Abstract
Background & Aims
CD26, a multifunctional transmembrane glycoprotein, is expressed in various cancers and functions as dipeptidyl peptidase 4 (DPP4). We investigated whether CD26 expression is associated with hepatocellular carcinoma (HCC) progression and whether DPP4 inhibitors exert antitumor effects against HCC.
Methods
CD26 expression was examined in 41 surgically resected HCC specimens. The effects of DPP4 inhibitors on HCC were examined by using HCC cell lines (Huh-7 and Li-7), xenograft tumors in nude mice, and a nonalcoholic steatohepatitis–related HCC mouse model.
Results
CD26 expression in HCC specimens was associated with increased serum DPP4 activity, as well as a more advanced stage, less tumor immunity, and poorer prognosis in HCC patients. The HCC cell lines and xenograft tumors exhibited CD26 expression and DPP4 activity. The DPP4 inhibitors did not exhibit antitumor effects in vitro, but natural killer (NK) and/or T-cell tumor accumulation suppressed growth of xenograft tumor and HCC in vivo. The antitumor effects of DPP4 inhibitors were abolished by the depletion of NK cells or the neutralization of CXCR3, a chemokine receptor on NK cells. EZ-TAXIScan, an optical horizontal chemotaxis apparatus, identified enhanced NK and T-cell chemotaxis by DPP4 inhibitors ex vivo in the presence of Huh-7 cells and the chemokine CXCL10, which binds to CXCR3. The DPP4 inhibitors prevented the biologically active form of CXCL10 from being truncated by Huh-7 cell DPP4 activity. DPP4 inhibitors also suppressed tumor angiogenesis.
Conclusions
These results provide a rationale for verifying whether DPP4 inhibitors clinically inhibit the progression of HCC or augment the antitumor effects of molecular-targeting drugs or immunotherapies against HCC.
Keywords: NK Cell, CD26, CXCL10, Tumor Immunity, TAXIScan, T Cell
Abbreviations used in this paper: anti-ASGM, anti-asialo GM1 antisera; CCK-8, Cell Counting Kit 8; DM, diabetes mellitus; DPP4, dipeptidyl peptidase 4; FBS, fetal bovine serum; HCC, hepatocellular carcinoma; HCV, hepatitis C virus; HPLC, high-performance liquid chromatography; IC50, inhibitory concentration of 50%; Ig, immunoglobulin; LDH, lactate dehydrogenase; MICA, MHC class I polypeptide-related sequence A; NAFLD, nonalcoholic fatty liver disease; NASH, nonalcoholic steatohepatitis; NK, natural killer; PBMC, peripheral blood mononuclear cell; PBS, phosphate-buffered saline; PTH, phenylthiohydantoin; SDS-PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis
Graphical abstract
Summary.
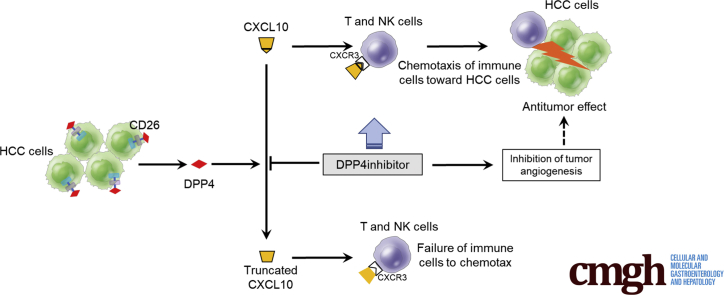
DPP4 inhibitors suppressed HCC development through the activated infiltration of lymphocytes into the xenograft tumors or liver tumors in mice. This effect was exerted by preventing the biologically active form of CXCL10 from being truncated by DPP4 activity.
Hepatocellular carcinoma (HCC) is the third greatest cause of cancer-related death in the world. A recent study from the United States found that although the overall rate of increasing HCC incidence has slowed from 2010 through 2012, the incidence rate is still increasing in subgroups such as men aged from 55 to 64 years, especially those born during the peak era of hepatitis C virus (HCV) infection.1 HCV infection has been a leading cause of HCC for the past several decades. However, the incidence of HCV-associated HCC is expected to decrease because of the high rate of HCV eradication via the rapid dissemination and use of direct-acting antivirals.
Alternatively, diabetes mellitus (DM), obesity, and nonalcoholic fatty liver disease (NAFLD) have recently emerged as new risk factors for HCC2; in particular, DM is a risk factor even after the successful treatment of chronic hepatitis C.3, 4 The American Diabetes Association and the American Cancer Society jointly published a consensus report on the association between DM and cancer,5 which was followed by a similar consensus report published by the joint committee of the Japan Diabetes Society and the Japanese Cancer Association.6 Notably, the relative risk of liver cancer development was highest among several cancer types in patients with DM.6 There is increasing interest in whether antidiabetic drugs have protective or promotive effects on HCC development and/or progression in these situations. Metformin has been shown to have protective effects on HCC development.7 However, although a meta-analysis of animal model studies showed that metformin significantly inhibited the growth of HCC,8 whether metformin has antitumor effects in patients with HCC remains to be elucidated. In clinical settings where no approved drugs for advanced HCC exist except for sorafenib and regorafenib, oral multikinase inhibitors, it is worth investigating antidiabetic drugs that have antitumor effects in HCC patients with DM.
Dipeptidyl peptidase 4 (DPP4) inhibitors were introduced in 2006 for the treatment of DM and are currently being used by more than 30 million people as antidiabetic medications, which highlights the safety profile of these drugs.9 There is still a lack of both basic and clinical investigations into whether DPP4 inhibitors have antitumor effects against HCC, probably because of the short time since their introduction into clinical settings. DPP4 is found as both a type II cell surface protein (CD26) and as a soluble protein lacking intracellular and transmembrane domains.10 DPP4 functions as a serine protease, selectively cleaving the N-terminal penultimate proline or alanine of proteins.11, 12 The prevailing and clinically relevant action of DPP4 is the degradation of endogenous glucagon-like peptide-1. On the other hand, the expression of CD26, also known as DPP4, in tumors has been shown to be correlated with clinicopathologic features such as tumor staging, degree of differentiation, and prognosis in patients with colorectal cancer.13 Although CD26 is expressed in HCC tissues as well,14 its clinical significance is obscure.
Several chemokines have been shown to be processed in vitro by DPP4.15 Particularly, the proinflammatory chemokine CXCL10 has been found to fail to induce immune cell chemotaxis when truncated in vitro.15 These results suggest that DPP4 inhibitors may have the potential to exert antitumor effects against HCC by modulating tumor immunity. Here we investigated whether DPP4 inhibitors have antitumor effects against HCC and the potential underlying mechanisms by using a mouse xenograft model of HCC and a nonalcoholic steatohepatitis (NASH)–related HCC mouse model. We found that DPP4 inhibitors prevented CXCL10 from being truncated by DPP4 activity and suppressed the growth of HCC tumors through the activation of natural killer (NK) cell and T-cell chemotaxis and the inhibition of tumor angiogenesis.
Results
CD26 Expression and Lymphocyte Infiltration in Tumor Tissue and Serum Dipeptidyl Peptidase 4 Activity of Hepatocellular Carcinoma Patients
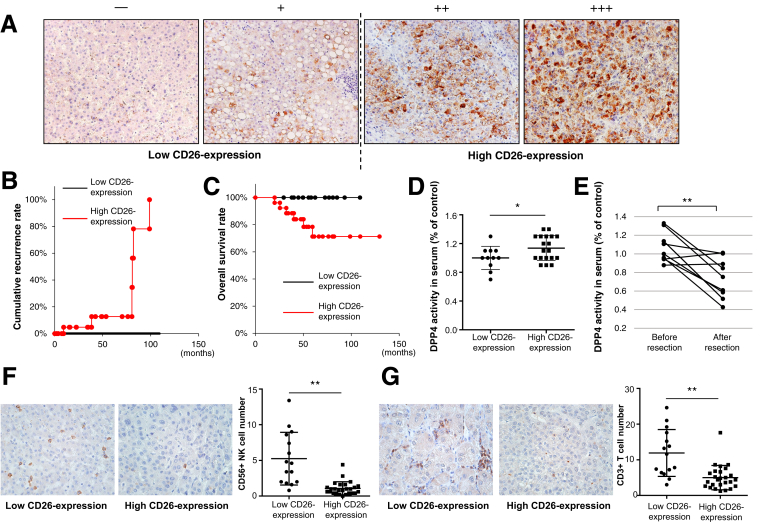
The etiology of the 41 HCC patients was HCV infection in 21 patients, hepatitis B virus infection in 11, alcoholic in 6, and unknown in 3. Immunohistologic staining for CD26 showed various degrees of expression of positive cells in HCC tissues, as shown in Figure 2A. We divided the 41 patients into 2 groups according to the degree of CD26+ cell expression as follows: the low CD26 expression group (− or + in Figure 2A, n = 15), with less than 33% of CD26+ cells, and the high CD26 expression group (++ or +++ in Figure 2A, n = 26), with greater than or equal to 33% of CD26+ cells. The high CD26 expression group showed higher alpha-fetoprotein levels, more advanced tumor stages, poorer histologic differentiation, more tumor cell infiltration into the tumor capsule, and more cell proliferation, as indicated by the Ki67 labeling index (Table 1). The cumulative recurrence rate of HCC was higher and the overall survival was shorter in the high CD26 expression group (P = .058, Figure 2B and C); however, neither difference was significant. Serum DPP4 activity was significantly higher in the high CD26 expression group (P < .05, Figure 2D). In addition, serum DPP4 activity significantly decreased after HCC resection (P = .01, Figure 2E). The number of infiltrated NK cells (CD56+) and T cells (CD3γ+) was significantly smaller in the high CD26 expression group (P < .05, Figure 2F and G). These results suggested that higher CD26 expression was associated with a more advanced tumor stage, poorer prognosis, and less tumor immunity in HCC patients and that CD26 expression in HCC tissues was reflected, in part, by serum DPP4 activity, although CD26 is expressed in many tissues other than the liver.16
Figure 2.
(A) Immunohistologic staining for CD26 in resected HCC tissues (original magnification, ×200). NIH image analysis software was used to quantify the mean percent area positive staining for CD26 for 5 randomly selected fields of view of digital images of each liver; the mean percent area was classified as follows: – (0%–16%), + (17%–32%), ++ (33%–66%), and +++ (≥67%). (B) Cumulative recurrence curve of patients with low CD26 expression (– or +, n = 15) and those with high CD26 expression (++ or +++, n = 26). P = .058. (C) Overall survival of patients with low CD26 expression (– or +, n = 15) and those with high CD26 expression (++ or +++, n = 26). P = .058. (D) Serum DPP4 activity before HCC resection in patients with low CD26 expression (– or +, n = 11) and those with high CD26 expression (++ or +++, n = 19). DPP4 activity was measured by using a DPP4 activity assay kit according to the manufacturer’s instructions. *P < .05. (E) Serum DPP4 activity before and 1–6 months after HCC resection in 9 patients with HCC. **P = .01. (F) and (G) Immunohistologic staining for CD56-positive cells (F) or CD3γ-positive cells (G) in resected HCC tissues (original magnification, ×400). NIH image analysis software was used to count the number of CD56-positive NK cells or CD3γ-positive T cells for 5 randomly selected fields of view of digital images of each liver. The number of intratumor NK cells or T cells in patients with low CD26 expression and those with high CD26 expression. **P < .01 (G).
Table 1.
