Abstract
Isotope labeling of biologically interesting proteins is a prerequisite for structural and dynamics studies by NMR spectroscopy. Many of these proteins require mammalian cofactors, chaperons, or posttranslational modifications such as myristoylation, glypiation, disulfide bond formation, or N- or O-linked glycosylation; and mammalian cells have the necessary machinery to produce them in their functional forms. Here, we describe recent advances in mammalian expression, including an efficient adenoviral vector-based system, for the production of isotopically labeled proteins. This system enables expression of mammalian proteins and their complexes, including proteins that require posttranslational modifications. We describe a roadmap to produce isotopically labeled 15N and 13C posttranslationally modified proteins, such as the outer domain of HIV-1 gp120, which has four disulfide bonds and 15 potential sites of N-linked glycosylation. These methods should allow NMR spectroscopic analysis of the structure and function of posttranslationally modified and secreted, cytoplasmic, or membrane-bound proteins.
1. INTRODUCTION
NMR spectroscopic characterization of proteins and protein domains generally requires the incorporation of the stable isotopes 15N, 13C, and/or 2H. Incorporating these isotopes is most easily done in prokaryotic systems; therefore, in most cases, full-length proteins or their individual domains are expressed and purified from prokaryotic expression systems, and a number of these methods are discussed in other chapters in this volume. Owing to structural genomics initiatives, large-scale expression trials have shown that only 10% of eukaryotic proteins can be expressed in their functional forms in Escherichia coli (Braun & LaBaer, 2003). This observation likely reflects the necessity of eukaryotic cofactors, chaperons, or posttranslational modifications such as myristoylation, disulfide bond formation, and glycosylation for proper folding and activity. Traditional eukaryotic systems such as insect, lower eukaryotes, and mammalian cells often produce low yields of the desired protein; in addition, these systems can be time consuming, prohibitively expensive, or both. Lower eukaryotes such as Dictyostelium discoideum (D. discoideum) have been used to produce isotopically enriched proteins in milligram quantities to allow resonance assignments from triple-resonance experiments (Cubeddu et al., 2000). D. discoideum has the potential to be an attractive expression system because isotopically enriched bacteria can be used as a nutrient source, thereby reducing the overall cost of isotopic labeling. However, heterologous expression of proteins in D. discoideum has its own challenges (Arya, Bhattacharya, & Saini, 2008); to date, there is only one published report describing successful production of an isotopically labeled protein using this system (Swarbrick et al., 2009). Thus, expression of isotopically enriched eukaryotic proteins in their stable, correctly folded form remains a bottleneck for spectroscopic characterization of many proteins. The development of large-scale mammalian transient expression systems along with advances in cell culture technologies have allowed milligram-to-gram-scale production of proteins suitable for structural analysis from Chinese hamster ovary (CHO) and human embryonic kidney (HEK) cells. Despite these advances, the absence of a suitable expression system than can incorporate labeling with high yield at a reasonable cost has stymied the production of isotopically enriched proteins suitable for NMR spectroscopy. We reported the adaptation of an efficient adenoviral vector-based mammalian expression system for the production of isotopically enriched cytoplasmic proteins and secreted glycoproteins (Sastry et al., 2011). This chapter gives a brief overview of mammalian expression systems and provides details for an adenoviral vector-based expression system through a case study of the human immunodeficiency virus type 1 (HIV-1) gp120 outer domain (OD) comprising 230 amino acids, 15 potential sites of N-linked glycosylation, and four disulfide bonds.
2. OVERVIEW OF MAMMALIAN EXPRESSION
Unlike prokaryotic systems, mammalian cells have the necessary cofactors, chaperons, and cellular machinery to produce correctly folded, posttranslationally modified functional proteins. Historically, heterologous expression of proteins using mammalian cells has proven difficult. However, the development of several eukaryotic expression systems—including yeast, D. discoideum, insect cells including Schneider cells, SF9, and silkworm-based baculovirus expression systems (Kato, Kajikawa, Maenaka, & Park, 2010; Unger & Peleg, 2012), and mammalian cells with their associated vectors which allow for the transient, inducible, or constitutive expression of a target gene (Kriz et al., 2010; Nettleship, Assenberg, Diprose, Rahman-Huq, & Owens, 2010; Trowitzsch, Klumpp, Thoma, Carralot, & Berger, 2011)—has allowed researchers to investigate the possibility of overexpressing proteins in eukaryotic systems. Large-scale protein production for therapeutic purposes generally uses constitutive or inducible expression from CHO, HEK, baby hamster kidney (BHK21), or human lymphoma (Namalwa) cells. Therefore, a number of research groups have succeeded in producing isotopically labeled proteins from mouse hybridoma or CHO stable cell lines using either a mixture of algal/bacterial hydrolysates (Hansen et al., 1992, 1994; Yamaguchi et al., 2006) or a mixture of labeled amino acids (Hinck et al., 1996; Lustbader et al., 1996; Shindo, Masuda, Takahashi, Arata, & Shimada, 2000; Wyss et al., 1995, 1993; Wyss, Dayie, & Wagner, 1997). However, these methods can be time consuming and prohibitively expensive leaving room for improvement in methodology (Table 1).
Table 1.
Mammalian Expression Systems Used to Obtain Isotopically Enriched Proteins
| Media | Cell Line | Protein Expression | 15N Media (mg/L) | 15N/13C Media (mg/L) |
|---|---|---|---|---|
| Algal and bacterial mixture of amino acidsa | Sp2/0 | Stable | 30 | 30 |
| Algal mixture of amino acidsb | CHO | Stable | 10 | 10 |
| CIL Bioexpress 6000 (15N/15N, 13C GKLQSTVW)c | HEK293 | Stable | 2 | 2.12 |
| Commercial Media (CIL) CGM6000d | A549 | Transient Adenoviral Expression | 50 | 43 |
Sastry et al. (2011). Adapted from Sastry et al. (2011) with permission from Springer.
