Abstract
Single-nucleotide polymorphisms (SNPs) located in the promoter region of the receptor for advanced glycation end products (RAGE) gene have been linked to the activity of RAGE. However, contrary to our expectation, we previously detected no correlation between SNPs within the RAGE promoter and ulcerative colitis (UC) risk in a case-control study. Here, we investigated the methylation of the RAGE promoter and analyzed the collective contribution of methylation and SNPs to UC risk. We found that RAGE promoter hypomethylation was more common in UC patients compared to controls (70% vs. 30%, respectively), as determined via bisulfite sequencing PCR (BSP) and methylation-specific PCR (MSP). Furthermore, we investigated the cooperativity of promoter methylation and SNPs and found that either of two SNPs (rs1800624 or rs1800625) and promoter methylation jointly contributed to UC risk (30 UC patients vs. 30 controls, P < 0.05). There was no correlation between UC risk and either methylation or SNPs when analyzed separately. This lack of correlation is likely due to promoter methylation repressing gene transcription, whereas SNPs in the RAGE promoter region activate RAGE transcription. We found that variant allele carriers with promoter hypomethylation were at an increased risk for UC (rs1800624, OR = 10, 95% CI: 1.641–60.21, P = 0.009; rs1800625, OR = 4.8, 95% CI: 1.074–21.447, P = 0.039). Furthermore, our data revealed that the RAGE mRNA levels in variant allele carriers with promoter hypomethylation were significantly higher compared to those with promoter hypermethylation (P < 0.05) as well as to those in wild-type allele individuals exhibiting promoter hypomethylation (P < 0.05). We therefore speculate that the methylation status and SNPs present in the RAGE promoter region alter RAGE transcription, thereby impacting UC risk. We also propose that the methylation status and RAGE promoter genotype could jointly serve as clinical biomarkers to assist in UC risk assessment.
Keywords: RAGE, Methylation, SNP, Inflammation, UC risk
Highlights
-
•
Promoter methylation and SNPs were detected in a Chinese UC population.
-
•
No correlation was detected in both methylation and SNPs with UC risk independently.
-
•
Methylation and SNPs may modulate RAGE transcription in opposite directions.
-
•
Combined effect of SNPs and methylation could be serve as biomarker for UC risk.
1. Introduction
Ulcerative colitis (UC) is an inflammatory bowel disease (IBD) that is thought to result from the complex interplay between genetic and environmental factors that results in dysregulation of the immune response [1]. Chronic inflammation in UC patients is associated with an increased incidence of colorectal cancer (CRC), the risk of which is directly correlated with the duration and extent of the inflammation [2], [3]. The cumulative CRC risk for long-standing UC patients is 5–10% at 20 years and 10–30% at 30 years [4], [5].
RAGE is a member of the immunoglobulin superfamily of cell surface molecules. It is a multi-ligand receptor that interacts with certain members of the S100/calgranulin family (e.g., S100A12 and S100B), high mobility group box-1 (HMGB1), and amyloid-β peptide (Aβ), among other factors. RAGE has been implicated in the amplification of proinflammatory responses via interactions with its ligands [6]. As a cell surface receptor, RAGE mediates rapid and sustained cellular activation through multiple intracellular signaling pathways that lead to the propagation of inflammatory responses [7].
Ordinarily, RAGE is constitutively expressed in the lung and at a low level in the intestinal epithelium [8]. However, during inflammation, its expression appears to be significantly upregulated in other cell types, including endothelial cells, epithelial cells and leukocytes [9]. In one model of sepsis, deletion of RAGE protected mice against sepsis in a manner that was dependent on the innate immune system. Furthermore, anti-RAGE antibody-treated animals were resistant to lethality in the cecal ligation and puncture (CLP) model of polymicrobial sepsis [10], demonstrating the role of RAGE in innate defense mechanisms. Furthermore, RAGE is known to be involved in other diseases that are characterized by inflammation, including diabetes, arthritis, sepsis, neurodegeneration and even cancer [10], [11], [12], [13].
Recent studies have revealed that RAGE expression is upregulated in the intestinal epithelium and colonic tissues of patients with active IBD [14]. Furthermore, a specific RAGE antibody was found to attenuate colitis in a mouse model [15]. Together, these findings suggest that RAGE plays an important role in regulating IBD-associated chronic inflammation, making the RAGE gene a candidate risk biomarker for IBD.