Clinical and Pathologic Comparison Between HCC Patients With Low and High CD26 Expression
| Factors | Low CD26 expression (n = 15) | High CD26 expression (n = 26) | P value |
|---|---|---|---|
| Age (y) | 64.7 ± 8.0 | 66.2 ± 8.5 | .515 |
| Gender (M/F) | 10/5 | 19/7 | .664 |
| Platelet count (×104/mL) | 13.8 ± 5.0 | 15.5 ± 5.1 | .401 |
| AST (IU/L) | 33.3 ± 15.6 | 45.1 ± 28.9 | .198 |
| ALT (IU/L) | 28.5 ± 19.9 | 39.4 ± 29.4 | .159 |
| γ-GT (IU/L) | 42.7 ± 22.6 | 77.9 ± 81.8 | .261 |
| Total bilirubin (mg/dL) | 0.75 ± 0.30 | 0.68 ± 0.31 | .388 |
| Albumin (g/dL) | 4.06 ± 0.44 | 3.98 ± 0.41 | .684 |
| Prothrombin time (%) | 96.7 ± 13.2 | 89.8 ± 12.7 | .160 |
| HbA1c (%) | 6.44 ± 0.95 | 6.49 ± 0.94 | .416 |
| Alpha-fetoprotein (ng/mL) | 19.3 ± 29.8 | 114.0 ± 187.8 | .012 |
| Des-γ-carboxy prothrombin (mAU/mL) | 425.1 ± 1319.0 | 508.9 ± 22,121.6 | .097 |
| Etiology (HBV/HCV/alcohol/other) | 7/7/0/1 | 4/14/6/2 | .071 |
| Non-tumor tissue (chronic hepatitis/cirrhosis) | 8/7 | 13/13 | .837 |
| Tumor size (mm) | 25.5 ± 15.0 | 29.9 ± 27.4 | .745 |
| Tumor number (multiple/single) | 2/13 | 7/19 | .311 |
| Portal vein invasion (+/−) | 2/13 | 8/18 | .211 |
| UICC staging (I/II/III) | 10/4/1 | 5/13/8 | .008 |
| Capsule formation of tumor (+/−) | 8/7 | 19/7 | .199 |
| Cancerous infiltration of capsule (+/−) | 3/12 | 14/12 | .034 |
| Differentiation (well/moderate/poor) | 9/5/1 | 0/22/4 | <.001 |
| Ki67 labeling index | 0.065 ± 0.048 | 0.189 ± 0.090 | <.001 |
ALT, alanine aminotransferase; AST, aspartate aminotransferase; γ-GT, gamma-glutamyl transferase; HbA1c, glycosylated hemoglobin; HBV, hepatitis B virus; UICC; Union for International Cancer Control.
Effects of Dipeptidyl Peptidase 4 Inhibitors on Liver Cancer Cell Lines Expressing CD26
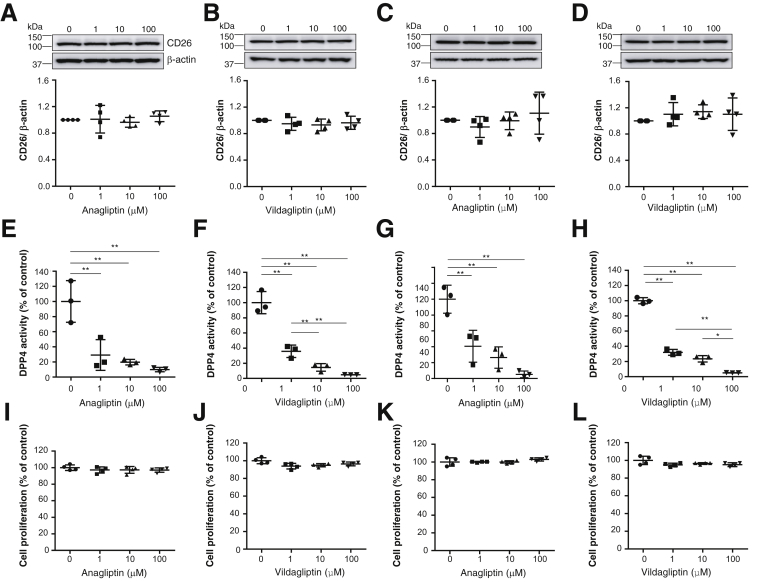
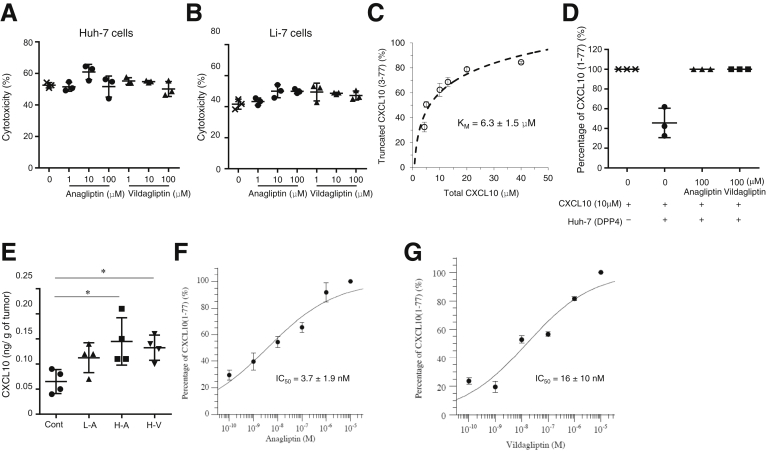
The results indicating that high HCC tissue CD26 expression was associated with a poorer prognosis and higher serum DPP4 activity suggested the potential of DPP4 as a therapeutic target for HCC. Therefore, we investigated whether DPP4 inhibitors exert antitumor effects against HCC. We confirmed the expression of CD26 in both the Huh-7 and Li-7 cell lines. The DPP4 inhibitors anagliptin and vildagliptin did not affect the expression of CD26 (Figure 3A–D), but they did suppress DPP4 activity in Huh-7 and Li-7 cells in a dose-dependent manner (Figure 3E–H). Anagliptin and vildagliptin did not affect Huh-7 and Li-7cell proliferation (Figure 3I–L). Anagliptin was also examined for effects on the cell cycle in Huh-7 cells, and the results showed that it did not affect the cell cycle (Figure 4A) or molecules regulating the transition from the G1 to the S phase, such as p21, p27kip1, pCDK2, and pRb (Figure 4B).
Figure 3.
(A) CD26 expression of Huh-7 cells in each anagliptin concentration group (n = 4). (B) CD26 expression of Huh-7 cells in each vildagliptin concentration group (n = 4). Immunoblots for CD26 were performed using Huh-7 cell lysates. (C) CD26 expression of Li-7 cells in each anagliptin concentration group (n = 4). (D) CD26 expression of Li-7 cells in each vildagliptin concentration group (n = 4). Immunoblots for CD26 were performed using Li-7 cell lysates. The amounts of protein were normalized to β-actin. (E) DPP4 activity of Huh-7 cells in each anagliptin concentration group (n = 3). (F) DPP4 activity of Huh-7 cells in each vildagliptin concentration group (n = 3). (G) DPP4 activity of Li-7 cells in each anagliptin concentration group (n = 3). (H) DPP4 activity of Li-7 cells in each vildagliptin concentration group (n = 3). DPP4 activity was measured using a DPP4 activity assay kit according to the manufacturer’s instructions. *P < .05, **P < .01. (I) Huh-7 cell proliferation in each anagliptin concentration group (n = 4). (J) Huh-7 cell proliferation in each vildagliptin concentration group (n = 4). (K) Li-7 cell proliferation in each anagliptin concentration group (n = 4). (L) Li-7 cell proliferation in each vildagliptin concentration group (n = 4). Cell proliferation was measured using a CCK-8 assay according to the manufacturer’s instructions.
Figure 4.
(A) Frequency of cells in the G0/G1, S, and G2/M phases of the cell cycle in each anagliptin concentration group (n = 3). Huh-7 cells were incubated with DNA dye (Hoechst 33342). Plates containing the cells were scanned with ImageXpress Micro Screening System. Classification of cell phases was based on the fluorescence intensity of the DNA dye. For the cell number and cell cycle analyses, the intensity of the integrated DNA dye was assessed using the cell-cycle application module. (B) Expression of p21, p27kip1, pCDK2, and pRb of Huh-7 cells in each anagliptin concentration group (n = 4). Immunoblots for p21, p27kip1, CDK2, and pRb were performed using Huh-7 cell lysates. Amounts of protein were normalized to β-actin. (C) Huh-7 cell xenograft tumors and growth curves in mice at 21 days after feeding in 3 groups (control diet, diet containing low dose of anagliptin [L-A] [0.7 g/kg diet], and diet containing high dose of anagliptin [H-A] [2 g/kg diet]) (n = 5 for each group). *P < .05 vs L-A, **P < .01 vs L-A, #P < .05 vs H-A, ##P < .01 vs H-A. (D) Li-7 cell xenograft tumors and growth curves in mice at 21 days feeding in the same 3 groups as described in (D) (n = 5 for each group). *P < .05 vs L-A, #P < .05 vs H-A, ##P < .01 vs H-A. (E) Huh-7 cell xenograft tumors and growth curves in mice at 21 days after feeding in 3 groups (control diet, diet containing high dose of anagliptin [2 g/kg diet], and diet containing high dose of vildagliptin [2 g/kg diet]) (n = 5 for each group). *P < .05, **P < .01 vs anagliptin group, #P < .05, ##P < .01 vs vildagliptin group.
Effects of Dipeptidyl Peptidase 4 Inhibitors on Xenograft Liver Tumors in Nude Mice
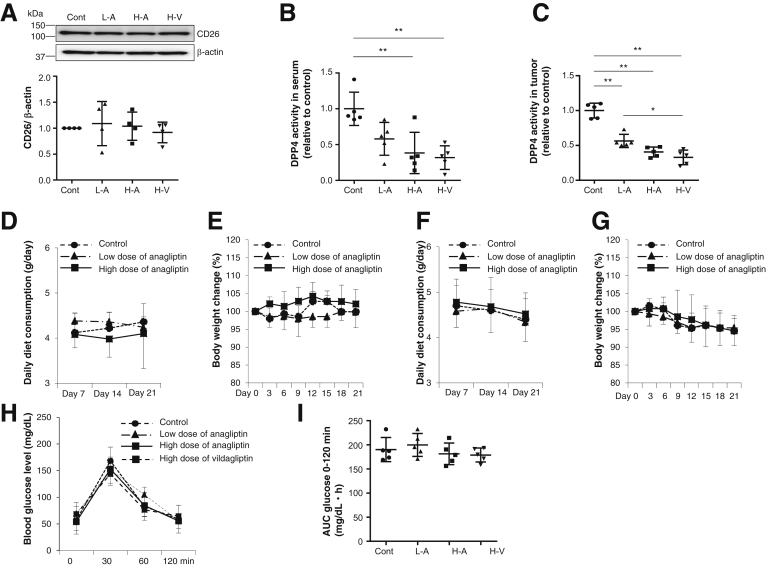
Although the DPP4 inhibitors did not affect cell proliferation or the cell cycle in vitro, anagliptin suppressed the growth of xenograft liver tumors in a dose-dependent manner (Figure 4C for Huh-7 cells, Figure 4D for Li-7 cells). Vildagliptin also suppressed the growth of xenograft liver tumors and did so to the same degree as anagliptin (Figure 4E). The DPP4 inhibitors did not affect the expression of CD26 in the xenograft tumors (Figure 5A) but did inhibit serum and tumor DPP4 activity in a dose-dependent manner (Figure 5B and C) at 21 days after the initiation of feeding. Daily diet consumption and body weight changes did not differ among the mice with Huh-7 or Li-7 cell xenografts fed the control diet and those fed an anagliptin-containing diet (Figure 5D and E for Huh-7 cells, Figure 5F and G for Li-7 cells). The levels of glucose, insulin, triglyceride, total cholesterol, and low-density lipoprotein cholesterol after fasting were similar among the 4 groups (the control, low anagliptin dose, high anagliptin dose, and high vildagliptin dose diets) at 21 days after the initiation of feeding (Table 2). We also compared the glucose tolerance of the xenograft mice fed the control diet and those fed an anagliptin-containing diet. The blood glucose levels and glucose areas under the curve between 0 and 120 minutes (area under the curve glucose 0–120 minutes) after the administration of 10 μL/g of body weight of a 15% glucose solution were similar among the 4 groups (Figure 5H and I).
Figure 5.
