Abstract
Angucyclines are one of the largest families of aromatic polyketides with various chemical structures and bioactivities. Decades of studies have made it easy for us to depict the picture of their early biosynthetic pathways. Two families of oxygenases, the FAD-dependent oxygenases and the ring opening oxygenases, contribute to the formation of some unique skeletons of atypical angucyclines. The FAD-dependent oxygenases involved in the biosynthetic gene clusters of typical angucyclines catalyze two hydroxylation reactions at C-12 and C-12b of prejadomycin, while their homolog JadH in jadomycin gene cluster catalyze the C-12 hydroxylation and 4a,12b-dehydration reactions of prejadomycin, which leads to the production of dehydrorabelomycin, a common intermediate during the biosynthesis of atypical angucyclines. Ring opening oxygenases of a unique family of oxygenases catalyze the oxidative C—C bond cleavage reaction of dehydrorabelomycin, followed by different rearrangement reactions, resulting in the formation of the various chemical skeletons of atypical angucyclines. These results suggested that the functional differentiation of these oxygenases could apparently enrich the sources of aromatic polyketides with greater structure diversities.
Keywords: Angucycline, Biosynthesis, Oxygenase, FAD-dependent monooxygenase, Ring opening oxygenase
1. Introduction
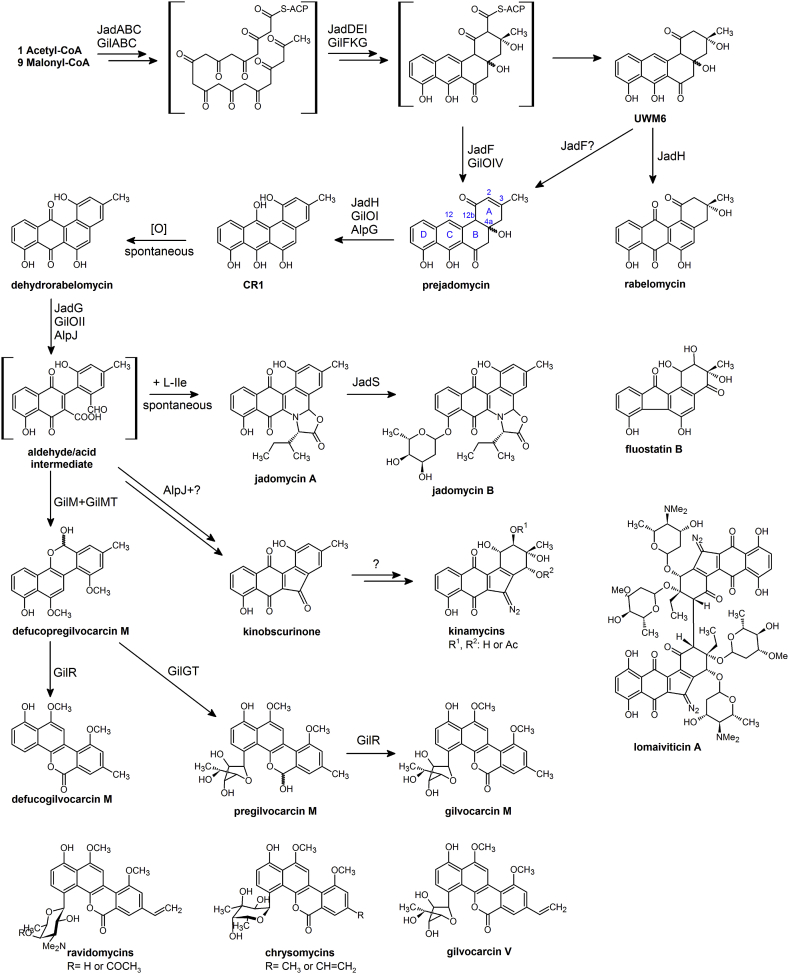
Angucyclines represent one of the largest families of aromatic polyketides, which show various bioactivities such as antibacterial, antitumor, antiviral and enzyme inhibitory activities [1]. Generally, the angucyclines have a benz[a]anthracene skeleton, which is biosynthesized by type II polyketide synthases (PKS). Briefly, a linear polyketide chain is assembled by Claisen condensation of short chain acyl-CoA (usually acetyl-CoA or its derivative malonyl-CoA), which is catalyzed by the minimal PKS, consisting of a heterodimer of ketosynthase α and β subunits and an acyl carrier protein (ACP) [[1], [2], [3]]. The nascent polyketide chain is then modified by accessory enzymes, including a C-9 ketoreductase, a steroidogenic acute regulatory protein related lipid-transfer (START) domain cyclase and a ferredoxin-like cyclase, to generate the key angucycline product UWM6 (see Fig. 1) [4,5]. Subsequent modification steps catalyzed by tailoring enzymes afford the hundreds of structurally diverse angucyclines that have been identified [1,6]. However, some aromatic polyketides without the typical benz[a]anthracene skeleton, for instance jadomycins [5,7,8], gilvocarcins [9,10], kinamycins [[11], [12], [13]], lomaiviticins [14,15] and fluostatins [16,17], have been proved to be biosynthesized via oxidative B-ring opening and rearrangement of intermediates with typical benz[a]anthracene skeleton instead (Fig. 1). Therefore, these natural products are defined as atypical angucyclines.
Fig. 1.
The biosynthetic pathways of atypical angucyclines.
Jadomycins, possessing a unique pentacyclic benz[b]oxazolophenanthridine ring system (Fig. 1), is synthesized by Streptomyces venezuelae ISP5230 (ATCC10712) under stress conditions [18,19]. The biosynthetic mechanism of the strange ring system remained obscure for years and was referred as a “black box” [20]. In 2005, Yang, Rohr and co-workers proved that UWM6 and prejadomycin (also known as 2,3-dehydro-UWM6) were biosynthetic intermediates of jadomycin, which harbored a typical benz[a]anthracene skeleton [5]. In vivo bioconversion experiments revealed that three oxygenases, including two FAD-dependent monooxygenases JadF and JadH, and a unique ring opening oxygenase JadG, could convert UWM6 and prejadomycin to jadomycin A, suggesting the involvement of these oxygenases in the formation the atypical skeleton of jadomycin [5].
Gilvocarcins are produced by Streptomyces griseoflavus Gö3592 and other Streptomycetes, which have a benzo[d]naphtha[1,2-b]pyran-6-one skeleton (Fig. 1) [10,21]. The gilvocarcin biosynthetic gene cluster was cloned in 2003 [21] and the pathway was elucidated clearly by Rohr and co-workers [10,20,[22], [23], [24], [25]]. Homologs of the three oxygenases in jadomycin biosynthetic cluster (JadF, JadH and JadG) were also found in the gilvocarcin biosynthetic cluster, GilOIV, GilOI, and GilOII, respectively [10,20,21]. Furthermore, Rohr and co-workers proved that the inactivation of gilOI could be complemented by jadH, as well as gilOIV which could be replaced by jadF when gilvocarcin cluster was expressed heterologously in S. lividans TK24 [20]. These results demonstrated that to a large extent the formation of the jadomycin and gilvocarcin skeletons might share a similar pathway.
