Abstract
Pancreatic cancers arise predominantly from ductal epithelial cells of the exocrine pancreas and are of the ductal adenocarcinoma histological subtype (PDAC). PDAC is an aggressive disease associated with a poor clinical prognosis, weakly effective therapeutic options, and a lack of early detection methods. Furthermore, the genetic and phenotypic heterogeneity of PDAC complicates efforts to identify universally efficacious therapies. PDACs commonly harbor activating mutations in the KRAS oncogene, which is a potent driver of tumor initiation and maintenance. Inactivating mutations in tumor suppressor genes such as CDKN2A/p16, TP53 and SMAD4 cooperate with KRAS mutations to cause aggressive PDAC tumor growth. PDAC can be classified into 3-4 molecular subtypes by global gene expression profiling. These subtypes can be distinguished by distinct molecular and phenotypic characteristics. This chapter will provide an overview of the current knowledge of PDAC pathogenesis at the genetic and molecular level as well as novel therapeutic opportunities to treat this highly aggressive disease.
1. INTRODUCTION
The pancreas is a glandular organ with both endocrine and exocrine function1,2. Its overall purpose is to maintain metabolic homeostasis by producing hormones that regulate blood glucose levels as well as enzymes that aid in digestion. The pancreas is derived from the embryonic foregut of the endodermal germ layer3. During embryonic development, two buds that ultimately give rise to the dorsal and ventral pancreas emerge from the foregut. As these buds expand, they are gradually repositioned over time until they come into contact and fuse together, forming the mature pancreas. Under the control of various developmental cues, pancreatic progenitor cells become acinar, endocrine, or ductal in function. Endocrine cells (α, β, and δ) secrete hormones like insulin, glucagon, and somatostatin into the circulatory system to modulate blood glucose levels1. This homeostatic function ensures that the metabolic demands of various tissues and organs are met. Acinar cells in the ducts secrete enzymes like trypsinogen, chymotrypsinogen, lipase, and amylase into the pancreatic duct2. These enzymes subsequently enter into the small intestine, where they aid in the digestion of various dietary macromolecules such as proteins, carbohydrates, and lipids.
Pancreatic dysfunction can lead to a number of common diseases including diabetes, pancreatitis, and cancer4-6. Diabetes is the most prevalent of these diseases. However, cancer of the pancreas is by far the deadliest, and its etiology is often linked to other pancreatic disorders, including diabetes. Pancreatic cancer is the fourth leading cause of cancer related deaths in the United States and is associated with a particularly poor prognosis7. Patients diagnosed with this disease exhibit a median overall survival of less than six months and a five-year survival rate of roughly 8%. The poor prognosis associated with pancreatic cancer is attributed in part to poor methods of early detection8. Patients often remain asymptomatic until the disease has disseminated throughout the body. Additionally, the therapeutics used to treat pancreatic cancer are relatively ineffective, as they fail to extend patient survival more than several months9. Overcoming these challenges will be critical in the future treatment of the disease, as it is expected to become the second leading cause of cancer related deaths in the United States by 203010.
Pancreatic cancers can arise from either endocrine or exocrine cells. Thus, endocrine and exocrine tumors can be distinguished by histological appearance. Endocrine tumors are relatively uncommon, and constitute less than 5% of all pancreatic cancers. They are associated with a median survival of 27 months and a 0.28-fold lower risk of mortality in comparison to the much more common pancreatic adenocarcinoma11. Endocrine tumors are commonly derived from pancreatic islet cells, and often produce constitutively high levels of pancreatic hormones. They can be further categorized into insulinomas, glucagonomas, and gastrinomas depending on their cell of origin and the hormones that they secrete. Pancreatic endocrine tumors can be readily diagnosed due to their excessive hormone secretion, which leads to dramatic symptoms such as hypoglycemia or necrolytic migratory erythema (skin rash)12. Pancreatic cancers derived from exocrine cells are much more common than endocrine tumors and can typically be classified into two histological subtypes. The pancreatic ductal adenocarcinoma (PDAC) subtype accounts for the majority of exocrine tumors and constitutes more than 90% of all pancreatic malignancies. PDACs are derived from epithelial cells that line the pancreatic duct and appear gland-like due to their origin11. These cancers frequently metastasize to the liver or lymph nodes13. Due to their lack of symptoms at the early stages of cancer development, PDACs are often diagnosed at a late stage, potentially after the cancer has already metastasized. As a result, anti-cancer therapeutics tend to be weakly effective due to the cancers having acquired strong cytoprotective mechanisms that promote drug resistance. Because of this aggressiveness and drug resistance, estimated median survival for PDAC can be as short as 4 months11.
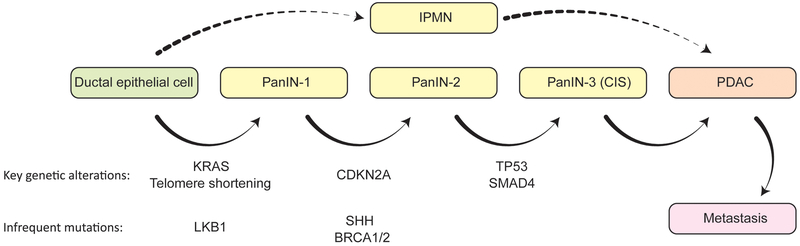
PDACs are preceded by the development of hyperplastic lesions known as pancreatic intraepithelial neoplasias (PanINs) and intraductal papillary mucinous neoplasms (IPMNs) that are precancerous and exhibit a propensity to develop into cancer (Fig. 1). IPMNs look like papillae (finger-like structures) that protrude into the pancreatic duct14. Mucinous tumors are the second most common histological subtype of pancreatic cancer, constituting less than 10% of cases. These tumors are usually much less invasive than adenocarcinomas at the time of diagnosis and have a 0.88-fold lower risk of mortality by comparison11. Mucinous tumors also originate from the pancreatic ductal epithelium, but secrete mucin, which can be seen in and around the cells, causing them to appear like they are ‘floating’15,16. There are many other subtypes of pancreatic cancer, such as those that arise from acinar cells, which are undifferentiated and resemble liver cancers. However, these subtypes are rare and will not be discussed in this chapter.
Figure 1. Disease Progression Model of Pancreatic Cancer.
Pancreatic cancer arises from two histological types of precursor lesions: PanINs and IPMNs. Through progressive stages of pathogenesis, molecular changes occur, leading to increasing degrees of nuclear and cytoskeletal abnormalities. Genetic alterations commonly observed in these lesions are indicated with respect to the stages in which they most often occur.
This chapter will focus on the molecular etiology of PDAC, the most prevalent form of pancreatic malignancy. The core genetic alterations that contribute to PDAC pathogenesis will be discussed. Furthermore, the molecular subtypes of PDAC will be presented with a focus on their cellular origin and the genetic alterations associated with them. The diverse and deregulated signaling pathways that contribute to PDAC pathogenesis will also be described in detail. To conclude, the molecular characteristics of PDAC will be discussed in relation to the current therapeutic strategies employed to manage the disease in the clinic and future approaches that may further improve patient prognosis.
2. GENETIC ALTERATIONS IN PANCREATIC CANCER
Whole exome-sequencing studies have revealed that PDAC is a molecularly heterogeneous disease characterized by four common genetic alterations: oncogenic KRAS mutation and inactivation of the tumor suppressors CDKN2A, TP53, and SMAD4 (Fig. 1, 2A)6. However, myriad additional genes are mutated in subsets of tumors, typically at a very low frequency (≤10%), with many of these mutations not occurring in a recurrent manner. Further analysis of these infrequent alterations has revealed that they converge on a relatively small number of pathways and cellular processes including KRAS, TGF-β, WNT, NOTCH, and Hedgehog signaling as well as S-phase entry, axonal guidance, chromatin remodeling, DNA repair, and RNA processing17,18. Understanding how context-dependent interactions between these various genetically altered pathways contribute to PDAC progression is a key goal in the development of more selective and efficacious therapeutic modalities to treat the disease.