(A) CD26 expression of Huh-7 cell xenograft tumors in mice in 4 groups (control, L-A, and H-A groups and the group fed diet containing high dose of vildagliptin [H-V] [2 g/kg diet]) (n = 5 for each group) at 21 days after feeding. Immunoblots for CD26 were performed using tumor tissue lysates. Amounts of protein were normalized to β-actin. (B) Serum DPP4 activity of mice in the 4 groups (n = 5 for each group). Data are expressed relative to data from the control group. **P < .01. (C) DPP4 activity in Huh-7 cell xenograft tumors of in the 4 groups (n = 5 for each group). Data are expressed relative to data from the control group. *P < .05, **P < .01. (D) Daily diet consumption of Huh-7 cell xenograft mice fed the control diet, those fed the diet containing low dose of anagliptin, and those fed the diet containing high dose of anagliptin (n = 5 for each group). (E) Body weight change of Huh-7 cell xenograft mice fed the control diet, those fed the diet containing low dose of anagliptin, and those fed the diet containing high dose of anagliptin (n = 5 for each group). (F) Daily diet consumption of Li-7 cell xenograft mice fed the control diet, those fed the diet containing low dose of anagliptin, and those fed the diet containing high dose of anagliptin (n = 5 for each group). (G) Body weight change of Li-7 cell xenograft mice fed the control diet, those fed the diet containing low dose of anagliptin, and those fed the diet containing high dose of anagliptin (n = 5 for each group). (H) Blood glucose levels at 0, 30, 60, and 120 minutes after administration of 10 μL/g of body weight of 15% glucose to Huh-7 cell xenograft mice fed the control diet, those fed the diet containing low dose of anagliptin, those fed the diet containing high dose of anagliptin, and those fed the diet containing high dose of vildagliptin (n = 5 for each group). (I) Blood glucose area under the curve between 0 and 120 minutes (AUC glucose 0–120 min) after administration of 10 μL/g of body weight of 15% glucose to Huh-7 cell xenograft mice fed the control diet, those fed the diet containing low dose of anagliptin, those fed the diet containing high dose of anagliptin, and those fed the diet containing high dose of vildagliptin (n = 5 for each group).
Table 2.
Serum Biochemical Parameters in 4 Groups of Mice at 21 Days After Initiation of Feeding
| Factors | Control | Low-dose anagliptina | High-dose anagliptinb | High-dose vildagliptinc | P value |
|---|---|---|---|---|---|
| Glucose (mg/dL) | 57.2 ± 16.9 | 61.8 ± 18.0 | 54.2 ± 16.0 | 68.6 ± 11.2 | .400 |
| Insulin (ng/mL) | 0.25 ± 0.14 | 0.24 ± 0.12 | 0.32 ± 0.24 | 0.29 ± 0.22 | .909 |
| Triglyceride (mg/dL) | 66.0 ± 47.4 | 60.0 ± 18.3 | 75.0 ± 51.3 | 76.8 ± 44.7 | .930 |
| Total cholesterol (mg/dL) | 96.6 ± 26.8 | 92.3 ± 7.9 | 98.0 ± 15.7 | 95.2 ± 29.4 | .983 |
| Low-density lipoprotein cholesterol (mg/dL) | 45.4 ± 12.2 | 38.3 ± 3.6 | 39.4 ± 12.7 | 49.6 ± 10.4 | .358 |
Mice fed diet containing low dose of anagliptin (0.7 g/kg diet).
Mice fed diet containing high dose of anagliptin (2 g/kg diet).
Mice fed diet containing high dose of vildagliptin (2 g/kg diet).
Effect of Anagliptin on Natural Killer Cell Infiltration Into Xenograft Liver Tumors
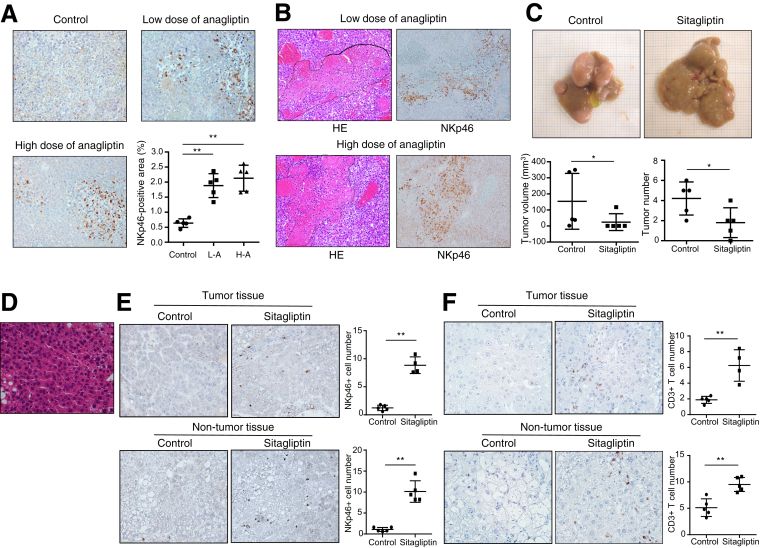
Nude mice lacking a thymus are unable to produce T cells but are able to produce NK cells.17 Because NK cells exhibit antitumor effects, we examined whether NK cells are associated with the effects of DPP4 inhibitors against xenograft liver tumors. Anagliptin significantly increased activated NK cell (NKp46+) infiltration into xenograft liver tumors in a dose-dependent manner (Figure 6A). In addition, the areas of activated NK cell infiltration almost completely coincided with necrotic areas of the xenograft liver tumors (Figure 6B). These results suggested that NK cells potentially play an important role in the effect of DPP4 inhibitors against xenograft liver tumors.
Figure 6.
(A) Immunohistologic staining for NKp46 in Huh-7 cell xenograft tumors of mice in 3 groups (n = 5 for each group) (original magnification, ×200). NIH image analysis software was used to quantify the mean percent area of positive staining for NKp46 for 5 randomly selected fields of view of digital images of each liver. **P < .01. (B) H&E and NKp46 staining of same tissue sections of Huh-7 cell xenograft tumors of mice in 2 groups (original magnification, ×200). Areas surrounded by dotted lines in the H&E images indicate necrotic areas in the tumor tissue. (C) The liver of STAM mice at 18 weeks of age showed multiple liver tumors, whereas the liver of STAM mice administered with sitagliptin showed significantly smaller and fewer liver tumors (n = 5 for each group). *P < .05. (D) Histology of liver tumor showed increased cellular density and/or markedly thickened trabeculae, which were compatible with HCC morphology (original magnification, ×400). (E) and (F) Immunohistologic staining for NKp46 (E) or CD3 (F) in liver tumors and non-tumor liver tissue of STAM mice in 2 groups (n = 5 for each group) (original magnification, ×200). NIH image analysis software was used to count the number of positive staining cells for NKp46 or CD3 for 5 randomly selected fields of view of digital images of each liver. **P < .01.
Effect of Sitagliptin on Tumor Development and Natural Killer Cell and T-Cell Infiltration in a Nonalcoholic Steatohepatitis–Related Hepatocellular Carcinoma Mouse Model
Nude mice are immunodeficient. Xenograft liver tumors in these mice may be insufficient to explore NK cell–mediated tumor biology because liver is a NK cell–rich organ. To overcome these weaknesses, we used a NASH-related HCC mouse model. STAM mice showed multiple large tumors in the liver at 18 weeks of age, but DPP4 inhibitor, sitagliptin, significantly suppressed both volume and number of liver tumors in STAM mice (Figure 6C). There was no significant difference in the incidence of liver tumors between STAM mice with sitagliptin (4/5, 80%) and those without (5/5, 100%). Histology of liver tumor showed increased cellular density and/or markedly thickened trabeculae, which were compatible with HCC morphology (Figure 6D). In addition, sitagliptin significantly increased activated NK cell (NKp46+) (Figure 6E) and CD3+ T cell (Figure 6F) infiltration into both tumor tissue and non-tumor tissue. Thus, antitumor effect of DPP4 inhibitors through activated NK cell and T-cell infiltration was confirmed in a NASH-related HCC mouse model in addition to NK cell infiltration in a xenograft mouse model, and this effect appeared to be a class effect.
Effect of Sitagliptin on Nonalcoholic Steatohepatitis–Related Parameters in a Nonalcoholic Steatohepatitis–Related Hepatocellular Carcinoma Mouse Model
DPP4 inhibitor has been shown to have anti-inflammatory and anti-steatotic activity in STAM mice.18 We examined the effects of sitagliptin on NASH-related parameters to ascertain whether those effects were involved in antitumor effect of sitagliptin in STAM mice. The results indicated that sitagliptin did not affect any biochemical and histologic NASH-related parameters (Table 3). Why sitagliptin did not affect serum insulin level was assumed to be due to damaged pancreatic beta cells by streptozotocin treatment in this mouse model.
Table 3.
Effects of Sitagliptin on Biochemical and Histologic NASH-Related Parameters
| Parameters | Control (n = 5) | Sitagliptin (n = 5) | P value |
|---|---|---|---|
| Biochemical parameters | |||
| Glucose (mg/dL) | 435.4 ± 96.8 | 365.4 ± 85.4 | .19 |
| HbA1c (%) | 6.0 ± 0.5 | 5.6 ± 0.8 | .36 |
| Insulin (ng/mL) | 0.17 ± 0.05 | 0.20 ± 0.07 | .55 |
| AST (IU/L) | 104.2 ± 35.6 | 84.0 ± 24.0 | .38 |
| ALT (IU/L) | 115.0 ± 44.1 | 98.4 ± 20.0 | .27 |
| Total cholesterol (mg/dL) | 152.4 ± 5.4 | 142.6 ± 26.1 | .92 |
| Triglyceride (mg/dL) | 409.8 ± 167.6 | 318.0 ± 92.2 | .38 |
| NAFLD activity score | |||
| Steatosis | 2.4 ± 0.9 | 1.6 ± 0.5 | .13 |
| Inflammation | 1.4 ± 0.5 | 1.0 ± 0.0 | .14 |
| Ballooning | 1.4 ± 0.9 | 1.0 ± 1.0 | .53 |
| NAFLD activity score | 5.2 ± 1.5 | 3.6 ± 1.5 | .11 |
ALT, alanine aminotransferase; AST, aspartate aminotransferase; HbA1c, glycosylated hemoglobin.
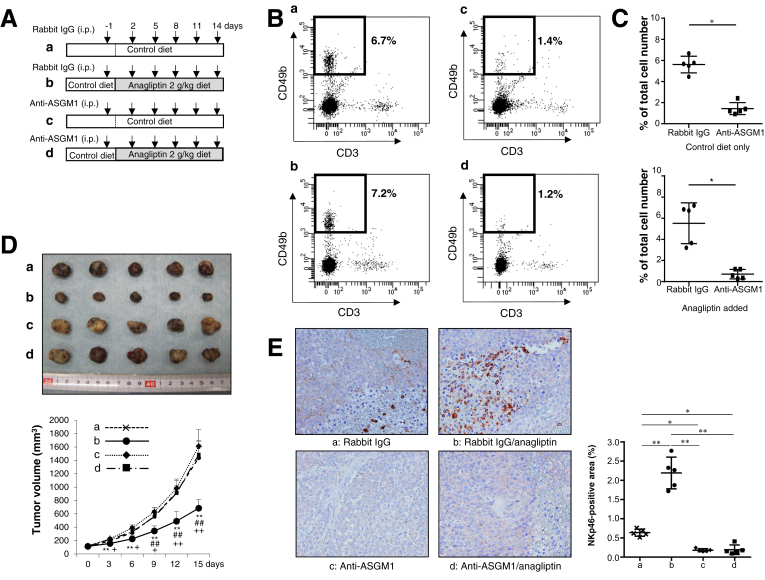
Natural Killer Cell Depletion Abrogates Antitumor Effects of Anagliptin
To clarify whether DPP4 inhibitors exert antitumor effects through NK cells, we assessed the effects of NK cell depletion (Figure 7A) on the antitumor effects of anagliptin. NK cells among spleen leukocytes were significantly depleted by anti-ASGM1 administration in both xenograft mice fed the control diet and those fed an anagliptin-containing diet (Figure 7B and C). Although anti-ASGM1 is known to deplete other unconventional T-cell populations such as NKT cells (CD3+CD49b+), neither anti-ASGM1 nor anagliptin affected NKT cell population (data not shown) because of extremely low frequency of NKT cells (0.02%–0.08%) in nude mice. Notably, the reduction in xenograft tumor growth induced by anagliptin was completely abrogated by depleting NK cells (Figure 7D). The immunohistochemical analysis also showed that the significantly increased NK cell infiltration into xenograft liver tumors induced by anagliptin was almost completely abolished by depleting NK cells (Figure 7E). These results indicate that NK cells are indispensable to the antitumor effects exerted by anagliptin in mice with xenograft liver tumors.