2.1. Transient Protein Expression in Mammalian Cells
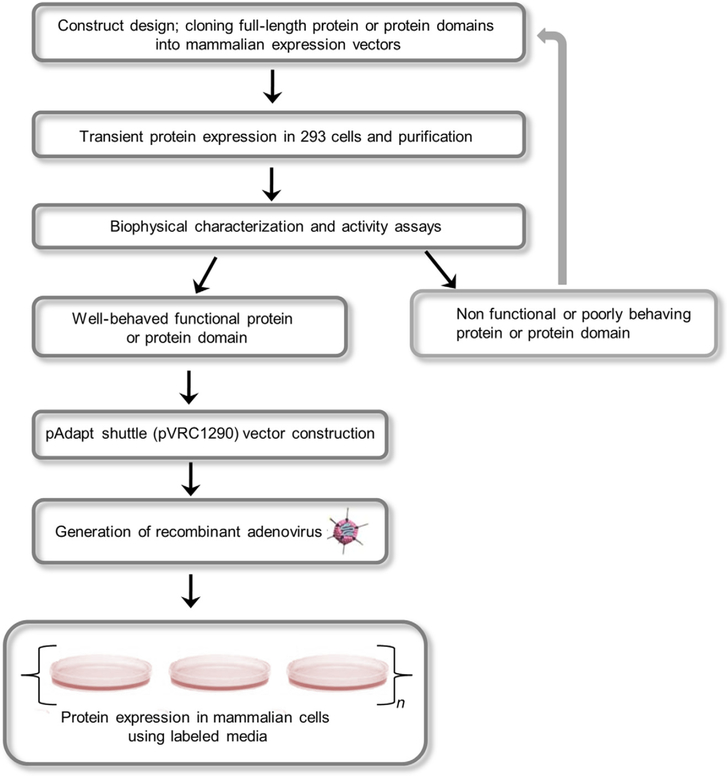
Protein production using transient transfection (Kingston, Chen, & Okayama, 2001; Potter & Heller, 2003) is an attractive alternative method and has been used routinely in the production of posttranslationally modified proteins including antibodies, glycoproteins, and membrane proteins. Transient transfection of HEK293 cells with polyethyleneimine (PEI)-mediated transfections has been used to obtain milligram-to-gram quantities of secreted proteins (Backliwal et al., 2008; Coleman et al., 1997). Transient transfection can be achieved in adherent as well as suspension cells (Geisse & Fux, 2009; Hopkins, Wall, & Esposito, 2012) to produce biologically important proteins that are functionally active and has been used extensively in biophysical and crystallographic studies (Kwon et al., 2015; Pancera et al., 2014; Zhou et al., 2015). A number of articles describing transient transfections and stable cell line development have been published (Longo, Kavran, Kim, & Leahy, 2013a, 2013b). Factors governing gene expression in eukaryotic cells are choice of expression vector, the presence of an optimized Kozak sequence upstream of the start codon, codon-optimization of your gene of interest and inclusion of targeting sequences such as signal peptide or endoplasmic reticulum localization sequences similar to requirements for prokaryotic expression systems discussed in preceding chapters. Protein expression is driven by a strong promoter such as the SV40 early promoter, the Rous sarcoma virus (RSV) promoter, or the cytomegalovirus (CMV) very early promoter (Xia et al., 2006). We provide a roadmap for overexpression and isotopic labeling of proteins from mammalian cells (Fig. 1) and discuss individual steps in the following sections.
Figure 1.
Schematic overview of isotopic labeling of proteins in mammalian cells. The gene of interest (GOI) is initially cloned into a suitable mammalian expression vector. The protein of interest is expressed using transient transfection and its functional and biophysical properties are analyzed. A construct either enters the adenoviral-based mammalian expression pathway to generate recombinant adenovirus followed by protein production or enters an iterative loop for further modifications to obtain an optimal construct.
2.1.1. Procedure for Transient Expression of Proteins and Protein Domains in Mammalian Cells
We outline a procedure for transient expression in this section and include relevant references and discuss details for production of isotopically enriched proteins using an adenoviral vector-based mammalian expression system with the HIV-1 gp120 OD as an example.
Design and clone the gene for the protein of interest (POI) in a mammalian expression vector of choice (Li et al., 2007; Van Craenenbroeck, Vanhoenacker, & Haegeman, 2000). A designed construct should include an upstream Kozak sequence such as gccgccA/GccATGG (Kozak, 1984, 1987), localization sequences particular to the protein (for example, a signal peptide for a secreted protein), a codon-optimized gene, and a poly A sequence 3’ to the gene of interest (GOI). For our case study, the HIV-1 gp120 OD was cloned into a CMV/R vector (pVRC8400) (Wu et al., 2006), protein expression was under the control of the CMV promoter. For easier tracking of an expressed protein, individual researchers may utilize C-terminal fusion of POI with green fluorescent protein (GFP) or yellow fluorescent protein. As with bacterial expression optimizing, the N- and C-termini may influence expression levels and protein stability. It is also highly recommended to generate varying lengths of a construct and test each for optimal levels of expression and ideally, function.
2.1.2. Protein Expression and Construct Selection
It is recommended to use high-quality endotoxin-free plasmid DNA preferably at 1μg/μl concentration to transfect HEK293T or HEK293FS cells. Small-scale expression to screen constructs can be performed using Lipofectamine, PEI, True Fect max, or 293 Fectin using established methods (Longo et al., 2013b; McLellan et al., 2013). The amount of DNA and the ratio of DNA to transfection reagent may need to be screened for optimal expression of the POI. Protein expression of the target gene can be monitored and confirmed by western blots or BioLayer Interferometry during the duration of expression. Once protein expression is confirmed, the crude supernatant or cell lysate can be further evaluated for the presence of a correctly folded POI by surface plasmon resonance (SPR) analysis (Raghavan & Bjorkman, 1995; Rich & Myszka, 2010), BioLayer Interferometry, ELISAs, or an equivalent technique using, for example, a structure-dependent antibody.
An individual researcher may also choose to characterize the expressed protein further using 1H–15N and 1H–13C HSQC spectra at natural abundance (Chen, Freedberg, & Keire, 2015) as protein expression from adherent cells using 15N, 15N/13C-labeled media is feasible although potentially prohibitively expensive. Yet, additional essential steps in obtaining isotopically labeled proteins from mammalian cells include choosing optimal growth media for your cell line and tracking cell viability during the duration of protein expression (Sastry et al., 2011). Furthermore, a researcher may choose to investigate suitable media and cell types in the searchable database (http://www.goodcellculture.com). Despite the advances in cell culture technology and the high yields obtained with transient transfection, lack of suitable labeling media for suspension cells complicates production and screening of labeled proteins for NMR spectroscopic measurements. As a result, we chose to adapt and optimize transient protein production using a mammalian expression system that utilizes mammalian viruses to deliver the GOI.