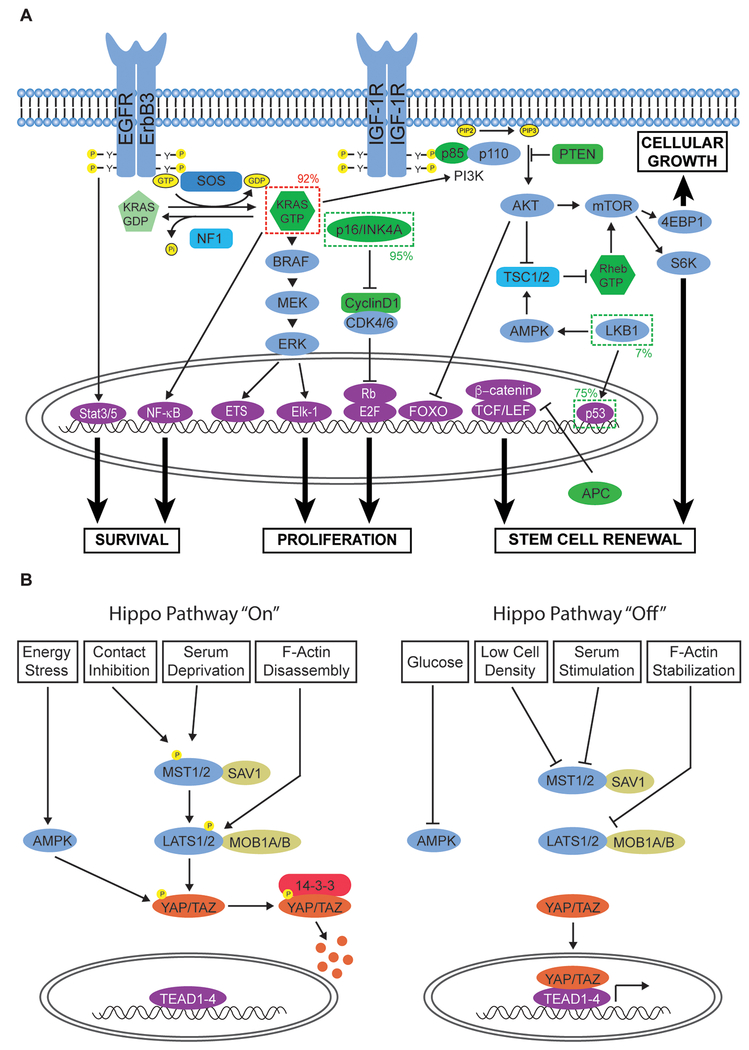
Figure 2. Deregulated Signaling Networks in Pancreatic Cancer.
A. RTK and cell cycle-regulatory signaling networks frequently altered in pancreatic cancer. Oncogenes exhibiting gain-of-function mutations are indicated by a dark dashed line. Tumor suppressor genes altered in the disease are indicated by a lighter dashed line. The frequencies at which these genes are altered are also included. Oncogenic KRAS mutation cooperates with the loss of various tumor suppressor genes to promote cellular proliferation, growth, survival, and stem cell renewal. B. Hippo signaling is frequently deregulated in pancreatic cancer. The ability of the pathway to restrict cell growth and induce apoptosis is mediated by a number of stimuli including cell density, glucose levels, serum levels, and cytoskeletal tension.
2.1. Oncogenic KRAS Mutations
Activating KRAS mutations are the defining genetic feature of PDAC progression and are found in approximately 92% of PDAC18. KRAS, a member of the RAS superfamily, encodes a small GTPase that regulates diverse cellular processes including cell proliferation, differentiation, survival, and migration (Fig. 2A)19. Under normal physiological conditions, KRAS cycles through a GTP-bound active state and a GDP-bound inactive state. The transition between these two states is modulated by guanine nucleotide exchange factors (GEFs; e.g. Sos1), which catalyze the exchange of GDP for GTP, and GTPase-activating proteins (GAPs; e.g. NF1), which enhance the intrinsically weak ability of KRAS to hydrolyze GTP20,21. In quiescent cells, KRAS is predominantly GDP-bound. However, upon growth factor stimulation, GEFs are able to bind to KRAS and promote its activation by catalyzing GDP-GTP exchange. In many solid cancers, KRAS or one of its closely related family members (HRAS and NRAS) undergo mutations that ultimately impair their ability to hydrolyze GTP. HRAS and NRAS mutations are typically not found in PDAC. Oncogenic RAS proteins are locked in an active state that results in constitutive stimulation of effector pathways capable of driving tumor development22. The major effector pathways downstream of active RAS are those mediated by RAF/MEK/ERK, PI3K and RAL-GEF.
KRAS point mutations typically result in a single amino acid substitution at one of three codons: G12, G13, or Q6121. All three substitutions occur in the catalytic domain of the GTPase. G12 and G13 mutations introduce a steric hindrance that blocks the formation of van der Waals interactions between KRAS and GAPs23. Q61 mutations disrupt the coordination of a water molecule required for GTP hydolysis24. Although all three mutations promote KRAS activation by impairing its GTPase activity, the frequencies at which they occur are vastly disparate. Substitutions at codon G12 (typically G12D or G12V) are the most prevalent (82%) followed by Q61 (14%) and G13 (<1%)21. Interestingly, the three common KRAS mutant alleles exhibit contrasting functional properties. Tumors harboring KRAS Q61 substitutions display reduced MAPK activity and are associated with a better prognosis than substitutions at other codons25. This suggests that different KRAS mutations promote tumorigenesis in distinct ways.
Activating KRAS mutations are the earliest genetic alteration in the development of almost all PDACs (Fig. 1), as they are prevalent in greater than 90% of low-grade precursor pancreatic intraepithelial neoplasia (PanIN) lesions26. PDAC is preceded by the emergence of PanIN lesions, which are classified into three stages (I-III) based on the degree of architectural disorganization and nuclear abnormalities of the tissue6. In vivo studies of genetically engineered mouse (GEM) models have demonstrated that PanINs readily develop as a consequence of KRAS-activating mutations but do not ultimately give rise to PDAC. Rather, the subsequent and combined inactivation of multiple tumor suppressor genes (i.e. CDK2NA, TP53, and SMAD4) is also required for malignant tumor progression6,27,28. These findings suggest that KRAS mutations alone are not sufficient to promote PDAC progression.
Although oncogenic KRAS promotes the initiation of PDAC development, its role in tumor maintenance is less clear. In a GEM PDAC model, pancreas-specific induction of KRASG12D expression coupled with loss of a single TP53 allele results in the dedifferentiation of normal epithelial cells and the development of PanINs29. Extinction of oncogenic KRAS leads to the regression of these lesions, suggesting that it is required for PanIN progression. In a subsequent study, pancreas-specific induction of KRASG12D expression coupled with the loss of one or both TP53 alleles led to the development of PDAC30. Similar to the effects observed in PanINs, extinction of oncogenic KRAS in these PDAC lesions led to robust tumor regression. Taken together these studies suggest that oncogenic KRAS plays an essential role in both tumor initiation and maintenance.
