Abstract
Four new metabolites, including three new oblongolides named C1, P1, and X1 (1-3) and 6-hydroxyphomodiol (10), along with eight known compounds – oblongolides B (4), C (5), D (6), O (7), P (8) and U (9), (3R,4aR,5S,6R)-6-hydroxy-5-methylramulosin (11), and (3R)-5-methylmellein (12) – were isolated from the endophytic fungal strain Phomopsis sp. XZ-01 of Camptotheca acuminate. Their structures were elucidated by spectroscopic analyses, including 1H- and 13C-NMR, 2D NMR (HSQC, HMBC, 1H-1H COSY and NOESY) and HR-FT-MS. Cytotoxic activities of these compounds were evaluated. Some of them showed weak selective activities.
Keywords: Camptotheca acuminate, endophytic fungus, Phomopsis sp. XZ-01, oblongolides, new metabolites
1. Introduction
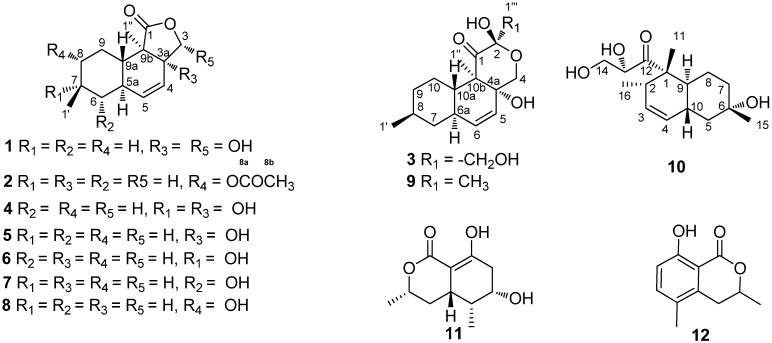
Endophytes, especially those found in medicinal plants, have drawn a lot of attention for the past few years as a rich and reliable source of bioactive and chemically novel compounds with huge medicinal and agricultural potential [1]. In the course of our exploration for bioactive or new chemical entities from the endophytic fungus of Camptotheca acuminate Decne (Cornaceae), numerous new compounds were obtained [2,3]. Continuous research on the secondary metabolisms of another endophytic fungus of Camptotheca acuminate (Phomopsis sp. XZ-01), led to the discovery of three new oblongolides C1 (1), P1 (2), and X1 (3), oblongolides B (4) [4], C (5) [4], D (6) [4], O (7) [3], P (8) [3] and U (9) [3], the new phomodiol 6-hydroxyphomodiol (10), (3R,4aR,5S,6R)-6-hydroxy-5-methyl-ramulosin (11) [5], and (3R)-5-methylmellein (12) [6]. In this paper, we report the isolation and structural elucidation of compounds 1-12 (Figure 1) and their anticancer activities.
Figure 1.
Structures of compounds 1-12.
2. Results and Discussion
We obtained oblongolide C1 (1) as white needles and determined it to have the molecular formula C14H20O4 by HR-FT-MS. The 13C-NMR, DEPT and HSQC spectra of compound 1 showed 14 carbon signals: two methyl groups, three methylene groups, three methine groups, one hemiacetal methine (δC 100.6), a disubstituted olefin (δC 137.8 and 124.2), an oxygenated quaternary carbon (δC 78.7), a lactone carbonyl (δC 176.6), and a quaternary carbon. The 1H-1H COSY correlations between H-4 and H-5, H-5 and H-5a, H-5a and H-6, H-5a and H-9a, H-6 and H-7, H-7 and H-1′, H-8 and H-7, H-8 and H-9, H-9a and H-9 established the structure of a 9-carbon moiety (Figure 2, in green). Key HMBC correlations from H-1″ to C-1, C-3a, C-9a and C-9b, from H-5 to C-3a, from H-4 to C-9b, and from H-3 to C-3a established the planar structure of 1. The relative configuration of 1 was deduced on the basis of NOESY spectroscopic data. The NOE correlations between H-7 and H-5a and between H-5a and H-1″ established the α-orientations of H-5a, H-7 and H-1″. NOESY cross-peaks from H-3 to H-9a and from H-9a to H-1′ indicated the β-orientations of H-3, H-9a and H-1′. A comparison of the 1H and 13C-NMR spectra of 1 with that of oblongolide C indicated that 1 was the 3α-hydroxy derivative of oblongolide C [4]. Therefore, we determined the structure of 1 to be 3α-hydroxyoblongolide C and it was named as oblongolide C1 for consistency with the literature [4].