2.2. Transient Protein Expression Using Mammalian Viruses
Mammalian viruses such as adenovirus, herpesviruses, poxviruses, papillomaviruses, and SV40 have been used to express heterologous proteins for almost four decades (Gluzman, Reichl, & Solnick, 1982; Howley, Sarver, & Law, 1983; Mulligan, Howard, & Berg, 1979; Southern & Berg, 1982; Warnock, Daigre, & Al-Rubeai, 2011). Adenoviruses used in our expression system are double-stranded DNA viruses (Berk, 1986) with a deletion of the E1 gene that renders it replication defective (Aoki, Barker, Danthinne, Imperiale, & Nabel, 1999; Sastry, Bewley, & Kwong, 2012). Recombinant adenovirus 5 (rAd5) initially binds cell surface coxsackie-adenovirus receptor (CAR), followed by an interaction with cellular integrins and subsequent internalization via receptor-mediated endocytosis. Adenovirus infection is epichromosomal in all known cells, and the replication-defective virus results in a very effective transfection system for introducing a functional gene into cells. Transfection with recombinant adenovirus is quick and simple, with a postinfection viability ~100%. Thus, a replication-defective recombinant adenovirus has the ability to transfer genetic material into cells with negligible apparent toxic effects. Viral promoters drive protein expression at the expense of production of cellular proteins and can increase the overall yield of the heterologous protein (Babich, Feldman, Nevins, Darnell, & Weinberger, 1983; Huang & Schneider, 1990). The following section focuses on the adenoviral expression system that we have adapted to produce isotopically enriched glycoproteins.
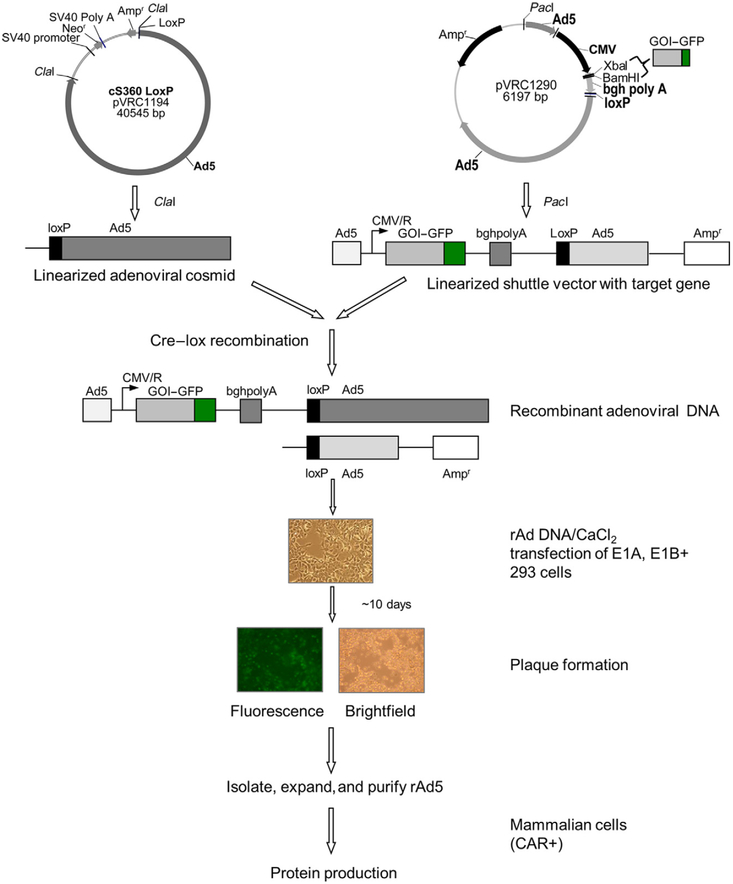
2.2.1. Construction of the Adenoviral Shuttle Vector
A schematic outline of the composition of an adenoviral shuttle vector is shown in Fig. 2. Once an optimal construct for the GOI is identified from the transient transfection study, the target gene is subcloned into pVRC1290 shuttle vector using suitable restriction sites such that the Kozak sequence, localization sequences, and purification tags are retained (Aoki et al., 1999; Sastry et al., 2012). Protein expression using this shuttle vector can once again be tested using small-scale transient transfection (McLellan et al., 2013) in HEK293 adherent cells or any other cell line of choice.
Figure 2.
Recombinant adenovirus as a tool to obtain isotopically labeled proteins. The target gene is cloned into a shuttle vector (pVRC1290) using the restriction sites (e.g., Xba1, BamH1) in the multiple cloning site; adenoviral cosmid DNA (pVRC1194) and shuttle vector are each linearized with ClaI and PacI, respectively, then recombined in vitro with Cre–Lox recombinase to obtain recombinant adenoviral genome (Aoki et al., 1999). The recombined adenoviral type 5 DNA is transfected into 293 adherent helper mammalian cells and recombinant adenovirus is isolated and purified using well-established methods. Target protein production is achieved by infecting CAR+ mammalian cells, such as A549 or CHO(CAR+).
2.2.2. Generation of Recombinant Adenoviral Genome Containing the GOI
A schematic outline of the generation of a recombinant adenoviral genome is shown in Fig. 2. The adenoviral cosmid (pVRC1194) consists of 9.2–100 mu (map units) of the adenoviral genome, with a deletion in the E1 region and a loxP site at 9.2 mu. The cosmid DNA (~26 kb) is digested with the restriction enzyme ClaI (New England Biolabs), and reaction is monitored by agarose gel electrophoresis. ClaI-cleaved cosmid DNA is purified using standard phenol/chloroform purification followed by ethanol precipitation. The shuttle plasmid (pVRC1290) containing the GOI–GFP or GOI is linearized with restriction enzyme PacI (New England Biolabs), reaction is monitored by agarose gel electrophoresis, and the linearized product is purified by phenol/chloroform purification (Moore & Dowhan, 2002; Voytas, 2001). Equimolar amounts of the shuttle plasmid and the adenoviral cosmid are recombined in vitro using Cre recombinase (New England Biolabs), recombination reaction is monitored using gel electrophoresis and upon completion, the recombinant adenoviral genome is purified by standard phenol/chloroform extraction and ethanol precipitation. The recombinant adenoviral DNA obtained from the Cre–Lox recombination reaction contains the GOI flanked by the adenoviral 5’ inverted terminal repeat (ITR), a 0- to 1-mu packaging signal, the bovine growth hormone polyadenylation signal (bghpolyA), and 9.2–100 mu of the adenoviral genome. These flanking sequences consist of DNA packaging sequences as well as sites for recombination with the rest of the viral DNA to reconstitute replication-defective adenoviruses. Generation of recombinant adenovirus can generally be a lengthy process. Therefore, prior to embarking on recombinant adenoviral production, traditional transient transfection into either HEK293 adherent or suspension cells is highly recommended in order to optimize the construct and perform functional analysis of the expressed protein.
