEM, 11965, Thermo Fisher Scientific) containing 1 mg/mL collagenase I (C9263-1G, Sigma-Aldrich) at 37°C for 90 minutes. Following centrifugation, residual tissue pieces were digested in 0.05% trypsin-EDTA (2530062, Thermo Fisher Scientific) at 37°C for 10 minutes and then with 1 U/mL Dispase (354235, BD Biosciences) and 100μg/mL DNase I (1010415901, Sigma-Aldrich) at 37°C for 10 minutes. Dissociated tumor cells were filtrated, rinsed and collected into a 5 mL round bottom tube with a 40 μm cell strainer cap (352235, BD Biosciences) with DPBS and pelleted by centrifugation at 1,500 rpm for 5 minutes at 4°C.
Flow Cytometry for Cell Surface Markers and Autophagy
Flow cytometry was performed using FACSCalibur, FACSCanto, or LSR II cytometers (BD Biosciences) and FlowJo software (Tree Star, Ashland, OR). Cells suspended in DPBS containing 1% bovine serum albumin (A2058, Sigma-Aldrich). 4',6-Diamidino-2-phenylindole (D1306, Thermo Fisher Scientific) was used to determine cell viability. Cells were subjected to flow cytometry for cell surface expression of CD44 as described13, 47 where cells were stained with APC-anti-CD44 (1:20; 31118; BD Biosciences) on ice for 30 minutes. AVs were determined with Cyto-ID fluorescent dye (ENZ-51031-K200, Enzo Life Sciences, Farmingdale, NY) as described13, 46 where cells were incubated with Cyto-ID at 1:1000 in DPBS containing 5% FBS and 1% bovine serum albumin at 37°C for 30 minutes. Unstained cells were utilized to establish the background fluorescence. The mean fluorescence in live cells was determined for each sample and is presented after subtraction of background fluorescence.
Statistical Analyses
Data were analyzed as indicated using the GraphPad Prism 7.0 software (GraphPad, La Jolla, CA). Equal variance across groups being compared was confirmed by Bartlett’s test for analysis of variance (ANOVA) or F test (Student’s t test). Statistical analysis of group differences was performed using the fisher’s exact test and done using SAS statistical software (SAS Institute, Cary, NC). P value of <.05 was considered significant.
Acknowledgements
The authors thank Ms Rie Tajiri for technical assistance in 3-dimensional organoid culture and morphological analyses at Kagoshima University and Mr Ben Rhoades and the staff of the Molecular Pathology and Imaging Core, Host-Microbial Analytic and Repository Core, Cell Culture/iPS Core and Flow Cytometry and Cell Sorting Facilities at the University of Pennsylvania for technical support. The authors thank members of the esophageal team at Department of Digestive Surgery, Breast and Thyroid Surgery of Kagoshima University, the Nakagawa lab, and the Rustgi lab for helpful discussions. Prasanna M Chandramouleeswaran is currently in the graduate program in Cell Biology, Physiology, and Metabolism at the University of Pennsylvania.
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This study was supported by the Grant-in-Aid for challenging Exploratory Research, Grant in Aid for Scientific Research B and Grant in Aid for Scientific Research C from the Ministry of Education, Culture, Sports, Science and Technology of Japan (17H04285 to SN; and 15K10108 to YK). This study was also supported by the following NIH Grants: P01CA098101 (HN, KT, KAW, VG, AJK, AB, JAD, AKR), U54CA163004 (HN, GGG, AKR), R01DK114436 (HN), R01AA026297 (HN), R01AA022986 (NGA), P30ES013508University of Pennsylvania Center of Excellence in Environmental Toxicology (HN and AKR), K01DK103953 (KAW), F32CA174176 (KAW), T32DK007066 (KAW), K08DK106444 (ABM), the American Cancer SocietyRP-10-033-01-CCE (AKR), NIH/NIDDKP30DK050306 Center of Molecular Studies in Digestive and Liver Diseases, The Molecular Pathology and Imaging, Molecular Biology/Gene Expression, Cell Culture/iPS and Mouse Core Facilities. KT is a recipient of the Japan Society for the Promotion of Science Postdoctoral Fellowship. VG is a recipient of the Fonds de Recherche du Québec – Santé Postdoctoral Fellowship (P-Giroux-27692 and P-Giroux-31601).
