Abstract
Background
Germline mutations in BAP1 have been associated with BAP1-Tumor Predisposition Syndrome (BAP1-TPDS), a predisposition to multiple tumors within a family that includes uveal melanoma (UM), cutaneous melanoma, malignant mesothelioma and renal cell carcinoma. Alternatively, somatic mutations in BAP1 in UM have been associated with high risk for metastasis. In this study, we compare the risk of metastasis in UM that carry germline versus somatic BAP1 mutations and mutation-negative tumors.
Methods
DNA extracted from 142 UM and matched blood samples was sequenced using Sanger or next generation sequencing to identify BAP1 gene mutations.
Results
Eleven of 142 UM (8%) carried germline BAP1 mutations, 43 (30%) had somatic mutations, and 88 (62%) were mutation-negative. All BAP1 mutations identified in blood samples were also present in the matched UM. There were 52 unique mutations in 54 tumors. All were pathogenic or likely pathogenic.
A comparison of tumors carrying somatic vs. germline mutations, or no mutations, showed a higher frequency of metastasis in tumors carrying somatic mutations: 74% vs. 36%, P=0.03 and 74% vs. 26% P<0.001, respectively. Tumors with a somatic mutation compared to mutation-negative had an older age of diagnosis of (61.8 vs. 52.2 years, P=0.002), and shorter time to metastasis (16 vs. 26 months, P=0.04). Kaplan-Meier analysis further showed that tumors with somatic (vs. germline) mutations demonstrated a greater metastatic risk (P=0.03). Cox multivariate analysis showed in addition to chromosome-3 monosomy and larger tumor diameter, the presence of BAP1 somatic, but not germline mutations, was significantly associated with risk of metastasis(P=0.02).
Personal or family history of BAP1-TPDS was available for 79 of the cases. All eight cases with germline mutations reported a history of BAP1-TPDS, which was significantly greater than what was observed in cases with somatic mutations (10 of 23, P=0.009) or mutation-negative cases (11 of 48, P<0.001).
Conclusions
Defining germline vs. somatic nature of BAP1 mutations in UM can inform the individual about both the risk of metastasis, and the time to metastasis, which are critically important outcomes for the individual. This information can also change the cascade screening and surveillance of family members.
Electronic supplementary material
The online version of this article (10.1186/s12885-018-5079-x) contains supplementary material, which is available to authorized users.
Keywords: uveal melanoma, ocular/eye cancer, BAP1, germline mutations, somatic mutations
Background
The BRCA1-associated protein 1 (BAP1) gene is located on chromosome 3p21 and encodes a nuclear localized, ubiquitin carboxy-terminal hydrolase tumor suppressor protein [1, 2]. BAP1 has been shown to bind to the BRCA1 protein enhancing BRCA1-mediated tumor suppression, and is involved in various biological processes including DNA damage response, regulation of the cell cycle and cell growth [2, 3]. Germline BAP1 mutations have been associated with hereditary predisposition to multiple different cancers that include uveal and cutaneous melanoma, malignant mesothelioma on exposure to asbestos, renal cell carcinoma and other cancer types, such as lung adenocarcinoma and meningioma, that are collectively referred to as BAP1 Tumor Predisposition Syndrome (BAP1-TPDS, OMIM #614327) [4, 5]. In vivo studies in melanocytic cells, have shown that BAP1 is involved in the maintenance of a melanocytic phenotype; the depletion of BAP1 protein levels result in dedifferentiation of cells and the acquisition of a more primitive, stem cell like phenotype [6]. It is well established that the BAP1 region of chromosome 3p is commonly deleted in several cancers including melanomas, breast and lung cancers, among others [1, 7].
Uveal melanoma (UM) is a rare malignant tumor of the eye with a frequency of 5.1 per million in the United States [8], and is the cancer most commonly associated with the BAP1-TPDS [9]. The prognosis for UM is poor in approximately 50% of cases primarily due to metastasis to the liver within a short time [10, 11]. Factors such as large tumor size, location and chromosome 3 monosomy [12–18], as well as a specific 12-gene expression pattern [19–21] have all been associated with an increased risk of metastasis. In addition, the presence of inactivating somatic [22–29] or germline [30] BAP1 mutations often in conjunction with chromosome 3 monosomy, loss of BAP1 expression, or lack of immunohistochemical staining, have all been associated with metastasizing UM.
Germline BAP1 mutations have been identified in approximately 2-5% of UM unselected for the presence of any high risk of hereditary cancers [30–33]. These studies reported that metastasis occurred more frequently in UM carrying germline mutations in the blood compared to BAP1 mutation-negative controls, but the differences were significant only in one study [30], possibly due to the small number of tumors with germline mutations. In studies where DNA from both tumor tissue and blood was sequenced to rule out the presence of germline mutations, the frequency of somatic BAP1 mutations was considerably higher than that observed in tumors with germline mutations, approaching 50% [23, 27, 34]. In other studies where somatic mutations were detected by BAP1 expression assays and/or negative immunohistochemical staining methods, as well as sequencing, the lack of BAP1 expression was also significantly associated with the risk of metastasis [22–29]. While these studies show a strong association between somatic BAP1 mutations and metastasis, the effect of germline changes remains unclear.
Most studies of BAP1 mutation status in UM have been carried out using enucleated UM which may not be representative of how UM are currently treated, especially smaller tumors that are treated with globe sparing procedures. In this study of BAP1 mutations in UM, the presence of germline or somatic mutations was determined using sequence analysis of DNA extracted from biopsies of both enucleated globes and fine needle aspirations (FNA) and from matched blood samples. Demographic and clinical characteristics of the tumors and the frequency of metastasis were compared among tumors with germline or somatic BAP1 mutations and mutation-negative UM.
Methods
UM cases
A total of 142 UM cases managed by the Ocular Oncology Service at Wills Eye Hospital, Thomas Jefferson University, Philadelphia, PA, USA between 1998 and 2016 were evaluated in this study of somatic and germline BAP1 mutations. Twenty-three tumor samples were obtained by solid open biopsy of enucleated UM and 119 by fine needle aspirate (FNA) biopsies. Matched peripheral blood samples were obtained from all 142 individuals. Data on demographic and clinical characteristics, metastatic status, location of metastasis and relevant follow-up times were obtained by retrospective chart reviews. Written informed consent was obtained for all individuals at the time of chromosome testing of the tumor DNA. This research was approved by the Institutional Review Board of the University of Pennsylvania, and is in accordance with the Declaration of Helsinki.
The 142 UM were initially collected from two cohorts. Cohort 1 cases (N=90) were chosen from a series of tumors diagnosed between 1998 and 2013 based on availability of matched tumor and blood samples. Cohort 2 consisted of 52 cases diagnosed between 2008 and 2016 for which BAP1 testing was requested, typically due to a personal and/or family history of cancer. The decision to combine the two sets of tumors into a single cohort for analysis was based on the finding that cohort membership was not a significant variable in either univariate (P=0.53) or multivariate (P=0.65) Cox regression analysis (Table 1). In addition, there was no significant difference between the two cohorts in the relative number of tumors carrying germline or somatic BAP1 mutations and mutation-negative tumors (P= 0.76), or in the number of cases with personal or family history of BAP1-TPDS cancers (P=0.35, see below and Additional file 1: Table S1). There was a significant difference in the time to metastasis (P=0.01) and in the number of metastases (P=0.003) between the two cohorts, possibly due to a longer follow-up time for the cohort 1 tumors (median=85 months, range 6-191) compared to 24 months (range 4-77 months, P<0.001, Additional file 1: Table S1) for cohort 2. However, taking both metastasis and follow-up time into account in a Kaplan-Meier analysis, there was no significant association between cohort membership and metastasis (P=0.52, Additional file 1: Figure S1).
