A primary means by which the intestine is protected against microbial onslaught is by production of a thick mucus gel that serves to physically separate the microbiota from epithelial cells. Accordingly, the outer regions of the mucus layer are heavily colonized by bacteria while the inner layer is nearly sterile.1 Alterations in intestinal microbiota composition are associated with various disease states, including inflammatory bowel disease, cancer, and metabolic syndrome. Although such associations generally have focused on the fecal microbiome, recent studies have highlighted the importance of mucosa-associated bacteria.2, 3 Moreover, recent confocal microscopy–based studies, in both mice and humans, suggest an outsize role for bacteria that penetrate the inner mucus layer, thereby encroaching on, and potentially inducing proinflammatory gene expression in intestinal epithelial cells.4 Thus, we sought a means to identify inner mucus bacterial species and examine how their relative abundance changed in response to dietary perturbations that influence inflammation.
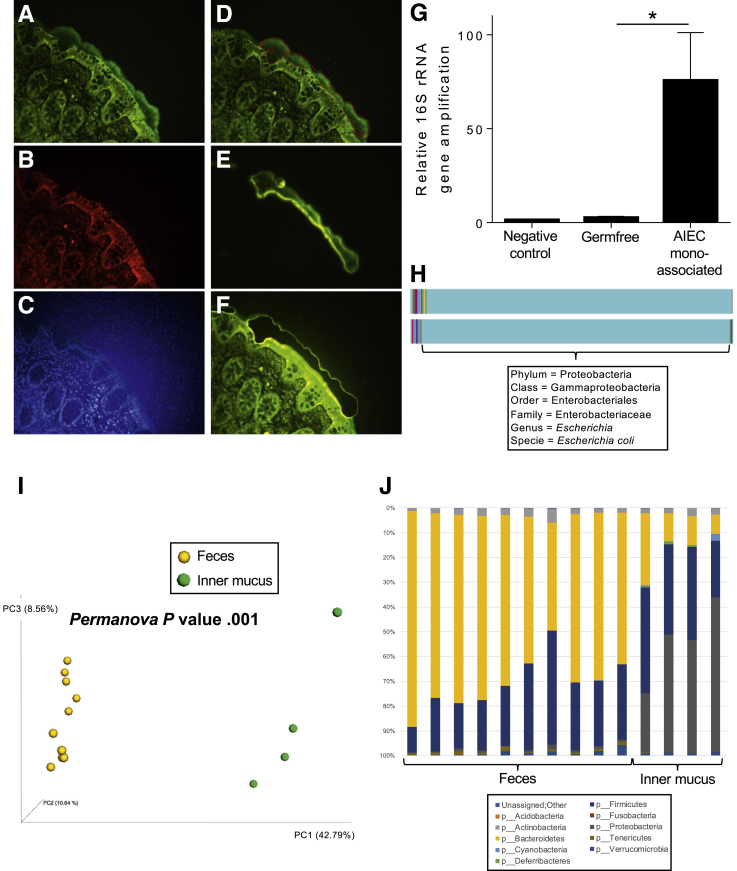
Laser capture microdissection (LCM) was used previously to interrogate mucosal- and crypt-associated microbial communities.5, 6, 7 Herein, as detailed in the Supplemental Methods section, we applied LCM to proximal human colon specimens fixed in Carnoy's solution, permitting a visually selective collection of fields of inner mucus from which bacterial DNA could be extracted and analyzed (Figure 1A–F). Application of DNA sequencing–based analysis of low biomass samples is prone to artifacts, resulting from trace bacterial DNA in samples themselves or analytical reagents. However, our approach yielded enough DNA that pre–polymerase chain reaction amplification, used in previous LCM-based microbiome analysis,5 was not needed, thus mitigating but not eliminating this concern. Hence, to validate our approach, we applied it to germ-free mice, which lack inner mucus bacteria, and mice mono-associated with adherent-invasive Escherichia coli. Although, as expected, some quantifiable and sequenceable bacterial DNA was present in inner mucus excised from germ-free mice (autoclaved chow contains dead bacteria), the level of bacterial DNA was much higher (50-fold) in adherent-invasive E coli mono-associated mice (Figure 1G) and, in these mice, the vast majority of sequences (>98%) were identified as E coli via the Greengenes database (Figure 1H). Such sequences were not detected in inner mucus collected from germ-free mice, suggesting that our method was indeed capable of characterizing inner mucus bacterial composition.
Figure 1.
Laser capture microdissection of mucus invaders. (A–F) Mucus layer staining on human colonic biopsy and LCM. (A) Muc2 (green). (B) Actin (red). (C) DNA/nucleus (blue, background owing to the use of PEN Membrane Frame Slides). (D) Selected area for microdissection. (E and F) After LCM, the inner mucus layer collected is transferred to the cap membrane and ready for DNA extraction. (G) 16S ribosomal RNA (rRNA) gene quantification after DNA extraction on unused cap membrane (negative control), and inner mucus microdissected from germ-free mice or E coli mono-associated mice. (H) Microbiota composition analysis of mucus invaders from E coli mono-associated mice. (I) Fecal and mucus-associated microbiota composition was analyzed by 16S rRNA gene Illumina sequencing. PCoA of the unweighted UniFrac distance is represented, with samples colored by sample type (feces and mucus). (J) Taxa summarization performed at the phylum level. Values are means ± SEM, N = 4–6. *P < .05 determined by a 1-way analysis of variance corrected for multiple comparisons with a Sidak post-test. AIEC, adherent-invasive E coli.