In contrast to observations using GEMMs, which bear similarities to subsets of human PDAC, alternative studies using human PDAC-derived cancer cell lines suggest that KRAS is dispensable in certain contexts31,32. Approximately half of human-derived PDAC cell lines readily undergo apoptosis following RNAi-mediated depletion of the GTPase, indicating a state of KRAS oncogene “dependency.” The discrepancy between these findings and those obtained in the GEMM studies may be attributed to a number of factors. First, pharmacological vulnerabilities in GEMMs do not often translate to clinical trials of human cancers, as has been observed with inhibitors of the MAPK and PI3K pathways20. This suggests that, although the same defined genetic mutations from human PDAC promote disease development in GEMMs, the resulting tumors likely display altered molecular characteristics compared to their human counterparts. Another potential explanation for the discrepancy between mouse and human cell line models is that some GEMM-derived tumors may be transiently dependent on KRAS during early stages of PDAC progression. In mouse xenografts, MAPK, PI3K, and RalGEF are all required for tumor initiation33. However, only the PI3K pathway is necessary for tumor maintenance, suggesting a reduced need for oncogenic KRAS. Furthermore, activation of PI3K/AKT signaling by the tumor microenvironment is able to rescue the loss of mutant KRAS. This suggests that KRAS may become dispensable in the later stages of PDAC progression if the PI3K pathway is activated. Such a context may be difficult to model in GEMMs due to the rapid disease onset that results in a severe “dependency” on mutant KRAS for tumor maintenance.
More recent studies in GEMMs have demonstrated that the need for KRAS can be bypassed by enhanced mitochondrial activity or Hippo pathway deregulation34-36. In both cases, depletion of the KRAS protein resulted in robust tumor regression, consistent with previous in vivo work. However, subpopulations of cells were able to drive tumor relapse. In the case of Hippo pathway deregulation, surviving cells expressed lower KRAS protein levels and were unresponsive to KRAS depletion. Furthermore, the relapsed tumors exhibited a more mesenchymal morphology, reminiscent of the phenotype observed in PDAC cell lines that were insensitive to KRAS depletion31. Similarly, surviving cells that bypassed KRAS dependence by increased oxidative phosphorylation were also more stem-like in morphology. Taken together, these studies demonstrate that KRAS may be dispensable in subsets of tumors or subsets of cells within a tumor that can bypass the dependence on oncogenic KRAS signaling. As discussed below, this poses a new challenge to developing effective therapies to treat PDAC by targeting KRAS or its downstream effectors.
2.2. Tumor Suppressor Genes
Tumor suppressor genes (TSGs) restrict cell proliferation in the presence of oncogenic driver mutations by inducing cell cycle arrest, apoptosis, or senescence. Hence, the functional inactivation of TSGs is essential for tumorigenesis37. A number of TSGs are functionally lost in PDAC, the three most common of which are CDKN2A, TP53, and SMAD4/DPC46. The inactivation of these genes occurs in a sequential manner following the appearance of oncogenic KRAS mutation in concordance with the multi-stage carcinogenesis model proposed by Vogelstein and colleagues (Fig. 1)38. Together, TSG alterations constitute a major driving force of PDAC development.
CDKN2A encodes the p16/INK4A protein, a cyclin-dependent kinase inhibitor responsible for blocking entry into S-phase of the cell cycle39. It is the earliest and most frequently altered TSG observed in PDAC. Approximately 95% of tumors exhibit functional loss of this gene as a consequence of intragenic mutation coupled with loss of the second allele (40%), homozygous deletion (40%), or promoter hypermethylation (15%)40,41. CDKN2A inactivation is typically detected in moderately advanced PanINs, prior to the development of PDAC42. CDKN2A loss is crucial in disease pathogenesis, as p16 induces senescence following the introduction of oncogenic KRAS43. For this reason, CDKN2A inactivation occurs immediately following the appearance of activating KRAS mutations and at a similar frequency to bypass the senescence response.
The CDKN2A locus also encodes the tumor suppressor ARF/p14, which is expressed via a distinct first exon that introduces an alternate reading frame in downstream exons shared with CDKN2A/p1639. The ARF/p14 protein induces growth arrest or apoptosis by inhibiting MDM2-dependent p53 proteolysis. However, the frequent inactivation of p53 in PDAC, which often occurs concomitantly with p14 loss, suggests that this mechanism may not be relevant to disease progression. Furthermore, p14 inactivation only occurs as a consequence of CDKN2A deletion (40%)40,41. Expression of this tumor suppressor is driven by an independent promoter that is unaffected by epigenetic changes influencing p16 transcription. Additionally, p14 appears to be spared by p16-inactivating mutations. These discrepancies indicate that loss of p16 is more critical in disease pathogenesis and that p14 loss is a byproduct. However, p14 may hinder the development of PDAC via p53-independent mechanisms, as it has been shown to repress ribosomal RNA processing, NF-κB transactivation, and c-Myc induced hyperproliferation.44-46 Furthermore, p14 can promote cell death by enhancing c-Myc-induced apoptosis as well proteasome-dependent degradation of the antiapoptotic transcriptional corepressor C-terminal binding proteins 1 and 2 (CtBP1/2)46,47. Thus, p14 may hinder PDAC development via multiple mechanisms in specific contexts.
Another major TSG in PDAC is TP53, which encodes the transcription factor p53. Functional loss of this gene has been observed in up to 75% of tumors as a consequence of missense mutation and loss of heterozygosity (LOH)40,48. Amino acid substitutions impair the ability of p53 to bind DNA, thus ablating its function as a transcription factor. As a consequence, mutant p53 is unable to induce the expression of genes that promote cell cycle arrest or apoptosis (e.g. CDKN1A/p21, BAX, NOXA, and PUMA) in response to cellular stress or DNA damage49. Inactivation of p53 is typically observed in advanced PanINs following the loss of CDKN2A43. At this stage of disease progression, the accumulation of DNA damage is believed to induce a selective pressure that necessitates the loss of p53 activity for the continued survival and proliferation of tumorigenic cells.
Emerging evidence suggests that TP53 mutations in PDAC may also contribute to the highly metastatic nature of the disease, thus defining a gain of function role for mutant p5350. In one study, a specific tumor-associated mutant form of p53 (p53R175H) was shown to bind the protein p73 and impair its ability to repress the expression of platelet-derived growth factor receptor β (PDGFRβ)51. The resulting upregulation of PDGFR expression promotes an autocrine signaling loop that enhances the motility, invasiveness, and metastatic potential of tumor cells in mice. Additional studies have demonstrated that inactivation of p53 in other cancer types also promotes metastasis by rendering the transcription factor incapable of inducing the expression of genes that counteract cell migration, epithelial-mesenchymal transition (EMT), and sternness52,53. Thus, mutation of p53 may promote metastasis in PDAC via PDGFR-independent mechanisms. Collectively, these studies highlight the complexity of genetic alterations in PDAC, as p53 functions as both a tumor suppressor and an enhancer of metastasis.
The low-frequency inactivation (<10%) of several additional TSGs has been observed in PDAC54. The most notable of these genes, STK11/LKB1, encodes a serine/threonine (S/T) kinase that regulates cell polarity and metabolism55. Loss of LKB1 as a consequence of germline mutation is most frequently associated with Peutz-Jeghers syndrome (PJS), a disease characterized by the development of benign polyps in the gastrointestinal tract. Individuals with PJS have a 93% cumulative lifetime risk of developing other malignancies, the most frequent of which is PDAC (36% lifetime risk)56. As both germline and somatic/sporadic LKBl mutations are found at low frequency in PDAC, LKB1 may play an important, context-specific role in disease pathogenesis57. One such context may be in the absence of p53 mutation, where LKB1 haploinsufficiency was shown to cooperate with KRASG12D in a GEM model to accelerate PDAC development by suppressing p21-dependent cell cycle arrest58. This limited but critical role highlights the potential importance of low frequency genetic alterations in subsets of PDAC.