Table 1.
Demographic and tumor characteristics for 142 UM and assessment of their association with metastasis evaluated by Cox univariate regression for each of the nine variables alone, and multivariate regression that included all variables described in the table
| Variables | All tumors N=142 (frequency) | Univariate regression | Multivariate regression | ||||
|---|---|---|---|---|---|---|---|
| P-value | Hazard ratio | 95% confidence intervals | P-value | Hazard ratio | 95% confidence intervals | ||
| Cohort | |||||||
| 1 | 90 (0.63) | reference | reference | ||||
| 2 | 52 (0.37) | 0.53 | 0.82 | 0.44-1.53 | 0.65 | 1.18 | 0.57-2.48 |
| Source of biopsied samplea | |||||||
| FNA, N=119 (0.84) | reference | reference | |||||
| BAP1 mutation negative | 73 (0.61) | ||||||
| BAP1 mutation positive | 46 (0.39) | ||||||
| Enucleated tumor, N= 23 (0.16) | 0.01 | 2.12 | 1.19-3.76 | 0.08 | 2.04 | 0.92-4.52 | |
| BAP1 mutation negative | 15 (0.65) | ||||||
| BAP1 mutation positive | 8 (0.35) | ||||||
| Age (years) | 0.04 | 1.02 | 1.00-1.04 | 0.03 | 1.02 | 1.00-1.05 | |
| Median | 56.7 | ||||||
| Mean±SD | 55.4±14.7 | ||||||
| Range | 14-88 | ||||||
| Sex | |||||||
| Male | 77 (0.54) | reference | reference | ||||
| Female | 65 (0.46) | 0.24 | 0.73 | 0.43-1.24 | 0.73 | 0.90 | 0.50-1.63 |
| Chromosome 3 | |||||||
| Disomy | 63 (0.44) | reference | reference | ||||
| Monosomy, partial monosomy (N=2), mosaic (N=5) | 79 (0.56) | <0.001 | 4.64 | 2.46-8.78 | 0.008 | 2.99 | 1.33-6.72 |
| Tumor diameter (mm) | <0.001 | 1.25 | 1.17-1.35 | <0.001 | 1.20 | 1.10-1.31 | |
| Median | 12.0 | ||||||
| Mean±SD | 12.5±4.1 | ||||||
| Range | 5.0-22.0 | ||||||
| Tumor thickness (mm) | <0.001 | 1.22 | 1.13-1.31 | 0.61 | 0.97 | 0.87-1.09 | |
| Median | 5.7 | ||||||
| Mean±SD | 6.2±3.3, | ||||||
| Range | 1.0-16.5 | ||||||
| Ciliary body involvement | |||||||
| Absent | 110 (0.78) | <0.001 | reference | reference | |||
| Present | 32 (0.22) | 3.05 | 1.79-5.20 | 0.15 | 1.54 | 0.85-2.80 | |
| BAP1 | |||||||
| Negative | 88 (0.62) | reference | reference | ||||
| Somatic | 43 (0.30) | <0.001 | 4.81 | 2.79-8.28 | 0.02 | 2.20 | 1.13-4.30 |
| Germline | 11 (0.08) | 0.33 | 1.70 | 0.59-4.91 | 0.81 | 0.87 | 0.28-2.72 |
aThere was no significant difference in the number of BAP1 mutation negative tumors and those carrying germline or somatic mutations UM biopsies from FNA samples compared to enucleated tumors (P=0.79, Fisher Exact test)
Figures is bold indicate significant P-values <0.05
A personal or family history of BAP1-TPDS syndrome, defined following the guidelines found in OMIM #614327 [4] and Pilarski et al [5], was available for a subset of the UM cases. Individuals having at least two BAP1-TPDS tumors (UM, cutaneous melanoma, malignant mesothelioma, or renal cell carcinoma) themselves, or one in the UM case plus at least one in a first or second degree relative were considered as indicative of BAP1-TPDS syndrome. Cases with non-specific cancer diagnoses were not included as BAP1-TPDS syndromic tumors,
BAP1 mutation screening and chromosome 3 copy number analysis
Genomic DNA was extracted from peripheral blood lymphocytes using the Gentra Puregene Blood Kit (Cat No.158489, Qiagen) following the manufacturer’s protocol, and from tumor samples as described previously [15, 35]. Sanger sequencing of DNA from matched peripheral blood lymphocytes was used to determine the presence of a germline mutation as noted by Abdel-Rahman et al [36]. DNA from enucleated tumor or FNA biopsies was sequenced to identify BAP1 mutations using either Sanger or next generation sequencing of all coding exons and adjacent intronic regions [37, 38]. Sequence alignment and variant calling was performed as described previously [37]. The 2015 ACMG guidelines were followed in determining the degree pathogenicity of all sequence variants [39].
Chromosome 3 copy number was determined by microsatellite analysis (N=51) or whole genome SNP array (Affymetrix Human 100K, SNP-5.0 or SNP-6.0 and Cytoscan HD genotyping arrays, Affymetrix, Santa Clara CA) [15, 35]. Tumors were categorized as either disomy-3 (N=63) or monosomy (N=79) that included 72 tumors with complete monosomy-3, two with partial monosomy and five with a mosaic monosomy 3 indicating tumor heterogeneity of cells with chromosome-3 disomy and monosomy.
Statistical analyses
Continuous variables (age, tumor diameter and thickness) were compared using a Mann-Whitney U-test, and discrete variables (source of tumor sample, cohort 1 or 2, metastasis status, sex, tumor location and chromosome 3 copy number) using Fisher Exact or chi-square tests (vassarstats.net). Cox univariate and multivariate proportional hazard regression was used to determine the association of clinical and tumor characteristics with metastasis. The Kaplan-Meier method was used to generate metastasis-free survival curves that were compared using a log-rank test. SPSS 24 (IBM, New York, NY) was used for survival analysis procedures. All tests were two-tailed and P-values less than 0.05 were considered statistically significant.
Results
UM individuals and tumor characteristics
Of the 142 UM characterized in this study, 54 (38%) carried a BAP1 gene mutation. Eleven (8%) of the mutations were germline and 43 (30%) were somatic. The remaining 88 (62%) were BAP1 mutation-negative. The eleven germline mutations were all retained in the matched UM samples. Thus, there were no cases where a germline mutation was lost in tumors, with or without monosomy-3.
Demographic and tumor characteristics for the 142 UM cases are shown in Table 1. The median age of all individuals was 56.7 years (range=14-88), and 77 (54%) were males. Overall, 63 (44%) of the tumors carried two copies of chromosome 3 (disomy), and 79 (56%) carried one copy of chromosome 3 (monosomy), partial monosomy or mosaic. The median tumor diameter and thickness was 12 mm (range=5.0-22.0) and 5.7 mm (range 1.0-16.5), respectively. The majority of the tumors had no ciliary body involvement (N=110, 78%). Most tumors were located only in the choroid (N=109, 77%), 26 (18%) were in the choroid plus ciliary body, three (2%) in the ciliary body, one (1%) in the iris, two (2%) in the iris plus ciliary body, and one (1%) in the iris plus ciliary body and choroid. Fifty-nine tumors (42%) metastasized within a median time of 19 months (range=0-107) while 83 (58%) did not metastasize during the observed follow-up period (median=56 months, range=4-191 months, Table 2). Thirty-six of 54 tumors with BAP1 mutations metastasized (67%) compared to only 23 of 88 (26%) mutation-negative tumors (P<0.001) within 2-107 months.
Table 2.