DNA methylation has been shown to regulate gene transcription and activation. The reciprocal relationship between promoter methylation and transcriptional activity of a gene has been widely documented [16], [17], [18], [19], [20], [21], [22], [23]. In previous studies, 3–6% of all cytosines were found to be methylated in normal human DNA [23]. Gene hypermethylation, especially in cases of cancer, is associated with restricted transcription of tumor suppressor genes. Specifically, aberrant methylation of CpG islands within the promoter regions of tumor suppressor genes contributes to many types of tumorigenesis [24], [25]. Chronic inflammation in UC patients was also shown to be tightly associated with DNA methylation. The methylation status of the ABCB1, CDH1, ESR1, GDNF, HPP1, and MYOD1 genes were demonstrated to be associated with the inflammatory status of UC patients [19]. Moreover, the methylation of the RUNX3, MINT1, and COX-2 promoters has also been confirmed to be a biomarker for the presence of colorectal dysplasia in patients with UC [26].
However, few studies have addressed the correlation between RAGE promoter methylation and UC risk. Previously, Wang (2016) found a significant correlation between RAGE single-nucleotide polymorphisms (G82S) and UC risk in a Chinese population [27]. However, no correlation was found between UC risk and two SNPs in the RAGE promoter region, which were expected to have an important effect on RAGE transcription in early stages of the disease. This finding led us to explore other mechanisms by which RAGE transcription is altered in UC patients. Because DNA methylation acts to regulate the patterns of gene transcription, and a CpG island has been predicted in the RAGE promoter region, we examined the methylation status of the RAGE promoter region in UC patients. We then analyzed the joint effect of methylation status and SNPs of the RAGE promoter region on UC risk.
2. Materials and methods
2.1. Sample preparation
Tissue samples were obtained via colon biopsy from 30 UC patients [mean age = 45.87 ± 13.45 years, 16 men (53.3%) and 14 women (46.7%)] and 30 age- and race-matched healthy controls [mean age 43.13 ± 12.62 years, 18 men (60%) and 12 women (40%)]. All samples were collected from the Affiliated Hospital of Guangdong Medical University in south China, and all participants were Chinese. IBD was diagnosed using conventional clinical, endoscopic, and histological criteria, and other inflammatory diseases were excluded. The healthy controls were asymptomatic with no history of malignancy and no personal or family history of IBD. Written informed consent was obtained from all patients, and the study was approved by the ethics committee of the Affiliated Hospital of Guangdong Medical University.
2.2. Genomic DNA extraction and bisulfite conversion
Genomic DNA was extracted from colon biopsy tissues using a tissue DNA extraction kit (Qiagen, Germany) according to the manufacturer's protocol. Genomic DNA was treated using the EpiTect Bisulfite Kit (Qiagen) according to the manufacturer's protocol to convert all unmethylated cytosines to uracils while leaving 5-methylcytosine residues unaltered. One microgram of genomic DNA was used for each conversion reaction.
2.3. SNP genotyping
High-resolution melting (HRM) was employed to genotype two SNPs (rs1800624, rs1800625) (Fig. 1) in the promoter region of the RAGE gene. The HRM genotyping protocol and the SNP-specific primers (including the internal temperature standard) were described previously [29].
Fig. 1.
CpG islands and SNPs in the 5′ UTR of the RAGE gene.
2.4. Methylation identification
A CpG island in the promoter region of the RAGE gene was predicted using software (MethPrimer, PUMCH, China) (Fig. 1). Bisulfite sequencing PCR (BSP) was used to determine the methylation status of the CpG island in the RAGE promoter region. The following primers were used: 5′GGGATATGATTTTTGGATAGAGG3′ (forward) and 5′CCAATCAATAATTCCCTAAAATAAC3′ (reverse). The BSP amplicon was a 189-bp fragment containing the CpG island of the RAGE promoter, which included 9 CpGs. The PCR amplicon was cloned into the pMD18-T vector (TaKaRa, China), and the resulting plasmid was transformed into E. coli strain DH5α. The amplicon sequence was then analyzed using an ABI 3730XL DNA sequencer (ABI, US).
Methylation-specific PCR (MSP) was used to verify the BSP result and to determine the methylation status of all samples. Three pairs of specific primers were used in the MSP analysis: one pair was targeted to the methylated sequence; one pair was targeted to the unmethylated sequence; and the last pair was for the same amplicon with untreated DNA template. All of the primers used in this study are listed in Table 1. Bisulfite-treated genomic DNA was used as template DNA in the MSP amplification and untreated genomic DNA was used as a control.
Table 1.