Several caretaker genes, which have tumor suppressor function, are also functionally lost in subsets of PDAC. Unlike CDKN2A and TP53, which are considered classical TSGs, caretaker genes do not directly regulate proliferation. Rather, their function is to maintain the integrity of the genome, preventing the accumulation of mutations that might otherwise promote tumor development. A number of caretaker genes are lost in PDAC including BRCA1, BRCA2, hMLH1, and hMSH26,54. Similar to LKB1, inactivation of these TSGs can occur as a consequence of germline or somatic mutation and is more commonly associated with other malignancies. However, they play an implicit role in disease pathogenesis.
2.3. TGF-β/SMAD4 Alterations
Alterations in the transforming growth factor-β (TGF-β) signaling pathway play context-dependent roles in PDAC pathogenesis, as the pathway can both induce apoptosis in some contexts and/or promote invasion and metastasis in others. The antiproliferative effect of TGF-β signaling depends on the activity of the transcriptional co-activator SMAD4/DCP4. SMAD4 mutations and homozygous deletions lead to accelerated tumor development in a KRASG12D GEM model of PDAC28. SMAD4 reintroduction in SMAD4-deficient GEMM-derived tumor cells results in apoptosis upon treatment with TGF-β, highlighting its tumor suppressive properties. Approximately 90% of PDAC cases exhibit loss of heterozygosity (LOH) for the SMAD4 locus59. Furthermore, SMAD4 biallelic inactivation has been observed in roughly 50% of tumors as a consequence of homozygous deletion or intragenic mutations 60. These genetic alterations typically occur in advanced PanINs following the loss of CDKN2A, making SMAD4 loss one of the final steps in tumor initiation43,61. Interestingly, SMAD4 loss predicts poorer patient prognosis compared to tumors with intact, wild-type SMAD4 expression62.
TGF-β is a known inducer of the developmental epithelial-to-mesenchymal transition (EMT) program, discussed in detail below. Upon EMT induction, TGF-β can also promote the activation of an apoptotic program, which is referred to as “lethal EMT.” It has recently been postulated that the tumor suppressor function of Smad4 in PDAC can be attributed to its involvement in the induction of lethal EMT63. In canonical TGF-β signaling, the TGF-β ligand binds to its receptor (TGFBR), resulting in activation of Smad2/3, which subsequently bind to Smad4. The resulting protein complex translocates to the nucleus and induces a Smad4-dependent transcriptional program that promotes EMT. As a consequence of this program, expression of the gastrointestinal-lineage master regulator Klf5 is repressed. Klf5 can cooperate with Sox4 to promote PDAC progression. However, loss of Klf5 expression dramatically alters the role of Sox4 to that of an apoptosis-inducer. This mechanism highlights the complex role of Smad4 in context-dependent tumor suppression by the TGF-β pathway in the pathogenesis of PDAC.
Although TGF-β signaling clearly plays a role in tumor suppression, the pathway can also drive invasion and metastasis. At present, the underlying mechanism behind this alternative role is unclear. It has been suggested that TGF-β promotes invasion and metastasis via the induction of EMT64. However, more recent work in lung and pancreatic cancers has demonstrated that EMT is dispensable for metastasis65,66. These conflicting studies highlight the need to better understand how TGF-β signaling contributes to PDAC progression. EMT is clearly an important phenotypic outcome of TGF-β mediated tumorigenic processes, as it can induce sternness and drug resistance67. However, other mechanisms downstream of TGF-β are likely to be involved in PDAC progression, including activation of the TGF-β activated kinase (TAK1), which activates anti-apoptotic signaling mechanisms68. Low-frequency mutations have been observed in the TGFBR1 and TGFBR2 TGF-β receptor subunits69. However, like SMAD4 inactivation, these alterations are most likely the result of a selective pressure to eliminate the tumor suppressor function of the pathway. Thus, the pro-metastatic functions of TGF-β remain intact in the vast majority of PDAC. Ultimately, these observations suggest that the TGF-β pathway is critical for PDAC pathogenesis, playing complex and context-dependent roles in disease progression.
2.4. Telomere Abnormalities
Telomeres are specialized nucleoprotein structures that maintain the integrity of the genome by protecting the ends of linear chromosomes. These structures gradually shorten, or erode, over time with each successive round of cell division due to inefficient telomere-directed replication. When telomeres become critically short, sister chromatids can fuse together at their ends, forming a bridge70,71. This linkage poses a problem during anaphase, as the chromatids are unable to properly separate. Thus, when fused sister chromatids break during segregation in mitosis, the gain or loss of chromosomal fragments can occur 72,73. This break-fusion-bridge (BFB) process occurs in a cyclical manner, promoting genomic instability and a selective pressure to eliminate anti-apoptotic pathways.
Telomere erosion has been observed in more than 90% of low-grade PanINs, suggesting that it is an early event in PDAC pathogenesis74. This genomic alteration would typically result in p53-induced senescence75. However, the frequent inactivation of p53 sustains continued tumor cell proliferation76. Robust telomere shortening precedes the loss of p53 and likely contributes to the selective pressure that results in its loss. Although telomeric loss can promote a genomically unstable state conducive to tumor formation, it may also be detrimental to disease progression if left unchecked, as the reactivation of telomerase has been observed in invasive PDAC77. This reactivation most likely occurs to prevent additional genomic alterations that would be catastrophic. Telomerase reactivation can promote tumor progression via induction of cellular immortalization, allowing for sustained telomere elongation during repeated rounds of DNA replication.
3. DEREGULATED EMT IN PANCREATIC CANCER
Epithelial cells are located at the surface of many tissues and organs that are derived from the endodermal and ectodermal embryonic germ layers. These cells form sheets that act as barriers against xenobiotic and pathogenic agents and serve specialized secretory functions in the intestine and the pancreas. Due to their location and function, epithelial cells exhibit a distinct apical-basolateral polarity created by macromolecular protein complexes at cell-cell contacts known as adherens and tight junctions. In contrast to epithelial cells, mesenchymal cells serve in anchoring or scaffolding roles and participate in early embryonic development, wound healing, and tissue repair. Mesenchymal cells lose apical-basolateral polarity as a consequence of the EMT transcriptional program. EMT is a developmental process in which the adherens and tight junctions of epithelial cells are degraded, resulting in a loss of cellular polarity and conversion to mesenchymal cells that are highly motile and invasive78. The key molecular changes associated with EMT are loss of epithelial protein marker expression, such as the adherens junction component E-cadherin, and gain of mesenchymal marker expression, including vimentin. During embryonic development, these cells can travel to distant sites and differentiate back into epithelial cells, known as mesenchymal-to-epithelial transition (MET), enabling tissue morphogenesis, tissue repair, and wound healing. EMT is a transcriptional program regulated by specific extracellular factors and cytokines, including TGF-β, Wnt, and Notch, resulting in the activation of signaling pathways such as NF-κB. Many of these factors and pathways are all commonly dysregulated in cancer79. Of note, cells within tumors displaying EMT properties have been identified using immunohistological (IHC) methods. Many parallels can be drawn between the processes of wound healing and tumorigenesis, since both involve the recruitment of mesenchymal stem cells and are associated with increased cellular invasiveness. It is believed that EMT is an important step in cancer invasion and metastasis, as cells exhibiting EMT-like characteristics are often observed at the invasive front of tumors80.