Another atypical angucycline kinamycins were isolated from Streptomyces murayamaensis in early 1970s [26,27], and their correct chemical structures were determined with a 5-diazobenzo[b]fluorene skeleton by Gould et al in 1994 [28]. Several biosynthetic intermediates or related derivatives were identified via bioconversion experiments, such as dehydrorabelomycin, kinobscurinone and prekinamycin [[29], [30], [31]]. The first identified kinamycin biosynthetic cluster in S. murayamaensis was proved to be partial by heterologous expression in S. lividans [32]. After that two new kinamycin clusters were found in Streptomyces ambofaciens and Streptomyces galtieri Sgt26 (the alp clusters) [[33], [34], [35], [36]], and the latter was proved as the completed kinamycin clusters via heterologous expression in Streptomyces albus J1074 [36]. Moreover, lomaiviticins were discovered in Micromonospora lomaivitiensis, surprisingly formed by a dimeric kinamycin-type skeleton [37]. Then lomaiviticins were reisolated in Salinispora pacifica strain DPJ-0019 [38] and the biosynthetic clusters were identified in several Salinispora strains as time went by [14,39,40].
Fluostatins were isolated in many actinomycetes strains [[41], [42], [43], [44], [45], [46]] and their biosynthetic clusters were identified [16,17,46] afterwards. Although the feeding experiment using 13C labelled acetate indicated that the benzo[a]fluorene skeleton of fluostatins was biosynthesized from a precursor with typical angucycline skeleton [17], the detailed reactions and enzymes involved were still not known.
These atypical angucyclines have different chemical skeletons; however, they are proved to share the same early biosynthetic pathway with the typical angucyclines, for example, landomycin, oviedomycin and gaudimycin [[47], [48], [49], [50], [51], [52]]. After decades of research, we can now depict the integrated picture of the biosynthetic pathways of these atypical angucyclines. Both typical and atypical angucyclines could be formed though similar early biosynthetic processes and shared two common intermediates, UWM6 and prejadomycin. The branch point of two types of angucyclines was prejadomycin, which was converted by different reactions catalyzed by a subfamily of FAD-dependent monooxygenases [48]. Dehydrorabelomycin was a common biosynthetic intermediate of atypical angucyclines, and it was the branch point mentioned appearing in the biosynthetic pathways of atypical ones. A unique oxygenase family, the AlpJ-family ring opening oxygenases, catalyzed the oxidative C—C bond cleavage reaction, followed by various rearrangement reactions and generated products with multifarious skeleton [7,9,12,13,53].
2. The functional diversities of FAD-dependent monooxygenases between atypical and typical angucyclines biosynthesis
FAD-dependent monooxygenases are widely presented in almost all biosynthetic gene clusters of aromatic polyketides, and the number of these enzymes and their sequence similarities have been reported closely related to the chemical structure of the final products [54]. All known biosynthetic gene clusters of angucyclines have at least two FAD-dependent monooxygenases, namely the M series and E series monooxygenases [54]. The M series FAD-dependent monooxygenases were proposed to catalyze the 2,3-dehydration reaction [25]. In the study about the enzymatic total synthesis of defucogilvocarcin M (a model compound with gilvocarcin-tyep skeleton), Rohr and co-workers tried various combinations of enzymes involved in the biosynthesis of angucyclines. They reported that combination of seven enzymes (ketosynthase α and β subunits GilA, GilB, ACP RavC, ketoreductase GilF, malonyl-CoA:ACP transacylase GilP, cyclases JadD and RavG) could convert acetyl-CoA and malonyl-CoA to rabelomycin and UWM6, while these enzymes incorporated with JadF could generate prejadomycin. Furthermore, prejadomycin could be converted to defucogilvocarcin M by combination of six post-PKS tailoring enzymes (including GilOI, GilOII, JadF, methyltransferases GilM and GilMT [23], oxidoreductase GilR [22] and necessary co-factors), but UWM6 could not be converted. Thus the authors concluded that UWM6 was a shunt product and JadF catalyzed the 2,3-dehydration of an ACP-tethered UWM6-like intermediate [25]. On the other hand, the E series monooxygenases catalyzed the C12-oxygenation reaction of prejadomycin, but the detailed reactions were different for atypical angucyclines (jadomycin) and typical angucycline (gaudimycin, urdamycin and landomycin) [8,48].
2.1. JadH was a bifunctional hydroxylase/dehydrase
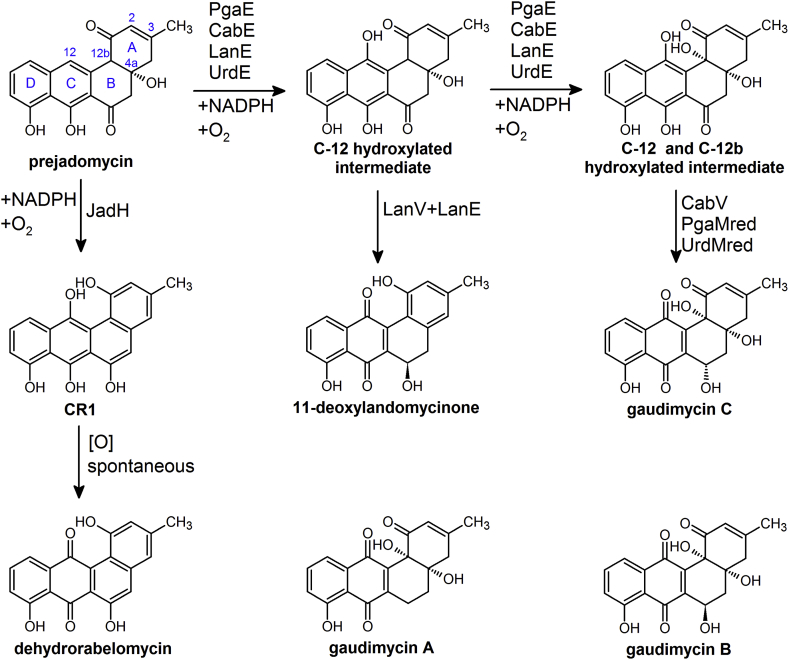
In vitro enzymatic assays revealed that JadH catalyzed the C-12 hydroxylation and 4a,12b-dehydration of prejadomycin and generated CR1 (Fig. 2), which could be converted to dehydrorabelomycin in the air spontaneously [8]. This result indicated that CR1 was the real product of JadH-catalyzed reaction, and JadH was a bifunctional hydroxylase/dehydrase.