Description of demographic and tumor variables with pairwise comparisons for 11 tumors with germline BAP1 mutations, 43 with BAP1 somatic mutations and 88 with no BAP1 mutations
| Variables | BAP1 germline | BAP1 somatic | BAP1 negative | Pairwise comparisons (P-value) | ||
|---|---|---|---|---|---|---|
| N=11 (0.08) (frequency) | N=43 (0.30) (frequency) | N=88 (0.62) (frequency) | Germline/Somatic | Germline/Negative | Somatic/Negative | |
| Metastases | ||||||
| Yes N=59 (0.42) | 4 (0.36) | 32 (0.74) | 23 (0.26) | 0.03 a | 0.72a | <0.001 a |
| No N=83 (0.58) | 7 (0.64) | 11 (0.26) | 65 (0.74) | |||
| Time to metastasis (months) (N=59) | N=4 (0.07) | N=32 (0.54) | N=23 (0.38) | 0.07b | 0.50b | 0.04 c |
| Median (19.0 months) | 37.5 | 16.0 | 26.0 | |||
| Mean±SD (26.4±20.9) | 35.0±15.8 | 23±22.5 | 29.7±19.2 | |||
| Range (0-107) | 16-49 | 2-45,90,107d | 0-84 | |||
| Follow-up time (months) in tumors with no metastasis (N=83) | N=7 (0.08) | N=11 (0.13) | N=65 (0.78) | 0.59c | 0.25c | 0.33c |
| Median (56.0 months) | 24 | 54.0 | 58.0 | |||
| Mean±SD (59.8±39.0) | 43.7±39.3 | 49.9±30.0 | 63.2±40.1 | |||
| Range (4-191) | 4-109 | 8-90 | 5-191 | |||
| Cohort | ||||||
| 1 | 6 (0.54) | 27 (0.63) | 57 (0.65) | 0.73a | 0.74a | 0.85a |
| 2 | 5 (0.46) | 16 (0.37) | 31 (0.35) | |||
| Source of biopsied sample | ||||||
| FNA | 10 (0.91) | 36 (0.84) | 73 (0.83) | 0.68a | 0.69a | 1.00a |
| Enucleated tumor | 1 (0.09) | 7 (0.16) | 15 (0.17) | |||
| BAP1-TPDS personal or family history (N=79 reports)e | ||||||
| Yes, syndromic tumors present (N=29, 0.37) | 8 (1.00) | 10 (0.44) | 11 (0.23) | 0.009 a | <0.001 a | 0.10 |
| No syndromic tumors (N=50, 0.63) | 0 | 13 (0.56) | 37 (0.77) | |||
| Age (years) | ||||||
| Median | 59.0 | 61.8 | 52.2 | 0.07c | 0.86c | 0.002 c |
| Mean±SD | 51.9±13.6 | 61.2±12.8 | 53.1±15.0, | |||
| Range | 22-67 | 28-88 | 14-84 | |||
| Sex | ||||||
| Male | 7 (0.64) | 25 (0.58) | 45 (0.51) | 1.00a | 0.53a | 0.46a |
| Female | 4 (0.36) | 18 (0.42) | 43 (0.49) | |||
| Chromosome 3 | ||||||
| Disomy | 1 (0.09) | 3 (0.07) | 59 (0.67) | 1.0a | <0.001 a | <0.001 a |
| Monosomy, partial monosomy (N=2), mosaic (N=5) | 10 (0.91) | 40 (0.93) | 26 (0.33) | |||
| Tumor diameter (mm) | ||||||
| Median | 14.0 | 14.0 | 11.5 | 0.70c | 0.23c | 0.003 c |
| Mean±SD | 13.4±4.4 | 13.9±3.9 | 11.7±4.0, | |||
| Range | 8.0-20.0 | 6.0-21.5 | 5.0-22.0 | |||
| Tumor thickness (mm) | ||||||
| Median | 6.6 | 7.0 | 5.0 | 0.41c | 0.51c | 0.002 c |
| Mean±SD | 6.5±3.6 | 7.5±3.5 | 5.6±2.9, | |||
| Range | 1.5-12.3 | 2.0-16.5 | 1.0-13.1 | |||
| Ciliary body involvement | ||||||
| Absent | 8 (0.73) | 28 (0.65) | 74 (0.84) | 0.73a | 0.40a | 0.02 a |
| Present | 3 (0.27) | 15 (0.35) | 14 (0.14) | |||
aAssociation test performed using two-tailed Fisher Exact or Chi-square tests
bReported P-values are calculated from a normal approximation of the Mann Whitney test statistic
cTests of means of quantitative variables performed using Mann-Whitney U tests
dThe time for metastasis for 30 of the tumors with somatic mutations was 2-45 months. The time to metastasis for the remaining two tumors was 90 and 107 months
eBAP1-TPDS, BAP1-tumor predisposition syndrome as defined in OMIM #614327 [4] and Pilarski et al [5]
Figures is bold indicate significant P-values <0.05
BAP1 status was determined for 119 (84%) FNA biopsies and 23 enucleated tumors (Table 1). There was a significant difference in the number of metastases depending on the source of the biopsied sample: 43 of 119 (36%) tumors biopsied from FNA metastasized compared to 16 of 23 from enucleated globes (70%, P=0.006). The source of the biopsied sample was also significantly associated with metastasis in a Cox univariate (P=0.01), but not in a multivariate analysis (0.08, Table 1). This is not surprising, however, given that some of the classic prognostic factors for UM metastasis, larger tumor diameter and thickness and tumor location which warrant enucleation, were significantly different between tumors sampled from FNAs or enucleated tumor samples (P=0.002, >0.001 and 0.02, respectively (data not shown). Importantly, there was no difference in the number of tumors carrying a germline or somatic mutation or mutation-negative tumors in enucleated tumors compared to FNA samples (P=0.79, Table 1 footnotea).
In a Cox univariate analysis, the presence of a somatic, but not a germline BAP1 mutation (hazard ratio (HR) =4.81, P<0.0001; HR=1.7, P=0.33, respectively, Table 1) was significantly associated with metastasis. Other tumor variables that were significant in the univariate regression analysis included tumor source (FNA or enucleated tumor, HR= 2.12; P=0.01), age HR=1.02, P=0.04), chromosome 3 monosomy, larger tumor diameter and thickness, and ciliary body involvement (HR=4.64, 1.25 and 1.22, respectively; all P≤0.001). Considering all variables in a multivariate analysis, the presence of a somatic BAP1 mutation (HR=2.20, P=0.02), age (HR=1.02, P=0.03) tumor diameter (HR=1.20, P<0.001), and chromosome 3 monosomy (HR=2.99, P=0.008) remained significant, while the presence of a germline mutation was not significant (HR=0.87, P=0.81).
BAP1 Gene Mutations
Fifty-two unique BAP1 mutations were identified in 54 tumors: 43 tumors (30%) carried somatic mutations, while 11 tumors (8%) carried germline mutations (Table 3). All mutations were defined as pathogenic or likely pathogenic. These included seven (13%) missense and 15 (28%) splice-site mutations, three (6%) in-frame deletions and 29 mutations causing premature truncation, including one large deletion encompassing exons 15-17 and the intervening introns. There was no significant difference in the frequencies of these categories of mutations found in UM with germline compared to somatic mutations (P=1.0), nor in tumors that metastasized compared to those that did not (P=0.53). Thirty-six (67%) of the mutations were in the catalytic UCH domain. One splice-site mutation, c.438-1A>G was present as a germline mutation in one tumor (UM-23) and as a somatic mutation in two tumors (UM-11 and UM-27). Thirty-three of these mutations were previously described in UM by Ewens et al [37] and 13 have been identified in UM by others (as noted in Table 3). Although the number of germline mutations was small (N=11), the locations of these were spread throughout the gene, similar to the distribution of somatic mutations (Table 3). Therefore, no new information was inferred from the location, or the associated functional domains, that would make the germline mutations have unique characteristic compared to somatic mutations.
Table 3.