EMT-associated signaling networks promote activation of the transcriptional repressors Snail, Slug, Zeb1, Zeb2/SIP1, and Twist 81. These factors bind to the promoter of E-cadherin and block transcription by promoting chromatin condensation via the activation of histone deacetylases and other corepressors. This results in the loss of adherens junctions as well as loss of cell polarity67. TGF-β is a cytokine that promotes EMT through its ability to drive Smad complex association with Zeb proteins, resulting in repression of E-cadherin expression82. TGF-β-mediated induction of EMT is accompanied by apoptosis and growth arrest, known as lethal EMT (see above). TGF-β can also induce transient activation of the RAS and PI3K/AKT pathways, which can help to block the apoptotic effects of the cytokine and produce a stable mesenchymal phenotype in cells83. Smad4, an important co-factor in TGF-β signal transduction, functions as a tumor suppressor in pancreatic cancer by promoting lethal EMT upon TGF-β stimulation.
In pancreatic cancer, as well as other malignancies, cancer stem cells (CSC) are multipotent or pluripotent progenitor cells in the tumor. CSCs can be identified and distinguished by high expression of cell surface markers including CD133 and CD44, which are typically not expressed on bulk tumor cells. They can self-renew as well as divide asymmetrically to give rise to differentiated cells. These characteristics allow CSCs to initiate or regenerate a tumor. CSCs are thought to be derived from existing progenitor cells or dedifferentiated cells within a tumor67. EMT is known to promote CSC-like properties in pancreatic cancer by inducing a CD44high and CD24low cell surface marker expression profile characteristic of cancer stem cells84. This observation suggests EMT plays a direct causal role in the emergence of CSCs. However, the mechanistic basis for the association between EMT and CSC induction remains to be fully elucidated.
Mesenchymal-like properties can render pancreatic cancers more resistant to anti-cancer therapeutics, especially cytotoxic agents that induce apoptotic cell death. When comparing mesenchymal-like cells in pancreatic cancer to epithelial-like cells, mesenchymal-like cells are more resistant to gemcitabine, 5-fluorouracil (5-FU), cisplatin, and epidermal growth factor receptor (EGFR) inhibitors as assessed by cellular growth and viability85. Furthermore, pancreatic cancer cells that are intrinsically resistant to gemcitabine express high levels of vimentin and low levels of E-cadherin, indicating that these drug resistant cells are more mesenchymal in nature86. As EMT has been associated with the emergence of CSCs, it has been noted that the use of cytotoxic agents such as gemcitabine can lead to an enrichment of CD44high, CD24low CSC-like cells84. Finally, EMT is also associated with resistance to adjuvant or neoadjuvant radiotherapy, as resistant cells express high levels of vimentin and low levels of E-cadherin. These drug-resistant cells express high levels of the stem cell markers Oct4, CD24, and CD133, further indicating that EMT is associated with the acquisition of stem-like properties87. In the context of oncogenic KRAS signaling, studies indicate that loss of dependence on oncogenic KRAS for survival significantly correlates with mesenchymal-like phenotypic characteristics in PDAC cell lines31. These studies are supported by findings that primary PDAC tumors can be classified into distinct molecular subtypes based on the expression of EMT markers. These subtypes have been designated classical, exocrine-like and quasi-mesenchymal. Of note, the cell lines with quasi-mesenchymal properties are weakly dependent on oncogenic KRAS to maintain viability32.
The clinical and pathophysiological significance of EMT in promoting metastasis and drug resistance remains controversial. A recent study has sought to tackle this question using EMT lineage tracing experiments in a spontaneous breast-to-lung metastasis model65. The study demonstrates that a small proportion of cells in primary tumors undergo EMT and that cells found in lung metastases are predominantly epithelial-like. Furthermore, blocking the EMT process via the overexpression of miR-200, a negative regulator of Zeb1, does not significantly impair the formation of distant lung metastases. These findings strongly suggest that EMT is not necessary for metastasis to occur efficiently. Using the same experimental conditions, the study also demonstrated that treatment of primary tumors with the chemotherapeutic agent cyclophosphamide results in the accumulation of mesenchymal-like cells which contribute more significantly to metastasis formation. Overexpression of miR-200 blocked this metastatic growth. Therefore, while EMT may not be required for metastasis under treatment naive conditions, drug resistant cells with EMT-like properties may emerge with an increased metastasis-forming ability. Such studies will be critical in addressing the key roles of EMT in driving cancer metastasis and drug resistance across a number of cancer types, including pancreatic cancer. Importantly, associations between the EMT program, the emergence of CSCs, cancer invasiveness, KRAS dependence, and drug resistance provide new therapeutic opportunities for pancreatic cancer treatment.
4. MOLECULAR SUBTYPE CLASSIFICATIONS OF PANCREATIC CANCER
PDACs harbor a number of recurrent genetic alterations, including activation of KRAS in addition to loss of TP53, SMAD4, and CDKN2A6. A multitude of other mutations occur to varying degrees in subsets of tumors, leading to dysregulation of cellular processes such as DNA damage repair, cell cycle regulation, TGF-β signaling, and chromatin modification18. However, some of the observed alterations may be “passenger” mutations that play minor roles in disease pathogenesis. Efforts to identify strategies to manage PDAC have been confounded by the molecular diversity of these aggressive tumors. Understanding the genetic variation in PDAC is now a major area of focus to develop more effective therapeutics. Although there are recurrent gene mutations, the transcriptional networks that are activated in tumors with similar genetic profiles can vary significantly as determined by global gene expression profiling. Using transcriptome data from RNA-seq analyses of primary PDACs, a recent study has generated a PDAC classification system of four major molecular subtypes. These are squamous (quasimesenchymal), pancreatic progenitor (classical), immunogenic, and aberrantly differentiated endocrine-exocrine (ADEX, exocrine-like)18,32. Of note, these subtypes recapitulate the major subtypes identified by Collison and colleagues, including those associated with EMT32.
The squamous PDAC subtype is characterized by high mesenchymal marker gene expression and has the worst prognosis in comparison to the other subtypes. This subtype primarily comprises PDACs that are histologically identified as adenosquamous carcinoma18,32. Squamous subtype tumors are enriched for TP53 mutations. They display activation of the p53 family member TP63ΔN and its associated transcriptional network. TP63ΔN regulates squamous epithelial cellular differentiation in contrast to columnar differentiation that is characteristic of pancreatic ductal epithelial cells. Thus squamous transdifferentiation in this molecular subtype is driven by an EMT-like program. Hypermethylation of pancreatic endodermal cell-fate determination genes, including PDX1, GATA6 and HNF1B, further contributes to the dedifferentiated and mesenchymal nature of these tumors. A number of other phenotypic characteristics can distinguish squamous subtype PDAC tumors including increased prevalence KDM6A mutations, which affect chromatin remodeling, inflammation, the hypoxia response, metabolic reprogramming, TGF-β signaling, MYC activation, and autophagy. Finally, cell lines derived from squamous subtype tumors are more sensitive to the cytotoxic effects of gemcitabine32.
Pancreatic progenitor subtype tumors are typically more epithelial-like in nature, as determined by high expression levels of epithelial marker genes, including adhesion-associated genes such as CDH1/E-cadherin18. This subtype bears molecular similarities to KRAS-dependent PDAC cell lines31. Furthermore, high expression levels of genes that contribute to early pancreatic development are prevalent (FOXA2/3, PDX2, MNX1, and GATA6). These genes are important for terminal differentiation to pancreatic ductal epithelial cells. For example, PDX2 induces differentiation of ductal, exocrine, and endocrine cells of the pancreas. Cellular processes that are characteristic of the pancreatic progenitor subtype are fatty acid oxidation, steroid hormone biosynthesis, drug metabolism, and O-linked glycosylation of mucins. In contrast to the squamous subtype, pancreatic progenitor subtype tumor-derived cell lines are more sensitive to the EGFR inhibitor erlotinib32. Interestingly, development of pancreatic progenitor subtype tumors is linked to maturity onset diabetes of the young (MODY)18.