Figure 2.
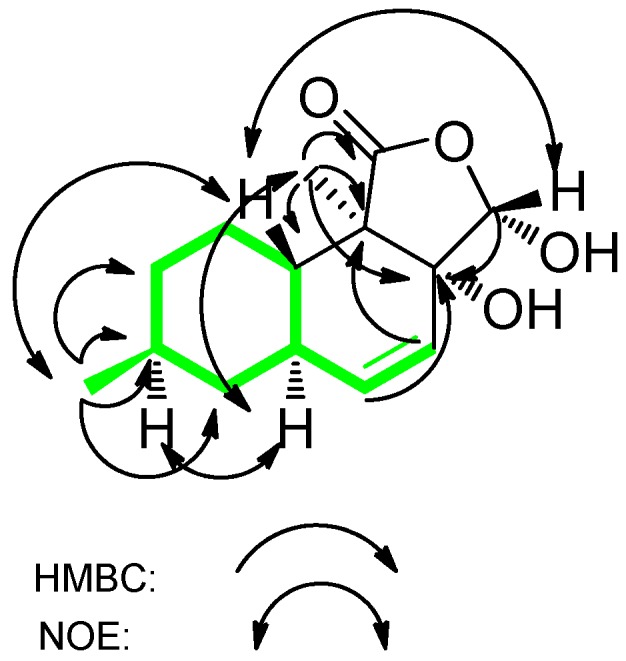
Key HMBC and NOE Correlations of compound 1.
Oblongolide P1 (2) was isolated as a white powder. The molecular formula C16H22O4 was deduced from HR-FT-MS and 13C-NMR. NMR data of 2 were similar to those of 1, except that the hemiacetal methine [δH 5.69 (1H, d, J = 11.5 Hz) and δC 100.6, CH-3], quaternary carbon (δC 78.7, C-3a) and methylene [δH 1.82 (1H, m), δH 0.91 (1H, m) and δC 34.6, CH-8] in 1 were replaced by oxymethylene [δH 4.44 (1H, t, J = 8.6 Hz), δH 3.85 (1H, dd, J = 10.9, 8.9 Hz) and δC 70.1, CH2-3], methine [δH 2.78 (1H, m) and δC 44.6, CH-3a], oxymethine [δH 4.54 (1H, dt, J = 10.8, 4.4 Hz) and δC 77.1, CH-8], and there was an acetyl group in 2. Key HMBC correlations from H-8 to C-8a, C-1′ and C-9a, from H-1′ to C-6, C-7 and C-8, from H-1″ to C-1, C-3a, C-9a and C-9b indicated the planar structure of 2. We determined the relative configuration of 2 by analysis of the NOESY spectrum. The NOE correlations between H-8 and H-1′, between H-8 and H-9a, between H-8 and H-9β, and between H-1′ and H-6β established the β-orientations of H-1′, H-8 and H-9a. The NOE correlations between H-3a and H-1″, between H-3α and H-3a and between H-1″ and H-5a indicated the α-orientations of H-1″, H-3a and H-5a. A comparison of the 1H- and 13C-NMR data of 2 with those of oblongolide P [3] revealed that these two compounds had similar structures, except that an acetyl group was attached to the C-8 hydroxyl group in 2. Therefore, we determined 2 to be 8-acetylobolngolide P and named it oblongolide P1.
Oblongolide X1 (3) was obtained as white oil. Its molecular formula, C16H24O5, was deduced on the basis of HR-FT-MS and 13C-NMR data. A comparison of the NMR data of 3 with those of known compound oblongolide X [7] indicated that 3 was a hydroxy-derivative of the latter. The HMBC correlations from H-1″′ to C-1 and C-2 located the hydroxyl substitution at C-2. The NOE correlations between H-10a and H-1′, between H-6a and H-8 and between H-6a and H-1″ determined the relative configuration of 3. Therefore, we determined 3 to be 1″′-hydroxyoblongolide X and named it oblongolide X1.