Contributor Information
Hiroshi Nakagawa, Email: nakagawh@pennmedicine.upenn.edu.
Anil K. Rustgi, Email: anil2@pennmedicine.upenn.edu.
Shoji Natsugoe, Email: natsugoe@m2.kufm.kagoshima-u.ac.jp.
References
- 1.Dotto G.P., Rustgi A.K. Squamous cell cancers: a unified perspective on biology and genetics. Cancer Cell. 2016;29:622–637. doi: 10.1016/j.ccell.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rustgi A.K., El-Serag H.B. Esophageal carcinoma. N Engl J Med. 2014;371:2499–2509. doi: 10.1056/NEJMra1314530. [DOI] [PubMed] [Google Scholar]
- 3.Ohashi S., Miyamoto S., Kikuchi O., Goto T., Amanuma Y., Muto M. Recent advances from basic and clinical studies of esophageal squamous cell carcinoma. Gastroenterology. 2015;149:1700–1715. doi: 10.1053/j.gastro.2015.08.054. [DOI] [PubMed] [Google Scholar]
- 4.Shapiro J., van Lanschot JJB, Hulshof M., van Hagen P., van Berge Henegouwen M.I., Wijnhoven B.P.L., van Laarhoven H.W.M., Nieuwenhuijzen G.A.P., Hospers G.A.P., Bonenkamp J.J., Cuesta M.A., Blaisse R.J.B., Busch O.R.C., Ten Kate F.J.W., Creemers G.M., Punt C.J.A., Plukker J.T.M., Verheul H.M.W., Bilgen E.J.S., van Dekken H., van der Sangen M.J.C., Rozema T., Biermann K., Beukema J.C., Piet A.H.M., van Rij C.M., Reinders J.G., Tilanus H.W., Steyerberg E.W., van der Gaast A., group Cs. Neoadjuvant chemoradiotherapy plus surgery versus surgery alone for oesophageal or junctional cancer (CROSS): long-term results of a randomised controlled trial. Lancet Oncol. 2015;16:1090–1098. doi: 10.1016/S1470-2045(15)00040-6. [DOI] [PubMed] [Google Scholar]
- 5.Domenge C., Hill C., Lefebvre J.L., De Raucourt D., Rhein B., Wibault P., Marandas P., Coche-Dequeant B., Stromboni-Luboinski M., Sancho-Garnier H., Luboinski B. French Groupe d'Etude des Tumeurs de la Tete et du C. Randomized trial of neoadjuvant chemotherapy in oropharyngeal carcinoma. French Groupe d'Etude des Tumeurs de la Tete et du Cou (GETTEC) Br J Cancer. 2000;83:1594–1598. doi: 10.1054/bjoc.2000.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh A., Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;29:4741–4751. doi: 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Apel A., Herr I., Schwarz H., Rodemann H.P., Mayer A. Blocked autophagy sensitizes resistant carcinoma cells to radiation therapy. Cancer Res. 2008;68:1485–1494. doi: 10.1158/0008-5472.CAN-07-0562. [DOI] [PubMed] [Google Scholar]
- 8.Sui X., Chen R., Wang Z., Huang Z., Kong N., Zhang M., Han W., Lou F., Yang J., Zhang Q., Wang X., He C., Pan H. Autophagy and chemotherapy resistance: a promising therapeutic target for cancer treatment. Cell Death Dis. 2013;4:e838. doi: 10.1038/cddis.2013.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao J.S., Li W.J., Ge D., Zhang P.J., Li J.J., Lu C.L., Ji X.D., Guan D.X., Gao H., Xu L.Y., Li E.M., Soukiasian H., Koeffler H.P., Wang X.F., Xie D. Tumor initiating cells in esophageal squamous cell carcinomas express high levels of CD44. PLoS One. 2011;6:e21419. doi: 10.1371/journal.pone.0021419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okamoto K., Ninomiya I., Ohbatake Y., Hirose A., Tsukada T., Nakanuma S., Sakai S., Kinoshita J., Makino I., Nakamura K., Hayashi H., Oyama K., Inokuchi M., Nakagawara H., Miyashita T., Hidehiro T., Takamura H., Fushida S., Ohta T. Expression status of CD44 and CD133 as a prognostic marker in esophageal squamous cell carcinoma treated with neoadjuvant chemotherapy followed by radical esophagectomy. Oncol Rep. 2016;36:3333–3342. doi: 10.3892/or.2016.5133. [DOI] [PubMed] [Google Scholar]