The aberrantly differentiated endocrine-exocrine (ADEX) tumor subtype is characterized by the simultaneous expression of transcriptional programs observed in the endocrine and exocrine pancreas. Both programs are typically activated in the later stages of normal organ development and differentiation in a mutually exclusive manner. Genes upregulated in ADEX tumors play a role in acinar and endocrine differentiation as well as regeneration and pancreatitis. Furthermore, a number of these genes are associated with KRAS activation. ADEX tumors comprise a subclass of the pancreatic progenitor tumor subtype and are histologically associated with rare acinar cell carcinomas18,32.
Immunogenic subtype tumors exhibit many of the same molecular characteristics observed in the pancreatic progenitor subtype but can be distinguished by the upregulation of various immune-related transcriptional programs. These programs are associated with B- and T-cell receptor signaling, Toll-like receptor signaling, antigen presentation, and acquired immune suppression through immune checkpoint pathways such as CTLA4 and PD1. Additionally, immunogenic subtype tumors exhibit a notable increase in infiltrating B- and T-cells. Tumors of this subtype display histological characteristics observed in mucinous non-cystic (colloid) adenocarcinomas and IPMN-derived carcinomas18. PDACs are generally non-responsive to new classes of anti-PD1 immunomodulatory checkpoint inhibitors, such as pembrolizumab. However, tumors of the immunogenic PDAC subtype may be more responsive to these immune checkpoint blockers either alone or in combination with other chemotherapeutic agents88.
5. DEREGULATED SIGNALING NETWORKS IN PANCREATIC CANCER
5.1. The EGFR-KRAS Network
Receptor tyrosine kinases (RTKs) are cell surface receptors for many growth factor ligands, including epidermal growth factor (EGF)89. RTK dysregulation plays a significant role in many cancers. Upon binding to a growth factor ligand, RTKs form homo- or heterodimers, bringing their intracellular kinase domains into close proximity. The intracellular receptor regions are transphosphorylated to create docking sites for SH2-domain containing adapter proteins and enzymes. This activates many downstream signaling cascades mediated by proteins including RAS and PI3K (Fig. 2A)90. While EGFR mutations are rare in pancreatic cancer, inhibition of receptor kinase activity with erlotinib is moderately effective for treating a subset of PDACs89. Since KRAS is so frequently mutated in pancreatic cancer, the activation of RAS-mediated signaling pathways is thought to be a major driver of disease pathogenesis. Other RAS isoform genes such as HRAS and NRAS are mutated infrequently in pancreatic cancer, suggesting that the KRAS locus provides a unique advantage in the context of PDAC pathogenesis. Although not mutated in PDAC, HRAS and NRAS proteins may participate in EGFR/KRAS signaling networks to promote tumorigenesis via protection against DNA damage-induced stress pathways91. As discussed above, KRAS mutations occur early in PanIN lesions. Given the high prevalence of KRAS mutations, it is not surprising that the KRAS oncoprotein is a potent driver of PDAC development92.
RAS GTPase-activating proteins (GAPs) catalyze the hydrolysis of GTP to GDP, resulting in the inactivation of RAS20,21. Thus, GAPs, such as the NF1 gene product, can serve as tumor suppressors, although they are not frequently mutated in PDAC. The GTP bound form of RAS undergoes a conformational change in which two “switch” regions (I and II) converge to form the effector binding domain. This domain forms biochemical interactions with effectors to either promote their allosteric activation or enable recruitment to the membrane, allowing for initiation of downstream signal transduction cascades. One of the most well studied classes of RAS effectors are RAF family serine/threonine (S/T) kinases, which initiate the MEK/ERK MAP kinase pathway through a cascade of sequential phosphorylation events19. This results in phosphorylation and nuclear localization of transcription factors, such as Elk-1, that drive cell proliferation, inflammatory signaling, differentiation, and cell survival. RAS can also activate the lipid kinase PI3K by associating with the p110 subunit of the PI3K complex. This complex is composed of a p110 catalytic subunit and a p85 regulatory subunit, which together regulate many key tumorigenic processes, including cell survival and proliferation. Activated RAS recruits PI3K to the plasma membrane and promotes its catalytic activity, which is to facilitate the conversion of the membrane phospholipid phosphatidylinositol-4,5-bisphosphate (PIP2) to phosphatidylinositol-3,4,5-triphosphate (PIP3). PIP3 serves as a binding site for proteins that contain a pleckstrin-homology (PH) domain. To inactivate the PI3K pathway, the lipid phosphatase PTEN hydrolyzes PIP3 to PIP2. PTEN is often dysregulated in late stages of pancreatic cancer resulting in hyperactivation of the PI3K pathway and an acceleration of PDAC development93. A primary effector of PI3K activation is the AKT S/T kinase, which is activated by sequential phosphorylation mediated by PDK1 and mTORC2. AKT suppresses apoptosis by promoting MDM2-dependent p53 proteolysis. It can also activate mTORC1, which phosphorylates 4EBP1 and ribosomal S6 kinase to promote CAP-dependent mRNA translation (Fig. 2A). The activation of mTORCI is a key feature of many PDACs that results in increased protein translation, stem cell renewal, proliferation, and inhibition of autophagy via ULK1 kinase activation94,95.
5.2. Hippo Signaling
The Hippo tumor suppressor pathway regulates organ size and maintains tissue homeostasis by restricting cell proliferation and, when necessary, inducing apoptosis96. The Hippo pathway is activated by a kinase cascade that was originally discovered via genetic screens aimed at identifying tumor suppressor genes in the fruit fly Drosophila melanogaster. Mutations in various Hippo pathway component genes, such as Hippo (Hpo) itself, Salvador (Sav), and Warts (Wts) results in increased organ size as well as tissue overgrowth. The evolutionary conservation of Hippo signaling has been established in mammals, where pathway deregulation is frequently observed in many solid cancers, including PDAC97,98.
The induction of Hippo signaling occurs in response to a number of stimuli including contact inhibition, serum deprivation, energy stress, and actin depolymerization (Figure 2B)99. These signals ultimately result in the phosphorylation and activation of large tumor suppressor kinases 1/2 (LATS1/2; Wts orthologs) by the MST1/2 (Hpo orthologs) kinases. Once activated, LATS1/2 phosphorylate the transcriptional co-activator Yes-associated protein (YAP) and its vertebrate-specific paralog transcriptional co-activator TAZ, encoded by the WWTR1 gene. Phosphorylation of YAP and TAZ promotes their association with 14-3-3 proteins, resulting in their cytoplasmic retention and subsequent degradation. In the absence of Hippo pathway activation, YAP and TAZ are able to translocate to the nucleus, where they interact with the TEAD family of transcription factors to induce the expression of genes associated with cell survival, proliferation, and migration. Thus, under normal physiological conditions in epithelial tissue such as the pancreatic duct, YAP/TAZ activity remains low.
Deregulation of Hippo signaling in PDAC is evidenced by increased YAP/TAZ protein levels and nuclear localization100-102. However, the underlying mechanisms accounting for increased YAP/TAZ activation remain an area of active investigation. Pan-cancer genomic analyses have revealed that mutations in the genes encoding Hippo pathway components occur at very low frequencies97,98. Given the oncogenic activity of YAP, this is a surprising finding103. The low frequency of Hippo pathway gene mutations in human cancer could indicate that that Hippo signaling is essential for both physiological and pathophysiological tissue homeostasis. This notion is supported by several studies in which the deletion of various Hippo pathway components in mice results in embryonic lethality104-108. Therefore, pathway output must be physiologically maintained within certain limits to ensure that cellular proliferation remains in check.