Fig. 2.
Reactions catalyzed by FAD-dependent monooxygenases and SDR family reductases in vitro.
Since dehydrorabelomycin was the common biosynthetic intermediate for atypical angucyclines (see below), and the homologs of JadH were conserved in their gene clusters, it was proposed that the catalytic activities of these JadH homologs were conserved in the biosynthesis of atypical angucyclines as well. In fact, CR1 and dehydrorabelomycin were observed products for FlsO2, a homolog of JadH in fluostatin biosynthetic cluster identified in Micromonospora rosaria SCSIO N160 [16].
2.2. PgaE catalyzed two hydroxylation reactions at C-12 and C-12b
In 2007, Metsä-Ketelä and co-workers reported the heterologous expression of two cryptic angucycline biosynthetic genes (designated as pga and cab clusters), which produced typical angucyclines gaudimycin A and B (see Fig. 2), respectively [54]. In the following years, the comprehensive studies about PgaE revealed that JadH and PgaE catalyzed different reactions of prejadomycin, although PgaE was considered as a homolog of JadH in the pga cluster [48,51,52].
In vitro enzymatic assays revealed that PgaE catalyzed two consecutive hydroxylation reactions of prejadomycin [52]. Together with CabV, a short-chain alcohol dehydrogenase/reductase (SDR) catalyzing the C-6 reduction of the product, PgaE could convert prejadomycin to gaudimycin C (Fig. 2). Parallel kinetic quantitation of prejadomycin and NADPH revealed that 2 moles of NADPH were consumed when 1 mole of prejadomycin was converted, indicating two separated monooxygenation reactions [52]. The authors only observed the intermediate generated by the first C-12 hydroxylation under controlled NADPH concentration, since the second C-12b hydroxylation reaction was inhibited by prejadomycin. However, the C-12 hydroxylated intermediate was too labile to be isolated [52].
2.3. The content-dependent regulation on the catalytic activities of FAD-dependent monooxygenases controlled the formation of various angucyclines
A comparative study about several FAD-dependent monooxygenases from typical angucycline biosynthetic clusters, such as UrdE (urdamycin), LanE (landomycin), PgaE and CabE (gaudimycins), together with JadH from the atypical angucycline jadomycin cluster, suggested interesting content-dependent regulation on the catalytic activities of these enzymes [48]. Enzymatic assays in vitro revealed CabE, UrdE and LanE also catalyzed two hydroxylation reactions of prejadomycin at C-12 and C-12b, turning out converting prejadomycin to gaudimycin C together with SDR family reductases (CabV or UrdMred, the reductase domain of UrdM) [48]. However, LanE and LanV (SDR family reductase in the landomycin cluster) converted prejadomycin to 11-deoxylandomycinone (Fig. 2), but not gaudimycin C [48]. As 11-deoxylandomycinone did not have a C-12b hydroxyl group, it was produced from the C-12 hydroxylated intermediate. Detailed kinetic analysis revealed that the formation of 11-deoxylandomycinone was in parallel with the conversion of prejadomycin, which demonstrated that the substrate of LanV was the C-12 hydroxylated intermediate, not the C-12 and C-12b double hydroxylated intermediate (Fig. 2) [48].
Given that different combinations of hydroxylases (PgaE, CabE, UrdE or LanE) and SDR family reductases (CabV, UrdMred or LanV) were tested for the conversion of prejadomycin, Metsä-Ketelä and co-workers made a conclusion that the products were largely determined by the SDR family reductases [48]. When CabV or UrdMred was used, prejadomycin would be transformed to gaudimycin C, while 11-deoxylandomycinone was produced when LanV participated. The authors suggested that these results could be explained by the relatively high substrate selectivity of LanV towards the C-12 hydroxylated intermediate, and presented an interesting example of biosynthetic context-dependent regulation as an essential cause of the structural diversities of the final products [48]. Notably, the authors also represented the potential of catalyzing the C-12b hydroxylation for JadH. When prejadomycin was utilized as substrate, JadH catalyzed the C-12 hydroxylation and 4a, 12b dehydration reactions with or without SDR family reductase. Nonetheless, JadH could convert the C-12 hydroxylated intermediate to gaudimycin C in the presence of either CabV, UrdMred or LanV [48]. These results indicated that JadH retained the catalytic activity of C-12b hydroxylation of the presumed ancestral enzymes [48].
3. The functional diversities of the ring opening oxygenases and the structural diversities of the atypical angucyclines
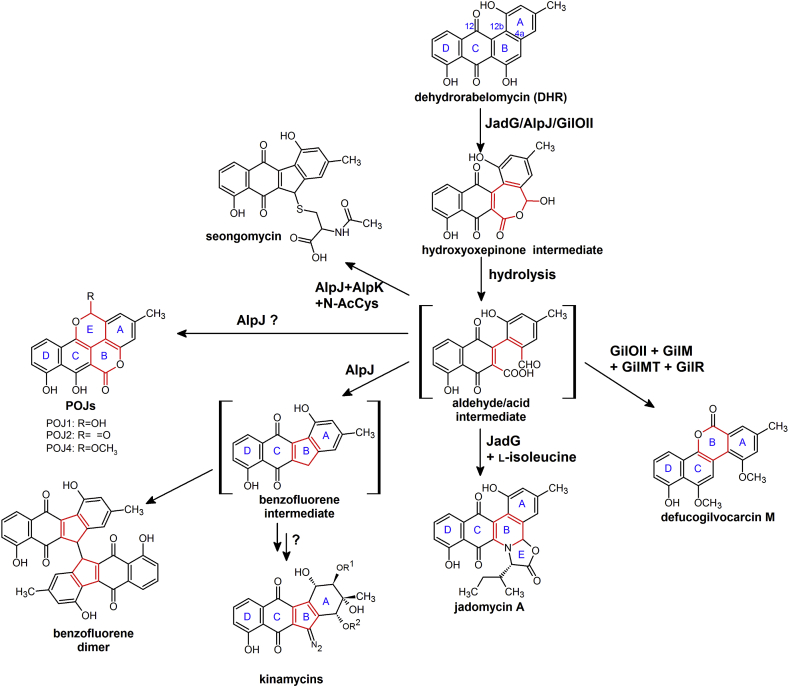
It is known that dehydrorabelomycin is determined as a common intermediate of atypical angucyclines [1,8,17,55]. A small family of oxygenases, ring-opening oxygenases, catalyze the oxidative B-ring opening of dehydrorabelomycin and the following rearrangement reactions [7,9,12,13], and other tailoring enzymes in subsequent steps contribute to the final structure of derivative products [9,23,35,36]. Sequence comparison and database searches demonstrate that these ring-opening oxygenases are found only in biosynthetic clusters of atypical angucyclines, strongly suggesting they played critical roles in the ring opening of atypical angucyclines [7,53]. JadG (jadomycin), GilOII (gilvocarcin), AlpJ (kinamycin) and other homologs are representative members of these particular ring-opening oxygenases family [53].