- 11.Sato F., Kubota Y., Natsuizaka M., Maehara O., Hatanaka Y., Marukawa K., Terashita K., Suda G., Ohnishi S., Shimizu Y., Komatsu Y., Ohashi S., Kagawa S., Kinugasa H., Whelan K.A., Nakagawa H., Sakamoto N. EGFR inhibitors prevent induction of cancer stem-like cells in esophageal squamous cell carcinoma by suppressing epithelial-mesenchymal transition. Cancer Biol Ther. 2015;16:933–940. doi: 10.1080/15384047.2015.1040959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Long A., Giroux V., Whelan K.A., Hamilton K.E., Tetreault M.P., Tanaka K., Lee J.S., Klein-Szanto A.J., Nakagawa H., Rustgi A.K. WNT10A promotes an invasive and self-renewing phenotype in esophageal squamous cell carcinoma. Carcinogenesis. 2015;36:598–606. doi: 10.1093/carcin/bgv025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whelan K.A., Chandramouleeswaran P.M., Tanaka K., Natsuizaka M., Guha M., Srinivasan S., Darling D.S., Kita Y., Natsugoe S., Winkler J.D., Klein-Szanto A.J., Amaravadi R.K., Avadhani N.G., Rustgi A.K., Nakagawa H. Autophagy supports generation of cells with high CD44 expression via modulation of oxidative stress and Parkin-mediated mitochondrial clearance. Oncogene. 2017;36:4843–4858. doi: 10.1038/onc.2017.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lancaster M.A., Knoblich J.A. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345:1247125. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- 15.Sato T., Clevers H. SnapShot: growing organoids from stem cells. Cell. 2015;161:1700–1700.e1. doi: 10.1016/j.cell.2015.06.028. [DOI] [PubMed] [Google Scholar]
- 16.Matano M., Date S., Shimokawa M., Takano A., Fujii M., Ohta Y., Watanabe T., Kanai T., Sato T. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat Med. 2015;21:256–262. doi: 10.1038/nm.3802. [DOI] [PubMed] [Google Scholar]
- 17.Sato T., Clevers H. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science. 2013;340:1190–1194. doi: 10.1126/science.1234852. [DOI] [PubMed] [Google Scholar]
- 18.van de Wetering M., Francies H.E., Francis J.M., Bounova G., Iorio F., Pronk A., van Houdt W., van Gorp J., Taylor-Weiner A., Kester L., McLaren-Douglas A., Blokker J., Jaksani S., Bartfeld S., Volckman R., van Sluis P., Li V.S., Seepo S., Sekhar Pedamallu C., Cibulskis K., Carter S.L., McKenna A., Lawrence M.S., Lichtenstein L., Stewart C., Koster J., Versteeg R., van Oudenaarden A., Saez-Rodriguez J., Vries R.G., Getz G., Wessels L., Stratton M.R., McDermott U., Meyerson M., Garnett M.J., Clevers H. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161:933–945. doi: 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boj S.F., Hwang C.I., Baker L.A., Chio I.I., Engle D.D., Corbo V., Jager M., Ponz-Sarvise M., Tiriac H., Spector M.S., Gracanin A., Oni T., Yu K.H., van Boxtel R., Huch M., Rivera K.D., Wilson J.P., Feigin M.E., Ohlund D., Handly-Santana A., Ardito-Abraham C.M., Ludwig M., Elyada E., Alagesan B., Biffi G., Yordanov G.N., Delcuze B., Creighton B., Wright K., Park Y., Morsink F.H., Molenaar I.Q., Borel Rinkes I.H., Cuppen E., Hao Y., Jin Y., Nijman I.J., Iacobuzio-Donahue C., Leach S.D., Pappin D.J., Hammell M., Klimstra D.S., Basturk O., Hruban R.H., Offerhaus G.J., Vries R.G., Clevers H., Tuveson D.A. Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160:324–338. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao D., Vela I., Sboner A., Iaquinta P.J., Karthaus W.R., Gopalan A., Dowling C., Wanjala J.N., Undvall E.A., Arora V.K., Wongvipat J., Kossai M., Ramazanoglu S., Barboza L.P., Di W., Cao Z., Zhang Q.F., Sirota I., Ran L., MacDonald T.Y., Beltran H., Mosquera J.M., Touijer K.A., Scardino P.T., Laudone V.P., Curtis K.R., Rathkopf D.E., Morris M.J., Danila D.C., Slovin S.F., Solomon S.B., Eastham J.A., Chi P., Carver B., Rubin M.A., Scher H.I., Clevers H., Sawyers C.L., Chen Y. Organoid cultures derived from patients with advanced prostate cancer. Cell. 2014;159:176–187. doi: 10.1016/j.cell.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sato T., Stange D.E., Ferrante M., Vries R.G., Van Es J.H., Van den Brink S., Van Houdt W.J., Pronk A., Van Gorp J., Siersema P.D., Clevers H. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 22.DeWard A.D., Cramer J., Lagasse E. Cellular heterogeneity in the mouse esophagus implicates the presence of a nonquiescent epithelial stem cell population. Cell Rep. 2014;9:701–711. doi: 10.1016/j.celrep.2014.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kasagi Y., Chandramouleeswaran P.M., Whelan K.A., Tanaka K., Giroux V., Sharma M., Wang J., Benitez A.J., DeMarshall M., Tobias J.W., Hamilton K.E., Falk G.W., Spergel J.M., Klein-Szanto A.J., Rustgi A.K., Muir A.B., Nakagawa H. The esophageal organoid system reveals functional interplay between notch and cytokines in reactive epithelial changes. Cell Mol Gastroenterol Hepatol. 2018;5:333–352. doi: 10.1016/j.jcmgh.2017.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whelan K.A., Muir A.B., Nakagawa H. Esophageal 3D culture systems as modeling tools in esophageal epithelial pathobiology and personalized medicine. Cell Mol Gastroenterol Hepatol. 2018;5:461–478. doi: 10.1016/j.jcmgh.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wagata T., Shibagaki I., Imamura M., Shimada Y., Toguchida J., Yandell D.W., Ikenaga M., Tobe T., Ishizaki K. Loss of 17p, mutation of the p53 gene, and overexpression of p53 protein in esophageal squamous cell carcinomas. Cancer Res. 1993;53:846–850. [PubMed] [Google Scholar]
- 26.Freed-Pastor W.A., Prives C. Mutant p53: one name, many proteins. Genes Dev. 2012;26:1268–1286. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kagawa S., Natsuizaka M., Whelan K.A., Facompre N., Naganuma S., Ohashi S., Kinugasa H., Egloff A.M., Basu D., Gimotty P.A., Klein-Szanto A.J., Bass A.J., Wong K.K., Diehl J.A., Rustgi A.K., Nakagawa H. Cellular senescence checkpoint function determines differential Notch1-dependent oncogenic and tumor-suppressor activities. Oncogene. 2015;34:2347–2359. doi: 10.1038/onc.2014.