Compound 10 had the molecular formula C16H26O4, as established by HR-FT-MS and 13C-NMR spectra. 1H- and 13C-NMR data of 10 were similar to those of phomodiol [8], except that the methine signal [δH 1.46 (1H, m), CH-6] was replaced by a quaternary carbon (δC 70.0, C-6). Key HMBC correlations from H-15 to C-5, C-6 and C-7, from H-11 to C-1, C-2, C-9 and C-12, from H-16 to C-1, C-2 and C-3, from H-4 to C-2, C-5 and C-9 and from H-10 to C-3, C-6 and C-8 indicated the planar structure of 10. The relative configuration of 10 was deduced on the basis of NOESY spectroscopic data. The NOE correlations between H-10 and H-11, between H-2 and H-11, between H-13 and H-11, between H-15 and H-10 and between H-9 and H-16 indicated β-orientation of the hydroxyl group (6-OH) and the α-orientation of the side chain attached to C-1. Therefore, the structure of 10 was determined. We named it 6-hydroxyphomodiol [8].
Besides the nine oblongolides, including three new ones, we isolated two more polyketides. We determined 11 to be (3R,4aR,5S,6R)-6-hydroxy-5-methylramulosin (11) [5] by a comparison of NMR data. This compound was previously isolated from a marine-derived fungus which was derived from the green alga Codium fragile [5]. The spectroscopic data of 12 were identical to those of the known compound (3R)-5-methylmellein, first isolated as the main phytotoxic metabolite of Fusicoccum amygdale [6].
Cytotoxicity
The results of cytotoxic tests of compounds 1-12 are shown in Table 4. They exhibited no significant activity against the three tested cancer cell lines.
Table 4.
Biological Activities of Compounds 1-12.
| Compound | Inhibitory rate(%) | ||
|---|---|---|---|
| HeLa | A549 | HepG2 | |
| Oblongolide C1 (1) | - | - | 18.01 ± 0.86 |
| Oblongolide P1 (2) | - | - | 28.59 ± 1.04 |
| Oblongolide X1 (3) | - | - | 27.89 ± 1.2 |
| Oblongolide B (4) | - | - | - |
| Oblongolide C (5) | - | 14.92 ± 0.86 | - |
| Oblongolide D (6) | 22.9 ± 0.78 | 13.82 ± 1.01 | - |
| Oblongolide O (7) | - | - | - |
| Oblongolide P (8) | - | - | - |
| Oblongolide U (9) | - | 18.76 ± 0.56 | 16.89 ± 1.01 |
| 6-Hydroxyphomodiol (10) | - | - | 23.86 ± 1.21 |
| (3R,4aR,5S,6R)-6-Hydroxy-5-methylramulosin (11) | - | - | - |
| (3R)-5-Methylmellein (12) | - | - | - |
3. Experimental
3.1. General
Optical rotations were measured with a Perkin-Elmer 341 automatic polarimeter in methanol. IR spectra were recorded on a Nicolet AVATAR 330FT spectrometer. NMR spectra were taken on a Bruker Avance III-600 NMR spectrometer with TMS as an internal standard. HR-FT-MS data were acquired by using En Apex ultra 7.0 FT-MS. TLC was carried out using glass-precoated silica gel GF254 (Qingdao) and visualized under UV light or by spraying with vanillin (contains H2SO4) ethanol reagent. Sephadex LH-20 (40-70 µm, Amersham Pharmacia Biotech AB, Uppsala, Sweden), silica gel (200-300mesh, Qingdao Marine Chemical, Inc., Qingdao, China), and lichroprep reversed-phase RP-18 silica gel (40-63 µm, Merck, Darmstadt, Germany) were used for column chromatography (CC).
3.2. Fungal Material
The fungus (XZ-01) was isolated from current-year twigs (8-12 × 1-2 cm, length × diameter) of Camptotheca acuminate collected from the Jiangshi Natural Reserve, Shaowu, Fujian, China. It was identified as a non-sporulating fungus by traditional morphology. A BLAST search result showed that the internal transcribed spaces (ITS) sequence of XZ-01 was highly homologous (98% percent similarity) to that of a Phomopsis species (BCC 9789 [GU086404]), indicating that XZ-01 belongs to this genus.
3.3. Fermentation and Extraction
XZ-01 was cultivated on potato dextrose agar at 28 °C. The agar blocks were chopped and transferred into Erlenmeyer flasks (10 × 3 L), each containing 1 L of potato dextrose broth (PDB), and then fermented at 28 °C on a rotary shaker (150 rpm) for 7d. The culture was filtered to separate broth and mycelia. The culture broth was extracted with EtOAc (6 × 10 L) for six times. The combined organic layer was concentrated under vacuum to afford 3.2 g of residue.