3.1. Ring-opening oxygenases catalyzed the oxidative C—C bond cleavage and rearrangement reactions of dehydrorabelomycin
For gilvocarcin biosynthesis, GilOII plays a pivotal role in the formation of gilvocarcin-type skeleton. In 2011, Rohr and co-workers reported that GilOII, GilM, GilMT and GilR could convert dehydrorabelomycin to defucogilvocarcin M, a model compound containing the same benzo[d]naphtha[1,2-b]pyran-6-one skeleton as other gilvocarcin derivatives (Fig. 3) [25]. Among the four enzymes, GilM and GilMT were proved to be O-methyltransferases, and GilR was identified as an FAD-dependent oxidoreductase which was responsible for oxidizing the hemiacetal of pregilvocarcins to the lactone of gilvocarcins [22,23]. A Baeyer-Villiger-type hydroxyoxepinone intermediate was isolated from the reaction mixture of GilOII and Fre (Escherichia coli flavin reductase [56,57]) using dehydrorabelomycin as substrate in the presence of the cofactors FAD and NADPH (Fig. 3). This hydroxyoxepinone intermediate could be converted to defucogilvocarcin M by GilOII, GilM, GilMT, GilR and Fre (with the supply of FAD and NADPH) [9]. However, the end product of the GilOII catalyzed reaction was not isolated.
Fig. 3.
Reactions catalyzed by ring opening oxygenases in vitro.
As for jadomycin biosynthesis, inactivation of JadG prevented the biosynthesis of jadomycin B and A, and then three compounds with an intact B-ring were accumulated, including dehydrorabelomycin and prejadomycin [58]. In vitro enzymatic analysis revealed that JadG catalyzed the ring opening and rearrangement of dehydrorabelomycin to form jadomycin A when l-isoleucine was supplied (Fig. 3) [7]. FMNH2 or FADH2 was required as cofactors, and it could be provided by either a cluster-situated flavin reductase JadY or the E. coli Fre. JadY was characterized as an NAD(P)H-dependent FMN/FAD reductase, with FMN as the preferred substrate [7]. Tan, Yang and co-workers reported that the transcription of jadY was tightly controlled by a cluster-situated regulator JadR*, to ensure a timely supply of cofactors and avoid unnecessary consumption of NAD(P)H [59].
For kinamycin biosynthesis, inactivation of AlpJ, the ring-opening oxygenase in the kinamycin cluster of S. ambofaciens, caused accumulation of dehydrorabelomycin, while the complement mutant (ΔΔalpJ::alpJ) produced a series of new compounds, POJ1, POJ2 and POJ4, all possessing a 7,8-dihydroxy-3-methyl-naphtho[2,3-c]chromen-6-one structure and regarded as rearrangement products of the B-ring opened aldehyde/acid intermediates (Fig. 3) [12]. Analysis in vitro presented that AlpJ (with FADH2 supplied by E. coli Fre) was qualified to convert dehydrorabelomycin to two B-ring contraction products: benzofluorene intermediate and its dimer (Fig. 3). Hence, these results highlighted that AlpJ could catalyze the oxidative ring opening and contraction of dehydrorabelomycin to generate kinamycin-type products [12].
Sequence comparison of the benzo[b]fluorene-producing gene clusters displayed a conserved pair of oxygenases (AlpK-AlpJ homologs), suggesting that AlpK was functionally associated with AlpJ. Inactivation of AlpK caused accumulation of PK1, a trimer of benzofluorene [12]. Heterologous expression in Streptomyces lividans TK24 and bioconversion using dehydrorabelomycin as substrate revealed that PK1 was produced by AlpJ solely, while co-expression of AlpJ and AlpK could convert dehydrorabelomycin to seongomycin (a shunt product of kinamycin biosynthesis) in vivo. While in vitro, researchers discovered that AlpK could replace Fre and offer cofactors for AlpJ. When N-acetylcysteine was added, AlpJ and AlpK turned dehydrorabelomycin to seongomycin in vitro. These results indicated that AlpK catalyzed the reaction following the ring-opening and rearrangement reactions in kinamycin biosynthesis [12]. Similar results were reported by Balskus and co-workers for AlpJ and KinO1 (the homolog of AlpK in S. murayamaensis) later [13]. They also found the formation of stealthin C via an intramolecular S—N-type Smiles rearrangement reaction for AlpJ, KinO1/Fre and l-cysteine with dehydrorabelomycin as substrate [13]. Stealthin C has an aminobenzo[b]fluorene skeleton, and has been proposed as a biosynthetic intermediate by Gould et al. [60]. However, recent study about the biosynthesis of fosfazinomycin and kinamycin revealed that the N—N bonds in the two natural products were formed via an acetylhydrazine biosynthetic synthon, which was carried on glutamic acid and would be transferred to a kinobscurinone-type substrate [61]. These results precluded the possibility of stealthin C as a biosynthetic intermediate of kinamycin.
3.2. The structure and evolution of ring-opening oxygenases
The crystal structure of AlpJ was determined by Liu, Yang and co-workers in 2016 [53]. AlpJ and other ring opening oxygenases showed weak sequence similarities with the well-known anthrone oxygenases, such as TcmH, ElmH and ActVA-Orf6, whereas they were doubled in length [[62], [63], [64]]. Both static light scattering and size-exclusion chromatography experiments indicated that AlpJ was a monomer in solution. The overall structure of AlpJ monomer resembled the dimeric structure of ferredoxin-like proteins, containing two similar domains. Structure comparison revealed that the monomeric AlpJ was a structural equivalent of the homodimer of ActVA-Orf6 [65]. Both the N- and C-terminal domains contained putative substrate binding pockets, which might form reasonable interactions with the substrate dehydrorabelomycin as revealed by molecular docking [53]. Mutations in the N-terminal domain (N60A and W64A), the C-terminal domain (W181A) and the domain interface (H50A and Y178A) significantly reduced the catalytic activity of AlpJ. These results made clear that the putative active sites in both the N- and C-terminal domain were vital for the catalytic activity of AlpJ, and they might function in a cooperative manner [53].