Specific primers used in this study.
| Primer | Product size | Length bp | Tm (°C) | 'C's | Sequence |
|---|---|---|---|---|---|
| F-N-RAGE | 106 | 25 | 59 | – | CCCCATGCATCCCCACCCCCGC |
| R-N-RAGE | 25 | 57 | – | ATCTGAGTCAGACACTCTCGTC | |
| F-M-RAGE | 112 | 25 | 55 | 15 | CGTTTTTATGTATTTTTATTTTCGT |
| R-M-RAGE | 25 | 55 | 4 | AATATCTAAATCAAACACTCTCGTC | |
| F-U-RAGE | 112 | 25 | 52 | 15 | TGTTTTTATGTATTTTTATTTTTGT |
| R-U-RAGE | 25 | 53 | 4 | AATATCTAAATCAAACACTCTCATC |
MSP products were resolved by electrophoresis on a 10% polyacrylamide gel (PAGE) and band patterns were analyzed independently by two individuals.
2.5. mRNA extraction and QPCR
Total mRNA extracted from colon biopsy tissues was reverse transcribed to cDNA using a reverse transcription kit (TaKaRa, Dalian, China). Quantitative real-time PCR (QPCR) was used to evaluate RAGE transcription in UC patients and in healthy controls. RAGE transcription levels were normalized to GAPDH transcription levels.
2.6. Statistical analysis
Allele frequencies and genotype distributions were determined as described previously. Hardy–Weinberg equilibrium was applied to evaluate allelic and genotypic distributions, and χ2 test and Fisher's exact test were used to compare the allele and genotype frequencies in patients versus controls. The odds ratio (OR) and 95% confidence interval (CI) were calculated to assess the relative risk of UC. The χ2 tests were performed using SPSS 19 software. The threshold for significance was set at P < 0.05 for all analyses.
3. Results
3.1. BSP
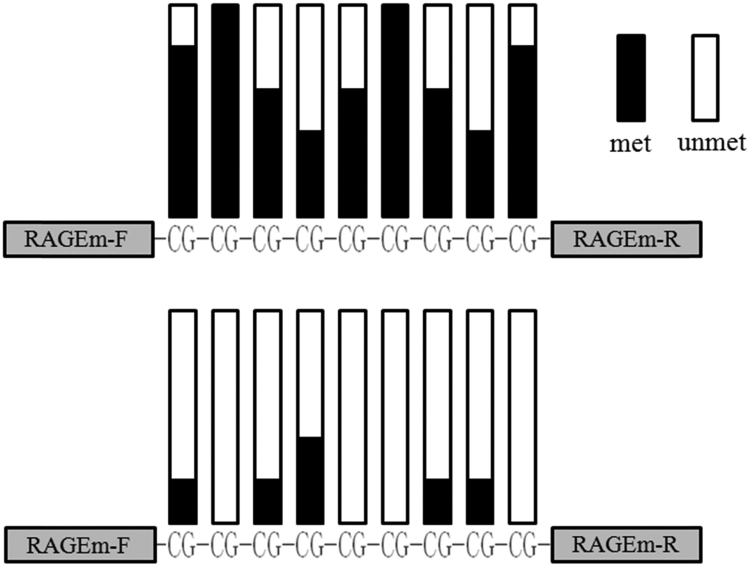
A CpG island 124 bp in length and containing 9 CpGs was predicted to exist in the RAGE promoter region using the online software MethPrimer (http://www.urogene.org/cgi-bin/methprimer/methprimer.cgi). The CpG island was located upstream of two SNPs (rs1800624 and rs1800625) in the promoter region. The BSP amplicon was 189 bp in length and contained the entire CpG island. After cloning the BSP products into competent cells, five bacterial colonies derived from each sample were sequenced using an ABI 3730XL DNA sequencer. The sequencing data revealed that most of the CpGs present in the RAGE promoter of healthy control samples were methylated, whereas very few were methylated in samples from UC patients (Fig. 2).
Fig. 2.
CpG island methylation status in the 5′ UTR of theRAGEgene. The black columns represent methylated CpGs and hollowcolumns represent unmethylated CpGs in the BSP amplicons. Five sets of sequencing data for each sample were stacked together to form this diagram. Above: methylation status of the healthy controls. Below: methylation status of the UC patients.
3.2. MSP
The MSP amplicons were 112 bp in length and were analyzed by PAGE. All reactions using the N-RAGE primers with untreated DNA template produced a positive band on the gel (Fig. 3a) while no positive bands were observed in reactions using the N-RAGE primers with untreated template DNA (Fig. 3c). The products generated in reaction using the M-RAGE primers with bisulfite-treated template DNA revealed differences between UC patients and healthy controls; a higher percentage of control samples produced positive bands compared to the UC patient samples (Fig. 3b). However, the products generated from the U-RAGE primers with bisulfite-treated template DNA revealed the opposite result, with a higher percentage of UC patient samples producing positive bands compared to the controls (Fig. 3d).
Fig. 3.