3.4. Isolation and Spectral Data
The crude extract was separated into fifteen fractions (1-15) by column chromatography on RP-18 silica gel, eluted by methanol/H2O (0:100, 30:70, 50:50, 70:30, and 100:0). Fraction 3 (100 mg) was subjected to silica gel CC (step gradient, elution with 0-10% MeOH in CHCl3) to afford eleven fractions (3-1-3-11). Fractions 3-11 (4.9 mg) were further separated by silica gel CC (step gradient, elution with 22.2-33.3% EtOAc in hexane) to yield 4 (2.3 mg). Fraction 5 (92.1 mg) was separated by Sephadex LH-20 (elution with 100% methanol) to give three subfractions (fraction 5-1–5-3). Fraction 5-2 (23.6 mg) was purified by silica gel CC (step gradient, 7.7-50% EtOAc in hexane) to produce fraction 5-2-1. Fraction 5-2-1 (3.7 mg) was separated by silica gel (eluted with 50% CHCl3 in petroleum ether) to afford 11 (2mg). Fraction 6 (225.8 mg) was fractionated by Sephadex LH-20 CC (elution with 100% MeOH) to provide nine fractions (6-1–6-9). Fraction 6-1 (28.8 mg) was further purified by silica gel CC (step gradient, 0-17% MeOH in CHCl3) to furnish 6 (11.5 mg), 8 (2.6 mg) and 10 (6.4 mg). Fraction 7 (247.1 mg) was subjected to Sephadex LH-20 CC (elution with 100% MeOH) to give 5 fractions (7-1–7-5). Fraction 7-4 (36.1 mg) was purified by silica gel CC (elution with CHCl3) to yield 7 (3.1 mg). Fraction 10 (109 mg) was fractionated by Sephadex LH-20 CC (elution with 100% MeOH) to provide two fractions (10-1–10-2). Fraction 10-1 (72 mg) was further purified by silica gel CC (step gradient, elution with 0-10% MeOH in CHCl3) to give two subfractions (10-1-1 and 10-1-2). Fraction 10-1-2 (11.7 mg) was separated by silica gel CC (elution with 100% CHCl3) to yield 3 (3.8 mg). Fraction 11 (318.3 mg) was separated by Sephadex LH-20 (elution with 100% MeOH) to provide five fraction (11-1–11-5). Fraction 11-5 (23.9 mg) was further purified by silica gel CC (elution with 10% CHCl3 in petroleum ether) to afford 12 (22.8 mg). Fraction 11-3 (99 mg) was separated by silica gel CC (step gradient, elution with 0-10% MeOH in CHCl3) to give 5 (34 mg) and 9 (2.3 mg). Fraction 12 (117 mg) was fractionated by Sephadex LH-20 CC (elution with 100% MeOH) to provide three fractions (12-1–12-3). Fraction 12-1 (12.8 mg) was further separated by silica gel CC (elution with 33.3% CHCl3 in petroleum ether) to yield 2 (7.4 mg). Fraction 9 (232 mg) was separated by Sephadex LH-20 (elution with 100% MeOH) to give two fractions (9-1–9-2). Fraction 9-2 (38 mg) was purified by silica gel CC (step gradient, 0-12.3% MeOH in CHCl3) to yield 1 (5.7 mg).
Oblongolide C-1 (1): White needles; [α]D20: – 22.6 (c 0.0072, MeOH). IR (KBr) νmax 2919, 2359, 1219, 772, 668 cm–1. 1H- and 13C-NMR: see Table 1; HR-FT-MS: m/z = 251.1281 [M − H]- (calcd. for C14H19O4, 251.1283, Temperature: 180, Resolution: 125,508).
Table 1.
1H- and 13C-NMR spectroscopic data of compounds 1 and 2 (1 and 2 at 600 MHz, CDCl3, chemical shift values are in ppm relative to TMS; multiplicity and J values (in Hz) are presented in parentheses.