Fifty-two unique germline and somatic BAP1 mutations identified in 54 UM
| ID | BAP1 mutation | Metastases | chr 3 | DNA change | Protein change | Predictred effect | Predicted pathogenic effecta | Previously reported mutations in UM | |
|---|---|---|---|---|---|---|---|---|---|
| UM_44 | germline | yes | monosomy | exon 1 | c.3G>A | p.Met1Ile | misense: start site lost | LP | [36] |
| UM_76 | somatic | yes | monosomy | intron 1 | c.38-1G>C | p.? | splicing | P | [36] |
| UM_9 | somatic | yes | monosomy | exon 2 | c.40_52del13 | p.Leu14SerfsTer54 | truncating | P | [36] |
| UM_1136 | germline | no | monosomy | exon 2 | c.58_59insTG | p.Glu20ValfsTer53 | truncating | P | |
| UM_113 | somatic | yes | monosomy | intron 2 | c.67+1G>A | p.? | splicing | P | [27, 36] |
| UM_1123 | somatic | yes | monosomy | intron 2 | c.67+2dupT | p.? | splicing | P | |
| UM_1080 | somatic | no | monosomy | intron 2/exon 3 | c.68-12_75del20 | p.? | splicing | P | |
| UM_802 | somatic | yes | monosomy | intron 2/exon 3 | c.68-16_69del18 | p.? | splicing | P | [36] |
| UM_584 | germline | no | monosomy | exon 3 | c.79delG | p.Val27CysfsTer45 | truncating | P | |
| UM_17 | somatic | yes | monosomy | exon 3 | c.82C>T | p.Gln28Ter | truncating | P | [36] |
| UM_1207 | somatic | no | monosomy | exon 3 | c.91_93delGAG | p.Glu31del | in-frame deletion | LP | |
| UM_1133 | germline | no | disomyb | intron 3 | c.122+1G>C | p.? | splicing | P | |
| UM_109 | germline | yes | monosomy | intron 3 | c.122+1G>T | p.? | splicing | P | [36] |
| UM_75 | somatic | no | monosomy | exon 4 | c.125_145del21 | p.Pro42_Phe48del | in-frame deletion | LP | [36] |
| UM_88 | somatic | yes | monosomy | exon 4 | c.145delC | p.Leu49CysfsTer23 | truncating | P | [36] |
| UM_877 | somatic | yes | monosomy | exon 4 | c.165_180del16 | p.Arg57SerfsTer10 | truncating | P | |
| UM_13 | somatic | yes | disomyb | exon 4 | c.178C>T | p.Arg60Ter | truncating | P | [36, 45] |
| UM_62 | somatic | yes | monosomy | exon 4 | c.202_227del26 | p.Asp68CysfsTer3 | truncating | P | [36] |
| UM_780 | somatic | yes | monosomy | exon 4/intron 4 | c.234del96 | p.? | splicing | P | |
| UM_119 | somatic | yes | monosomy | exon 4 | c.253C>T | p.Gln85Ter | truncating | P | [36] |
| UM_106 | somatic | yes | monosomy | exon 4 | c.254A>C | p.Gln85Pro | missense | P | [36] |
| UM_1126 | somatic | no | monosomy | exon 5 | c.295_312del18 | p.Val99_Ser104del | in-frame deletion | LP | |
| UM_1046 | somatic | no | monosomy | exon5/intron5 | c.370_375+12del18 | p.? | splicing | P | |
| UM_1208 | somatic | yes | monosomy | intron 5/exon 6 | c.376-20_383del28 | p.? | splicing | P | |
| UM_56 | somatic | yes | monosomy | exon 6 | c.422A>G | p.His141Arg | missense | LP | [36, 46] |
| UM_23 | germline | yes | monosomy | intron 6 | c.438-2A>G | p.? | splicing | P | [36] |
| UM_11 | somatic | no | disomyb | intron 6 | c.438-2A>G | p.? | splicing | P | |
| UM_27 | somatic | yes | monosomy | intron 6 | c.438-2A>G | p.? | splicing | P | |
| UM_58 | germline | no | monosomy | exon 7 | c.458_459delCT | p.Pro153ArgfsTer7 | truncating | P | [9, 36] |
| UM_114 | somatic | yes | monosomy | exon 7 | c.497_509del13 | p.Glu166ValfsTer17 | truncating | P | [36] |
| UM_51 | somatic | no | monosomy | exon 7 | c.506A>C | p.His169Pro | missense | P | [36] |
| UM_1118 | somatic | no | monosomy | exon 7 | c.524C>G | p.Pro175Arg | missense | LP | [27] |
| UM_1086 | somatic | yes | monosomy | intron 7/exon 8 | c.581-2_591del13 | p.? | splicing | P | |
| UM_60 | somatic | yes | monosomy | exon 8 | c.588G>A | p.Trp196Ter | truncating | P | [23, 47] |
| UM_1334 | germline | no | mosaic | exon 8 | c.619delC | p.Arg207GlyfsTer24 | truncating | P | |
| UM_105 | somatic | yes | monosomy | intron 8 | c.659+1G>A | p.? | splicing | P | [27, 36] |
| UM_35 | somatic | yes | monosomy | exon 9 | c.723T>A | p.Tyr241Ter | truncating | P | [36, 48:c.723T>C] |
| UM_107 | somatic | yes | monosomy | exon 9 | c.781C>T | p.Gln261Ter | truncating | P | [36, 49] rs772448753c |
| UM_46 | somatic | yes | monosomy | exon 10 | c.904_905insT | p.Pro302LeufsTer5 | truncating | P | [36] |
| UM_1029 | somatic | yes | monosomy | exon 12 | c.1134_1143del10_insAA | p.Ala379ArgfsTer16 | truncating | P | |
| UM_115 | somatic | yes | monosomy | exon 12 | c.1153C>T | p.Arg385Ter | truncating | P | [32, 36, 44, 50] |
| UM_55 | somatic | no | monosomy | exon 12 | c.1175_1182delAGCAGTAC | p.Gln392LeufsTer3 | truncating | P | [36] |
| UM_863 | somatic | no | monosomy | exon 12 | c.1192G>T | p.Glu398Ter | truncating | P | |
| UM_950 | germline | yes | mosaic | exon 12 | c.1203dupT | p.Glu402Ter | truncating | P | |
| UM_69 | somatic | yes | monosomy | exon 12 | c.1217_1220delAGGA | p.Glu406ValfsTer23 | truncating | P | [36] |
| UM_1333 | germline | no | mosaic | exon 13 | c.1695dupT | p.Glu566Ter | truncating | P | |
| UM_48 | somatic | no | monosomy | exon 13 | c.1729G>C | p.Glu577Gln | missense | P | [36] |
| UM_61 | somatic | yes | monosomy | exon 14 | c.1881C>G | p.Tyr627Ter | truncating | P | [36] |
| UM_74 | germline | no | monosomy | exon 14 | c.1882_1885delTCAC | p.Ser628ProfsTer8 | truncating | P | [29, 36, 51] |
| UM_708 | somatic | no | disomyb | intron 14-3'UTR | c.1890+38_2573del | p.Glu631Ter | truncating (3 exon deletion) | P | |
| UM_1113 | somatic | yes | monosomy | exon 15 | c.1926_1951del 26 | p.Ile643GlyfsTer12 | truncating | P | |
| UM_104 | somatic | yes | monosomy | exon 15 | c.1932_1948deldel17 | p.Asn645GlnfsTer13 | truncating | P | [36] |
| UM_804 | somatic | yes | monosomy | exon 16 | c.1986_1989delTGAT | p.Ile662MetfsTer29 | truncating | P | [36] |
| UM_108 | somatic | yes | monosomy | exon 16 | c.2015A>G | p.Asp672Gly | missense | LP | [23, 36] |
aPredicted pathogenic effect based on ACMG guidelines [39] P, pathogenic; LP, likely pathogenic
bindicates UM with chromosome 3 disomy and carrying a germline or somatic BAP1 mutation
**Population frequency rs772448753: 8.2E-06
Characterizations of UM with and without BAP1 mutations
Clinical and tumor characteristics of UM carrying BAP1 germline or somatic mutations and mutation-negative tumors, as well as pairwise comparisons of the three groups are presented in Table 2. There was no significant difference in the cohort, source of biopsy sample, or sex of the individuals among the three categories of mutations. The age at diagnosis of individuals with somatic mutations was significantly older (61.8 years, range=28-88) than individuals with mutation-negative tumors (median=52.2 years, range=14-84 years, P=0.002) and approached significance compared to individuals with a tumor carrying a germline mutation (median=59.0 years, range=22-67, P=0.07).