2.2.3. Generation of Adenoviruses Containing GOI
Recombinant adenoviral DNA obtained from the Cre–Lox reaction is transfected into helper 293 cells (Fig. 2) using calcium phosphate transfection methodology. HEK293 cells are transfected with varying amounts of rAd5 DNA and monitored over a time course of nearly 10 days. HEK293 cell line with its indigenous E1 protein complements the absence of E1 gene in the recombinant adenoviral DNA (Graham, Smiley, Russell, & Nairn, 1977) and allows for the production of recombinant adenovirus. If the recombination reaction and calcium phosphate-mediated transfection are successful, rAd5 viruses are released via lysis of HEK293 cells resulting in a near circular clearing in the monolayer of cells. This region of visible lysis seen clearly in the case of a domain of HIV-1 gp120-GFP construct in Fig. 2 is known as a plaque and is indicative of virus formation. Upon plaque formation cells are harvested, the recombinant crude virus is isolated and protein expression of the desired gene tested by infecting A549 (CAR+) lung carcinoma cells. Once protein expression is confirmed, the crude virus preparation is used to infect low-passage HEK293 cells to produce recombinant adenovirus as described previously (Sastry et al., 2012). Briefly, HEK293 cells are seeded at 2.0 × 107 cells per 15-cm plate 24 h prior to infection. Cells are infected with the crude virus. Nearly 30 h postinfection, cells and culture media are collected and spun down at ~300 × g for 10 min, the cell pellet is washed twice with phosphate-buffered saline (PBS) resuspended in 10 mM Tris–HCl, pH 8.0. Recombinant virus is released by a series of freeze–thaw cycles, and the virus is purified using well-established CsCl gradient centrifugation methodology (Chillon & Alemany, 2011; Duffy, O’Doherty, O’Brien, & Strappe, 2005; Moore & Dowhan, 2002; Tan, Li, Jiang, & Ma, 2006). The purified adenovirus is quantitated, aliquoted aseptically, and stored at −20 °C until needed for protein expression.
3. PROTEIN EXPRESSION
Once the pure recombinant adenovirus has been obtained, small-scale protein expression can be initiated in six-well plates. This is necessary prior to embarking on large-scale protein production to assess the expression conditions as well as proper folding and activity of the desired protein. In our case study of HIV-1 gp120 OD, the glycoprotein is secreted into the culture medium. A researcher may choose to direct expression of a POI into the secretory pathway; however, attention must be paid to monitor inadvertent glycosylation of a cytoplasmic protein when it is directed into the Golgi secretory pathway. Additionally, expression of glycoproteins from mammalian cells in contrast to insect or yeast hosts (Jenkins, Parekh, & James, 1996) can result in heterogeneous glycosylation due to variation in the occupancy as well as the nature of glycan. The glycosylation pattern obtained from mammalian cells can be high mannose, hybrid, or complex in nature, and this may have an impact on stability and folding as well as in vivo functional activity (Weigel & Yik, 2002). Thus, a researcher may choose to design and direct POI production based on the source of a recombinant gene. In our case study, we used a combination of kifunensine, a potent inhibitor of α-mannosidase I (Elbein, Tropea, Mitchell, & Kaushal, 1990) as well as swainsonine a potent inhibitor of α-mannosidase II (Elbein, 1991) to obtain endoglycosidase H-sensitive high-mannose glycans (Kong et al., 2010; Magnelli, Bielik, & Guthrie, 2012).
Small-scale protein expression using the adenoviral expression system can be performed in six-well plates using A549 adherent cells as described previously (Sastry et al., 2012). Typically, cells are seeded at0.8 × 106 cell/well in a six-well plate on Day 1 in fresh Dulbecco’s minimal eagle medium (DMEM) containing 10% heat-inactivated-dialyzed fetal bovine serum (FBS), 1% penicillin/streptomycin. Cells are allowed to grow overnight at 37 °C and 5% CO2. The following day (Day 2), media is replaced with labeled 15N, 15N/13C CGM6000, or fresh DMEM containing 10% heat-inactivated-dialyzed FBS and 1% penicillin/streptomycin. A549 cells are then infected with rAd5 containing the GOI to a final concentration of 2500 particles/cell. Protein expression of a cytoplasmic or secreted protein is monitored 72–96 h postinfection by either harvesting cells or the culture media, respectively, and testing for the presence of the protein using ELISAs, SPR, BioLayer Interferometry or an equivalent technique.
Large-scale production of isotopically enriched glycoprotein is performed using the protocol described previously (Sastry et al., 2011, 2012) and purified using multistep affinity chromatography followed by size-exclusion chromatography. For cytoplasmic proteins, culture supernatant is removed, and adherent A549 cells are harvested following treatment with Trypsin-EDTA. Cells are pelleted at 300 × g, washed twice with PBS, and subsequently lysed with cell lysis buffer (cell signaling) and the POI purified by the method of choice.
3.1. Selective Labeling of Specific Amino Acids
CHO cell lines are most commonly used for production of therapeutic reagents. As a result, protein production for structural studies was focused on obtaining labeled material from stable CHO and HEK293 cell lines. Due to the exorbitant cost of labeled media and low yields, researches interested in studying biologically interesting proteins by NMR spectroscopy turned to partial (Werner et al., 2008) or amino acid type-specific labeling (Anglister, Frey, & McConnell, 1984; Arata, Kato, Takahashi, & Shimada, 1994; Klein-Seetharaman et al., 2002, 2004) of proteins from mammalian expression systems. Insect cells have proven to be a good source for obtaining specifically labeled proteins (Gossert et al., 2011). The adenoviral expression system is particularly suitable to obtaining proteins with specific amino acids labeled. We demonstrated this for 15N glycine and 15N valine (Sastry et al., 2012). Herein, we extend the methodology to incorporating 15N/13C proline. The methodology to produce OD selectively labeled with 15N/13C proline is similar to that described previously and in Section 3. CGM-6750-CUSTOM media containing 15N/13C-labeled proline and 15N labels in all other amino acids and media components can be used to produce protein specifically labeled with 15N/13C proline. We estimate yields of 33 mg/L of pure glycosylated his-tagged 15N/13C proline-enriched OD protein, thus an NMR sample can be obtained from only 200–300 mL culture. These yields balance out the high cost of the labeled media and allow for the production of selectively labeled proteins.