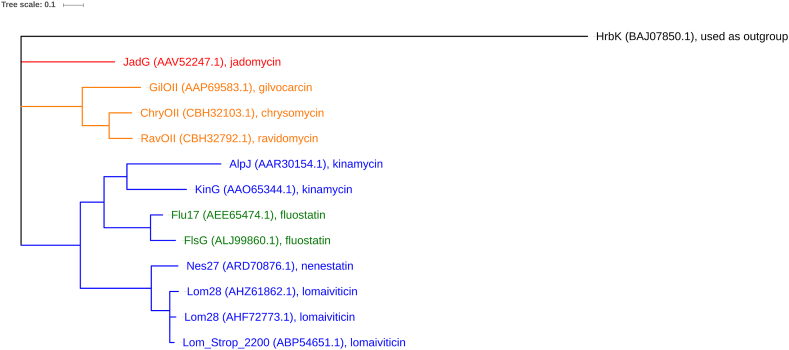
The homologs of ring opening oxygenases (200–250 residues) were found mainly in the biosynthetic gene clusters of atypical angucyclines, including AlpJ and KinG (kinamycin) [32,34,36], Lom28 and its homologs (lomaiviticin) [14,15,39], Nes27 (nenestatin) [66], Flu17, FlsG, and Fluo6 (fluostatin) [16,17,46], GilOII (gilvocarcin) [21], ChryOII (chrysomycin), RavOII (ravidomycin) [67], and JadG (jadomycin) [68], etc. The only exception so far was HrbK in the gene cluster of the typical angucycline hatomarubigin, whose catalytic function was still unknown [69]. Phylogenetic analysis certified these ring opening oxygenases formed a unique protein family, which involved three subclades (see Fig. 4) [53]. Each subclade of ring opening oxygenases is correlated with a family of atypical angucyclines sharing the same chemical skeleton, for instance the subclade of JadG for jadomycin skeleton, the subclade of GilOII, ChryOII and RavOII for gilvocarcin-type skeleton, and a large subclade including ring opening oxygenases from kinamycin, lomaiviticin, nenestatin and fluostatin biosynthetic gene clusters. It was notable that the ring opening oxygenases from the fluostatin clusters were in the same subclade of those from the kinamycin and lomaiviticin clusters, although the skeletons of their final products were different [53]. This interesting discovery indicates that the fluostatin-type skeleton is possibly generated via the rearrangement of a kinamycin-type structure [16,70].
Fig. 4.
Phylogenetic analysis of ring opening oxygenases involved in known atypical angucycline biosynthetic gene clusters. The tree was generated by MEGA7 using the Le_Gascuel_2008 method and the maximum likelihood algorithm. Bar, 1.0 substitutions per amino acid position. Red, product with jadomycin-type skeleton; orange, products with gilvocarcin-type skeleton; blue, products with kinamycin-type skeleton; green, products with fluostatin-type skeleton. HrbK doesn't catalyze the ring opening reaction as indicated by the chemical structure of hatomarubigins, and shows weak sequence similarities with the ring opening oxygenases, thus it is used as an outgroup. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4. Conclusion
It is widely acknowledged that aromatic polyketides are usually biosynthesized by type II PKS, and that tailoring enzymes contribute to the formation of their final structures [6]. Case studies about the biosynthesis of angucyclines have revealed that functional diverse oxygenases are responsible for the chemical diversity of these natural products.
Although PgaE, CabE, UrdE, LanE and JadH, FlsO2 belonged to the same subfamily of FAD-dependent monooxygenase, they catalyzed different reactions of prejadomycin [5,8,16,48,52]. Among them JadH and FlsO2 catalyzed the C-12 hydroxylation and 4a,12b-dehydration reactions of prejadomycin and produced dehydrorabelomycin, namely the common intermediate of atypical angucycline biosynthesis [8,16]. On the contrary, PgaE and its homologs catalyzing the C-12 and C-12b hydroxylation reactions of prejadomycin, lead to the formation of several typical angucyclines with C-12b hydroxyl group [48,52]. Similar diversification appears to have occurred for the ring opening oxygenases. Although these oxygenases, GilOII, JadG and AlpJ, for instance, belonged to this unique oxygenase family, they catalyzed three types of reaction of dehydrorabelomycin and formed final products with the corresponding structures [7,9,12]. These results suggested that the functional differentiation of the oxygenases were one of the considerable sources of the structure diversities of natural products.
The comparative studies about the FAD-dependent monooxygenases also revealed the biosynthetic context-dependent regulation on their catalytic activities [48]. Although PgaE, CabE, UrdE and LanE, could catalyze the consecutive C-12 and C-12b hydroxylation reactions, the end products were controlled by the following SDR family reductases such as CabV and LanV. Due to the different substrate affinities of the reductases towards various biosynthetic intermediates, the second C-12b hydroxylation might happen or not happen at all, which caused the formation of gaudimycin and landomycin-like products, respectively [48]. Thus, the substrate affinities of the tailoring enzymes contributed to the structure diversities of the natural products as well.
Taken together, the post-PKS tailoring enzymes, especially the FAD-dependent monooxygenase family and the ring opening oxygenase family, offered interesting examples for the study about the enzyme evolution and functional differentiation, which also contributed to the discovery of natural products with novel chemical skeletons.