| No. | 1 | 2 | ||
|---|---|---|---|---|
| δH | δC | δH | δC | |
| 1 | 176.6 | 179.4 | ||
| 3α | 4.44 (t, 8.6) | 70.1 | ||
| 3β | 5.69 (d, 11.5) | 100.6 | 3.85 (dd, 8.9, 10.9) | 77.1 |
| 3a | 78.7 | 2.78 (m) | 44.6 | |
| 4 | 5.53 (dd, 10.2, 2.8) | 124.2 | 5.62 (dd, 12.8, 2.5) | 122.2 |
| 5 | 5.79 (d, 9.9) | 137.8 | 5.65 (d, 12.8) | 133.0 |
| 5a | 2.03 (m) | 36.3 | 1.97 (m) | 35.4 |
| 6α | 0.84 (q, 12.4) | 41.0 | 1.00 (m) | 39.0 |
| 6β | 1.90 (m) | 41.0 | 1.94 (m) | 39.0 |
| 7 | 1.50 (m) | 32.7 | 1.68 (m) | 37.4 |
| 8α | 0.91 (m) | 34.6 | ||
| 8β | 1.82 (m) | 34.6 | 4.54 (dt, 4.4, 10.8) | 77.1 |
| 9α | 1.34 (dd, 12.4, 3.1) | 25.6 | 1.38 (q, 12.2) | 31.1 |
| 9β | 1.79 (m) | 25.6 | 2.10 (m) | 31.1 |
| 9a | 1.49 (m) | 44.1 | 1.50 (m) | 37.3 |
| 9b | 51.5 | 42.8 | ||
| 1′ | 0.93 (d, 6.6) | 22.2 | 0.94 (d, 6.5) | 18.0 |
| 1″ | 1.18 (s) | 9.5 | 1.16 (s) | 16.1 |
| 8a | 170.5 | |||
| 8b | 2.06 (s) | 21.1 | ||
Oblongolide O-1 (2): White powder; [α]D20: – 72.2(c 0.0025, MeOH). IR (KBr) νmax 3344, 2922, 1588, 1383, 772 cm–1. 1H- and 13C-NMR: see Table 1; HR-FT-MS: m/z = 301.1418 [M + Na]+ (calcd. for C16H22O4Na, 301.1416, Temperature: 180, Resolution: 14,100).
Oblongolide X-1 (3): White oil; [α]D20: – 21.7(c 0.0056, MeOH). IR (KBr) νmax 3422, 1583, 773, 685 cm–1. 1H- and 13C-NMR: see Table 2; HR-FT-MS: m/z = 295.1541 [M − H]- (calcd. for C16H21O5, 295.1545, Temperature: 180, Resolution: 106,466).
Table 2.
1H- and 13C-NMR spectroscopic data of compound 3 (3 at 600 MHz, CDCl3, chemical shift values are in ppm relative to TMS; multiplicity and J values (in Hz) are presented in parentheses.
| No. | 3 | |
|---|---|---|
| δH | δC | |
| 1 | - | 207.0 |
| 2 | - | 94.9 |
| 4α | 3.57 (d, 12.4) | 66.1 |
| 4β | 4.63 (d, 12.4) | 66.1 |
| 4a | 78.2 | |
| 5 | 5.36 (dd, 10.1, 2.8) | 126.9 |
| 6 | 5.68 (dd, 10.1, 1.6) | 136.2 |
| 6a | 1.93 (m) | 37.9 |
| 7α | 1.86 (m) | 41.1 |
| 7β | 0.89 (m) | 41.1 |
| 8 | 1.49 (m) | 33.0 |
| 9α | 1.77 (m) | 34.8 |
| 9β | 1.03 (m) | 34.8 |
| 10α | 1.26 (m) | 26.8 |
| 10β | 1.23 (m) | 26.8 |
| 10a | 2.33 (ddd, 11.5, 10.6 3.0) | 43.8 |
| 10b | 55.4 | |
| 1′ | 0.93 (d, 6.5) | 22.3 |
| 1‴ | 1.09 (s) | 10.4 |
| 1‴ | 3.61 (d, 11.9) | 65.2 |
| 1‴ β | 3.95 (d, 11.9) | 65.2 |
6-Hydroxyphomodiol (10): Transparent oil; [α]D20: + 43.3(c 0.002, MeOH). IR (KBr) νmax 2365, 1223, 771 cm–1. 1H- and 13C-NMR: see Table 3; HR-FT-MS: m/z = 305.1736 [M + Na]- (calcd. for C16H26NaO4, 305.1729, Temperature: 180, Resolution: 36,000).
Table 3.
1H- and 13C-NMR spectroscopic data of compound 10 (600 MHz, in CDCl3, chemical shift values are in ppm relative to TMS; multiplicity and J values (in Hz) are presented in parentheses.