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Natsuizaka M., Whelan K.A., Kagawa S., Tanaka K., Giroux V., Chandramouleeswaran P.M., Long A., Sahu V., Darling D.S., Que J., Yang Y., Katz J.P., Wileyto E.P., Basu D., Kita Y., Natsugoe S., Naganuma S., Klein-Szanto A.J., Diehl J.A., Bass A.J., Wong K.K., Rustgi A.K., Nakagawa H. Interplay between Notch1 and Notch3 promotes EMT and tumor initiation in squamous cell carcinoma. Nat Commun. 2017;8:1758. doi: 10.1038/s41467-017-01500-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimada Y., Imamura M., Wagata T., Yamaguchi N., Tobe T. Characterization of 21 newly established esophageal cancer cell lines. Cancer. 1992;69:277–284. doi: 10.1002/1097-0142(19920115)69:2<277::aid-cncr2820690202>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 30.Facompre N.D., Harmeyer K.M., Sole X., Kabraji S., Belden Z., Sahu V., Whelan K., Tanaka K., Weinstein G.S., Montone K.T., Roesch A., Gimotty P.A., Herlyn M., Rustgi A.K., Nakagawa H., Ramaswamy S., Basu D. JARID1B enables transit between distinct states of the stem-like cell population in oral cancers. Cancer Res. 2016;76:5538–5549. doi: 10.1158/0008-5472.CAN-15-3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giroux V., Lento A.A., Islam M., Pitarresi J.R., Kharbanda A., Hamilton K.E., Whelan K.A., Long A., Rhoades B., Tang Q., Nakagawa H., Lengner C.J., Bass A.J., Wileyto E.P., Klein-Szanto A.J., Wang T.C., Rustgi A.K. Long-lived keratin 15+ esophageal progenitor cells contribute to homeostasis and regeneration. J Clin Invest. 2017;127:2378–2391. doi: 10.1172/JCI88941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kalabis J., Oyama K., Okawa T., Nakagawa H., Michaylira C.Z., Stairs D.B., Figueiredo J.L., Mahmood U., Diehl J.A., Herlyn M., Rustgi A.K. A subpopulation of mouse esophageal basal cells has properties of stem cells with the capacity for self-renewal and lineage specification. J Clin Invest. 2008;118:3860–3869. doi: 10.1172/JCI35012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta J., Nebreda A.R. Roles of p38alpha mitogen-activated protein kinase in mouse models of inflammatory diseases and cancer. FEBS J. 2015;282:1841–1857. doi: 10.1111/febs.13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hisha H., Tanaka T., Kanno S., Tokuyama Y., Komai Y., Ohe S., Yanai H., Omachi T., Ueno H. Establishment of a novel lingual organoid culture system: generation of organoids having mature keratinized epithelium from adult epithelial stem cells. Sci Rep. 2013;3:3224. doi: 10.1038/srep03224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Casale F., Canaparo R., Serpe L., Muntoni E., Pepa C.D., Costa M., Mairone L., Zara G.P., Fornari G., Eandi M. Plasma concentrations of 5-fluorouracil and its metabolites in colon cancer patients. Pharmacol Res. 2004;50:173–179. doi: 10.1016/j.phrs.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 36.Kikuchi O., Ohashi S., Nakai Y., Nakagawa S., Matsuoka K., Kobunai T., Takechi T., Amanuma Y., Yoshioka M., Ida T., Yamamoto Y., Okuno Y., Miyamoto S., Nakagawa H., Matsubara K., Chiba T., Muto M. Novel 5-fluorouracil-resistant human esophageal squamous cell carcinoma cells with dihydropyrimidine dehydrogenase overexpression. Am J Cancer Res. 2015;5:2431–2440. [PMC free article] [PubMed] [Google Scholar]