4. NMR CHARACTERIZATION OF EXPRESSED PROTEIN
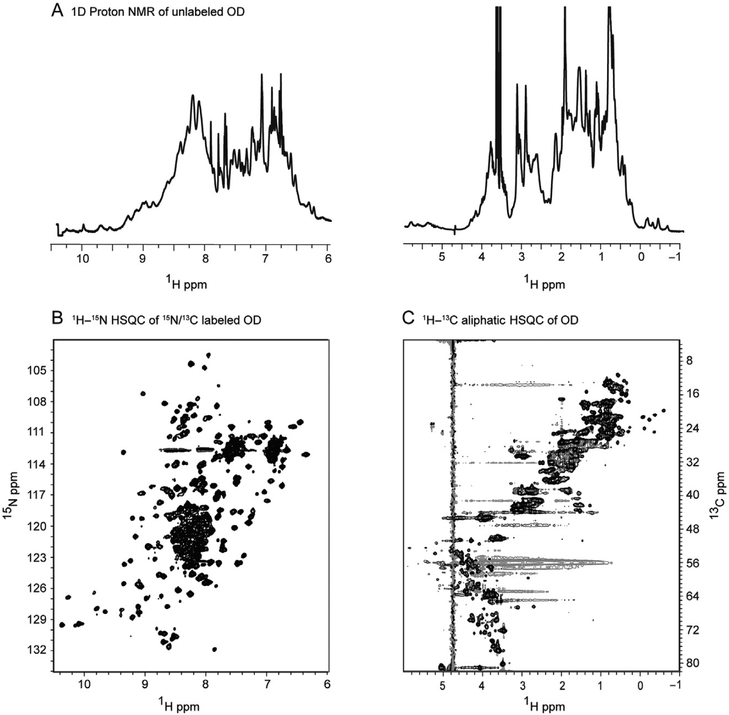
Initially, an unlabeled sample of a full-length protein or a protein domain of interest expressed either transiently or by the adenoviral-induced expression can be characterized using 1D or 2D NMR spectroscopy. Thus, in our example, the HIV-1 gp120 OD obtained from the adenovirus expression system exhibits a one-dimensional 1H NMR spectrum (Fig. 3A) that is well dispersed with well resolved, upfield-shifted methyl protons as well as a nicely dispersed amide region, indicative of a folded protein. The 1H–15N HSQC spectra of the OD are also of very high quality and exhibits resolved backbone and side-chain amides (Fig. 3B). The 1H–13C HSQC (Fig. 3C) exhibits very good chemical shift dispersion along with upfield-shifted methyl resonances, indicative of a structured protein.
Figure 3.
HIV-1 gp120 outer domain (OD) expressed and purified from the adenoviral expression system is functionally active and well-folded protein. One-dimensional proton spectra of unlabeled deglycosylated HIV-1 gp120 outer domain acquired at 600 MHz and 25 °C are shown in (A). 1H–15N and 1H–13C HSQC of 15N/13C-labeled outer domain acquired at 900 MHz and 25 °C are shown in (B) and (C), respectively. Reproduced from Sastry et al. (2011) with permission from Springer.
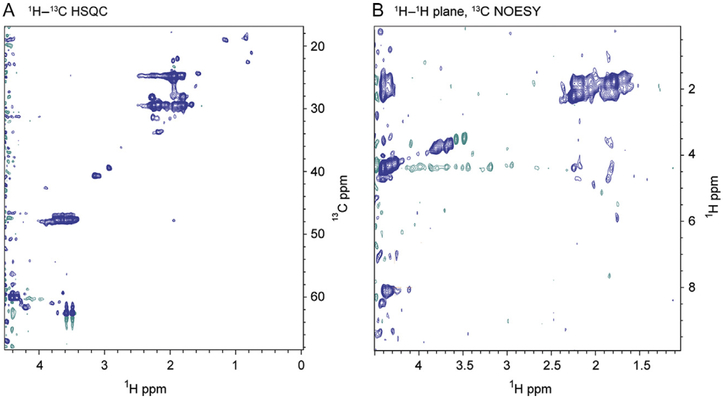
To assess selective 15N/13C proline incorporation necessary for assignment of backbone and side-chain atoms, two data sets were recorded. In Fig. 4A, a high-quality 1H–13C aliphatic HSQC spectrum acquired at 900 MHz and 25 °C shows selective incorporation of isotopically labeled pro-line residues. The 1H–1H plane of a 1H–13C 3D NOESY spectrum at a Cα chemical shift of 60 ppm also shows nOes to amide residues in the 8–9 ppm range, demonstrating conclusively that the adenoviral expression system provides sufficient isotope enrichment to allow acquisition of triple-resonance and NOESY-type experiments to obtain full backbone and side-chain resonance assignments. Selective labeling of specific amino acids allows the study of specific interactions and enables confirmation of assignments in extremely crowded regions of 3D spectra. Selectively labeled OD spectra enriched with 15N/13C proline (Fig. 4A and B) along with our previously published data provide further proof that the adenoviral expression system is suitable for obtaining isotopically enriched proteins for structural characterization of proteins and protein complexes by NMR spectroscopy.
Figure 4.
Single amino acid labeling of HIV-1 gp120 OD from the adenoviral expression system is feasible. (A) 1H–13C HSQC spectra of HIV-1 gp120 OD selectively labeled with 15N/13C proline acquired at 900 MHz and 25 °C. (B) 1H–13C NOESY HSQC spectra of HIV-1 gp120 OD selectively labeled with 15N/13C proline in a 15N background acquired at 900 MHz and 25 °C. An 1H–1H NOESY plane at Cα 60.0 ppm is shown. The 1H–13C aliphatic HSQC and NOESY of selectively labeled outer domain spectrum demonstrate that the adenoviral system can be used to obtain selectively labeled proteins.
5. CONCLUSIONS
Isotope labeling of biologically interesting proteins is a prerequisite for structural and dynamics studies by NMR spectroscopy. Despite the technological advances in NMR spectroscopy, difficulties in obtaining uniformly and specifically labeled protein samples of posttranslationally modified proteins have limited the breadth and scope of solution NMR measurements. In this chapter, we described a mammalian expression system that is based on the delivery of transgenes using mammalian viruses with nearly 100% efficiency and low toxicity. The high level of protein expression offsets the cost of the labeled media, and the versatility of the expression system should allow solution NMR studies of difficult to express proteins and their complexes.