Acknowledgment
This work was supported by National Natural Science Foundation of China (Grants: 31670800, 31470176, and 31130001) and Ministry of Science and Technology of China (Grants: 2014CB910400).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
References
- 1.Kharel M.K., Pahari P., Shepherd M.D., Tibrewal N., Nybo S.E., Shaaban K.A. Angucyclines: biosynthesis, mode-of-action, new natural products, and synthesis. Nat Prod Rep. 2012;29:264–325. doi: 10.1039/c1np00068c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Das A., Khosla C. Biosynthesis of aromatic polyketides in bacteria. Acc Chem Res. 2009;42:631–639. doi: 10.1021/ar8002249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhan J. Biosynthesis of bacterial aromatic polyketides. Curr Top Med Chem. 2009;9:1598–1610. doi: 10.2174/156802609789941906. [DOI] [PubMed] [Google Scholar]
- 4.Kulowski K., Wendt-Pienkowski E., Han L., Yang K., Vining L.C., Hutchinson C.R. Functional characterization of the jadI gene as a cyclase forming angucyclinones. J Am Chem Soc. 1999;121:1786–1794. [Google Scholar]
- 5.Chen Y., Wang C., Greenwell L., Rix U., Hoffmeister D., Vining L.C. Functional analyses of oxygenases in jadomycin biosynthesis and identification of JadH as a bifunctional oxygenase/dehydrase. J Biol Chem. 2005;280:22508–22514. doi: 10.1074/jbc.M414229200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olano C., Méndez C., Salas J.A. Post-PKS tailoring steps in natural product-producing actinomycetes from the perspective of combinatorial biosynthesis. Nat Prod Rep. 2010;27:571–616. doi: 10.1039/b911956f. [DOI] [PubMed] [Google Scholar]
- 7.Fan K., Pan G., Peng X., Zheng J., Gao W., Wang J. Identification of JadG as the B ring opening oxygenase catalyzing the oxidative C-C bond cleavage reaction in jadomycin biosynthesis. Chem Biol. 2012;19:1381–1390. doi: 10.1016/j.chembiol.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 8.Chen Y., Fan K., He Y., Xu X., Peng Y., Yu T. Characterization of JadH as an FAD- and NAD(P)H-dependent bifunctional hydroxylase/dehydrase in jadomycin biosynthesis. ChemBioChem. 2010;11:1055–1060. doi: 10.1002/cbic.201000178. [DOI] [PubMed] [Google Scholar]
- 9.Tibrewal N., Pahari P., Wang G., Kharel M.K., Morris C., Downey T. Baeyer-Villiger C-C bond cleavage reaction in gilvocarcin and jadomycin biosynthesis. J Am Chem Soc. 2012;134:18181–18184. doi: 10.1021/ja3081154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu T., Fischer C., Beninga C., Rohr J. Oxidative rearrangement processes in the biosynthesis of gilvocarcin V. J Am Chem Soc. 2004;126:12262–12263. doi: 10.1021/ja0467521. [DOI] [PubMed] [Google Scholar]
- 11.Seaton P.J., Gould S.J. New products related to kinamycin from Streptomyces murayamaensis. II. Structures of pre-kinamycin, keto-anhydrokinamycin, and kinamycins E and F. J Antibiot. 1989;42:189–197. doi: 10.7164/antibiotics.42.189. [DOI] [PubMed] [Google Scholar]
- 12.Wang B., Ren J., Li L., Guo F., Pan G., Ai G. Kinamycin biosynthesis employs a conserved pair of oxidases for B-ring contraction. Chem Commun. 2015;51:8845–8848. doi: 10.1039/c5cc01986a. [DOI] [PubMed] [Google Scholar]
- 13.Wang P., Hong G.J., Wilson M.R., Balskus E.P. Production of stealthin C involves an S-N-type Smiles rearrangement. J Am Chem Soc. 2017;139:2864–2867. doi: 10.1021/jacs.6b10586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janso J.E., Haltli B.A., Eustáquio A.S., Kulowski K., Waldman A.J., Zha L. Discovery of the lomaiviticin biosynthetic gene cluster in Salinispora pacifica. Tetrahedron. 2014;70:4156–4164. doi: 10.1016/j.tet.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woo C.M., Gholap S.L., Herzon S.B. Insights into lomaiviticin biosynthesis. Isolation and structure elucidation of (-)-homoseongomycin. J Nat Prod. 2013;76:1238–1241. doi: 10.1021/np400355h. [DOI] [PubMed] [Google Scholar]
- 16.Yang C., Huang C., Zhang W., Zhu Y., Zhang C. Heterologous expression of fluostatin gene cluster leads to a bioactive heterodimer. Org Lett. 2015;17:5324–5327. doi: 10.1021/acs.orglett.5b02683. [DOI] [PubMed] [Google Scholar]
- 17.Feng Z., Kim J.H., Brady S.F. Fluostatins produced by the heterologous expression of a TAR reassembled environmental DNA derived type II PKS gene cluster. J Am Chem Soc. 2010;132 doi: 10.1021/ja104550p. 11902–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doull J.L., Singh A.K., Hoare M., Ayer S.W. Conditions for the production of jadomycin B by Streptomyces venezuelae ISP5230: effects of heat shock, ethanol treatment and phage infection. J Ind Microbiol. 1994;13:120–125. doi: 10.1007/BF01584109. [DOI] [PubMed] [Google Scholar]
- 19.Doull J.L., Ayer S.W., Singh A.K., Thibault P. Production of a novel polyketide antibiotic, jadomycin B, by Streptomyces venezuelae following heat shock. J Antibiot. 1993;46:869–871. doi: 10.7164/antibiotics.46.869. [DOI] [PubMed] [Google Scholar]
- 20.Kharel M.K., Zhu L., Liu T., Rohr J. Multi-oxygenase complexes of the gilvocarcin and jadomycin biosynthesis. J Am Chem Soc. 2007;129:3780–3781. doi: 10.1021/ja0680515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fischer C., Lipata F., Rohr J. The complete gene cluster of the antitumor agent gilvocarcin V and its implication for the biosynthesis of the gilvocarcins. J Am Chem Soc. 2003;125:7818–7819. doi: 10.1021/ja034781q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kharel M.K., Pahari P., Lian H., Rohr J. GilR, an unusual lactone-forming enzyme involved in gilvocarcin biosynthesis. ChemBioChem. 2009;10:1305–1308. doi: 10.1002/cbic.200900130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tibrewal N., Downey T.E., Van Lanen S.G., Ul Sharif E., O'Doherty G.A., Rohr J. Roles of the synergistic reductive O-methyltransferase GilM and of O-methyltransferase GilMT in the gilvocarcin biosynthetic pathway. J Am Chem Soc. 2012;134:12402–12405. doi: 10.1021/ja305113d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu T., Kharel M.K., Zhu L., Bright S.A., Mattingly C., Adams V.R. Inactivation of the ketoreductase gilU gene of the gilvocarcin biosynthetic gene cluster yields new analogues with partly improved biological activity. ChemBioChem. 2009;10:278–286. doi: 10.1002/cbic.200800348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pahari P., Kharel M.K., Shepherd M.D., van Lanen S.G., Rohr J. Enzymatic total synthesis of defucogilvocarcin M and its implications for gilvocarcin biosynthesis. Angew Chem Int Ed Engl. 2012;51:1216–1220. doi: 10.1002/anie.201105882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Itŏ S., Matsuya T., Omura S., Otani M., Nakagawa A. A new antibiotic, kinamycin. J Antibiot. 1970;23:315–317. doi: 10.7164/antibiotics.23.315. [DOI] [PubMed] [Google Scholar]