The mechanisms that control YAP/TAZ activation in PDAC have yet to be fully elucidated. One possible mechanism is loss of LATS1/2 expression as a consequence of promoter hypermethylation. Epigenetic regulation of the Hippo pathway, which has been documented in both colon and breast cancers109,110, functionally disconnects YAP/TAZ activity from upstream pathway induction. Another alteration that may account for the elevated YAP/TAZ levels in PDAC is gene amplification. YAP and TAZ gene copy number gains have been documented in several different cancer types97,111. In such contexts, increased YAP/TAZ protein levels result in constitutive activation even when the Hippo pathway is intact and active.
Recent studies of YAP function in GEMMs suggest that it plays a key role in PDAC initiation and maintenance. Ectopic expression of YAP in the developing mouse pancreas results in the appearance of metaplasia and impaired differentiation of both the endocrine and exocrine compartments112,113. However, increased organ size and tumor formation were not observed. Subsequent work in Kras and Kras/Tp53-mutant mice demonstrated that YAP is essential for the progression of neoplasia to PDAC102. Furthermore, YAP gene amplification is observed in some cases following loss of oncogenic KRAS expression in GEMMs and cancer cell lines, leading to increased cell survival and tumor recurrence35,36. In these studies, YAP activity can bypass the requirement for sustained KRAS signaling, conferring a loss of KRAS oncogene dependence. Furthermore, YAP can become activated as a consequence of MAPK signaling to promote survival and proliferation. Hence, the loss of KRAS expression in PDAC GEM models initially results in robust tumor regression. However, a subpopulation of cells with amplification and overexpression of YAP, is able to repopulate the tumor, leading to disease relapse. Collectively, these studies suggest that Hippo pathway deregulation plays a critical role in PDAC progression as well as dependence on the KRAS-MAPK pathway. Thus, modulation of YAP and/or TAZ activity represents an attractive opportunity for therapeutic intervention.
5.3. Inflammation
Innate immune responses and inflammation have been associated with cancer etiology in many contexts, and pancreatic cancer is no exception. Recent studies using KRAS-driven GEM models have highlighted the role of acute pancreatitis-associated inflammation in driving the progression of PanIN lesions to full-blown PDAC114. Some studies estimate that up to 50% of the PDAC tumor cell mass can be composed of stromal and immune cells recruited to the tumor via paracrine cytokine signaling115. Furthermore, oncogenic KRAS signaling can promote the development of the inflammatory microenvironment found in PDAC tumors. It is hypothesized that inflammation promotes PDAC initiation and progression through several different mechanisms. Firstly, it promotes cell survival and proliferation through inflammatory mediators that activate transcription factors responsible for anti-apoptotic signaling as well as cancer cell invasion and metastasis. Examples of such proinflammatory, prosurvival transcription factors include STAT3, the AP-1 complex (Jun/Fos), and NF-κB. KRAS induces IL-6 and IL-11 cytokine expression and secretion, leading to STAT3 transcriptional activation. KRAS also promotes NF-κB signaling via induction of the MAPK pathway, leading to increased TNFα and IL-1 levels, which drive a positive feedback loop for NF-κB activation. Reciprocally, NF-κB can enhance RAS activity through a RAS-NF-κB-cyclooxygenase-2 positive feedback loop68. Thus, inflammatory cytokines and RAS signaling are intimately linked. Secondly, proinflammatory cytokines, including TNFα, IL-6, and IL-1, can promote metastasis by inducing EMT as well as the acquisition of cancer stem cell-like traits115. These mechanisms, coupled with the ability of NF-κB to activate Notch and other oncogenic pathways, lead to accelerated PDAC development. Finally, inflammation-associated cytokines can impair immunosurveillance of tumor cells due to an increase in immune cell subsets that have immunosuppressive properties, including regulatory T-cells (T-regs) and myeloid-derived suppressor cells (MDSCs). These cell types can negatively regulate the numbers and functional activation of cytotoxic CD8+ T-cells, possibly via increased expression of granulocyte-macrophage colony-stimulating factor (GM-CSF)116,117.
There are likely to be a number of additional and diverse mechanisms by which inflammation promotes cancer. Inflammation can cause cellular damage, for example, via the accumulation of reactive oxygen species (ROS) that promote oxidative DNA damage115. This may lead to increased genetic evolution in PDAC development as tumor cells acquire somatic mutations that confer selective advantages to promote growth and survival. When cells acquire an activating oncogenic mutation (e.g. KRAS), a phenomenon known as oncogene induced senescence (OIS) is triggered as a consequence of CDKN2A/p16 induction. This senescence response can be suppressed by inflammatory signaling, representing a mechanism by which inflammation promotes cancer progression. Taken together, inflammation clearly plays a critical role in immunosuppression, KRAS pathway modulation, and cancer metastasis. Thus, targeting inflammatory pathways could represent an attractive avenue for therapeutic intervention.
5.4. Autophagy
Autophagy is a well-characterized metabolic, homeostatic process by which cellular constituents, such as proteins and organelles, are degraded and recycled to meet cellular demands under conditions of nutrient deprivation or stress. Such conditions occur during tumorigenesis, and indeed, constitutive activation of autophagy has been observed in many tumors including PDAC118,119. Three types of autophagy are typically described: macroautophagy, microautophagy, and chaperonin-mediated autophagy. Macroautophagy, henceforth referred to simply as autophagy, is the primary pathway, as well as the most significant in PDAC. During nutrient deprivation, misfolded and non-essential proteins, as well as organelles, are sequestered into a lipid bilayer known as a phagophore. This gives rise to a double-membrane structure known as the autophagosome, which subsequently fuses with a lysosome, forming an autophagolysosome, or autolysosome. The presence of degradative enzymes combined with low pH within autolysosomes cause the breakdown of cellular macromolecules. Amino acids and other building blocks are then recycled, allowing for cellular homeostasis to be maintained. In normal pancreatic tissue, there is a basal level of autophagy that serves to maintain homeostasis118.
The role of autophagy in pancreatic cancer is complicated and has yet to be fully elucidated. PDACs typically exhibit high basal levels of autophagic activity, including increased number and size of both autophagosomes and autolysosomes, when compared to normal pancreatic tissue120. It is hypothesized that tumor cells gain a selective advantage when autophagy is activated, as it may allow them to cope with the stresses resulting from high rates of cell division as well as nutrient deprivation, which can occur from hypoxic conditions in poorly vascularized regions of the tumor121. Therefore, autophagy can drive the survival of PDAC cells under these conditions. Chloroquine, which inhibits autophagy by blocking the formation of autolysosomes, potently suppresses the growth of some PDAC cell lines. When autophagy is inhibited with chloroquine, or by genetic ablation of the key autophagy regulator Atg-7, Kras-induced progression of PanIN to PDAC is blocked as a consequence of cell death, growth arrest, or senescence122. However, when combined with TP53 deletion, Atg-7 loss enhances, rather than suppresses, Kras-driven PDAC progression123. These findings highlight the highly context-dependent role of autophagy in PDAC pathogenesis, which depends on TP53 status and possibly other tumor suppressor gene mutations. Lastly, autophagy could allow PDAC cells to cope with the deleterious effects of chemotherapeutics and radiation therapy. For example, treatment of the PDAC cell line PANC-1 with gemcitabine or 5-fluorouracil induces autophagy121. Interestingly, the combination of autophagy inhibitors, such as chloroquine, with chemotherapy or radiation therapy greatly enhances cytotoxicity in PDAC cell lines.