CpG island methylation status detection by MSP. a: PAGE product of the N-RAGE primers with untreated DNA. b: PAGE product of the M-RAGE primers with bisulfite treated DNA. c: PAGE product of the N-RAGE primers with bisulfite treated DNA. d: PAGE product of the U-RAGE primers with bisulfite treated DNA.
3.3. Joint analysis of RAGE promoter methylation and SNP genotype/allele frequency
We analyzed the SNP genotype/allele frequencies combined with the methylation status of the RAGE promoter region and their collective association with UC risk. After carrying out the Harvey-Weinberg equilibrium calculation, no deviations from HWE were observed in the control population. However, the data revealed that rs1800624 or rs1800625 variant allele carriers exhibiting hypomethylation in the RAGE promoter region were at significant UC risk (rs1800624, OR = 2.5, 95% CI: 1.241–5.037, P = 0.009; rs1800625, OR = 2.118, 95% CI: 0.972–4.614, P = 0.039) (Table 2, Table 3).
Table 2.
Association of rs1800624 genotype/allele, promoter methylation and UC risk.
| genotype/allele+M | UC | Control | OR | 95%CI | P | N |
|---|---|---|---|---|---|---|
| TT+M+ | 12 | 20 | – | |||
| TT+M- | 17 | 12 | 0.424 | 0.151–1.185 | 0.127 | 61 |
| TA+AA+M+ | 7 | 15 | 1.286 | 0.408–4.051 | 0.775 | 54 |
| TA+AA+M- | 24 | 11 | 0.275 | 0.100–0.765 | 0.015 | 67 |
| T+M+ | 29 | 53 | – | |||
| T+M- | 53 | 34 | 0.351 | 0.188–0.656 | 0.001 | 169 |
| A+M+ | 9 | 17 | 1.034 | 0.409–2.609 | 1 | 108 |
| A+M- | 29 | 12 | 0.226 | 0.101–0.509 | < 0.001 | 123 |
genotype/allele+M: genotype/allele and methylation status of RAGE. M+: methylation positive, M-: methylation negative.
Table 3.
Association of rs1800625 genotype/allele, promoter methylation and UC risk.
| genotype/allele+M | UC | Control | OR | 95%CI | P | N |
|---|---|---|---|---|---|---|
| TT+M+ | 10 | 16 | – | |||
| TT+M- | 18 | 11 | 0.382 | 0.128–1.135 | 0.108 | 55 |
| TC+CC+M+ | 8 | 18 | 1.406 | 0.446–4.432 | 0.771 | 52 |
| TC+CC+M- | 24 | 13 | 0.339 | 0.120–0.957 | 0.045 | 63 |
| T+M+ | 28 | 48 | – | |||
| T+M- | 58 | 44 | 0.443 | 0.241–0.814 | 0.010 | 178 |
| C+M+ | 8 | 20 | 1.458 | 0.568–3.745 | 0.492 | 104 |
| C+M- | 26 | 14 | 0.314 | 0.141–0.699 | 0.006 | 116 |
In addition, hypomethylation was significantly more prevalent among UC patients than healthy controls (70% vs. 30%). Further analyses revealed that the combination of −374A (rs1800624) and hypomethylation in the RAGE promoter region behaved as a UC risk factor (OR = 2.75, 95% CI: 1.718–4.402, P < 0.001) (Table 2). The combination of -429C (rs1800625) and hypomethylation in the RAGE promoter region also behaves as a UC risk factor (OR = 2.361, 95% CI: 1.378–4.046, P = 0.002) (Table 3). After analyze the distribution frequency of genotype/allele and methylation status in different genders of UC patients, no significant difference between the sexes was detected (Table 4).
Table 4.
Distribution of RAGE genotype/allele and promoter methylation frequencies between different genders of UC patients.