Figure 7.
(A) Nude mice were intraperitoneally injected with 50 μL rabbit anti-ASGM1 or rabbit IgG (isotype control) 6 times during the course of 15 days. (a) Mice fed the control diet and injected with rabbit IgG. (b) Mice fed the diet containing anagliptin (2 g/kg diet) and injected with rabbit IgG. (c) Mice fed the control diet and injected with anti-ASGM1. (d) Mice fed the diet containing anagliptin (2 g/kg diet) and injected with anti-ASGM1. (B) Flow cytometric analysis of CD49b+CD3– NK cells among spleen leukocytes of mice in the 4 groups [a, b, c, and d in (A)]. Areas surrounded by bold lines indicate CD49b+CD3– NK cells, and adjacent figures indicate percentages of CD49b+CD3– NK cells among spleen leukocytes. (C) Data represent the mean of flow cytometric analyses repeated 4 times in each group and compared between groups a and c or between groups b and d. *P < .05. (D) Huh-7 cell xenograft tumors and growth curves in mice at 15 days after initial injection of rabbit IgG or anti-ASGM1 in the 4 groups [a, b, c, and d in (A)] (n = 5 for each group). **P < .01 vs group c, ##P < .01 vs group d, +P < .05, ++P < .01 vs group a. (E) Immunohistologic staining for NKp46 in Huh-7 cell xenograft tumors of mice in the 4 groups [a, b, c, and d in (A)] (n = 5 for each group) (original magnification, ×200). NIH image analysis software was used to quantify the mean percent area of positive staining for NKp46 in 5 randomly selected fields of view of digital images of each liver tumor. *P < .05, **P < .01.
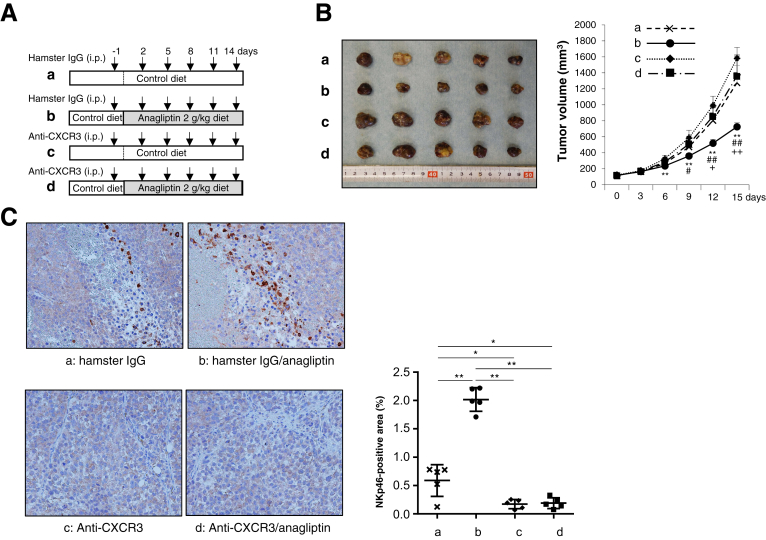
Defective Natural Killer Cell Trafficking Abrogates Antitumor Effects of Anagliptin
To exert antitumor effects, NK cells need to be mobilized from the bone marrow and subsequently recruited from the peripheral blood into tumor tissues. NK cell accumulation in tumor tissue has been shown to be dependent on the chemokine receptor CXCR3,19 which binds to the structurally and functionally related chemokines CXCL9, CXCL10, and CXCL11.20 We inhibited the binding of CXCR3 to chemokines by using an anti-CXCR3 neutralizing antibody, and we investigated whether defective NK-cell trafficking abrogates the antitumor effects of anagliptin. Xenograft mice fed the control diet and those fed an anagliptin-containing diet were intraperitoneally injected with either the anti-CXCR3 antibody or hamster IgG (isotype control) 6 times during the course of 15 days, as shown in Figure 8A. The administration of anti-CXCR3 abrogated the reduction in xenograft tumor growth induced by anagliptin (Figure 8B) and abolished NK cell accumulation in xenograft liver tumors (Figure 8C). These results suggest that the antitumor effects of anagliptin are dependent on NK-cell migration into tumor tissue.
Figure 8.
(A) Nude mice were intraperitoneally injected with 200 μg anti-CXCR3 or hamster IgG (isotype control) 6 times during the course of 15 days. (a) Mice fed the control diet and injected with hamster IgG. (b) Mice fed the diet containing anagliptin (2 g/kg diet) and injected with hamster IgG. (c) Mice fed the control diet and injected with anti-CXCR3. (d) Mice fed the diet containing anagliptin (2 g/kg diet) and injected with anti-CXCR3. (B) Huh-7 cell xenograft tumors and growth curves in mice at 15 days after initial injection of hamster IgG or anti-CXCR3 in the 4 groups [a, b, c, and d in (A)] (n = 5 for each group). **P < .01 vs group c, #P < .05, ##P < .01 vs group d, +P < .05, ++P < .01 vs group a. (C) Immunohistologic staining for NKp46 in Huh-7 cell xenograft tumors of mice in the 4 groups (n = 5 for each group) (original magnification, ×200). NIH image analysis software was used to quantify the mean percent area of positive staining for NKp46 in 5 randomly selected fields of view of digital images of each liver tumor. *P < .05, **P < .01.
Antitumor Effects of Dipeptidyl Peptidase 4 Inhibitors Are Dependent on the Chemokine CXCL10
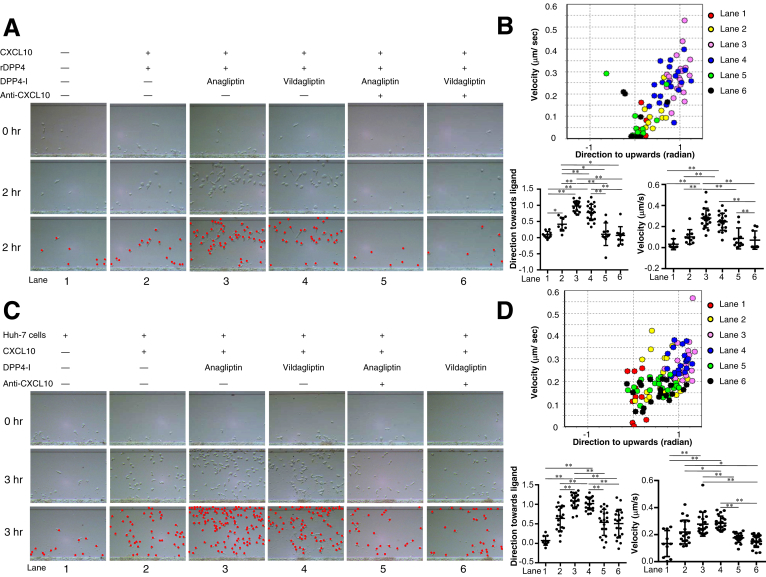
Several chemokines have been shown to be processed in vitro by DPP4,15 which is capable of enzymatically removing the first 2 amino acids from a protein that possesses a proline or alanine in the penultimate N-terminal position.11 Among these chemokines, the proinflammatory chemokine CXCL10 has been shown to be readily truncated in vitro15; this truncated form becomes an antagonist that engages its receptor, CXCR3, but does not induce chemotaxis.21 We hypothesized that DPP4 inhibitors may exhibit antitumor effects by preserving the CXCR3-CXCL10 axis for chemotaxis via protecting the biologically active form of CXCL10. Therefore, we assessed the effects of DPP4 inhibitors on the chemotaxis of NK cells in vitro in the presence of CXCL10 by using EZ-TAXIScan (ECI Inc, Kawasaki, Japan), a cell mobility analysis device. DPP4 inhibitors (anagliptin and vildagliptin) significantly enhanced the mobility of NK cells in the presence of recombinant DPP4 and CXCL10, and these effects were canceled by the addition of anti-CXCL10 (Figure 9A and B). To reproduce the truncation of CXCL10 by DPP4 activity in tumor cells, we cultured Huh-7 cells expressing CD26 with 10 μmol/L CXCL10 for 8 hours in the presence or absence of DPP4 inhibitors, and we then examined the mobility of NK cells and T cells by using this culture supernatant as a chemoattractant. The results showed a significant increase in the NK cell and T cell mobility induced by the DPP4 inhibitors (Figure 9C–F). We also confirmed that the DPP4 inhibitors had no effect on the cytotoxicity of NK cells against Huh-7 cells and Li-7 cells (Figure 10A and B). These results suggest that DPP4 inhibitors exert antitumor effects by enhancing NK cell and T-cell chemotaxis, most likely through the protection of the biologically active form of CXCL10.
Figure 9.
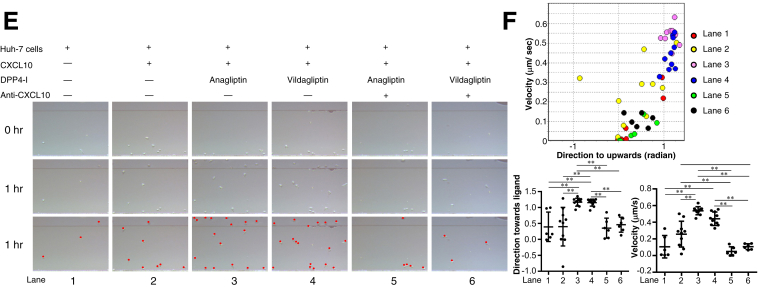
(A) NK cell migration in 260-μm-long microchannel. After aligning the NK cells (1 × 103 cells/well) on the edge of the microchannel (bottom in each panel), medium alone (lane 1), medium containing CXCL10 and recombinant DPP4 (rDPP4) (lane 2), medium containing CXCL10, rDPP4, and anagliptin (lane 3), medium containing CXCL10, rDPP4, and vildagliptin (lane 4), medium containing CXCL10, rDPP4, anagliptin, and anti-CXCL10 (lane 5), or medium containing CXCL10, rDPP4, vildagliptin, and anti-CXCL10 (lane 6) was injected into the compartment opposite the NK cell–containing compartment, thus forming a concentration gradient of CXCL10 in each channel from top to bottom in each panel (refer to Figure 1A). NK cells in the images (each panel in the middle line) are drawn as red dots in each panel in the third line. (B) Migration of 20 NK cells randomly selected from those in each lane (except for 10 NK cells in lane 1) was analyzed using TAXIScan Analyzer 2 software. To obtain statistical data of cell migration, median values of velocity and directionality (direction toward ligands) of each cell within an experimental period were calculated from the migratory pathway data obtained from time-lapse images, and the data are expressed using velocity-directionality plots. *P < .05, **P < .01. (C) NK cell migration analyzed in the same manner as described in (A), except for injected medium into the compartment opposite the NK cell–containing compartment (lane 1, culture supernatant of Huh-7 cells alone; lane 2, culture supernatant of Huh-7 cells with CXCL10; lane 3, culture supernatant of Huh-7 cells with CXCL10 + anagliptin; lane 4, culture supernatant of Huh-7 cells with CXCL10 + vildagliptin; lane 5, culture supernatant of Huh-7 cells with CXCL10 + anagliptin + anti-CXCL10; lane 6, culture supernatant of Huh-7 cells with CXCL10 + vildagliptin + anti-CXCL10). NK cells in the images (each panel in the middle line) are drawn as red dots in each panel in the third line. (D) Migration of 20 NK cells randomly selected from those in each lane (except for 10 NK cells in lanes 1, 2, 5, and 6) was analyzed using TAXIScan Analyzer 2 software. Median velocity and directionality values of each cell within an experimental period and the statistical data of cell migration were calculated in the same manner as described in (B). *P < .05, **P < .01. (E) CD8+ T-cell migration analyzed in the same manner as described in (C). CD8+T cells in the images (each panel in the middle line) are drawn as red dots in each panel in the third line. (F) Migration of 10 CD8+ T cells randomly selected from those in each lane (except for 6 CD8+T cells in lanes 1, 5, and 6) was analyzed using TAXIScan Analyzer 2 software. Median velocity and directionality values of each cell within an experimental period and the statistical data of cell migration were calculated in the same manner as described in (B). *P < .05, **P < .01.