- 37.Edge S.B., Compton C.C. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 38.Natsugoe S., Okumura H., Matsumoto M., Uchikado Y., Setoyama T., Yokomakura N., Ishigami S., Owaki T., Aikou T. Randomized controlled study on preoperative chemoradiotherapy followed by surgery versus surgery alone for esophageal squamous cell cancer in a single institution. Dis Esophagus. 2006;19:468–472. doi: 10.1111/j.1442-2050.2006.00615.x. [DOI] [PubMed] [Google Scholar]
- 39.Eisenhauer E.A., Therasse P., Bogaerts J., Schwartz L.H., Sargent D., Ford R., Dancey J., Arbuck S., Gwyther S., Mooney M., Rubinstein L., Shankar L., Dodd L., Kaplan R., Lacombe D., Verweij J. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 40.Society. JE Japanese Classification of Esophageal Cancer, 10th Edition: part Ⅰ. Esophagus. 2009;6:1–25. [Google Scholar]
- 41.Society J.E. Japanese Classification of Esophageal cancer, 10th edition: parts Ⅱ and Ⅲ. Esophagus. 2009;6:71–94. [Google Scholar]
- 42.Nakagawa H., Zukerberg L., Togawa K., Meltzer S.J., Nishihara T., Rustgi A.K. Human cyclin D1 oncogene and esophageal squamous cell carcinoma. Cancer. 1995;76:541–549. doi: 10.1002/1097-0142(19950815)76:4<541::aid-cncr2820760402>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 43.Nishihira T., Hashimoto Y., Katayama M., Mori S., Kuroki T. Molecular and cellular features of esophageal cancer cells. J Cancer Res Clin Oncol. 1993;119:441–449. doi: 10.1007/BF01215923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohashi S., Kikuchi O., Tsurumaki M., Nakai Y., Kasai H., Horimatsu T., Miyamoto S., Shimizu A., Chiba T., Muto M. Preclinical validation of talaporfin sodium-mediated photodynamic therapy for esophageal squamous cell carcinoma. PLoS One. 2014;9:e103126. doi: 10.1371/journal.pone.0103126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kikuchi O., Ohashi S., Horibe T., Kohno M., Nakai Y., Miyamoto S., Chiba T., Muto M., Kawakami K. Novel EGFR-targeted strategy with hybrid peptide against oesophageal squamous cell carcinoma. Sci Rep. 2016;6:22452. doi: 10.1038/srep22452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whelan K.A., Merves J.F., Giroux V., Tanaka K., Guo A., Chandramouleeswaran P.M., Benitez A.J., Dods K., Que J., Masterson J.C., Fernando S.D., Godwin B.C., Klein-Szanto A.J., Chikwava K., Ruchelli E.D., Hamilton K.E., Muir A.B., Wang M.L., Furuta G.T., Falk G.W., Spergel J.M., Nakagawa H. Autophagy mediates epithelial cytoprotection in eosinophilic oesophagitis. Gut. 2017;66:1197–1207. doi: 10.1136/gutjnl-2015-310341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kinugasa H., Whelan K.A., Tanaka K., Natsuizaka M., Long A., Guo A., Chang S., Kagawa S., Srinivasan S., Guha M., Yamamoto K., St Clair D.K., Avadhani N.G., Diehl J.A., Nakagawa H. Mitochondrial SOD2 regulates epithelial-mesenchymal transition and cell populations defined by differential CD44 expression. Oncogene. 2015;34:5229–5239. doi: 10.1038/onc.2014.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Japan Esophageal S. Japanese Classification of Esophageal Cancer, 11th Edition: part I. Esophagus. 2017;14:1–36. doi: 10.1007/s10388-016-0551-7. [DOI] [PMC free article] [PubMed] [Google Scholar]