| Genotype/allele & methylation | Male | Female | OR | 95% | P | N |
|---|---|---|---|---|---|---|
| rs1800624 | ||||||
| TT+M+ | 8 | 4 | – | |||
| TT+M- | 12 | 10 | 1.667 | 0.385–7.209 | 0.717 | 34 |
| TA+AA+M+ | 4 | 2 | 1 | 0.125–7.995 | 1 | 18 |
| TA+AA+M- | 12 | 8 | 1.333 | 0.298–5.957 | 1 | 32 |
| T+M+ | 19 | 10 | – | – | – | – |
| T+M- | 32 | 26 | 1.509 | 0.603–3.776 | 0.493 | 90 |
| A+M+ | 6 | 2 | 0.76 | 0.124–4.643 | 1 | 36 |
| A+M- | 16 | 10 | 1.221 | 0.393–3.799 | 0.778 | 52 |
| rs1800625 | ||||||
| TT+M+ | 6 | 4 | – | |||
| TT+M- | 10 | 8 | 1.2 | 0.250–5.768 | 1 | 28 |
| TC+CC+M+ | 6 | 2 | 0.5 | 0.065–3.845 | 0.638 | 18 |
| TC+CC+M- | 14 | 10 | 1.071 | 0.238–4.816 | 1 | 34 |
| T+M+ | 16 | 10 | – | |||
| T+M- | 31 | 24 | 1.239 | 0.478–3.213 | 0.81 | 81 |
| C+M+ | 8 | 2 | 0.4 | 0.070–2.277 | 0.438 | 36 |
| C+M- | 17 | 12 | 1.129 | 0.383–3.332 | 1 | 55 |
RAGE mRNA levels were also analyzed for correlation with promoter SNP genotypes and methylation. In general, individuals carrying variant alleles of either rs1800624 or rs1800625 (TA+AA or TC+CC, respectively) showed higher RAGE mRNA levels compared to individuals carrying the homozygous wild-type allele (TT) (Fig. 4). Similarly, individuals with hypomethylation in the RAGE promoter region exhibited higher average RAGE mRNA levels compared to those with hypermethylation. Further analyses revealed significant correlation between the RAGE mRNA level and −374 TA/AA+M- carriers (P < 0.05) (Fig. 4a), as well as between the mRNA level and −429 TC/CC+M- carriers (P < 0.05) (Fig. 4b).
Fig. 4.
Association of promoter SNPs, methylation status and RAGE mRNA level. a: RAGE mRNA levels in patients with different rs1800624 genotypes and methylation status. b: RAGE mRNA levels in patients with different rs1800625 genotypes and methylation status.
4. Discussion
Recently, a growing body of evidence has suggested that there is a correlation between DNA methylation and the development of UC [28], [29]. Lin (2012) identified 11 IBD-associated CpG sites, 14 CD-specific CpG sites, and 24 UC-specific CpG sites that exhibit methylation changes in B cells and found that these CpG sites were distributed among several genes that play important roles in the immune and inflammatory responses [18]. Nevertheless, few studies have focused on the correlation between the methylation status of the RAGE gene and UC risk, although the role of the RAGE gene in immune and inflammatory responses is well documented.
Accumulation studies have highlighted the possibility that SNPs within key domains of the RAGE gene may influence its function, and individuals with certain genotypes or alleles may be predisposed to heightened disease risk [8], [9]. Nevertheless, there is long-standing debate over whether SNPs in the RAGE promoter region can alter RAGE transcription. Hudson et al. [30] demonstrated that the −374A polymorphism results in a threefold increase in transcriptional activity of the RAGE promoter compared with the -374T allele (P < 0.001) as assessed by chloramphenicol acetyl transferase (CAT) reporter gene transcription assays, and −374A/T allele carriers exhibited significantly altered nuclear protein binding activities. The same study also found that the incidence of the −429 C allele was increased in the retinopathy group (P < 0.05). Däbritz (2011) reported that the rs1800624 polymorphism in the RAGE promoter facilitates transcription of the gene and may, to some degree, protect against the development of strictures in CD patients [31]. Wang (2014) reported that the CD risk associated with the rs1800624 mutant allele was markedly decreased [32].
Conflicting results have also been reported. Picheth (2007) reported that the −429T>C, rather than the -374T>A, RAGE promoter polymorphism is associated with type 1 diabetes in a Brazilian population, and a 2-fold increase in the -429C allele frequency was observed among diabetic subjects compared with nondiabetic subjects [33]. Similarly, Peng et al. (2009) found that Chinese patients with type 2 diabetes mellitus who carry the C allele of -429T/C and G allele of G82S exhibited significantly higher sRAGE levels, while no such association was found in patients with different combinations of the -374T/A allele [34].
One preliminary case-control study found a correlation between the G82S allele in RAGE and UC risk in a Chinese population. However, no correlation between polymorphisms in the RAGE promoter region and UC risk were observed [27]. In the present study, we investigated the RAGE promoter methylation status and found no significant correlation between promoter methylation and UC risk. However, a significant correlation was found when considering both the methylation status and genotype of the RAGE promoter with respect to UC risk. These analyses revealed that individuals with a variant allele of rs1800624 or rs1800625 and hypomethylation were at significantly increased UC risk.