6. MATERIALS
We report materials used for selective 15N/13C labeling of Proline residues. Materials for summarized experiments can be found in the respective primary publications (Aoki et al., 1999; Sastry et al., 2011). pVRC1194 and pVRC1290 vectors are available upon request.
Specifically labeled 15N/13C proline-CGM6750 CUSTOM media, in which all other amino acids are uniformly 15N labeled while other components are unlabeled, were obtained from Cambridge Isotope Laboratories, Inc. (Andover, MA) (CIL). High-glucose-containing DMEM with Hepes and NaHCO3 was obtained from Life Technologies. Kifunensine and swainsonine were obtained from Enzo Life Sciences. Nickel-NTA was obtained from Qiagen Inc. HEK293 and A549 cells were obtained from american tissue culture collection (ATCC).
ACKNOWLEDGMENTS
We thank the NMR staff at the New York Structural Biology Consortium for assistance with instrumentation. We also thank the members of the Structural Biology Section and Structural Bioinformatics Section at the Vaccine Research Center for insightful comments and discussions. Support for this work was provided by the Intramural Program of the NIH (NIAID and NIDDK). 900 MHz spectrometers were purchased with funds from NIH, USA, the Keck Foundation, New York State, and the NYC Economic Development Corporation.
ABBREVIATIONS
- ATCC
American Type Culture Collection
- BGHpA
bovine growth hormone polyadenylation signal
- BHK21
baby-hamster-kidney cell line
- CAR
coxsackie-adenovirus receptor
- CHO
Chinese hamster ovary cells
- CMV
cytomegalovirus
- DMEM
Dulbecco’s minimal eagle media
- FBS
dialyzed fetal bovine serum
- HEK
human embryonic kidney cells
- HIV-1
human immunodeficiency virus type 1
- PBS
phosphate-buffered saline
- PEI
polyethyleneimine
- SPR
surface plasmon resonance
REFERENCES
- Anglister J, Frey T, & McConnell HM (1984). Magnetic resonance of a monoclonal anti-spin-label antibody. Biochemistry, 23(6), 1138–1142. [DOI] [PubMed] [Google Scholar]
- Aoki K, Barker C, Danthinne X, Imperiale MJ, & Nabel GJ (1999). Efficient generation of recombinant adenoviral vectors by Cre-lox recombination in vitro. Molecular Medicine, 5(4), 224–231. [PMC free article] [PubMed] [Google Scholar]
- Arata Y, Kato K, Takahashi H, & Shimada I (1994). Nuclear magnetic resonance study of antibodies: A multinuclear approach. Methods in Enzymology, 239, 440–464. [DOI] [PubMed] [Google Scholar]
- Arya R, Bhattacharya A, & Saini KS (2008). Dictyostelium discoideum—A promising expression system for the production of eukaryotic proteins. The FASEB Journal, 22(12), 4055–4066. [DOI] [PubMed] [Google Scholar]
- Babich A, Feldman LT, Nevins JR, Darnell JE Jr., & Weinberger C (1983). Effect of adenovirus on metabolism of specific host mRNAs: Transport control and specific translational discrimination. Molecular and Cellular Biology, 3(7), 1212–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backliwal G, Hildinger M, Chenuet S, Wulhfard S, De Jesus M, & Wurm FM (2008). Rational vector design and multi-pathway modulation of HEK 293E cells yield recombinant antibody titers exceeding 1 g/l by transient transfection under serum-free conditions. Nucleic Acids Research, 36(15), e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk AJ (1986). Adenovirus promoters and E1A transactivation. Annual Review of Genetics,20, 45–79 (Review). [DOI] [PubMed] [Google Scholar]
- Braun P, & LaBaer J (2003). High throughput protein production for functional proteomics. Trends in Biotechnology, 21(9), 383–388. [DOI] [PubMed] [Google Scholar]
- Chen K, Freedberg DI, & Keire DA (2015). NMR profiling of biomolecules at natural abundance using 2D 1H-15N and 1H-13C multiplicity-separated (MS) HSQC spectra. Journal of Magnetic Resonance, 251, 65–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chillon M, & Alemany R (2011). Methods to construct recombinant adenovirus vectors. Methods in Molecular Biology, 737, 117–138. [DOI] [PubMed] [Google Scholar]
- Coleman TA, Parmelee D, Thotakura NR, Nguyen N, Bürgin M, Gentz S, et al. (1997). Production and purification of novel secreted human proteins. Gene, 190(1), 163–171. [DOI] [PubMed] [Google Scholar]
- Cubeddu L, Moss CX, Swarbrick JD, Gooley AA, Williams KL, Curmi PM, et al. (2000). Dictyostelium discoideum as expression host: isotopic labeling of a recombinant glycoprotein for NMR studies. Protein Expression and Purification, 19(3), 335–342. [DOI] [PubMed] [Google Scholar]
- Duffy AM, O’Doherty AM, O’Brien T, & Strappe PM (2005). Purification of adenovirus and adeno-associated virus: Comparison of novel membrane-based technology to conventional techniques. Gene Therapy, 12(S1), S62–S72. [DOI] [PubMed] [Google Scholar]
- Elbein AD (1991). Glycosidase inhibitors: Inhibitors of N-linked oligosaccharide processing. The FASEB Journal, 5(15), 3055–3063. [DOI] [PubMed] [Google Scholar]
- Elbein AD, Tropea JE, Mitchell M, & Kaushal GP (1990). Kifunensine, a potent inhibitor of the glycoprotein processing mannosidase I. Journal of Biological Chemistry, 265(26), 15599–15605. [PubMed] [Google Scholar]
- Geisse S, & Fux C (2009). Chapter 15 recombinant protein production by transient gene transfer into mammalian cells In Richard RB & Murray PD (Eds.), Methods in enzymology: Vol. 463 (pp. 223–238). San Diego: Academic Press. [DOI] [PubMed] [Google Scholar]
- Gluzman Y, Reichl H, & Solnick D (1982). Helper-free adenovirus type-5 vectors In Gluzman Y (Ed.), Eukaryotic viral vectors (pp. 187–192). Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. [Google Scholar]
- Gossert A, Hinniger A, Gutmann S, Jahnke W, Strauss A, & Fernández C (2011). A simple protocol for amino acid type selective isotope labeling in insect cells with improved yields and high reproducibility. Journal of Biomolecular NMR, 51(4), 449–456. [DOI] [PubMed] [Google Scholar]
- Graham FL, Smiley J, Russell WC, & Nairn R (1977). Characteristics of a human cell line transformed by DNA from human adenovirus type 5. Journal of General Virology, 36(1), 59–72. [DOI] [PubMed] [Google Scholar]