There was a significant difference in the comparison of chromosome 3 monosomy in mutation-negative tumors compared to tumors with either germline or somatic mutations. Only 26 (33%) mutation-negative tumors carried monosomy 3, while ten (91%) tumors with germline mutations and 40 (93%) with somatic mutations were chromosome 3 monosomy (P<0.001 in both cases). A comparison of chromosome 3 copy number between tumors with germline vs. somatic mutations (91% vs. 93%) was not significant (P=1.0, Table 2). There were four tumors with BAP1 mutations and chromosome 3 disomy (marked with superscript b in Table 3). These would suggest that chromosome 3 loss was not a necessary consequence of BAP1 loss. The one germline mutation was a splice-site mutation, intron 3, c.122+1G>C. The three somatic mutations included one splice-site mutation (intron 6, c.438-2A>G) and two truncating mutations: exon 4, c.178C>T (p.Arg60X) and a large deletion of exons 15-17 and the intervening introns. The tumor carrying the exon 4, c.178C>T (p.Arg60X) truncating mutation metastasized at 11 months, but the other three tumors had not metastasized within 24 months (c.122+1G>C) or within at least 65 months (c.438-2A>G and the large three exon deletion).
Tumors with BAP1 somatic mutations had a significantly larger median diameter (14.0 mm, range=6.0-21.5) and thickness (7.0 mm, range=2.0-16.5) compared to mutation negative tumors (median diameter=11.5 mm, range=5.0-22.0; thickness=5.0 mm, range=0.0-13.1, P=0.003 and P=0.002 respectively, Table 2). However, there was no significant difference in the tumor diameter or thickness between BAP1 germline tumors and tumors with either somatic mutations (P=0.70 and P=0.41, respectively), or BAP1-negative tumors (P=0.23 and P=0.51, respectively). Table 2 shows that there was also a significant difference in ciliary body involvement of tumors carrying a somatic mutation, where the tumor was more likely to have ciliary body involvement (33%) compared to BAP1-negative tumors (14%, P=0.04), but not compared with those with a germline mutation (27%, P=1.0).
Tumors carrying BAP1 somatic mutations metastasized significantly more often (32 of 43, 74%) compared to those with either BAP1 germline mutations (4 of 11, 36%, P=0.03) or to BAP1 mutation-negative tumors (23 of 88, 26%, P<0.001, Table 2). However, there was no significant difference in the frequency of metastasis between tumors with germline mutations and BAP1-negative tumors (P=0.72). This finding was corroborated by the multivariate regression analyses shown in Table 1 indicating that somatic, but not germline BAP1 mutations, were significantly associated with metastasis (P=0.03 and 0.70, respectively). Kaplan-Meier analysis (Figure 1) also showed that tumors with a BAP1 somatic mutation had a significantly poorer metastatic outcome compared to those with germline mutations (P=0.03) or mutation-negative tumors (P<0.001), but the difference between tumors with germline mutations and BAP1 mutation-negative tumors was not significant (P=0.23). The time to metastasis was also significantly shorter in the tumors with somatic mutations (median=16.0 months, range=2-107) compared to BAP1-negative tumors (median=26 months, range=0-84 months, P=0.04, Table 2).
Fig. 1.
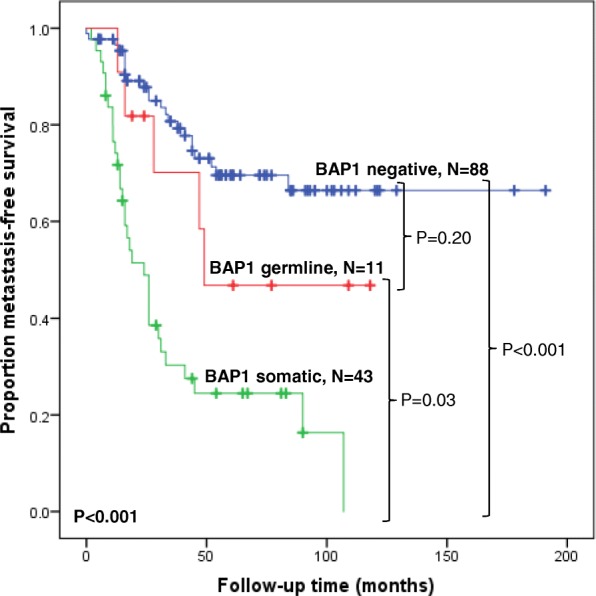
Kaplan-Meier curves showing metastasis-free survival following diagnosis for 142 UM stratified by BAP1 mutation status
A positive personal and/or family history of BAP1-TPDS tumors was reported for 29 (37%) of the 79 cases that provided information of other cancers in themselves or their families (Table 2). There were eight cases that carried a germline mutation with information on personal or familial cancers; all had a positive history of BAP1-TPDS. Comparisons of BAP1-TPDS tumors in UM cases with a germline (8, 100%) vs. somatic mutation (10, 44%) or germline vs. mutation-negative (11, 23%) were significant (P=0.009 and P<0.001, respectively), while a comparison of those with somatic mutations was not significantly different from those with mutation-negative tumors (P=0.10)
Discussion
Inactivating somatic mutations in the BAP1 gene in UM were first identified and associated with metastatic disease in 2010 [23]. Subsequent to this original publication, there have been numerous studies characterizing BAP1 somatic [22, 24–29, 40–43] and germline [9, 30, 32, 33] mutations in UM. However, to our knowledge, this is the first report directly comparing risk of metastasis in tumors carrying germline or somatic BAP1 mutations and mutation-negative tumors. In this study, the presence of somatic mutations was strictly defined as those found in tumors, and not in matched blood samples. A second important difference in this study is that, compared to most other reports where UM tumors analyzed for BAP1 mutations were comprised primarily of samples biopsied from larger, enucleated tumors, 84% of the samples analyzed in this study were from FNA biopsies. We found no significant difference in the frequency of somatic and germline mutations in biopsies from FNAs compared to enucleated tumors. Since the majority of UM are currently being treated by globe sparing procedures, it was important to determine whether the consequences of carrying a BAP1 mutation identified in larger enucleated tumors could be applied to smaller UM biopsied by FNA sampling.
It is well established that tumor characteristics such as tumor diameter, ciliary body involvement and chromosome 3 monosomy are all associated with poor prognosis as measured by the development of metastases within 48 months after the primary is treated [12–18]. It has also been shown that that these same variables are significantly associated with BAP1 mutation status in tumors with somatic or germline mutations [24, 25, 28, 30]. In this study, multivariate regression analysis showed that in addition to these classic tumor variables, the presence of somatic, but not germline, BAP1 mutations was significantly associated with metastasis (HR=2.20, P=0.02; HR=0.87, P=0.81, respectively). Kaplan-Meier analysis further showed that tumors with somatic mutations were associated with the poorest metastatic outcome, while there was no significant difference between tumors carrying a germline mutation and mutation-negative tumors. Furthermore, the time to metastasis was seven months shorter for individuals whose tumors carried somatic mutations compared to mutation-negative tumors (P=0.04) suggesting a more aggressive phenotype. Thus, in a previous publication [37] we showed that tumors with any mutation in the BAP1 gene, in combination with chromosome 3-monosomy, have the highest risk of metastasis (HR=11.5). In this manuscript we show that monosomy 3 plus a somatic mutation in the BAP1 gene, as opposed to a germline mutation, have a significantly higher risk of metastasis (P=0.01, H.R.=2.81) and a shorter time to metastasis compared to the ones with a germline mutation.