Most studies suggest that methylation within a promoter region downregulates gene transcription, whereas SNPs in the RAGE promoter region are largely thought to increase RAGE transcription. The CpG island that we identified in the RAGE promoter region was predicted to contain binding sites for 12 transcription factors (dissimilarity margin ≤ 5%), as assessed using the promo database (http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/). Among these, the sites for 3 transcription factors, namely, YY1 [T00915], GATA-1 [T00306], and c-Jun [T00133], were identical in sequence to the known binding sites for these proteins. We speculate that CpG methylation in this region may affect regulation by these transcription factors thereby alter RAGE expression. However, this potential mechanism needs to be experimentally verified. Moreover, unlike the G82S SNP located in the V-type immunoglobulin domain of the receptor's extracellular region, which could directly affect protein function [35], SNPs located in the promoter region are more likely to alter RAGE transcription. Thus, it is conceivable that no association would be found when promoter methylation and polymorphisms are independently assessed for their correlation with UC risk. Our analyses of the relationships between methylation, SNPs, and RAGE transcriptional levels support this notion.
Our results suggest that RAGE promoter methylation in conjunction with promoter genotype may be a candidate biomarker for the identification of patients at high risk for UC. For the first time, our analyses have linked the combinatorial effect of RAGE promoter SNPs and methylation status to UC risk within a Chinese population.
We assessed RAGE promoter methylation by MSP in a fraction of our samples. The occurrence of RAGE methylation in the inflammatory rectal mucosa was defined by the presence or absence of a band as assessed by PAGE. There are limits to the sensitivity of MSP detection; therefore, alternative methylation detection methods, such as pyrosequencing, may provide more accurate data. We investigated the relationship between RAGE promoter methylation, SNPs and UC risk using a case-control study. The case-control design is suitable for the study of rare diseases such as UC, and the association of multiple factors with a disease can be studied simultaneously. However, case-control studies only acquire information regarding disease risk and not morbidity, as opposed to cohort studies. Furthermore, the small population size involved in this study, as well as conceivable links between DNA methylation and additional factors such as environment and lifestyle, indicate that further verification with larger sample sizes and diverse populations is warranted.
In addition, a soluble form of RAGE (sRAGE) derived from both alternative splicing of the RAGE gene and cleavage of membrane-bound RAGE [36], [37], [38], [39] exists in circulation. The sRAGE protein may play a protective role in chronic inflammation due to its ability to block RAGE ligands. Thus, while RAGE signaling promotes inflammation [6], sRAGE is capable of neutralizing ligands in circulation, exerting a protective effect against inflammation [36], [37], [38], [39]. The regulatory mechanism mediating this phenomenon is not yet clear. However, Blocking RAGE has been demonstrated to be effective in preventing colitis in an animal model of the disease [40].
Nevertheless, further investigation is necessary to determine whether the RAGE pathway is a viable target for UC therapeutics in humans. The combined effect of methylation status and polymorphisms in the promoter region of RAGE may facilitate use of the RAGE pathway as a biomarker for clinical UC risk detection.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (81600445, 81302246), the Medical Scientific Research Foundation of Guangdong Province (No. A2016522), the Guangdong Medical University Research Fund (2XJ14006, 2XJ14040) and the Doctoral Research Fund of Affiliated Hospital of Guangdong Medical College (528B20150012).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2018.11.001
Contributor Information
Yulan Zhou, Email: zyl092078@163.com.
Lingli Zhang, Email: semy518@126.com.
Transparency document. Supplementary material
Supplementary material
References
- 1.Cho J.H. The genetics and immunopathogenesis of inflammatory bowel disease. Nat. Rev. Immunol. 2008;8:458–466. doi: 10.1038/nri2340. [DOI] [PubMed] [Google Scholar]