- Hansen AP, Petros AM, Mazar AP, Pederson TM, Rueter A, & Fesik SW (1992). A practical method for uniform isotopic labeling of recombinant proteins in mammalian cells. Biochemistry, 31(51), 12713–12718. [DOI] [PubMed] [Google Scholar]
- Hansen AP, Petros AM, Meadows RP, Nettesheim DG, Mazar AP, Olejniczak ET, et al. (1994). Solution structure of the amino-terminal fragment of urokinase-type plasminogen activator. Biochemistry, 33(16), 4847–4864. [DOI] [PubMed] [Google Scholar]
- Hinck AP, Archer SJ, Qian SW, Roberts AB, Sporn MB, Weatherbee JA, et al. (1996). Transforming growth factor β1: Three-dimensional structure in solution and comparison with the X-ray structure of transforming growth factor β2†,‡. Biochemistry, 35(26), 8517–8534. [DOI] [PubMed] [Google Scholar]
- Hopkins RF, Wall VE, & Esposito D (2012). Optimizing transient recombinant protein expression in mammalian cells In Hartley JL (Ed.), Protein expression in mammalian cells: Vol. 801 (pp. 251–268). New York: Humana Press. [DOI] [PubMed] [Google Scholar]
- Howley PM, Sarver N, & Law M-F (1983). Eukaryotic cloning vectors derived from bovine papillomavirus DNA In Wu L. G. K. M. Ray (Ed.), Methods in enzymology: Vol. 101 (pp. 387–402): Academic Press. [DOI] [PubMed] [Google Scholar]
- Huang JT, & Schneider RJ (1990). Adenovirus inhibition of cellular protein synthesis is prevented by the drug 2-aminopurine. Proceedings of the National Academy of Sciences of the United States of America, 87(18), 7115–7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins N, Parekh RB, & James DC (1996). Getting the glycosylation right: Implications for the biotechnology industry. Nature Biotechnology, 14(8), 975–981. [DOI] [PubMed] [Google Scholar]
- Kato T, Kajikawa M, Maenaka K, & Park E (2010). Silkworm expression system as a platform technology in life science. Applied Microbiology and Biotechnology, 85(3), 459–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston RE, Chen CA, & Okayama H (2001). Calcium phosphate transfection In Current protocols in immunology: Vol 31 (pp. 10.13.1–10.13.9). New York, NY: John Wiley & Sons, Inc. [DOI] [PubMed] [Google Scholar]
- Klein-Seetharaman J, Reeves PJ, Loewen MC, Getmanova EV, Chung J, Schwalbe H, et al. (2002). Solution NMR spectroscopy of [α−15N]lysine-labeled rhodopsin: The single peak observed in both conventional and TROSY-type HSQC spectra is ascribed to Lys-339 in the carboxyl-terminal peptide sequence. Proceedings of the National Academy of Sciences of the United States of America, 99(6), 3452–3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein-Seetharaman J, Yanamala NVK, Javeed F, Reeves PJ, Getmanova EV, Loewen MC, et al. (2004). Differential dynamics in the G protein-coupled receptor rhodopsin revealed by solution NMR. Proceedings of the National Academy of Sciences of the United States of America, 101(10), 3409–3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Sheppard NC, Stewart-Jones GBE, Robson CL, Chen H, Xu X, et al. (2010). Expression-system-dependent modulation of HIV-1 envelope glycoprotein antigenicity and immunogenicity. Journal of Molecular Biology, 403(1), 131–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M (1984). Point mutations close to the AUG initiator codon affect the efficiency of translation of rat preproinsulin in vivo. Nature, 308(5956), 241–246. http://dx.doi.org/10.1038/308241a0. [DOI] [PubMed] [Google Scholar]
- Kozak M (1987). An analysis of 5’-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Research, 15(20), 8125–8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriz A, Schmid K, Baumgartner N, Ziegler U, Berger I, Ballmer-Hofer K, et al. (2010). A plasmid-based multigene expression system for mammalian cells. Nature Communications, 1, 120. [DOI] [PubMed] [Google Scholar]
- Kwon YD, Pancera M, Acharya P, Georgiev IS, Crooks ET, Gorman J, et al. (2015). Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nature Structural & Molecular Biology, 22(7), 522–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Menzel C, Meier D, Zhang C, Dübel S, & Jostock T (2007). A comparative study of different vector designs for the mammalian expression of recombinant IgG antibodies. Journal of Immunological Methods, 318(1–2), 113–124. [DOI] [PubMed] [Google Scholar]
- Longo PA, Kavran JM, Kim M-S, & Leahy DJ (2013a). Generating mammalian stable cell lines by electroporation In Lorsch J (Ed.), Methods in Enzymology: Vol. 529 (pp. 209–226). Waltham, MA: Academic Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo PA, Kavran JM, Kim M-S, & Leahy DJ (2013b). Transient mammalian cell transfection with Polyethylenimine (PEI) In Lorsch J (Ed.), Methods in Enzymology: Vol. 529 (pp. 227–240). Waltham, MA: Academic Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustbader JW, Birken S, Pollak S, Pound A, Chait BT, Mirza UA, et al. (1996). Expression of human chorionic gonadotropin uniformly labeled with NMR isotpes in Chinese hamster ovary cells: An advance toward rapid determination of glycoprotein structures. Journal of Biomolecular NMR, 7(4), 295–304. [DOI] [PubMed] [Google Scholar]
- Magnelli P, Bielik A, & Guthrie E (2012). Identification and characterization of protein glycosylation using specific endo- and exoglycosidases In Hartley JL (Ed.), Protein expression in mammalian cells: Vol. 801 (pp. 189–211). New York: Humana Press. [DOI] [PubMed] [Google Scholar]
- McLellan JS, Chen M, Leung S, Graepel KW, Du X, Yang Y, et al. (2013). Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science (New York, N.Y.), 340(6136), 1113–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore D, & Dowhan D (2002). Purification and concentration of DNA from aqueous solutions Current protocols in molecular biology: Vol. 59 (pp. 2.1.1–2.1.10). New York, NY: John Wiley & Sons, Inc. [DOI] [PubMed] [Google Scholar]