Given that somatic BAP1 mutations appear to have a highly significant association with poor metastatic prognosis, it is surprising that the age of diagnosis of the individuals whose tumors carried somatic mutations was significantly older (median=61.8 years) compared to those with mutation-negative tumors (median=52.2 years, P=0.002). Previous studies have also reported that tumors with somatic mutations were diagnosed at least 8 years later than mutation-negative controls [24, 27, 29], but the significance of this observation is not clear.
Among the 54 tumors with BAP1 mutations, there were four with chromosome 3 disomy. It cannot be ruled out that although these tumors appeared to be disomy-3, small deletions not detectable on the SNP arrays leading to partial monosomy or heterogeneity of the tumor sample could be responsible for this finding. Alternatively, in some cases it is possible the during tumor evolution, BAP1 mutation on one copy of chromosome 3 could precede the loss of the other copy of chromosome 3. While several studies have found total concordance between the presence BAP1 mutations and monosomy 3 [23, 27, 40], others using sequencing, gene expression, and/or immunostaining methods have identified BAP1 mutations in a small number of tumors with disomy 3 [24, 25, 28, 44].
One interesting observation in this study was the presence of BAP1-TPDS history in 44% of somatic cases and 23% of mutation negative tumors. The data on BAP1-TPDS was based on self-reported personal and family history of cancer. We classified individuals as having BAP1-TPDS if they carried at least two relevant tumors in their personal or family history. Since it is self-reported, the information can be imprecise, and there is a possibility for over/under representation of the BAP1-TPDS cases as well.
While the presence of germline BAP1 mutations in cancers associated with BAP1-TPSD is well documented, the role of somatic mutations in this syndrome is not well studied. It is possible that the presence of TPDS in our cohort of UM cases that have a somatic BAP1 mutation or are mutation-negative could be interpreted as evidence for the existence of a second type of the syndrome associated with mutations in a second gene different from BAP1.
A major strength of this study is that it includes UM sampled from enucleated tumors and FNAs with both somatic and germline BAP1 mutations. However, it is limited by the small number of tumors with germline mutations. It is important to confirm these findings in a larger cohort of UM.
Conclusions
This study identified significant differences in the risk of metastasis for individuals whose tumors carry somatic vs. germline BAP1 mutations. While overall 36 of 59 (61%) metastasizing tumors carried a BAP1 mutation, only 7% carried germline mutations, compared to 54% with somatic mutations. From the viewpoint of counseling individuals with UM, there are different implications depending on whether the mutation is classified as germline or somatic. The most relevant information for the individuals with UM is the risk of metastasis, as well as an estimate of metastasis-free survival. The presence of germline mutations can provide the individual with information concerning the risk of development of other cancers themselves or in other family members, while information about the presence of somatic mutations is relevant to their individual risk of developing metastases within a shorter time period. Thus it is important to determine whether a tumor carries a BAP1 mutation, and if positive, to also evaluate a matched blood sample to establish whether the mutation is germline or somatic. In addition, this information can also change the cascade screening and surveillance of the family members.
Additional file
Table S1. Comparison of demographic and tumor variables of two cohorts of 142 UM. Description of demographic and tumor variables with pairwise comparisons for 142 divided into two cohorts depending on whether tumors were selected from archived samples for which both tumor and blood samples were available (cohort 1, N=90) or tumors for which BAP1 sequencing was specifically requested (cohort 2, N=52). References: 1. OMIM: Tumor predisposition syndrome; TPDS. https://www.omim.org/entry/614327. 2. Pilarski R, Rai K, Cebulla C, Abdel-Rahman M.BAP1 Tumor Predisposition Syndrome. 2016 Oct 13 In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, Amemiya A, editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993-2018. Available from: https://www.ncbi.nlm.nih.gov/books/NBK390611/. Figure S1. Kaplan-Meier analysis comparing metastasis-free survival of two cohorts of 142 UM. Kaplan-Meier curves showing metastasis-free survival following treatment for 142 UM stratified by cohort, as described in the legend for Additional file 1: Table S1. (DOCX 81 kb)
Acknowledgements
The authors wish to thank Jessica Ebrahimzadeh for her service in reviewing case charts.
Funding
This work was supported in part by a grant from the National Cancer Institute (5R21CA181935) to AG, and internal Research and Development funds of the Genetic Diagnostic Laboratory.
Availability of data and materials
The datasets analyzed for this study are available from the corresponding author upon reasonable request.
Abbreviations
- BAP1
BRCA1-associated protein 1
- BAP1-TPDS
BAP1 tumor predisposition syndrome
- FNA
Fine needle aspirate
- GDL
Genetic Diagnostic Laboratory, University of Pennsylvania, Perelman School of Medicine
- UM
Uveal melanoma
- vs
Versus
Authors’ contributions
KGE participated in data collection and analysis and manuscript preparation; EL contributed to statistical analysis and manuscript review; J R-Y was responsible for BAP1 sequence data and analysis; CLS provided all case samples and access to charts, participated in the design of the project and review of the manuscript; AG conceived the study and was involved in all aspects of data collection, analysis and manuscript preparation. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Written informed consent was obtained at the time of testing the tumor samples from all individuals. This research was approved by the Institutional Review Board of the University of Pennsylvania, and is in accordance with the Declaration of Helsinki.
Consent for publication
Not applicable
Competing interests
The authors declare they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
K. G. Ewens, Email: ewens@pennmedicine.upenn.edu
E. Lalonde, Email: elalonde@pennmedicine.upenn.edu
J. Richards-Yutz, Email: jarichar@pennmedicine.upenn.edu
C. L. Shields, Email: carolshields@gmail.com
A. Ganguly, Phone: 215-898-3122, Email: ganguly@pennmedicine.upenn.edu
References
- 1.Jensen DE, Proctor M, Marquis ST, Gardner HP, Ha SI, Chodosh LA, et al. BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene. 1998;16(9):1097–1112. doi: 10.1038/sj.onc.1201861. [DOI] [PubMed] [Google Scholar]
- 2.Jensen DE, Rauscher FJ. BAP1, a candidate tumor suppressor protein that interacts with BRCA1. Ann N Y Acad Sci. 1999;886(1):191–194. doi: 10.1111/j.1749-6632.1999.tb09414.x. [DOI] [PubMed] [Google Scholar]
- 3.Ventii KH, Devi NS, Friedrich KL, Chernova TA, Tighiouart M, Van Meir EG, et al. BRCA1-associated protein-1 is a tumor suppressor that requires deubiquitinating activity and nuclear localization. Can Res. 2008;68(17):6953–6962. doi: 10.1158/0008-5472.CAN-08-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.OMIM: Tumor predisposition syndrome; TPDS. https://www.omim.org/entry/614327.