- 2.Gyde S.N., Prior P., Allan R.N. Colorectal cancer in ulcerative colitis: a cohort study of primary referrals from three centres. Gut. 1988;29(2):206–217. doi: 10.1136/gut.29.2.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Isbell G., Levin B. Ulcerative colitis and colon cancer. Gastroenterol. Clin. N. Am. 1988;17(4):773–791. [PubMed] [Google Scholar]
- 4.Ekbom A., Helmick C., Zack M. Ulcerative colitis and colorectal cancer. A population-based study. N. Engl. J. Med. 1990;323(18):1228–1233. doi: 10.1056/NEJM199011013231802. [DOI] [PubMed] [Google Scholar]
- 5.Pia A., Rainer P., Michael G. Epigenetic control of the E-cadherin gene (CDH1) by CpG methylation in colectomy samples of patients with ulcerative colitis. Genes Chromosomes Cancer. 2002;35(2):121–126. doi: 10.1002/gcc.10101. [DOI] [PubMed] [Google Scholar]
- 6.Hofmann M.A., Drury S., Fu C. RAGE mediates a novel proinflammatory axis: a central cell surface receptor for S100/calgranulin polypeptides. Cell. 1999;97:889–901. doi: 10.1016/s0092-8674(00)80801-6. [DOI] [PubMed] [Google Scholar]
- 7.Sparvero L.J., Asafu-Adjei D., Rui K. RAGE (Receptor for Advanced Glycation Endproducts), RAGE ligands, and their role in cancer and inflammation. J. Transl. Med. 2009;7(1):17. doi: 10.1186/1479-5876-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng C., Tsuneyama K., Kominami R., Shinohara H., Sakurai S., Yonekura H., Watanabe T., Takano Y., Yamamoto H., Yamamoto Y. Expression profiling of endogenous secretory receptor for advanced glycation end products in human organs. Mod. Pathol. 2005;18:1385–1396. doi: 10.1038/modpathol.3800450. [DOI] [PubMed] [Google Scholar]
- 9.Bierhaus A., Humpert P.M., Morcos M. Understanding RAGE, the receptor for advanced glycation end products. J. Mol. Med. 2005;83(11):876. doi: 10.1007/s00109-005-0688-7. [DOI] [PubMed] [Google Scholar]
- 10.Lutterloh E.C., Opal S.M., Pittman D.D. Inhibition of the RAGE products increases survival in experimental models of severe sepsis and systemic infection.[J] Crit. Care. 2007;11(6):R122. doi: 10.1186/cc6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Zoelen M.A., Schmidt A.M., Florquin S. Receptor for advanced glycation end products facilitates host defense during Escherichia coliinduced abdominal sepsis in mice. J. Infect. Dis. 2009;200:765–773. doi: 10.1086/604730. [DOI] [PubMed] [Google Scholar]
- 12.Chavakis T., Bierhaus A., Al-Fakhri N. The pattern recognition receptor (RAGE) is a counterreceptor for leukocyte integrins: a novel pathway for inflammatory cell recruitment. J. Exp. Med. 2003;198(10):1507–1515. doi: 10.1084/jem.20030800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi F.Y., Ramasamy R., Schmidt A.M. Receptor for age (RAGE) & its ligands – cast into leading roles in diabetes & the inflammatory response. J. Mol. Med. 2009;87(3):235–247. doi: 10.1007/s00109-009-0439-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zen K., Chen C.X., Chen Y.T. Receptor for advanced glycation endproducts mediates neutrophil migration across intestinal epithelium. J. Immunol. 2007;178(4):2483–2490. doi: 10.4049/jimmunol.178.4.2483. [DOI] [PubMed] [Google Scholar]
- 15.Geetha S., Jonamani N., Bernd W. Carboxylated N-glycans on RAGE promote S100A12 binding and signaling[J] J. Cell. Biochem. 2010;110(3):645–659. doi: 10.1002/jcb.22575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones P.A., Laird P.W. Cancer-epigenetics comes of age. Nat. Genet. 1999;21(2):163. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 17.Arisawa T., Tahara T., Shibata T. Promoter hypomethylation of protease-activated receptor 2 associated with carcinogenesis in the stomach. J. Gastroenterol. Hepatol. 2007;22(6):943–948. doi: 10.1111/j.1440-1746.2007.04847.x. [DOI] [PubMed] [Google Scholar]
- 18.Lin Z., Hegarty J.P., Yu W. Identification of disease-associated DNA methylation in B cells from Crohn's disease and ulcerative colitis patients. Dig. Dis. Sci. 2012;57(12):3145–3153. doi: 10.1007/s10620-012-2288-z. [DOI] [PubMed] [Google Scholar]
- 19.Saito S., Kato J., Hiraoka S. DNA methylation of colon mucosa in ulcerative colitis patients: correlation with inflammatory status. Inflamm. Bowel Dis. 2011;17(9):1955–1965. doi: 10.1002/ibd.21573. [DOI] [PubMed] [Google Scholar]
- 20.Herman J.G., Baylin S.B. Gene silencing in cancer in association with promoter hypermethylation. N. Engl. J. Med. 2003;349(21):2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 21.Feinberg A.P., Tycko B. The history of cancer epigenetics. Nat. Rev. Cancer. 2004;4(2):143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 22.Gunnar E., Nils W. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429(6990):457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 23.Esteller M. Aberrant DNA methylation as a cancer-inducing mechanism. J. Annual Rev.Pharmacol. Toxicol. 2005;45(45):629–656. doi: 10.1146/annurev.pharmtox.45.120403.095832. [DOI] [PubMed] [Google Scholar]