| No. | 10 | |
|---|---|---|
| δH | δC | |
| 1 | – | 5.15 |
| 2 | 2.18 (m) | 39.5 |
| 3 | 5.58 (ddd, 9.9, 4.9, 2.6) | 130.2 |
| 4 | 5.36 (d, 10.0) | 129.0 |
| 5α | 1.28 (m) | 45.5 |
| 5β | 1.75 (m) | 45.5 |
| 6 | – | 70.0 |
| 7α | 1.56 (dt, 13.6, 4.4) | 39.4 |
| 7β | 1.69 (dd, 14.1, 3.0) | 39.4 |
| 8α | 1.09 (brs) | 22.8 |
| 8β | 1.32 (m) | 22.8 |
| 9 | 1.79 (m) | 40.5 |
| 10 | 2.22 (m) | 33.0 |
| 11 | 1.35 (s) | 16.7 |
| 12 | – | 214.0 |
| 13 | 4.52 (brs) | 75.7 |
| 14 | 4.03 (dd, 11.8, 3.6), 3.79 (dd, 11.7, 4.7) | 63.3 |
| 15 | 1.27 (s) | 31.6 |
| 16 | 0.84 (d, 7.0) | 18.7 |
3.5. Biological Assay
Cancer cell lines were derived from the cell bank of The Chinese Academy of Sciences. Cells were seeded at a density of 5 × 103/100 µL medium in 96-well microtiter plate and treated with the compounds at the concentration of 20 µg/mL. Viable cells were incubated with MTT (5 mg/mL) for 4 h and formazan precipitate was dissolved in 100 µL DMSO and the absorbance at 490 nm was measured by Multimode Detector DTX880 (Beckman Coulter).
4. Conclusions
Four new compounds, oblongolides C1 (1), P1 (2), X1 (3), 6-hydroxyphomodiol (10), together with eight known compounds were isolated from the endophytic fungus Phomopsis sp. XZ-01. oblongolides C1 (1), P1 (2), X1 (3), and 6-hydroxyphomodiol (10) showed modest selective activities against HepG2 cancer cell lines. Oblongolide C (5) exhibited minor selective activity against A549.
Footnotes
Sample Availability: Samples of the compounds 1-12 are available from the authors.
References
- 1.Tan R.X., Zou W.X.B. Endophytes: a rich source of functional metabolites. Nat. Prod. Rep. 2001;18:448–459. doi: 10.1039/b100918o. [DOI] [PubMed] [Google Scholar]
- 2.Tan Q.F., Yan X.F., Lin X., Huang Y.J., Zheng Z.H., Song S.Y., Lu C.H., Shen Y.M. Chemical constituents of the endophytic fungal strain Phomopsis sp NXZ-05 of Camptotheca acuminate. Helv. Chim. Acta. 2007;90:1811–1817. doi: 10.1002/hlca.200790190. [DOI] [Google Scholar]
- 3.Lin T., Lin X., Lu C., Hu Z., Huang W., Huang Y., Shen Y. Secondary Metabolites of Phomopsis sp.XZ-26, an Endophytic Fungus from Camptotheca acuminate. Eur. J. Org. Chem. 2009;2009:2975–2982. doi: 10.1002/ejoc.200801021. [DOI] [Google Scholar]
- 4.Dai J., Krohn K., Gehle D., Kock I., Flörke U., Aust H.J., Draeger S., Schulz B., Rheinheimer J. New Oblongolides Isolated from the Endophytic Fungus Phomopsis sp. from Melilotus dentata from the Shores of the Baltic Sea. Eur. J. Org. Chem. 2005;2005:4009–4016. [Google Scholar]
- 5.Elbeih A.A., Kato H., Ohta T., Tsukamoto S. (3R,4aR,5S,6R)-6-Hydroxy-5-methylramulosin: a New Ramulosin Derivative from a Marine-Derived Sterile Mycelium. Chem. Pharm. Bull. 2007;55:953–954. doi: 10.1248/cpb.55.953. [DOI] [PubMed] [Google Scholar]
- 6.Ballio A., Barcellona S., Santurban B. 5-Methylmellein, a new natural dihydroisocoumarin. Tetrahedron Lett. 1966;7:3723–3726. doi: 10.1016/S0040-4039(01)99954-X. [DOI] [Google Scholar]
- 7.Bunyapaiboonsri T., Yoiprommarat S., Srikitikulchai P., Srichomthong K., Lumyong S. Oblongolides from the Endophytic Fungus Phomopsis sp. BCC9789. J. Nat. Prod. 2010;73:55–59. doi: 10.1021/np900650c. [DOI] [PubMed] [Google Scholar]
- 8.Horn W.S., Schwartz R.E., Simmonds M.S.J., Blaney W.M. Isolation and Characterization of Phomodiol, a New Antifungal from Phomopsis. Tetrahedron Lett. 1994;35:6037–6040. doi: 10.1016/0040-4039(94)88068-9. [DOI] [Google Scholar]