- 5.Pilarski R, Rai K, Cebulla C, Abdel-Rahman M. BAP1 Tumor Predisposition Syndrome. 2016 Oct 13 In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, Amemiya A, editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993-2018. Available from: https://www.ncbi.nlm.nih.gov/books/NBK390611/. [PubMed]
- 6.Matatall KA, Agapova OA, Onken MD, Worley LA, Bowcock AM, Harbour JW. BAP1 deficiency causes loss of melanocytic cell identity in uveal melanoma. BMC Cancer. 2013; 13:371-371. [DOI] [PMC free article] [PubMed]
- 7.Angeloni D. Molecular analysis of deletions in human chromosome 3p21 and the role of resident cancer genes in disease. Brief Funct Genomics. 2007;6(1):19–39. doi: 10.1093/bfgp/elm007. [DOI] [PubMed] [Google Scholar]
- 8.Singh AD, Turell ME, Topham AK. Uveal Melanoma: Trends in incidence, treatment, and survival. Ophthalmology. 2011;118(9):1881–1885. doi: 10.1016/j.ophtha.2011.01.040. [DOI] [PubMed] [Google Scholar]
- 9.Rai K, Pilarski R, Cebulla CM, Abdel-Rahman MH. Comprehensive review of BAP1 tumor predisposition syndrome with report of two new cases. Clin Genet. 2016;89(3):285–294. doi: 10.1111/cge.12630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kujala E, Mäkitie T, Kivelä T. Very long-term prognosis of patients with malignant uveal melanoma. Invest Ophthalmol Vis Sci. 2003;44(11):4651–4659. doi: 10.1167/iovs.03-0538. [DOI] [PubMed] [Google Scholar]
- 11.Lorigan JG, Wallace S, Mavligit GM. The prevalence and location of metastases from ocular melanoma: imaging study in 110 patients. Am J Roentgenol. 1991;157(6):1279–1281. doi: 10.2214/ajr.157.6.1950883. [DOI] [PubMed] [Google Scholar]
- 12.Aalto Y, Eriksson L, Seregard S, Larsson O, Knuutila S. Concomitant loss of chromosome 3 and whole arm losses and gains of chromosome 1, 6, or 8 in metastasizing primary uveal melanoma. Invest Ophthalmol Vis Sci. 2001;42(2):313–317. [PubMed] [Google Scholar]
- 13.Damato B, Coupland SE. A reappraisal of the significance of largest basal diameter of posterior uveal melanoma. Eye. 2009;23(12):2152–2162. doi: 10.1038/eye.2009.235. [DOI] [PubMed] [Google Scholar]
- 14.Damato B, Dopierala J, Klaasen A, van Dijk M, Sibbring J, Coupland SE. Multiplex ligation-dependent probe amplification of uveal melanoma: correlation with metastatic death. Invest Ophthalmol Vis Sci. 2009;50(7):3048–3055. doi: 10.1167/iovs.08-3165. [DOI] [PubMed] [Google Scholar]
- 15.Ewens KG, Kanetsky PA, Richards-Yutz J, Al-Dahmash S, De Luca MC, Bianciotto CG, et al. Genomic profile of 320 uveal melanoma cases: Chromosome 8p-loss and metastatic outcome. Invest Ophthalmol Vis Sci. 2013;54(8):5721–5729. doi: 10.1167/iovs.13-12195. [DOI] [PubMed] [Google Scholar]
- 16.Shields CL, Furuta M, Thangappan A, Nagori S, Mashayekhi A, Lally DR, et al. Metastasis of uveal melanoma millimeter-by-millimeter in 8033 consecutive eyes. Arch Ophthalmol. 2009;127(8):989–998. doi: 10.1001/archophthalmol.2009.208. [DOI] [PubMed] [Google Scholar]
- 17.Shields CL, Say EAT, Hasanreisoglu M, Saktanasate J, Lawson BM, Landy JE, et al. Personalized prognosis of uveal melanoma based on cytogenetic profile in 1059 patients over an 8-year period: The 2017 Harry S. Gradle Lecture. Ophthalmology. 2017;124(10):1523–1531. doi: 10.1016/j.ophtha.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 18.Shields CL, Say EAT, Hasanreisoglu M, Saktanasate J, Lawson BM, Landy JE, et al. Cytogenetic abnormalities in uveal melanoma based on tumor features and size in 1059 patients: The 2016 W. Richard Green Lecture. Ophthalmology. 2017;124(5):609–618. doi: 10.1016/j.ophtha.2016.12.026. [DOI] [PubMed] [Google Scholar]
- 19.Onken MD, Worley LA, Ehlers JP, Harbour JW. Gene expression profiling in uveal melanoma reveals two molecular classes and predicts metastatic death. Can Res. 2004;64(20):7205–7209. doi: 10.1158/0008-5472.CAN-04-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Onken MD, Worley LA, Tuscan MD, Harbour JW. An accurate, clinically feasible multi-gene expression assay for predicting metastasis in uveal melanoma. J Mol Diagn. 2010;12(4):461–468. doi: 10.2353/jmoldx.2010.090220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Worley LA, Onken MD, Person E, Robirds D, Branson J, Char DH, et al. Transcriptomic versus chromosomal prognostic markers and clinical outcome in uveal melanoma. Clin Can Res. 2007;13(5):1466–1471. doi: 10.1158/1078-0432.CCR-06-2401. [DOI] [PubMed] [Google Scholar]
- 22.Decatur CL, Ong E, Garg N, Anbunathan H, Bowcock AM, Field MG, et al. Driver mutations in uveal melanoma: Associations with gene expression profile and patient outcomes. JAMA Ophthalmol. 2016;134(7):728–733. doi: 10.1001/jamaophthalmol.2016.0903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harbour JW, Onken MD, Roberson EDO, Duan S, Cao L, Worley LA, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330(6009):1410–1413. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalirai H, Dodson A, Faqir S, Damato BE, Coupland SE. Lack of BAP1 protein expression in uveal melanoma is associated with increased metastatic risk and has utility in routine prognostic testing. Br J Cancer. 2014;111(7):1373–1380. doi: 10.1038/bjc.2014.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koopmans AE, Verdijk RM, Brouwer RW, van den Bosch TP, van den Berg MM. Vaarwater et al. Clinical significance of immunohistochemistry for detection of BAP1 mutations in uveal melanoma. Mod Pathol. 2014;27(10):1321–1330. doi: 10.1038/modpathol.2014.43. [DOI] [PubMed] [Google Scholar]
- 26.Shah AA, David Bourne T, Murali R. BAP1 protein loss by immunohistochemistry: A potentially useful tool for prognostic prediction in patients with uveal melanoma. Pathology. 2013;45(7):651–656. doi: 10.1097/PAT.0000000000000002. [DOI] [PubMed] [Google Scholar]
- 27.van de Nes JA, Nelles J, Kreis S, Metz CH, Hager T, Lohmann DR, et al. Comparing the prognostic value of BAP1 mutation pattern, chromosome 3 status, and BAP1 immunohistochemistry in uveal melanoma. Am J Surg Pathol. 2016;40(6):796–805. doi: 10.1097/PAS.0000000000000645. [DOI] [PubMed] [Google Scholar]
- 28.van Essen TH, van Pelt SI, Versluis M, Bronkhorst IHG, van Duinen SG, Marinkovic M, et al. Prognostic parameters in uveal melanoma and their association with BAP1 expression. Br J Ophthalmol. 2014;98(12):1738–1743. doi: 10.1136/bjophthalmol-2014-305047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yavuzyigitoglu S, Koopmans AE, Verdijk RM, Vaarwater J, Eussen B, van Bodegom A, et al. Uveal melanomas with SF3B1 mutations: A distinct subclass associated with late-onset metastases. Ophthalmology. 2016;123(5):1118–1128. doi: 10.1016/j.ophtha.2016.01.023. [DOI] [PubMed] [Google Scholar]
- 30.Gupta MP, Lane AM, DeAngelis MM, Mayne K, Crabtree M, Gragoudas ES, et al. Clinical characteristics of uveal melanoma in patients with germline BAP1 mutations. JAMA Ophthalmol. 2015;133(8):881–887. doi: 10.1001/jamaophthalmol.2015.1119. [DOI] [PubMed] [Google Scholar]