Figure 10.
(A) Cytotoxicity of NK cells against Huh-7 cells in each anagliptin or vildagliptin concentration group (n = 3). NK cells (effector cells, E) were added to each culture of Huh-7 or Li-7 cells (target cells, T) at E/T ratio of 50:1 in the absence or presence of various concentrations (1, 10, or 100 μmol/L) of anagliptin or vildagliptin. NK cell cytotoxicity was assessed using LDH Cytotoxicity Detection Kit according to the manufacturer’s instructions. (B) Cytotoxicity of NK cells against Li-7 cells in each anagliptin or vildagliptin concentration group (n = 3). Cytotoxicity was assayed in the same manner as described for Huh-7 cells. (C) Kinetic property of DPP4 expressed by Huh-7 cells. Huh-7 cells expressing CD26 (1 × 105 cells/well) were incubated with various concentrations of recombinant CXCL10 (4.3, 5.0, 10, 13, 20, and 40 μmol/L) for 8 hours. KM is the Michaelis-Menten constant. Levels of intact CXCL10 (1-77) and truncated CXCL10 (3-77) were quantified using immunoprecipitation, Edman degradation, and HPLC. (D) Inhibitory effect of anagliptin or vildagliptin on CXCL10 truncation by Huh-7 cell DPP4 activity. (E) Concentration of CXCL10 in Huh-7 cell xenograft tumors of mice fed the control diet, mice fed the diet containing low dose of anagliptin (L-A), mice fed the diet containing high dose of anagliptin (H-A), and mice fed the diet containing high dose of vildagliptin (H-V) (n = 4 for each group). This CXCL10 is referred to herein as total CXCL10 and was detected by conventional assays measuring both intact and truncated CXCL10 levels. *P < .05 vs control. (F) The IC50 value of anagliptin was determined with KaleidaGraph version 4.0 (Synergy Software) using a 4-parameter curve fit formula: y = 100/[1 + (x/a)b], where y = the percentage of intact CXCL10 (%), x = the concentration of DPP4 inhibitors, and a = the estimated IC50 value. (G) The IC50 value of vildagliptin was determined in the same manner as for anagliptin.
Dipeptidyl Peptidase 4 Inhibitors Prevent the Biologically Active Form of CXCL10 From Being Truncated by Dipeptidyl Peptidase 4
To know whether DPP4 inhibitors actually prevent the biologically active (intact) form of CXCL10 from being truncated by DPP4 and consequently enhance NK cell chemotaxis, we quantified the concentrations of intact CXCL10 (1-77) and truncated CXCL10 (3-77) by using immunoprecipitation, Edman degradation,22 and HPLC. The proportion of truncated CXCL10 was calculated by using the following formula:
Proportion of truncated CXCL10 (%) = Truncated CXCL10 (pmol) / [Intact CXCL10 (pmol) + Truncated CXCL10 (pmol)].
When Huh-7 cells (1×105 cells/well) were incubated with various concentrations of CXCL10 for 8 hours, the kinetic property of DPP4 expressed on Huh-7 cells was examined (Figure 10C). The Michaelis-Menten constant (KM) of DPP4 was 6.3 ± 1.5 μmol/L. This was similar to the KM (4.0 ± 1.0 μmol/L) of soluble DPP4 reported by Lambeir et al,15 suggesting the validity of our quantification for intact CXCL10 and truncated CXCL10. We next examined the inhibitory effect of DPP4 inhibitors on the truncation of CXCL10 by using the same cell culture supernatant as was used in the NK-cell mobility assay, where Huh-7 cells were cultured with CXCL10 in the absence or presence of 100 μmol/L anagliptin or vildagliptin. The DPP4 inhibitors almost completely suppressed the truncation of CXCL10, whereas approximately 60% of CXCL10 was truncated in the absence of DPP4 inhibitors (Figure 10D). These results clearly indicate that NK cell and T-cell chemotaxis is enhanced through the prevention of CXCL10 truncation by DPP4 inhibitors. In addition, the concentration of CXCL10 in tumor tissues was significantly greater in mice fed a diet with a high dose of anagliptin or vildagliptin than in those fed the control diet, even though this CXCL10 is referred to herein as total CXCL10 and was detected by conventional assays measuring both intact and truncated CXCL10 levels (Figure 10E) Finally, we determined the IC50 values of the DPP4 inhibitors (Figure 10F and G), which were 3.7 ± 1.9 nmol/L and 16 ± 10 nmol/L for anagliptin and vildagliptin, respectively. Of note, the IC50 values found in this study were not so different from previously reported values (3.3 nmol/L for anagliptin and 2.5 nmol/L for vildagliptin).23
Effects of Dipeptidyl Peptidase 4 Inhibitors on Tumor Angiogenesis
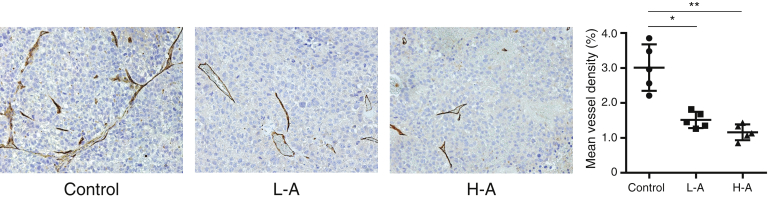
A recent study has highlighted the ability of DPP4 inhibitors to interfere with high fat diet–induced HCC development through the inhibition of tumor angiogenesis.24 Therefore, we examined whether DPP4 inhibitor inhibited angiogenesis in xenograft liver tumors. Anagliptin significantly suppressed CD34-positive vessel density (Figure 11) in xenograft liver tumors. These results suggested that antitumor effect of DPP4 inhibitors potentially involve their ability to suppress tumor angiogenesis.
Figure 11.
Immunohistologic staining for CD34 in Huh-7 cell xenograft tumors of mice in 3 groups (n = 5 for each group) (original magnification, ×200). Mean vascular density was determined by mean percent area positive staining for CD34 for 5 randomly selected fields of view of digital images of each liver using NIH image analysis software. L-A, low dose of anagliptin; H-A, high dose of anagliptin. *P < .05, **P < .01.
Discussion
The present study suggested that CD26 expression in HCC tissues was associated with a more advanced tumor stage, less tumor immunity, and poorer prognosis in HCC patients, and these results are consistent with the prognostic significance of CD26 in patients with colorectal cancer.13 We also found significantly increased serum DPP4 activity in patients with high CD26 expression HCC and a decrease in serum DPP4 activity after HCC resection. These results may be explained in part by a soluble form of CD26 derived from a breakdown product of membrane CD26 on HCC cells because the soluble form of CD26 has DPP4 activity25 and was found to be derived from cell surface CD26 in pulse-chase experiments.26
DPP4 can cleave many chemokines and cytokines possessing a proline or alanine in the penultimate N-terminal position11, 12; this cleavage influences the regulation of immune cell migration.12 Although we chose nude mice for use in an HCC cell xenograft model, this model might be a double-edged sword in terms of investigating the in vivo effects of DPP4 inhibitors on HCC. Because nude mice can produce NK cells but not T cells,2 they are suitable for clarifying the pure role of NK cell migration in tumor immunity against HCC but not for assessing T-cell migration or its role in tumor immunity. Considering NK cell immunity against liver tumors, another critical point is that liver is a NK cell–rich organ. In this regard we confirmed anti-tumor effect of DPP4 inhibitors through NK cell and T-cell infiltration in STAM mice liver. In addition, we used sitagliptin as a DPP4 inhibitor in this model because we wanted to know whether antitumor effect of DPP4 inhibitors is class effect. Several studies have provided evidence that high levels of tumor-infiltrating NK cells are correlated with a good prognosis and increased patient survival.27, 28 NK cells are particularly important for the elimination of tumors with reduced or absent MHC class I expression that evade CD8+ T cell–mediated killing.28 Recently, genetic variants of MHC class I polypeptide-related sequence A (MICA) have been demonstrated to be associated with HCV-associated HCC.29 MICA is a membrane protein that acts as a ligand for NKG2D expressed on NK cells to activate antitumor effects through NK cells. Thus, the role of NK cells in tumor immunity against HCC has attracted much attention.
The expression of chemokine receptors by human NK cells is a matter of debate. Among many chemokine receptors, CXCR3, CXCR4, and CX3CR1 have been reported to be expressed by 2 major human NK cell subsets (CD56dim CD16+ and CD56bright CD16–) and unfractionated NK cells.30 Mouse NK cells express CXCR3 as well.19, 31 Interferon-γ and the expression of CXCR3 by NK cells have been shown to be prerequisites for NK cell infiltration into tumor tissue in mice.19 CXCR3 binds the structurally and functionally related chemokines CXCL9, CXCL10, and CXCL11.20 On the other hand, only CXCL10 has a proline in the penultimate N-terminal position in both humans and mice, although human CXCL9 and CXCL11 have a proline in the same position.32 The first 2 amino acids of these chemokines are enzymatically truncated by DPP4.11 The truncated form of these chemokines becomes an antagonist that engages its receptor, CXCR3, but does not induce chemotaxis.21 On the basis of these results from previous reports, we investigated the effects of DPP4 inhibitors on NK cell chemotaxis, focusing on the CXCR3-CXCL10 axis because we used mice for assessing NK cell accumulation in tumors and human NK cells for assessing NK cell chemotaxis. However, CXCL9 and CXCL11 have also been demonstrated to be truncated by DPP4 activity15; therefore, DPP4 inhibitors may enhance NK cell chemotaxis through the CXCR3-CXCL9 and/or CXCR3-CXCL11 axis. One limitation of this study is that we did not examine the truncation of CXCL9 and/or CXCL11 by DPP4 or their restoration by DPP4 inhibitors.
Regarding the methodology presented herein, the optically accessible chemotaxis assay using the EZ-TAXIScan device should be emphasized. This chemotaxis assay enables the real-time optical observation of NK cell and T-cell chemotaxis and the quantification of cell migration during a fixed time. When Huh-7 cells (1×105 cells/well) were incubated with 10 μmol/L recombinant CXCL10 in the absence of a DPP4 inhibitor, approximately 60% of the CXCL10 was truncated, whereas CXCL10 was hardly truncated in the presence of 100 μmol/L DPP4 inhibitor (Figure 10D). Because we used these culture supernatants in the chemotaxis assay as chemoattractants containing approximately 40% or 100% of intact CXCL10, the differences in the concentrations of intact CXCL10 as a chemoattractant were estimated to lead to differences in the cumulative number of migrated cells between lane 2 and lanes 3 or 4, as shown in Figure 9C and E, as well as differences in migration velocity and direction toward the ligands, as shown in Figure 9D and F. Although the real-time chemotaxis of NK cells could not be evaluated in the HCC xenograft model, we could assess the extent to which NK cell chemotaxis was disrupted because of the truncation of CXCL10 caused by HCC cell DPP4 activity in vitro. These results suggest a real-time relationship between CXCL10 degradation due to HCC cells and disrupted NK cell and T-cell chemotaxis.
There have been several lines of evidence indicating that truncated CXCL10 (3-77) acts as an antagonist of intact CXCL10 (1-77) in clinical settings, resulting in the inability of patients to eradicate acute33 and chronic34 HCV in infections most likely because of the inhibition of antiviral innate and adaptive host immunity. In addition, DPP4 inhibition has been demonstrated to enhance lymphocyte trafficking through the prevention of CXCL10 truncation, improving both naturally occurring tumor immunity and immunotherapies in mouse melanoma models.12 Although our quantitative assay for assessing intact and truncated CXCL10 levels differs from the enzyme-linked immunosorbent assay–based Luminex assay used in previous studies,12, 33, 34 the validity of our method was verified by the observation of DPP4 KM values and DPP4 inhibitor IC50 values comparable with those of previous studies.15, 23 However, the limitation of our quantitative assay was its failure to measure intact and truncated CXCL10 concentrations in vivo, because the serum concentrations of both CXCL10 forms in mice were assumed to be too low for quantification by our method. Although it remains unknown why a high DPP4 inhibitor dose significantly increased the CXCL10 concentration in xenograft liver tumors, even though this CXCL10 was referred to herein as total CXCL10 and was detected by conventional assays measuring both intact and truncated CXCL10 levels, these results are not contradictory to those showing that DPP4 inhibitors significantly increased activated NK cell infiltration into xenograft liver tumors.