The application of our method to fecal and inner mucus samples from conventional mice, followed by principle coordinate analysis (PCoA) of unweighted UniFrac distances, showed profound differences between fecal and inner mucus bacteria composition (Figure 1I and Supplementary Figure 1). Such distinctness between fecal and inner mucus bacterial communities was evident at the phyla level as well, with inner mucus communities comprising 20%–60% Proteobacteria and a concomitantly marked reduction in Bacteroidetes (Figure 1J and Supplementary Figure 1). Moreover, the inner mucus microbiome showed a moderately higher level of species (α)-diversity compared with fecal samples (Figure 2C and D), suggesting a complex microbial ecosystem is harbored.
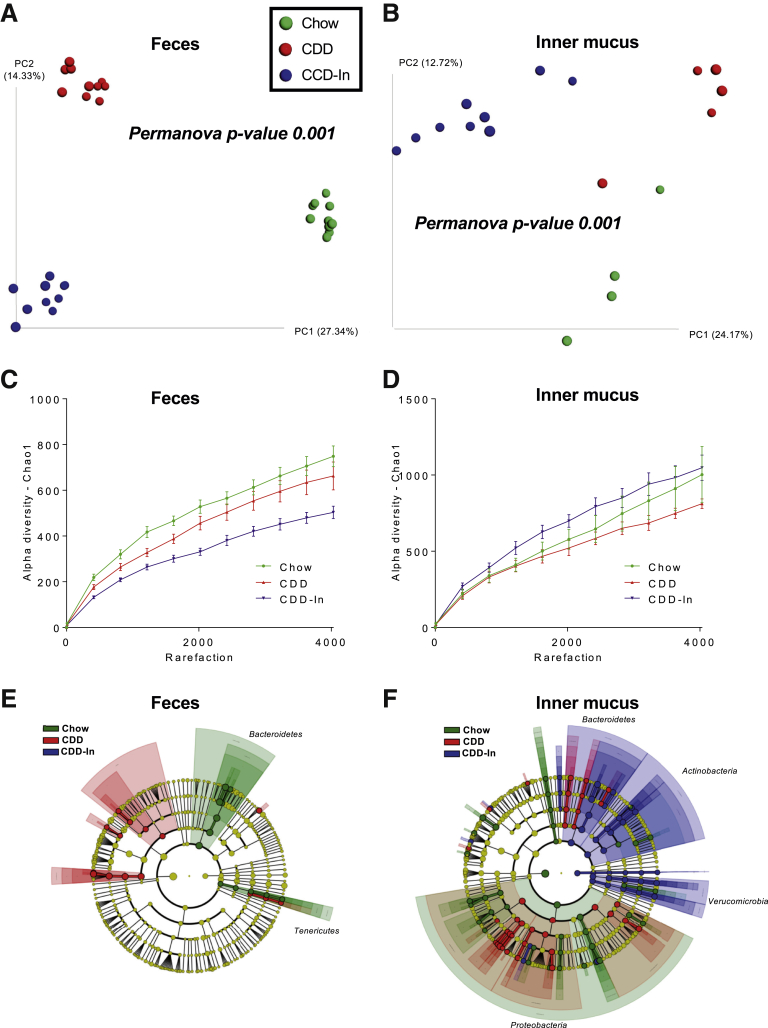
Figure 2.
Impact of purified diet and inulin supplementation on fecal and mucus-associated microbiota composition. Fecal and mucus-associated microbiota composition was analyzed by 16S ribosomal RNA gene Illumina sequencing. (A) PCoA of the unweighted UniFrac distance in fecal samples colored by experimental group (Chow, CDD, and CDD-In). (B) PCoA of the unweighted UniFrac distance in inner mucus samples colored by experimental group (Chow, CDD, and CDD-In). (C) α-Diversity in fecal samples using the chao1 measure. (D) α-Diversity in inner mucus samples using the chao1 measure. (E and F) Linear discriminant analysis effect size was used to investigate bacterial members that drive differences between the Chow, CDD, and CDD-In groups in fecal and inner mucus samples. (E) Taxonomic cladogram obtained from linear discriminant analysis effect size analysis of fecal 16S ribosomal RNA genes sequencing, highlighting taxa significantly altered in 1 group compared with the other (linear discriminant analysis score > 2.0). (F) Taxonomic cladogram obtained from linear discriminant analysis effect size of inner mucus-associated 16S ribosomal RNA gene sequencing, highlighting taxa significantly altered in 1 group compared with the other (linear discriminant analysis score > 2.0).