- 31.Aoude LG, Vajdic CM, Kricker A, Armstrong B, Hayward NK. Prevalence of germline BAP1 mutation in a population-based sample of uveal melanoma cases. Pigment Cell Melanoma Res. 2013;26(2):278–279. doi: 10.1111/pcmr.12046. [DOI] [PubMed] [Google Scholar]
- 32.Njauw C-NJ, Kim I, Piris A, Gabree M, Taylor M, Lane AM, et al. Germline BAP1 inactivation is preferentially associated with metastatic ocular melanoma and cutaneous-ocular melanoma families. PLoS ONE. 2012;7(4):e35295. doi: 10.1371/journal.pone.0035295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turunen JA, Markkinen S, Wilska R, Saarinen S, Raivio V, Täll M, Lehesjoki A-E, et al. BAP1 germline mutations in finnish patients with uveal melanoma. Ophthalmology. 2016;123(5):1112–1117. doi: 10.1016/j.ophtha.2016.01.008. [DOI] [PubMed] [Google Scholar]
- 34.Field MG, Durante MA, Anbunathan H, Cai LZ, Decatur CL, Bowcock AM, et al. Punctuated evolution of canonical genomic aberrations in uveal melanoma. Nature Commun. 2018;9(1):116. doi: 10.1038/s41467-017-02428-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shields CL, Ganguly A, Materin MA, Teixeira L, Mashayekhi A, Swanson LA, et al. Chromosome 3 analysis of uveal melanoma using fine-needle aspiration biopsy at the time of plaque radiotherapy in 140 consecutive cases: the Deborah Iverson, MD. Lectureship. Arch Ophthalmol. 2007;125(8):1017–1024. doi: 10.1001/archopht.125.8.1017. [DOI] [PubMed] [Google Scholar]
- 36.Abdel-Rahman MH, Rai K, Pilarski R, Davidorf FH, Cebulla CM. Germline BAP1 mutations misreported as somatic based on tumor-only testing. Familial Cancer. 2016;15(2):327–330. doi: 10.1007/s10689-016-9865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ewens KG, Kanetsky PA, Richards-Yutz J, Purrazzella J, Shields CL, Ganguly T, et al. Chromosome 3 status combined with BAP1 and EIF1AX mutation profiles are associated with metastasis in uveal melanoma. Invest Ophthalmol Vis Sci. 2014;55(8):5160–5167. doi: 10.1167/iovs.14-14550. [DOI] [PubMed] [Google Scholar]
- 38.Chen Z, Moran K, Richards-Yutz J, Toorens E, Gerhart D, Ganguly T, et al. Enhanced sensitivity for detection of low-level germline mosaic RB1 mutations in sporadic retinoblastoma cases using deep semiconductor sequencing. Hum Mut. 2014;35(3):384–391. doi: 10.1002/humu.22488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and Guidelines for the Interpretation of Sequence Variants: A Joint Consensus. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dono M, Angelini G, Cecconi M, Amaro A, Esposito AI, Mirisola V, et al. Mutation frequencies of GNAQ, GNA11, BAP1, SF3B1, EIF1AX and TERT in uveal melanoma: detection of an activating mutation in the TERT gene promoter in a single case of uveal melanoma. Br J Cancer. 2014;110(4):1058–1065. doi: 10.1038/bjc.2013.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johansson P, Aoude LG, Wadt K, Glasson WJ, Warrier SK, Hewitt AW, et al. Deep sequencing of uveal melanoma identifies a recurrent mutation in PLCB4. Oncotarget. 2016;7(4):4624–4631. doi: 10.18632/oncotarget.6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robertson AG, Shih J, Yau C, Gibb EA, Oba J, Mungall KL, et al. Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell. 2017;32(2):204–220. doi: 10.1016/j.ccell.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Staby KM, Gravdal K, Mørk SJ, Heegaard S, Vintermyr OK, Krohn J. Prognostic impact of chromosomal aberrations and GNAQ, GNA11 and BAP1 mutations in uveal melanoma. Acta Ophthalmol. 2018;96(1):31–38. doi: 10.1111/aos.13452. [DOI] [PubMed] [Google Scholar]
- 44.Martin M, Maszhofer L, Temming P, Rahmann S, Metz C, Bornfeld N, et al. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat Genet. 2013;45(8):933–936. doi: 10.1038/ng.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wadt KA, Aoude LG, Johansson P, Solinas A, Pritchard A, Crainic O, et al. A recurrent germline BAP1 mutation and extension of the BAP1 tumor predisposition spectrum to include basal cell carcinoma. Clin Genet. 2015;88(3):267–272. doi: 10.1111/cge.12501. [DOI] [PubMed] [Google Scholar]
- 46.van Engen-van Grunsven ACH, Baar MP, Pfundt R, Rijntjes J, Küsters-Vandevelde HVN, Delbecq A-L, et al. Whole-genome copy-number analysis identifies new leads for chromosomal aberrations involved in the oncogenesis and metastastic behavior of uveal melanomas. Melanoma Res. 2015;25(3):200–209. doi: 10.1097/CMR.0000000000000152. [DOI] [PubMed] [Google Scholar]
- 47.Popova T, Hebert L, Jacquemin V, Gad S, Caux-Moncoutier V, Dubois-d’Enghien C, et al. Germline BAP1 mutations predispose to renal cell carcinomas. Am J Hum Genet. 2013;92(6):974–980. doi: 10.1016/j.ajhg.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheung M, Talarchek J, Schindeler K, Saraiva E, Penney LS, Ludman M, et al. Further evidence for germline BAP1 mutations predisposing to melanoma and malignant mesothelioma. Cancer Genet. 2013;206(5):206–210. doi: 10.1016/j.cancergen.2013.05.018. [DOI] [PubMed] [Google Scholar]
- 49.Laurent C, Gentien D, Piperno-Neumann S, Némati F, Nicolas A, Tesson B, et al. Patient-derived xenografts recapitulate molecular features of human uveal melanomas. Mol Oncology. 2013;7(3):625–636. doi: 10.1016/j.molonc.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wiesner T, Obenauf AC, Murali R, Fried I, Griewank KG, Ulz P, et al. Germline mutations in BAP1 predispose to melanocytic tumors. Nat Genet. 2011;43(10):1018–1021. doi: 10.1038/ng.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pilarski R, Cebulla CM, Massengill JB, Rai K, Rich T, Strong L, et al. Expanding the clinical phenotype of hereditary BAP1 cancer predisposition syndrome, reporting three new cases. Genes Chromosomes Cancer. 2014;53(2):177–182. doi: 10.1002/gcc.22129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Comparison of demographic and tumor variables of two cohorts of 142 UM. Description of demographic and tumor variables with pairwise comparisons for 142 divided into two cohorts depending on whether tumors were selected from archived samples for which both tumor and blood samples were available (cohort 1, N=90) or tumors for which BAP1 sequencing was specifically requested (cohort 2, N=52). References: 1. OMIM: Tumor predisposition syndrome; TPDS. https://www.omim.org/entry/614327. 2. Pilarski R, Rai K, Cebulla C, Abdel-Rahman M.BAP1 Tumor Predisposition Syndrome. 2016 Oct 13 In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, Amemiya A, editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993-2018. Available from: https://www.ncbi.nlm.nih.gov/books/NBK390611/. Figure S1. Kaplan-Meier analysis comparing metastasis-free survival of two cohorts of 142 UM. Kaplan-Meier curves showing metastasis-free survival following treatment for 142 UM stratified by cohort, as described in the legend for Additional file 1: Table S1. (DOCX 81 kb)
Data Availability Statement
The datasets analyzed for this study are available from the corresponding author upon reasonable request.