- 27.Hata T., Omura S., Iwai Y., Nakagawa A., Otani M. A new antibiotic, kinamycin: fermentation, isolation, purification and properties. J Antibiot. 1971;24:353–359. doi: 10.7164/antibiotics.24.353. [DOI] [PubMed] [Google Scholar]
- 28.Gould S.J., Tamayo N., Melville C.R., Cone M.C. Revised structures for the kinamycin antibiotics: 5-diazobenzo[b]fluorenes rather than benzo[b]carbazole cyanamides. J Am Chem Soc. 1994;116:2207–2208. [Google Scholar]
- 29.Cone M.C., Seaton P.J., Halley K.A., Gould S.J. New products related to kinamycin from Streptomyces murayamaensis. I. Taxonomy, production, isolation and biological properties. J Antibiot. 1989;42:179–188. doi: 10.7164/antibiotics.42.179. [DOI] [PubMed] [Google Scholar]
- 30.Gould S.J., Melville C.R.C. Kinamycin biosytheesis. Synthesis, detection, and incorporation of kinobscurinone, a benzo[b]fluorenone. Bioorg Med Chem Lett. 1995;5:51–54. [Google Scholar]
- 31.Gould S.J. Biosynthesis of the kinamycins. Chem Rev. 1997;97:2499–2510. doi: 10.1021/cr9600215. [DOI] [PubMed] [Google Scholar]
- 32.Gould S.J., Hong S.-T., Carney J.R. Cloning and heterologous expression of gene from the kinamycin biosynthesis pathway of Strptomyces murayamaensis. J Antibiot. 1998;51:50–57. doi: 10.7164/antibiotics.51.50. [DOI] [PubMed] [Google Scholar]
- 33.Bunet R., Mendes M.V., Rouhier N., Pang X., Hotel L., Leblond P. Regulation of the synthesis of the angucyclinone antibiotic alpomycin in Streptomyces ambofaciens by the autoregulator receptor AlpZ and its specific ligand. J Bacteriol. 2008;190:3293–3305. doi: 10.1128/JB.01989-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pang X., Aigle B., Girardet J., Mangenot S., Pernodet J., Decaris B. Functional angucycline-like antibiotic gene cluster in the terminal inverted repeats of the Streptomyces ambofaciens linear chromosome. Antimicrob Agents Chemother. 2004;48:575–588. doi: 10.1128/AAC.48.2.575-588.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang B., Guo F., Ren J., Ai G., Aigle B., Fan K. Identification of Alp1U and Lom6 as epoxy hydrolases and implications for kinamycin and lomaiviticin biosynthesis. Nat Commun. 2015;6:7674. doi: 10.1038/ncomms8674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu X., Liu D., Xu M., Tao M., Bai L., Deng Z. Reconstitution of kinamycin biosynthesis within the heterologous host Streptomyces albus J1074. J Nat Prod. 2018;81:72–77. doi: 10.1021/acs.jnatprod.7b00652. [DOI] [PubMed] [Google Scholar]
- 37.He H., Ding W.D., Bernan V.S., Richardson A.D., Ireland C.M., Greenstein M. Lomaiviticins A and B, potent antitumor antibiotics from Micromonospora lomaivitiensis. J Am Chem Soc. 2001;123:5362–5363. doi: 10.1021/ja010129o. [DOI] [PubMed] [Google Scholar]
- 38.Woo C.M., Beizer N.E., Janso J.E., Herzon S.B. Isolation of lomaiviticins C-E, transformation of lomaiviticin C to lomaiviticin A, complete structure elucidation of lomaiviticin A, and structure-activity analyses. J Am Chem Soc. 2012;134:15285–15288. doi: 10.1021/ja3074984. [DOI] [PubMed] [Google Scholar]
- 39.Kersten R.D., Lane A.L., Nett M., Richter T.K.S., Duggan B.M., Dorrestein P.C. Bioactivity-guided genome mining reveals the lomaiviticin biosynthetic gene cluster in Salinispora tropica. ChemBioChem. 2013;14:955–962. doi: 10.1002/cbic.201300147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crüsemann M., O'Neill E.C., Larson C.B., Melnik A.V., Floros D.J., da Silva R.R. Prioritizing natural product diversity in a collection of 146 bacterial strains based on growth and extraction protocols. J Nat Prod. 2017;80:588–597. doi: 10.1021/acs.jnatprod.6b00722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akiyama T., Harada S., Kojima F., Takahashi Y., Imada C., Okami Y. Fluostatins A and B, new inhibitors of dipeptidyl peptidase III, produced by Streptomyces sp. TA-3391. I. Taxonomy of producing strain, production, isolation, physico-chemical properties and biological properties. J Antibiot. 1998;51:553–559. doi: 10.7164/antibiotics.51.553. [DOI] [PubMed] [Google Scholar]
- 42.Akiyama T., Nakamura K.T., Takahashi Y., Naganawa H., Muraoka Y., Aoyagi T. Fluostatins A and B, new inhibitors of dipeptidyl peptidase III, produced by Streptomyces sp. TA-3391. II. Structure determination. J Antibiot. 1998;51:586–588. doi: 10.7164/antibiotics.51.586. [DOI] [PubMed] [Google Scholar]
- 43.Schneider K., Nicholson G., Ströbele M., Baur S., Niehaus J., Fiedler H. The structures of fluostatins C, D and E, novel members of the fluostatin family. J Antibiot. 2006;59:105–109. doi: 10.1038/ja.2006.15. [DOI] [PubMed] [Google Scholar]
- 44.Baur S., Niehaus J., Karagouni A.D., Katsifas E.A., Chalkou K., Meintanis C. Fluostatins C-E, novel members of the fluostatin family produced by Streptomyces strain Acta 1383. J Antibiot. 2006;59:293–297. doi: 10.1038/ja.2006.41. [DOI] [PubMed] [Google Scholar]
- 45.Zhang W., Liu Z., Li S., Lu Y., Chen Y., Zhang H. Fluostatins I-K from the south China sea-derived Micromonospora rosaria SCSIO N160. J Nat Prod. 2012;75:1937–1943. doi: 10.1021/np300505y. [DOI] [PubMed] [Google Scholar]
- 46.Jin J., Yang X., Liu T., Xiao H., Wang G., Zhou M. Fluostatins M-Q featuring a 6-5-6-6 ring skeleton and high oxidized A-Rings from marine Streptomyces sp. PKU-MA00045. Mar Drugs. 2018;16:87. doi: 10.3390/md16030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Westrich L., Domann S., Faust B., Bedford D., Hopwood D.A., Bechthold A. Cloning and characterization of a gene cluster from Streptomyces cyanogenus S136 probably involved in landomycin biosynthesis. FEMS Microbiol Lett. 1999;170:381–387. doi: 10.1111/j.1574-6968.1999.tb13398.x. [DOI] [PubMed] [Google Scholar]