- Mulligan RC, Howard BH, & Berg P (1979). Synthesis of rabbit [beta]-globin in cultured monkey kidney cells following infection with a SV40 [beta]-globin recombinant genome. Nature, 277(5692), 108–114. [DOI] [PubMed] [Google Scholar]
- Nettleship JE, Assenberg R, Diprose JM, Rahman-Huq N, & Owens RJ (2010). Recent advances in the production of proteins in insect and mammalian cells for structural biology. Journal of Structural Biology, 172(1), 55–65. [DOI] [PubMed] [Google Scholar]
- Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, et al. (2014). Structure and immune recognition of trimeric prefusion HIV-1 Env. Nature, 514(7523), 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter H, & Heller R (2003). Transfection by electroporation In Current protocols in molecular biology: Vol. 62 (pp. 9.3.1–9.3.6). New York, NY: John Wiley & Sons, Inc. [DOI] [PubMed] [Google Scholar]
- Raghavan M, & Bjorkman PJ (1995). BIAcore: A microchip-based system for analyzing the formation of macromolecular complexes. Structure, 3(4), 331–333. [DOI] [PubMed] [Google Scholar]
- Rich RL, & Myszka DG (2010). Grading the commercial optical biosensor literature—Class of 2008: ‘The Mighty Binders’. Journal of Molecular Recognition, 23(1), 1–64. [DOI] [PubMed] [Google Scholar]
- Sastry M, Bewley C, & Kwong P (2012). Mammalian expression of isotopically labeled proteins for NMR spectroscopy In Atreya HS (Ed.), Isotope labeling in biomolecular NMR: Vol. 992 (pp. 197–211). The Netherlands: Springer. [DOI] [PubMed] [Google Scholar]
- Sastry M, Xu L, Georgiev I, Bewley C, Nabel G, & Kwong P (2011). Mammalian production of an isotopically enriched outer domain of the HIV-1 gp120 glycoprotein for NMR spectroscopy. Journal of Biomolecular NMR, 50(3), 197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo K, Masuda K, Takahashi H, Arata Y, & Shimada I (2000). Letter to the Editor: Backbone 1H, 13C, and 15 N resonance assignments of the anti-dansyl antibody Fv fragment. Journal of Biomolecular NMR, 17(4), 357–358. [DOI] [PubMed] [Google Scholar]
- Southern P, & Berg P (1982). Transformation of mammalian cell to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. Journal of Molecular and Applied Genetics, 1, 327–341. [PubMed] [Google Scholar]
- Swarbrick J, Cubeddu L, Ball G, Curmi PG, Gooley A, Williams K, et al. (2009). NMR assignment of prespore specific antigen—A cell surface adhesion glycoprotein from Dictyostelium discoideum. Biomolecular NMR Assignments, 3(1), 1–3. [DOI] [PubMed] [Google Scholar]
- Tan R, Li C, Jiang S, & Ma L (2006). A novel and simple method for construction of recombinant adenoviruses. Nucleic Acids Research, 34(12), e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowitzsch S, Klumpp M, Thoma R, Carralot J-P, & Berger I (2011). Light it up: Highly efficient multigene delivery in mammalian cells. BioEssays, 33(12), 946–955. [DOI] [PubMed] [Google Scholar]
- Unger T, & Peleg Y (2012). Recombinant protein expression in the baculovirus-infected insect cell system In Zanders ED (Ed.), Chemical genomics and proteomics: Vol. 800 (pp. 187–199). New York: Humana Press. [DOI] [PubMed] [Google Scholar]
- Van Craenenbroeck K, Vanhoenacker P, & Haegeman G (2000). Episomal vectors for gene expression in mammalian cells. European Journal of Biochemistry, 267(18), 5665–5678. [DOI] [PubMed] [Google Scholar]
- Voytas D (2001). Agarose gel electrophoresis Current protocols in molecular biology: Vol. 51(pp. 2.5A.1–2.5A.9). New York, NY: John Wiley & Sons, Inc. [DOI] [PubMed] [Google Scholar]
- Warnock JN, Daigre C, & Al-Rubeai M (2011). Introduction to viral vectors. Methods in Molecular Biology, 737, 1–25. [DOI] [PubMed] [Google Scholar]
- Weigel PH, & Yik JHN (2002). Glycans as endocytosis signals: The cases of the asialoglycoprotein and hyaluronan/chondroitin sulfate receptors. Biochimica et Biophysica Acta (BBA)—General Subjects, 1572(2–3), 341–363. [DOI] [PubMed] [Google Scholar]
- Werner K, Richter C, Klein-Seetharaman J, & Schwalbe H (2008). Isotope labeling of mammalian GPCRs in HEK293 cells and characterization of the C-terminus of bovine rhodopsin by high resolution liquid NMR spectroscopy. Journal of Biomolecular NMR, 40(1), 49–53. [DOI] [PubMed] [Google Scholar]
- Wu L, Yang Z-Y, Xu L, Welcher B, Winfrey S, Shao Y, et al. (2006). Cross-clade recognition and neutralization by the V3 region from clade C human immunodeficiency virus-1 envelope. Vaccine, 24(23), 4995–5002. [DOI] [PubMed] [Google Scholar]
- Wyss DF, Choi J, Li J, Knoppers M, Willis K, Arulanandam A, et al. (1995). Conformation and function of the N-linked glycan in the adhesion domain of human CD2. Science, 269(5228), 1273–1278. [DOI] [PubMed] [Google Scholar]
- Wyss DF, Dayie KT, & Wagner G (1997). The counterreceptor binding site of human CD2 exhibits an extended surface patch with multiple conformations fluctuating with millisecond to microsecond motions. Protein Science, 6(3), 534–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss DF, Withka JM, Knoppers MH, Sterne KA, Recny MA, & Wagner G (1993). Proton resonance assignments and secondary structure of the 13.6 kDa glycosylated adhesion domain of human CD2. Biochemistry, 32(41), 10995–11006. [DOI] [PubMed] [Google Scholar]
- Xia W, Bringmann P, McClary J, Jones PP, Manzana W, Zhu Y, et al. (2006). High levels of protein expression using different mammalian CMV promoters in several cell lines. Protein Expression and Purification, 45(1), 115–124. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Nishimura M, Nagano M, Yagi H, Sasakawa H, Uchida K, et al. (2006). Glycoform-dependent conformational alteration of the Fc region of human immunoglobulin G1 as revealed by NMR spectroscopy. Biochimica et Biophysica Acta (BBA)—General Subjects, 1760(4), 693–700. [DOI] [PubMed] [Google Scholar]
- Zhou T, Lynch RM, Chen L, Acharya P, Wu X, Doria-Rose NA, et al. (2015). Structural repertoire of HIV-1-neutralizing antibodies targeting the CD4 supersite in 14 donors. Cell, 161(6), 1280–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]