DPP4 inhibitors suppressed angiogenesis in xenograft liver tumors, which potentially contribute to antitumor effect of DPP4 inhibitors. We did not investigate the mechanisms underlying this effect because we focused on the enhancement of tumor immunity by DPP4 inhibitors in the present study. Chemokine ligand 2 has been shown to mediate the angiogenic effect of DPP4.24 DPP4 inhibitors may suppress angiogenesis through inactivation of chemokine ligand 2, although further studies are required to clarify this matter.
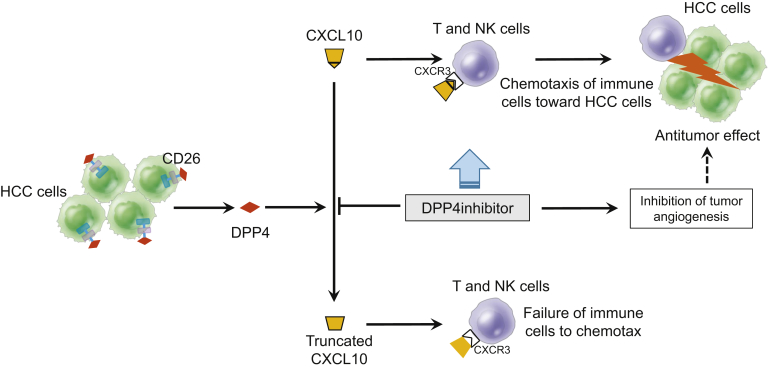
In summary, CD26 expressed in HCC tissues contributed to increased serum DPP4 activity in HCC patients. As shown in Figure 12, DPP4 inhibitors prevented CXCL10 from being truncated by DPP4 activity, activated NK cell and T-cell chemotaxis through preservation of the CXCR3-CXCL10 axis, and consequently suppressed the growth of xenograft liver tumors in nude mice and orthotopic HCC in STAM mice. DPP4 inhibitors also suppressed tumor angiogenesis. These results provide a rationale for verifying whether DPP4 inhibitors clinically inhibit the progression of HCC or augment the antitumor effects of molecular-targeting drugs and immunotherapies against HCC.
Figure 12.
Schematic diagram depicting mechanisms by which DPP4 inhibitors prevent CXCL10 truncation by DPP4 activity, activate NK cell and T-cell chemotaxis through preservation of the CXCR3-CXCL10 axis, inhibit tumor angiogenesis, and consequently suppress tumor growth in a mouse xenograft model of HCC and a NASH-related HCC mouse model.
Materials and Methods
Patients and Clinical Liver and Serum Samples
HCC specimens were obtained from 41 patients who had undergone surgical resection for HCC at the Kawasaki Medical School Hospital between 2006 and 2014. For the assessment of DPP4 activity, serum samples were obtained from 9 patients before and 1–6 months after the operation. All patients were not prescribed DPP4 inhibitors and were followed up after the operation. The overall survival was defined as the period between the diagnosis of HCC and either death or the last visit to the outpatient clinic up to November 20, 2016. HCC stage was classified by using the Union for International Cancer Control tumor node metastasis classification system.35 The expression of CD26, CD56+ NK cell, CD3+ T cell, and Ki67 in tumors was determined immunohistochemically. The study protocol conformed to the 1975 Helsinki Declaration and was approved by the Institutional Research Ethics Committee (Admission No: 1694-1 and 2503-1). The need for informed consent was waived by the Research Ethics Committee because the study was retrospective, and some patients had already died.
Animals and Experimental Design
Eight-week-old male nude mice (BALB/c-nu/nu) received subcutaneous injections of Huh-7 or Li-7 cells. When the xenograft tumors reached a size of 100–150 mm3, the mice were fed a diet containing a low dose of anagliptin (0.7 g/kg diet) (donated by Sanwa Kagaku Kenkyusho), a diet containing a high dose of anagliptin (2 g/kg diet), a diet containing a high dose of vildagliptin (2 g/kg diet) (Heat Biochem, Shanghai, China), or a control diet for 15–21 days. Tumor size (V) was calculated by using the following formula: V (mm3) = width (mm)2 × length (mm) / 2. Dietary intake, body weight, and tumor size were measured every 3 days. Four to 5 mice in each group were killed at 15–21 days after the commencement of feeding by carbon dioxide asphyxiation, as approved by the Panel on Euthanasia of the American Veterinary Association. After the mice were killed, tumor tissue and blood samples were collected for the determination of biochemical parameters, CD26 expression, DPP4 activity, and NK cell infiltration. An oral glucose tolerance test was performed at 2 weeks after the commencement of feeding. Briefly, after the mice fasted overnight, 10 μL/g body weight of a 15% glucose solution was administered by gastric intubation, and blood glucose was measured at 0, 30, 60, and 120 minutes after administration.
We also used a NASH-related HCC mouse model (STAM mouse; SMC Laboratories, Inc, Tokyo, Japan). The mice were developed as described previously.36 Briefly, 2-day-old male C57BL/6J mice were injected with streptozotocin (200 μg/mouse) and fed a high-fat diet (HFD-32; Clea, Tokyo, Japan) from the age of 4 weeks, which was followed by development of NASH and HCC at 8 and 18 weeks of age, respectively. Eight-week-old-mice were orally administered with either sitagliptin (30 mg/kg/day) (donated by MSD K.K.) or distilled water for 10 weeks. Five mice in each group were killed, and their liver tumor size was calculated as was done for nude mice with xenograft tumors. Blood samples and tumor tissues were collected for the biochemical and histologic assessment and the determination of NK cell and T-cell infiltration. Noncancerous liver histology was assessed on the basis of NAFLD activity score.37 The mice were bred and maintained according to the guidelines approved by the Institutional Animal Care Use Committee.
Cell Culture
Huh-7 and Li-7 hepatoma cells were obtained from the Cell Resource Center for Biomedical Research of the Institute of Development, Aging and Cancer at Tohoku University. These cells were cultured in Dulbecco modified Eagle medium containing 10% fetal bovine serum (FBS) at 37°C in a humidified atmosphere with 5% CO2.
Histopathologic Procedures
Tumor tissues and/or non-tumor tissues from patients and mice were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) or 10% buffered formalin and embedded in paraffin for histologic analysis. In addition to undergoing H&E staining, the liver sections were incubated with 1:200 dilution of a rabbit anti-CD26 monoclonal antibody (Abcam, Cambridge, MA), a rabbit anti-Ki67 polyclonal antibody (clone MIB-1; Abcam), a rabbit anti-CD3 monoclonal antibody (GeneTex, Inc, Irvine, CA), a rabbit anti-CD3γ polyclonal antibody (Proteintech, Rosemont, IL), a rabbit anti-CD56 polyclonal antibody (Proteintech), a rabbit anti-CD34 monoclonal antibody (Abcam), and a goat anti-NKp46 antibody (R&D Systems, Minneapolis, MN) at 4°C overnight. After incubation with avidin conjugated peroxidase (Vecstatin Elite ABC; Vector Laboratories, Burlingame, CA), the tissue sections were developed in 3,3’-diaminobenzidine tetrahydrochloride (Sigma Chemical Co, St Louis, MO). National Institutes of Health image analysis software was used to quantify the mean percent area or the mean cell number of positive staining for CD26, CD3, CD3γ, CD56, CD34, or NKp46 for 5 randomly selected fields of view of digital images of each xenograft tumor or liver tumor. The proportion of CD26-positive cells was classified as follows: – (0%–16%), + (17%–32%), ++ (33%–66%), and +++ (≥67%). The labeling index for Ki67-positive nuclei was determined by a random evaluation of 1000 cells selected from several areas. Mean vascular density was determined by the mean percent area positive staining for CD34.
Measurement of Biochemical Parameters
Blood glucose levels were measured with a glucometer (Life Scan, West Chester, PA). Serum insulin levels were measured by using a mouse insulin enzyme-linked immunosorbent assay kit (Morinaga, Yokohama, Japan) according to the manufacturer’s instructions. The concentrations of triglyceride, cholesterol, and low-density lipoprotein cholesterol were determined by using a SPOTCHEM EZ SP-4430 system (Arkray Inc, Kyoto, Japan).
Immunoblotting
Cells were harvested by using trypsin/ethylenediamine tetraacetic acid, washed in PBS, and resuspended in cell lysis buffer (Cell Signaling Technology, Danvers, MA) containing a 1% protease inhibitor mixture (Sigma-Aldrich, St Louis, MO) and 100 mmol/L phenylmethylsulfonyl fluoride. Tissue samples were also lysed in the same lysis buffer. Lysates of cultured cells or tumor tissues were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). The proteins were transferred to polyvinylidene difluoride membranes (Pall Corporation, New York, NY) and blocked for 1 hour at room temperature with 5% bovine serum albumin and 0.1% Tween-20 in Tris-buffered saline. The samples were subsequently incubated overnight at 4°C with a rabbit anti-CD26 monoclonal antibody (Abcam), a rabbit anti-p21 monoclonal antibody (Cell Signaling Technology), a rabbit anti-p27-kip1 monoclonal antibody (Cell Signaling Technology), a rabbit anti-phospho-CDK2 (Thr 160) polyclonal antibody (Cell Signaling Technology), or a rabbit anti-phospho-Rb (Ser807/811) monoclonal antibody (Cell Signaling Technology). The transferred proteins were also blocked for 1 hour at room temperature with 5% milk and 0.1% Tween-20 in Tris-buffered saline and were subsequently incubated for 1 hour at room temperature with a rabbit anti-β-actin monoclonal antibody (Cell Signaling Technology). Immunoreactive proteins were detected by using an enhanced chemiluminescence reagent (Western Lightning Plus-ECL; Perkin-Elmer, Waltham, MA).
Dipeptidyl Peptidase 4 Activity
DPP4 activity in the serum and the lysates of cells and tumor tissues was measured by using a DPP4-activity assay kit (BioVision, San Francisco, CA) according to the manufacturer’s instructions.
Cell Proliferation Analysis
Cell proliferation was measured by using a Cell Counting Kit-8 (CCK-8) (Dojindo Laboratories, Kumamoto, Japan) assay according to the manufacturer’s instructions. Briefly, 5 × 103 Huh-7 and Li-7 cells were incubated with different concentrations of anagliptin or vildagliptin (0, 1, 10, 100 μmol/L) in each well of a 96-well plate for 24 hours. CCK-8 solution was then added to each well. After 2 hours of incubation, sample absorbance at 450 nm was measured by using a FLUO star OPTIMA system (BMG Labtechnologies, Offenburg, Germany).
Cell Cycle Analysis
After 24 hours of in vitro treatment with anagliptin or vildagliptin, Huh-7 cells were fixed with 4% formalin, incubated for 30 minutes with DNA dye (Hoechst 33342), and rinsed with PBS. Plates containing the cells were scanned with an ImageXpress Micro Screening System (Molecular Devices, Tokyo, Japan). Cell phase classification was based on the fluorescence intensity of the DNA dye. For the cell number and cell cycle analyses, the intensity of the integrated DNA dye was assessed by using the cell-cycle application module (MetaXpress; Molecular Devices), as previously described.38
CXCL10 Concentration in Tumor Tissue
CXCL10 concentrations in xenograft tumor lysates were measured by using a mouse CXCL10 enzyme-linked immunosorbent assay kit (R&D Systems, Inc) according to the manufacturer’s instructions.