Next, we examined the extent to which bacterial composition of inner mucus might be impacted by 2 compositionally defined diets (CDDs) known to impact fecal microbiome composition. Mice were fed grain-based chow, control CDD that lacks fermentable fiber and results in microbiota encroachment, or CDD enriched with inulin fiber (CDD-In) that restores a normal microbiota localization (Supplementary Table 1).8, 9 Each condition used 2 cages of mice, thus reducing the risk of cage-clustering artifacts. In accord with our previous work, CDD caused a stark shift in fecal microbiome composition, which was shifted further in mice fed CDD-In, when using PCoA of both unweighted and weighted UniFrac distances (Figure 2A and Supplementary Figure 2A). These diets also caused clear, albeit slightly less stark, shifts in the composition of the inner mucus microbiome (Figure 2B and Supplementary Figure 2B). However, a combined PCoA plot of fecal and inner mucus samples illustrates that these marked differences in fecal microbiomes between mice fed these different diets were, in fact, of only moderate magnitude relative to the dramatic differences between fecal and inner mucus microbiomes of mice consuming the same diet (Supplementary Figure 1), further highlighting the distinctness between the fecal and inner mucus microbiome. Perhaps more importantly, although both CDD and CDD-In induced changes in fecal and inner mucus microbiome, the taxa that were impacted differed markedly between these sites. Changes at the phyla level included an expansion of Tenericutes and Actinobacteria in inner mucus of mice fed CDD-In. Moreover, when viewed at the family level, both diets induced changes in the fecal microbiome, but the extent of change was markedly greater and different in inner mucus samples (Supplementary Figure 3B). More generally, although CDD-In led to a marked loss of α-diversity in the fecal microbiome, this parameter was highest in the inner mucus in mice fed this diet (Figure 2C and D). Accordingly, use of the linear discriminant analysis effect size tool showed a panel of diet-specific changes, including a marked expansion of numerous taxa in mice fed CDD-In that were not observed in fecal samples (Figure 2E and F).
Although defining the extent to which changes in inner mucus microbiome impact host phenotype will require further investigation, we speculate that some of the specific alterations observed herein may be functionally important. For example, CDD-fed mice showed higher levels of Rickettsiales order (phyla Alphaproteobacteria) (Supplementary Figure 4C), which are known endosymbionts of eukaryotic cells and thus can be envisaged to play a role in linking microbiota encroachment to low-grade inflammation and altered metabolism, which was previously observed in these mice.9 Conversely, CDD-In, which abrogates low-grade inflammation and associated parameters of metabolic syndrome, enriched inner mucus Akkermansia species, which are known to be mucus-associated and generally considered health-promoting. Moreover, CDD-In feeding enriched the inner mucus levels of health-associated Bifidobacterium and Lactococcus species, while decreasing inflammation-associated Gammaproteobacteria (Supplementary Figure 4C). These diet-induced alterations were not observed in fecal samples, which highlights the potential importance of examining inner mucus bacteria.
In conclusion, we herein report the development of a technique to analyze the inner mucus microbiome. Our validation of this technique used samples of proximal mouse colon, which is more penetrable to bacteria than distal mouse and human colon.10 Nonetheless, we have preliminarily found the technique yields seemingly reasonable results when applied to small pinch human colon biopsy specimens. We believe that the inner mucus microbiome plays a pivotal role in health and disease and hope this report will spur investigation of this microbial ecosystem and how it impacts its host.
Footnotes
Author contributions Benoit Chassaing and Andrew T. Gewirtz conceived the project, designed the experiments, interpreted the results, and wrote the manuscript; Benoit Chassaing performed experiments and analysis.
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported by National Institutes of Health grants DK099071 and DK083890 (A.T.G.); a Career Development Award from the Crohn's and Colitis Foundation and an Innovator Award from the Kenneth Rainin Foundation (B.C.).
Supplementary Material
References
- 1.Johansson M.E. Proc Natl Acad Sci U S A. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gevers D. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Cruz P. J Gastroenterol Hepatol. 2015;30:268–278. doi: 10.1111/jgh.12694. [DOI] [PubMed] [Google Scholar]
- 4.Bretin A. Am J Physiol Endocrinol Metab. 2018;315:E1–E6. doi: 10.1152/ajpendo.00014.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y. Appl Microbiol Biotechnol. 2010;88:1333–1342. doi: 10.1007/s00253-010-2921-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richard M.L. Gut Microbes. 2018;9:131–142. doi: 10.1080/19490976.2017.1379637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pedron T. mBio. 2012;3(3) doi: 10.1128/mBio.00116-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chassaing B. Am J Physiol Gastrointest Liver Physiol. 2015;309:G528–G541. doi: 10.1152/ajpgi.00172.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zou J. Cell Host Microbe. 2018;23:41–53 e4. doi: 10.1016/j.chom.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ermund A. Am J Physiol Gastrointest Liver Physiol. 2013;305:G341–G347. doi: 10.1152/ajpgi.00046.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.