- 24.Baylin S.B., Herman J.G., Graff J.R. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv. Cancer Res. 1998;72:141–196. [PubMed] [Google Scholar]
- 25.Kanai Y. Genome‐wide DNA methylation profiles in precancerous conditions and cancers. Cancer Sci. 2010;101(1):36–45. doi: 10.1111/j.1349-7006.2009.01383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garritypark M.M., Loftus E.V.J., Sandborn W.J. Methylation status of genes in non-neoplastic mucosa from patients with ulcerative colitis-associated colorectal cancer. Am. J. Gastroenterol. 2010;105(7):1610–1619. doi: 10.1038/ajg.2010.22. [DOI] [PubMed] [Google Scholar]
- 27.Wang J., Zeng J., Wang H. Genetic polymorphisms of RAGE and risk of ulcerative colitis in a Chinese population. Immunol. Lett. 2016;170(2):88–94. doi: 10.1016/j.imlet.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 28.Tahara T., Shibata T., Nakamura M. Effect of MDR1 gene promoter methylation in patients with ulcerative colitis. Int. J. Mol. Med. 2009;23(4):521–527. doi: 10.3892/ijmm_00000160. [DOI] [PubMed] [Google Scholar]
- 29.Tahara T., Shibata T., Nakamura M. Promoter methylation of protease-activated receptor (PAR2) is associated with severe clinical phenotypes of ulcerative colitis (UC) Clin. Exp. Med. 2009;9(2):125–130. doi: 10.1007/s10238-008-0025-x. [DOI] [PubMed] [Google Scholar]
- 30.Hudson B.I., Stickland M.H., Futers T.S. Effects of novel polymorphisms in the RAGE gene on transcriptional regulation and their association with diabetic retinopathy. Diabetes. 2001;50(6):1505–1511. doi: 10.2337/diabetes.50.6.1505. [DOI] [PubMed] [Google Scholar]
- 31.Däbritz J., Friedrichs F., Weinhage T. The functional -374T/A polymorphism of the receptor for advanced glycation end products may modulate Crohn's disease. Gastroenterology. 2011;300(5):G823–G832. doi: 10.1152/ajpgi.00115.2010. [DOI] [PubMed] [Google Scholar]
- 32.Wang Z.T., Hu J.J., Fan R. RAGE gene three polymorphisms with Crohn's disease susceptibility in Chinese Han population. World J. Gastroenterol. 2014;20(9):2397. doi: 10.3748/wjg.v20.i9.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Picheth G., Heidemann M., Pedrosa F.O. The -429 T>C polymorphism of the receptor for advanced glycation end products (RAGE) is associated with type 1 diabetes in a Brazilian population. Clin. Chim. Acta. 2007;383(1):163–164. doi: 10.1016/j.cca.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 34.Peng W.H., Lu L., Wang L.J. RAGE gene polymorphisms are associated with circulating levels of endogenous secretory RAGE but not with coronary artery disease in Chinese patients with type 2 diabetes mellitus. Arch. Med. Res. 2009;40(5):393–398. doi: 10.1016/j.arcmed.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 35.Hofmann M.A., Drury S., Hudson B.I. RAGE and arthritis: the G82S polymorphism amplifies the inflammatory response. Genes Immunity. 2002;3(3):123–135. doi: 10.1038/sj.gene.6363861. [DOI] [PubMed] [Google Scholar]
- 36.Kalea A.Z., Reiniger N., Yang H. Alternative splicing of the murine receptor for advanced glycation end-products (RAGE) gene. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2009;23(6):1766–1774. doi: 10.1096/fj.08-117739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohe K., Watanabe T., Harada S. Regulation of alternative splicing of the receptor for advanced glycation endproducts (RAGE) through G-rich cis-elements and heterogenous nuclear ribonucleoprotein H. J. Biochem. 2010;147(5):651–659. doi: 10.1093/jb/mvp207. [DOI] [PubMed] [Google Scholar]
- 38.Raucci A., Cugusi S., Antonelli A., Barabino S.M., Monti L., Bierhaus A., Reiss K., Saftig P., Bianchi M.E. A soluble form of the receptor for advanced glycation endproducts (RAGE) is produced by proteolytic cleavage of the membrane-bound form by the sheddase a disintegrin and metalloprotease 10 (ADAM10) FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2008;22(10):3716–3727. doi: 10.1096/fj.08-109033. [DOI] [PubMed] [Google Scholar]
- 39.Zhang L., Bukulin M., Kojro E. Receptor for advanced glycation end products is subjected to protein ectodomain shedding by metalloproteinases. J. Biol. Chem. 2008;283(51):35507. doi: 10.1074/jbc.M806948200. [DOI] [PubMed] [Google Scholar]
- 40.Davé S.H., Tilstra J.S., Matsuoka K. Ethyl pyruvate decreases HMGB1 release and ameliorates murine colitis. J. Leukoc. Biol. 2009;86(3):633–643. doi: 10.1189/jlb.1008662. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material