- 48.Patrikainen P., Kallio P., Fan K., Klika K.D., Shaaban K.A., Mäntsälä P. Tailoring enzymes involved in the biosynthesis of angucyclines contain latent context-dependent catalytic activities. Chem Biol. 2012;19:647–655. doi: 10.1016/j.chembiol.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lombó F., Abdelfattah M.S., Braña A.F., Salas J.A., Rohr J., Méndez C. Elucidation of oxygenation steps during oviedomycin biosynthesis and generation of derivatives with increased antitumor activity. ChemBioChem. 2009;10:296–303. doi: 10.1002/cbic.200800425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lombó F., Braña A.F., Salas J.A., Méndez C. Genetic organization of the biosynthetic gene cluster for the antitumor angucycline oviedomycin in Streptomyces antibioticus ATCC 11891. ChemBioChem. 2004;5:1181–1187. doi: 10.1002/cbic.200400073. [DOI] [PubMed] [Google Scholar]
- 51.Kallio P., Patrikainen P., Belogurov G.A., Mäntsälä P., Yang K., Niemi J. Tracing the evolution of angucyclinone monooxygenases: structural determinants for C-12b hydroxylation and substrate inhibition in PgaE. Biochemistry. 2013;52:4507–4516. doi: 10.1021/bi400381s. [DOI] [PubMed] [Google Scholar]
- 52.Kallio P., Patrikainen P., Suomela J.-P., Mäntsälä P., Metsä-Ketelä M., Niemi J. Flavoprotein hydroxylase PgaE catalyzes two consecutive oxygen-dependent tailoring reactions in angucycline biosynthesis. Biochemistry. 2011;50:5535–5543. doi: 10.1021/bi200600k. [DOI] [PubMed] [Google Scholar]
- 53.Pan G., Gao X., Fan K., Liu J., Meng B., Gao J. Structure and function of a C-C bond cleaving oxygenase in atypical angucycline biosynthesis. ACS Chem Biol. 2017;12:142–152. doi: 10.1021/acschembio.6b00621. [DOI] [PubMed] [Google Scholar]
- 54.Palmu K., Ishida K., Mäntsälä P., Hertweck C., Metsä-Ketelä M. Artificial reconstruction of two cryptic angucycline antibiotic biosynthetic pathways. ChemBioChem. 2007;8:1577–1584. doi: 10.1002/cbic.200700140. [DOI] [PubMed] [Google Scholar]
- 55.Seaton P.J., Gould S.J. Kinamycin biosynthesis. Derivation by excision of an acetate unit from a single-chain decaketide intermediate. J Am Chem Soc. 1987;109:5282–5284. [Google Scholar]
- 56.Lin S., Van Lanen S.G., Shen B. Regiospecific chlorination of (S)-beta-tyrosyl-S-Carrier protein catalyzed by SgcC3 in the biosynthesis of the enediyne antitumor antibiotic C-1027. J Am Chem Soc. 2007;129:12432–12438. doi: 10.1021/ja072311g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spyrou G., Haggård-Ljungquist E., Krook M., Jörnvall H., Nilsson E., Reichard P. Characterization of the flavin reductase gene (fre) of Escherichia coli and construction of a plasmid for overproduction of the enzyme. J Bacteriol. 1991;173:3673–3679. doi: 10.1128/jb.173.12.3673-3679.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rix U., Wang C., Chen Y., Lipata F.M., Remsing Rix L.L., Greenwell L.M. The oxidative ring cleavage in jadomycin biosynthesis: a multistep oxygenation cascade in a biosynthetic black box. Chembiochem. 2005;6:838–845. doi: 10.1002/cbic.200400395. [DOI] [PubMed] [Google Scholar]
- 59.Zhang Y., Pan G., Zou Z., Fan K., Yang K., Tan H. JadR*-mediated feed-forward regulation of cofactor supply in jadomycin biosynthesis. Mol Microbiol. 2013;90:884–897. doi: 10.1111/mmi.12406. [DOI] [PubMed] [Google Scholar]
- 60.Gould S.J., Melville C.R., Cone M.C., Chen J., Carney J.R. Kinamycin biosynthesis. synthesis, isolation, and incorporation of stealthin C, an aminobenzo[b]fluorene. J Org Chem. 1997;62:320–324. doi: 10.1021/jo961486y. [DOI] [PubMed] [Google Scholar]
- 61.Wang K.-K.A., Ng T.L., Wang P., Huang Z., Balskus E.P., van der Donk W.A. Glutamic acid is a Carrier for hydrazine during the biosyntheses of fosfazinomycin and kinamycin. Nat Commun. 2018;9:3687. doi: 10.1038/s41467-018-06083-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shen B., Hutchinson C.R. Tetracenomycin F1 monooxygenase: oxidation of a naphthacenone to a naphthacenequinone in the biosynthesis of tetracenomycin C in Streptomyces glaucescens. Biochemistry. 1993;32:6656–6663. doi: 10.1021/bi00077a019. [DOI] [PubMed] [Google Scholar]
- 63.Rafanan E.R., Le L., Zhao L., Decker H., Shen B. Cloning, sequencing, and heterologous expression of the elmGHIJ genes involved in the biosynthesis of the polyketide antibiotic elloramycin from Streptomyces olivaceus Tü2353. J Nat Prod. 2001;64:444–449. doi: 10.1021/np010007+. [DOI] [PubMed] [Google Scholar]
- 64.Kendrew S.G., Hopwood D.A., Marsh E.N. Identification of a monooxygenase from Streptomyces coelicolor A3(2) involved in biosynthesis of actinorhodin: purification and characterization of the recombinant enzyme. J Bacteriol. 1997;179:4305–4310. doi: 10.1128/jb.179.13.4305-4310.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sciara G., Kendrew S.G., Miele A.E., Marsh N.G., Federici L., Malatesta F. The structure of ActVA-Orf6, a novel type of monooxygenase involved in actinorhodin biosynthesis. EMBO J. 2003;22:205–215. doi: 10.1093/emboj/cdg031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jiang X., Zhang Q., Zhu Y., Nie F., Wu Z., Yang C. Isolation, structure elucidation and biosynthesis of benzo[b]fluorene nenestatin A from deep-sea derived Micromonospora echinospora SCSIO 04089. Tetrahedron. 2017;73:3585–3590. [Google Scholar]
- 67.Kharel M.K., Nybo S.E., Shepherd M.D., Rohr J. Cloning and characterization of the ravidomycin and chrysomycin biosynthetic gene clusters. ChemBioChem. 2010;11:523–532. doi: 10.1002/cbic.200900673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Han L., Yang K., Ramalingam E., Mosher R.H., Vining L.C. Cloning and characterization of polyketide synthase genes for jadomycin B biosynthesis in Streptomyces venezuelae ISP5230. Microbiology. 1994;140:3379–3389. doi: 10.1099/13500872-140-12-3379. [DOI] [PubMed] [Google Scholar]
- 69.Kawasaki T., Hirashima R., Maruta T., Sato H., Maeda A., Yamada Y. Cloning and characterization of a gene cluster for hatomarubigin biosynthesis in Streptomyces sp. strain 2238-SVT4. Appl Environ Microbiol. 2010;76:4201–4206. doi: 10.1128/AEM.00668-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Proteau P.J., Li Y., Chen J., Williamson R.T., Gould S.J., Laufer R.S. Isoprekinamycin is a diazobenzo[a]fluorene rather than a diazobenzo[b]fluorene. J Am Chem Soc. 2000;122:8325–8326. [Google Scholar]