Natural Killer Cell Depletion in Mice
Nude mice were intraperitoneally injected with 50 μL rabbit anti-asialo GM1 antisera (anti-ASGM1) (Wako Pure Chemicals, Osaka, Japan) or rabbit immunoglobulin (Ig) G (isotype control) 6 times during the course of 15 days. The frequency of NK cells in the spleen leukocytes was examined by staining with anti-CD49b (DX5)-PE and anti-CD3-APC (Miltenyi Biotec, Gladbach, Germany) and by using FACS flow cytometer (BD FACS Aria III; BD Biosciences, San Jose, CA). The infiltration of NK cells in the tumor tissues was immunohistochemically assessed by staining with a goat anti-NKp46 polyclonal antibody.
Isolation of Mouse Spleen Leukocytes
The mouse spleens were processed with a cell strainer in 10 mL cold PBS containing 1% FBS. After the digested tissue was washed and centrifuged, the supernatant was discarded, and red blood cells were lysed by using an RBC Lysis Buffer (BioLegend, San Diego, CA). The purified leukocytes were subjected to flow cytometry for the quantification of NK cell frequency.
Neutralization of Chemokine Receptor CXCR3
To neutralize CXCR3 on NK cells, nude mice were intraperitoneally injected with 200 μg anti-CXCR3 antibody (Bio X cell, West Lebanon, NH) or hamster IgG (isotype control) 6 times during the course of 15 days. NK cell infiltration into tumor tissues was immunohistochemically assessed by staining with a goat anti-NKp46 polyclonal antibody.
Natural Killer Cell and T-Cell Isolation
Peripheral blood mononuclear cells (PBMCs) from 50 mL blood collected from healthy human adult volunteers were isolated by using Lymphoprep (AXIS-SHIELD, Oslo, Norway) according to the manufacturer’s instructions. NK cells were isolated from purified PBMCs by using an NK cell isolation kit (Miltenyi Biotech), activated by interleukin 2, and suspended in RPMI 1640 containing 1% FBS. CD8+ T cells were similarly isolated by using a CD8+ T-cell isolation kit (Miltenyi Biotech) except for activation by interleukin 2. We confirmed that these preparations contained more than 95% of CD56+/CD3– NK cells or CD8+ T cells by using a FACS flow cytometer (FACSCanto II; BD Biosciences). Purified NK cells or CD8+ T cells were then resuspended in assay buffer.
Analysis of Natural Killer Cell Cytotoxicity
NK cells (effector cells, E) were added to cultures of Huh-7 or Li-7 cells (target cells, T) at an E/T ratio of 50:1 in the absence or presence of various concentrations (1, 10, or 100 μmol/L) of anagliptin or vildagliptin. NK cell cytotoxicity was assessed by using a lactate dehydrogenase (LDH) Cytotoxicity Detection Kit (Takara Bio, Kusatsu, Japan) according to the manufacturer’s instructions. The amount of LDH released into the culture supernatant after 4 hours was determined by using a FLUO star OPTIMA system (BMG Labtechnologies). The percent specific cytotoxicity was calculated as (experimental release − spontaneous release) / (maximum release − spontaneous release). The mean cytotoxicity of the 2 cultures per E:T was also calculated.
Chemotaxis Assay of Natural Killer Cells and T Cells
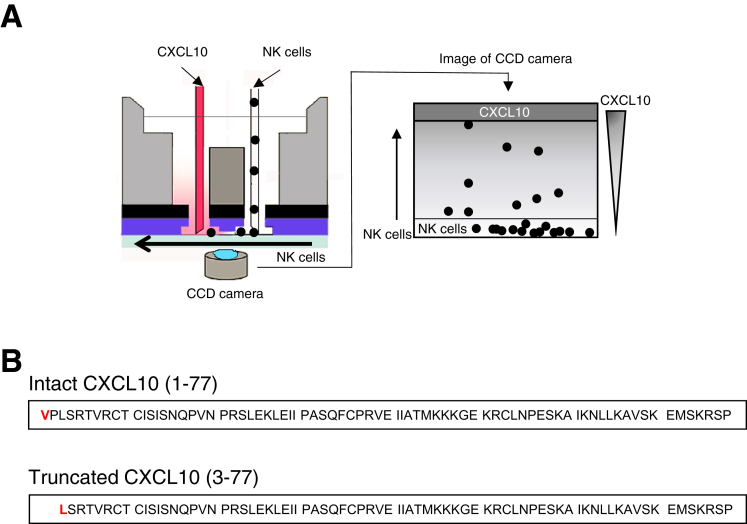
A suspension of purified CD56+/CD3– NK cells (1 × 103 cells/well) or CD8+ T cells (5 × 104 cells/well) was exposed to a chemokine concentration gradient in the EZ-TAXIScan device (Effector Cell Institute, Tokyo, Japan) (Figure 1A), as previously described.39 Chips 4 μm in depth were used for this study. The chemokine concentration gradient that formed from the chemokine injection side to the cell injection side of the microchannel was generated by adding either a mixture of 1 μmol/L recombinant human DPP4 (R&D Systems) and 10 μmol/L recombinant CXCL10 (PeproTech, Rocky Hill, NJ) or the culture supernatant of Huh-7 cells (1 × 105 cells/mL) with 10 μmol/L recombinant CXCL10, either in the presence or absence of a DPP4 inhibitor (100 μmol/L anagliptin or vildagliptin) and 2 μg/μL mouse anti-human CXCL10 monoclonal antibody (anti-CXCL10; R&D Systems). The migration of NK cells or CD8+ T cells in the microchannel was traced with time-lapse intervals by using a charge-coupled device camera and analyzed by using TAXIScan Analyzer 2 software, as previously reported.22 To obtain statistical data regarding cell migration, the median values of velocity and directionality of each cell within an experimental period were calculated from the migratory pathway data obtained from the time-lapse images; the data are expressed by using velocity-directionality plots.
Figure 1.
Schematic diagram depicting the EZ-TAXIScan device (left panel). (A) Concentration gradient of CXCL10 in the channel forms from the chemokine-injection side (top) to the cell-injection side (bottom) (right panel). (B) Amino acid sequence of intact CXCL10 (1-77) and truncated CXCL10 (3-77).
Quantification of Intact CXCL10 (1-77) and Truncated CXCL10 (3-77) Concentrations
Huh-7 cells expressing CD26 (1 × 105 cells/well) were incubated with various concentrations of recombinant CXCL10 (4.3, 5.0, 10, 13, 20, or 40 μmol/L) for 8 hours. Subsequently the CXCL10 peptides, including intact CXCL10 (1-77) and truncated CXCL10 (3-77), in the culture supernatant were immunoprecipitated with Dynabeads bound to anti-CXCL10 and eluted by using Dynabeads Protein G (ThermoFisher Scientific, Waltham, MA) according to the manufacturer’s instructions. The eluted CXCL10 was subjected to SDS-PAGE. The bands corresponding to CXCL10 peptides were cut out from the gel, homogenized, and resuspended in a mixture of 0.1% trifluoroacetic acid and acetonitrile (1:1). The N-terminal amino acid of intact CXCL10 is valine (Val, V), whereas that of truncated CXCL10 is leucine (Leu, L) because of the removal of the first 2 amino acids by DPP4 (Figure 1B). We quantified both intact and truncated CXCL10 levels on the basis of the difference in N-terminal amino acids between both peptides by using Edman degradation.40 In brief, N-terminal amino acids were labeled with fluorescein isothiocyanate, and this derivative of the terminal amino acids was cleaved as a thiazolinone derivative under acidic conditions. The thiazolinone amino acid was then converted to the more stable phenylthiohydantoin (PTH)-amino acid derivative. PTH-Val and PTH-Leu were identified by using high-performance liquid chromatography (HPLC), and their respective levels were quantified by using a standard curve constructed with a known PTH-amino acid standard solution. Thus, the Michaelis-Menten constant (KM) of CXCL10 for DPP4 was determined. Finally, we measured the percentage of intact CXCL10 in the culture supernatant of Huh-7 cells with 10 μmol/L recombinant CXCL10 in the presence of various concentrations of a DPP4 inhibitor. The percentage of intact CXCL10 was calculated by using the following formula: proportion of intact CXCL10 (%) = intact CXCL10 (pmol) / [intact CXCL10 (pmol) + truncated CXCL10 (pmol)]. The inhibitory concentration of 50% value of each DPP4 inhibitor was determined with KaleidaGraph version 4.0 (Synergy Software) by using a 4-parameter curve fit formula: y = 100/[1 + (x/a)b], where y is the percentage of intact CXCL10 (%), x is the concentration of DPP4 inhibitors, and a is the estimated inhibitory concentration of 50% value.
Statistics
Quantitative values are expressed as the mean ± standard deviation. Two groups among the multiple groups were compared by using the rank-based, Kruskal-Wallis analysis of variance test, followed by Scheffe’s test. Data of 2 groups were compared by the Student t test for continuous variables. The cumulative recurrence rate and overall survival were calculated by using the Kaplan-Meier method, and differences among the groups were analyzed by using the log-rank test. A P value less than .05 was considered significant.
Acknowledgments
The authors thank Prof Heiichiro Udono for critical discussion and suggestion for this manuscript and Chizuho Nagato, Nanami Mizukawa, and Hiroshi Ishii for their technical support with the experiments and mouse breeding.
Footnotes
Author contributions S.H.: study design, data acquisition, analysis and interpretation, drafting the manuscript, statistical analysis, funding acquisition; A.Y.: data acquisition, analysis and interpretation, critical revision of the manuscript for important intellectual content, technical support; T.K.: data acquisition, analysis and interpretation, critical revision of the manuscript for important intellectual content, material support; K.K.: critical revision of the manuscript for important intellectual content, material support; M.G.: data analysis and interpretation, critical revision of the manuscript for important intellectual content, material support; K.S.: technical support, critical revision of the manuscript for important intellectual content; Y.H.: critical revision of the manuscript for important intellectual content; Y.T.: critical revision of the manuscript for important intellectual content; F.K.: critical revision of the manuscript for important intellectual content; T.T.: critical revision of the manuscript for important intellectual content; K.H.: study supervision, study concept and design, data analysis and interpretation, drafting the manuscript, funding acquisition.
Conflicts of interest These authors disclose the following: Keisuke Hino received support from Sanwa Kagaku Kenkyusho; and Moritaka Goto is an employee of Sanwa Kagaku Kenkyusho, Co, Ltd. The remaining authors disclose no conflicts.
FundingKeisuke Hino was supported by a Grant-in-aid for Scientific Research (B) (26293179) from the Japan Society for the Promotion of Science and the Research Program on Hepatitis from the Japan Agency for Medical Research and Development (18fk0210016h0003). Sohji Nishina was supported by a Grant-in-aid for Young Scientists (B) (16K19374) from the Japan Society for the Promotion of Science and Kawasaki Medical School Research Project Grant P2. Akira Yamauchi was supported by a Grant-in-aid for Scientific Research (JP15K10201) from the Japan Society for the Promotion of Science.
References
- 1.White D.L., Thrift A.P., Kanwal F., Davila J., El-Serag H.B. Incidence of hepatocellular carcinoma in all 50 United States, from 2000 through 2012. Gastroenterology. 2017;152:812–820 e5. doi: 10.1053/j.gastro.2016.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singal A.G., EI-Serag H.B. Hepatocellular carcinoma from epidemiology to prevention: translating knowledge into practice. Clin Gastroenterol Hepatol. 2015;13:2140–2151. doi: 10.1016/j.cgh.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hedenstierna M., Nangarhari A., Weiland O., Aleman S. Diabetes and cirrhosis are risk factors for hepatocellular carcinoma after successful treatment of chronic hepatitis C. Clin Infect Dis. 2016;63:723–729. doi: 10.1093/cid/ciw362. [DOI] [PubMed] [Google Scholar]
- 4.EI-Serag H.B., Kanwal F., Richardson P., Kramer J. Risk of hepatocellular carcinoma after sustained virological response in veterans with hepatitis C virus infection. Hepatology. 2016;64:130–137. doi: 10.1002/hep.28535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giovannucci E., Harlan D., Archer M.C., Bergenstal R.M., Gapstur S.M., Habel L.A., Pollak M., Regensteiner J.G., Yee D. Diabetes and cancer. Diabetes Care. 2010;33 doi: 10.2337/dc10-0666. 1674–Cancer85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kasuga M., Ueki K., Tajima N., Noda M., Ohashi K., Noto H., Goto A., Ogawa W., Sakai R., Tsugane S., Hamajima N., Nakagama H., Tajima K., Miyazono K., Imai K. Report of the Japan Diabetes Society/Japanese Cancer Association joint committee on diabetes and cancer. Cancer Sci. 2013;104:965–976. doi: 10.1111/cas.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh S., Singh P.P., Singh A.G., Murad M.H., Sanchen W. Anti-diabetic medication and the risk of hepatocellular cancer: a systematic review and meta-analysis. Am J Gastroenterol. 2013;108:881–891. doi: 10.1038/ajg.2013.5. [DOI] [PubMed] [Google Scholar]
- 8.Li J., Hernanda P.Y., Bramer W.M., Peppelenbosch M.P., van Luijk J., Pan Q. Anti-tumor effects of metformin in animal models of hepatocellular carcinoma: a systemic review and meta-analysis. PLoS One. 2015;10:e0127967. doi: 10.1371/journal.pone.0127967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karagiannis T., Boura P., Tsapas A. Safety of dipeptidyl peptidase 4 inhibitors: a perspective review. Ther Adv Drug Saf. 2014;5:138–146. doi: 10.1177/2042098614523031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lambeir A.M., Durinx C., Scharpe S., De Meester I. Dipeptidyl peptidase IV from bench to bedside: an update on structural properties, functions, and clinical aspects of the enzyme DPP IV. Crit Rev Clin Lab Sci. 2003;40:209–294. doi: 10.1080/713609354. [DOI] [PubMed] [Google Scholar]
- 11.Bongers J., Lambros T., Ahmad M., Heimer E.P. Kinetics of dipeptidyl peptidase IV proteolysis of growth hormone-releasing factor and analogs. Biochim Biophys Acta. 1992;1122:147–153. doi: 10.1016/0167-4838(92)90317-7. [DOI] [PubMed] [Google Scholar]
- 12.Barreira da Silva R., Laird M.E., Yatim N., Fiette L., Ingersoll M.A., Albert M.L. Dipeptidylpeptidase 4 inhibition enhances lymphocyte trafficking, improving both naturally occurring tumor immunity and immunotherapy. Nat Immunol. 2015;16:850–858. doi: 10.1038/ni.3201. [DOI] [PubMed] [Google Scholar]
- 13.Lam C.S., Cheung A.H., Wong S.K., Wan T.M., Ng L., Chow A.K., Cheng N.S., Pak R.C., Li H.S., Man J.H., Yau T.C., Lo O.S., Poon J.T., Pang R.W., Law W.L. Prognostic significance of CD26 in patients with colorectal cancer. PLoS One. 2014;9:e98582. doi: 10.1371/journal.pone.0098582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawaguchi T., Kodama T., Hikita H., Makino Y., Saito Y., Tanaka S., Shimizu S., Sakamori R., Miyagi T., Wada H., Nagano H., Hiramatsu N., Tatsumi T., Takehara T. Synthetic lethal interaction of combined CD26 and Bcl-xL inhibition is a powerful anticancer therapy against hepatocellular carcinoma. Hepatol Res. 2015;45:1023–1033. doi: 10.1111/hepr.12434. [DOI] [PubMed] [Google Scholar]
- 15.Lambeir A.M., Proost P., Durinx C., Bal G., Senten K., Augustyns K., Scharpe S., Van Damme J., De Meester I. Kinetic investigation of chemokine truncation by CD26/dipeptidyl peptidase IV reveals a striking selectivity within the chemokine family. J Biol Chem. 2001;276:29839–29845. doi: 10.1074/jbc.M103106200. [DOI] [PubMed] [Google Scholar]
- 16.Gorrell M.D., Gysbers V., McCaughan G.W. CD26: a multifunctional integral membrane and secreted protein of activated lymphocytes. Scand J Immunol. 2001;54:249–264. doi: 10.1046/j.1365-3083.2001.00984.x. [DOI] [PubMed] [Google Scholar]
- 17.Fraker L.D., Halter S.A., Forbes J.T. Effects of orally administered retinol on natural killer cell activity in wild type BALB/c and congenitally athymic BALB/c mice. Cancer Immunol Immunother. 1986;21:114–118. doi: 10.1007/BF00199858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klein T., Fujii M., Sandel J., Shibazaki Y., Wakamatsu K., Mark M., Yoneyama H. Linagliptin alleviates hepatic steatosis and inflammation in a mouse model of non-alcoholic steatohepatitis. Med Mol Morphol. 2014;47:137–149. doi: 10.1007/s00795-013-0053-9. [DOI] [PubMed] [Google Scholar]
- 19.Wendel M., Galani I.E., Suri-Payer E., Cerwenka A. Natural killer cell accumulation in tumors is dependent on IFN-γ and CXCR3 ligands. Cancer Res. 2008;68:8437–8445. doi: 10.1158/0008-5472.CAN-08-1440. [DOI] [PubMed] [Google Scholar]
- 20.Clark-Lewis I., Mattioli I., Gong J.H., Loetscher P. Structure-function relationship between the human chemokine receptor CXCR3 and its ligands. J Biol Chem. 2003;278:289–295. doi: 10.1074/jbc.M209470200. [DOI] [PubMed] [Google Scholar]
- 21.Proost P., Schutyser E., Menten P., Struyf S., Wuyts A., Opdenakker G., Detheux M., Parmentier M., Durinx C., Lambeir A.M., Neyts J., Liekens S., Maudgal P.C., Billiau A., Van Damme J. Amino-terminal truncation of CXCR3 agonists impairs receptor signaling and lymphocyte chemotaxis, while preserving antiangiogenic properties. Blood. 2001;98:3554–3561. doi: 10.1182/blood.v98.13.3554. [DOI] [PubMed] [Google Scholar]
- 22.Yamauchi A., Degawa-Yamauchi M., Kuribayashi F., Kanegasaki S., Tsuchiya T. Systematic single cell analysis of migration and morphological changes of human neutrophils over stimulus concentration gradients. J Immunol Methods. 2014;404:59–70. doi: 10.1016/j.jim.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 23.Watanabe Y.S., Yasuda Y., Kojima Y., Okada S., Motoyama T., Takahashi R., Oka M. Anagliptin, a potent dipeptidyl peptidase IV inhibitor: its single-crystal structure and enzyme interactions. J Enzyme Inhib Med Chem. 2015;30:981–988. doi: 10.3109/14756366.2014.1002402. [DOI] [PubMed] [Google Scholar]
- 24.Qin C.J., Zhao L.H., Zhou X., Zhang H.L., Wen W., Tang L., Zeng M., Wang M.D., Fu G.B., Huang S., Huang W.J., Yang Y., Bao Z.J., Zhou W.P., Wang H.Y., Yan H.X. Inhibition of dipeptidyl peptidase IV prevents high fat diet-induced liver cancer angiogenesis by downregulating chemokine ligand 2. Cancer Lett. 2018;420:26–37. doi: 10.1016/j.canlet.2018.01.064. [DOI] [PubMed] [Google Scholar]
- 25.Durinx C., Lambeir A.M., Bosmans E., Falmagne J.B., Berghmans R., Haemers A., Scharpe S., De Meester I. Molecular characterization of dipeptidyl peptidase activity in serum: soluble CD26/dipeptidyl peptidase IV is responsible for the release of X-Pro dipeptides. Eur J Biochem. 2000;267:5608–5613. doi: 10.1046/j.1432-1327.2000.01634.x. [DOI] [PubMed] [Google Scholar]
- 26.Hong W.J., Piazza G.A., Hixson D.C., Doyle D. Expression of enzymatically active rat dipeptidyl peptidase IV in Chinese hamster ovary cells after transfection. Biochemistry. 1989;28:8474–8479. doi: 10.1021/bi00447a030. [DOI] [PubMed] [Google Scholar]
- 27.Ishigami S., Natsugoe S., Takuda K., Nakajo A., Che X., Iwashige H., Aridome K., Hokita S., Aikou T. Prognostic value of intratumoral natural killer cells in gastric carcinoma. Cancer. 2000;88:577–583. [PubMed] [Google Scholar]
- 28.Marincola F.M., Jaffee E.M., Hicklin D.J., Ferrone S. Escape of human solid tumors from T-cell recognition: molecular mechanisms and functional significance. Adv Immunol. 2000;74:181–273. doi: 10.1016/s0065-2776(08)60911-6. [DOI] [PubMed] [Google Scholar]
- 29.Kumar V., Kato N., Urabe Y., Takahashi A., Muroyama R., Hosono N., Otsuka M., Tateishi R., Omata M., Nakagawa H., Koike K., Kamatani N., Kubo M., Nakamura Y., Matsuda K. Genome-wide association study identifies a susceptibility locus for HCV-induced hepatocellular carcinoma. Nat Genet. 2011;43:455–458. doi: 10.1038/ng.809. [DOI] [PubMed] [Google Scholar]
- 30.Robertson M.J. Role of chemokines in the biology of natural killer cells. J Leukoc Biol. 2002;71:173–183. [PubMed] [Google Scholar]
- 31.Marquartdt N., Wilk E., Pokoyski C., Schmidt R.E., Jacobs R. Murine CXCR3+CD27brightNK cells resemble the human CD56brightNK-cell population. Eur J Immunol. 2010;40:1428–1439. doi: 10.1002/eji.200940056. [DOI] [PubMed] [Google Scholar]
- 32.Ou X., O’Leary H.A., Broxmeyer H.E. Implications of DPP4 modification of proteins that regulate stem/progenitor and more mature cell types. Blood. 2013;122:161–169. doi: 10.1182/blood-2013-02-487470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riva Antonio, Laird M., Gasrouge A., Ambrozaitis A., Williams R., Naoumov N.V., Albert M.L., Chokshi S. Truncated CXCL10 is associated with failure to achieve spontaneous clearance of acute hepatitis C infection. Hepatology. 2014;60:487–496. doi: 10.1002/hep.27139. [DOI] [PubMed] [Google Scholar]
- 34.Gasrouge A., Decalf J., Ahloulay M., Lababidi C., Mansour H., Vallet-Pichard A., Mallet V., Mottez E., Mapes J., Fontanet A., Pol S., Albert M.L. Evidence for an antagonist form of the chemokine CXCL10 in patients chronically infected with HCV. J Clin Invest. 2011;121:308–317. doi: 10.1172/JCI40594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sobin L.H., Gospodarowicz M.K., Witterkind C. 7th ed. Wiley-Blackwell; Oxford: 2010. TNM classification of malignant tumours. [Google Scholar]
- 36.Fujii M., Shibazaki Y., Wakamatsu K., Honda Y., Kawauchi Y., Suzuki K., Arumugam S., Watanabe K., Ichida T., Asakura H., Yoneyama H. A murine model for non-alcoholic steatohepatitis showing evidence of association between diabetes and hepatocellular carcinoma. Med Mol Morphol. 2013;46:141–152. doi: 10.1007/s00795-013-0016-1. [DOI] [PubMed] [Google Scholar]
- 37.Kleiner D.E., Brunt E.M., Van Natta M., Behling C., Contos M.J., Cummings O.W., Ferrell L.D., Liu Y.C., Torbenson M.S., Unalp-Arida A., Yeh M., McCullough A.J., Sanyal A.J. Nonalcoholic Steatohepatitis Clinical Research Network: design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 38.Kawase T., Yasui Y., Nishina S., Hara Y., Yanatori I., Tomiyama Y., Nakashima Y., Yoshida K., Kishi F., Nakamura M., Hino K. Fibroblast activation protein-α-expressing fibroblast promote the progression of pancreatic ductal adenocarcinoma. BMC Gastroenterol. 2015;15:109. doi: 10.1186/s12876-015-0340-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanegasaki S., Nomura Y., Nitta N., Akiyama S., Tamatani T., Goshoh Y., Yoshida T., Sato T., Kikuchi Y. A novel optic assay for the quantitative measurement of chemotaxis. J Immunol Methods. 2003;282:1–11. doi: 10.1016/j.jim.2003.07.008. [DOI] [PubMed] [Google Scholar]
- 40.Edman P. Method for determination of the amino acid sequence in peptides. Acta Chem Scand. 1950;4:283–293. [Google Scholar]