6. CURRENT AND FUTURE THERAPEUTIC STRATEGIES FOR PANCREATIC CANCER
PDACs are notoriously difficult to treat for a number of reasons9. Most patients with PDAC are often asymptomatic, and diagnoses are not usually made until after the tumors have become metastatic. Currently, there are few effective therapeutic options for PDAC patients. The only “curative” treatment is surgical resection, but its success rate in patients with operable tumors is low, with a 5 year survival rate of only 20%, a 60% rate of relapse within 6 months, and an overall relapse rate of more than 80%. Due to the typically late diagnosis, many patients are not candidates for surgical resection. In contrast, chemotherapy has marginal, but measureable, effects on overall survival in PDAC patients. The efficacy of chemoradiation as a PDAC therapeutic regimen remains unclear when compared to chemotherapy alone. With current PDAC therapeutic options, overall survival of 5 years or greater is estimated to be less than 5%, and these rates have not changed significantly in the past 30 years. Understanding PDAC etiology and pathogenesis at the detailed molecular level as a means to developing better therapeutics to treat the disease remains a pressing goal. PDAC is a complex disease with multiple stages that will respond to different sets of anti-cancer agents. The complexity of this disease is further highlighted by recent studies using a PDAC GEM model demonstrating that cells from PanIN lesions can metastasize. Pancreatic epithelial cells are able to disseminate from the pancreas at an early stage of the disease, when a primary lesion is not yet detectable. These cells are capable of seeding in the liver and potentially other distant sites. Therefore, therapeutics may be more successful if they are designed to treat pancreatic cancer as a systemic disease rather than localized one124. This could explain the low rate of success with local treatment, as cancer cells may be present at distal sites such as the liver or the lymph nodes. It is clear that new approaches to treat PDAC must be developed. A number of innovative avenues for therapeutic intervention are currently being evaluated in basic science and clinical studies9.
For PDAC patients with resectable lesions, the standard of care involves surgical resection and adjuvant chemotherapy with gemcitabine or 5-fluorouracil (5-FU). Both agents are pyrimidine analogs that block various stages of DNA replication, leading to cell cycle arrest and, in some cases, apoptosis. These conventional cytotoxic agents preferentially target rapidly dividing cells and can lead to modest tumor regression or growth suppression. In metastatic disease, the standard of care is a drug regimen that consists of gemcitabine, FOLFIRINOX, or nab-paclitaxel. FOLFIRINOX is a combination chemotherapy made up of folinic acid (leucovorin), 5-FU, irinotecan, and oxaliplatin. These agents collectively interfere with DNA replication and transcription. Paclitaxel, on the other hand, binds to, and stabilizes, microtubules, preventing their disassembly and ultimately blocking progression through mitosis. Nab-paclitaxel is an albumin-bound form of the drug that has increased bioavailability. As these drugs indiscriminately target all rapidly dividing cells, many side-effects and dose-limiting toxicities are associated with their utilization. For this reason, conventional cytotoxic chemotherapeutics provide only marginal increases in median patient survival on the order of weeks or months9. To improve the survival benefit conferred by these agents, many ongoing studies are investigating optimal dosing regimens and combinations for treating different stages of pancreatic cancer. Clinical trials involving chemoradiotherapy have returned inconsistent results, and the treatment remains controversial. This is possibly due to the lack of biomarkers to determine which patients will be responsive to radiation treatment. A newer strategy involves gemcitabine in combination with the EGFR tyrosine kinase inhibitor erlotinib, which results in a modest survival benefit of 12 days compared to gemcitabine alone. Thus, erlotinib is the only FDA-approved targeted therapy available for treatment of pancreatic cancer. Some patients respond much more favorably to erlotinib than others, suggesting that there could be biomarkers to identify PDAC patients who will likely benefit most from anti-EGFR therapies such as erlotinib. A recent study found that TP53-wild-type tumors may be more sensitive to EGFR inhibition. Thus, identifying the right patient population for a particular targeted therapy, in the interests of precision medicine, remains a key goal of PDAC therapeutics9.
Effective targeted therapeutics and precision medicine-based approaches for PDAC have yet to be identified. One approach under investigation is to take advantage of the tumor specific environment. Due to the high density of fibrous connective tissue that is characteristic of pancreatic cancer, drug delivery to tumors is severely impaired. Hyaluronic acid is an extracellular matrix component found surrounding tumors that presents a physical barrier for drug delivery. The degradation of hyaluronic acid via hyaluronidase may enhance drug delivery. PEGPH20 is a PEGylated form of hyaluronidase that is currently being tested and shows prolonged survival of tumor-bearing mice when given in combination with gemcitabine125. DNA damaging agents such as TH-302 (evofosfamide) can take advantage of the hypoxic environment in pancreatic tumors to increase their specificity. This drug is derived from a nitrogen mustard that becomes activated under hypoxic conditions and releases its active form, dibromoisophosphoramide mustard (Br-IDM), a DNA alkylating agent. In combination with gemcitabine, TH-302 provides a 6 month survival benefit compared to 3.6 months with gemcitabine alone126,127. Drug modifications that provide improved delivery are also being evaluated, such as nab-paclitaxel or nanoliposomal formulations of irinotecan (MM-398). These modifications can increase the plasma half-life of drugs and increase the availability of their active metabolites. MM-398 provides a 2-month survival advantage when given in combination with 5-FU and folinic acid128. To identify accurate biomarkers of response to particular agents, DNA sequencing of PDACs for sensitizing genetic alterations represents a major step in advancing precision medicines to treat the disease. Some examples currently under investigation include SMAD4 for chemoradiotherapy, STK11 for mTOR inhibitors, and the genes PALB2, ATM, and BRCA2 for DNA damaging agents9.
Another avenue for therapeutic intervention could be to exploit oncogene “addiction”129. As mutant KRAS is a key linchpin in PDAC pathogenesis, it remains a major therapeutic target, albeit a stubborn one. Thus far, there has been little success in targeting the RAS-related proteins, due to their high affinity for GTP and the abundance of GTP in the cell, which prevent access to the protein active site. Nonetheless, many alternate methods of RAS inhibition are currently being investigated. The RAS protein must undergo several post-translational modifications, including a farnesylation step, before the protein is functional. Blocking farnesylation of the protein is one potential method to inhibit the RAS pathway, but results have been disappointing thus far because KRAS can be alternatively isoprenylated with a geranylgeranyl group. After the RAS protein has been translated, it must be transported to the cell membrane, a process that requires the PDEδ protein. Thus, the PDEδ inhibitor Deltarasin has been developed, which can block RAS membrane translocation. This prevents the downstream activation of ERK, leading to suppression of KRAS driven PDAC cell proliferation and viability. Although directly inhibiting KRAS has proven to be difficult, allosteric, covalent-modifying inhibitors that stabilize the GDP bound form of the protein have been identified for the G12C isoform commonly found in non-small cell lung cancer (NSCLC). It is possible that a similar agent could be identified for the most common isoform found in pancreatic cancer, KRAS G12D. As direct inhibition of KRAS remains a challenge, downstream inhibition of the RAF-MEK-ERK cascade and/or PI3K/mTOR has become an active area of investigation9,19. Lastly, synthetic lethality is another approach that attempts to identify genetic interactors with mutant KRAS that cooperate to promote PDAC tumor cell survival129. Thus far, genome-wide KRAS synthetic lethality siRNA screens have failed to yield promising candidate therapeutic targets, perhaps due to the complexity and molecular heterogeneity of oncogenic KRAS mutant PDAC tumors.
Altered cellular metabolism is yet another area being studied to develop PDAC therapeutics. In the tumor microenvironment, dense desmoplastic regions surround the tumor, leading to hypoxia and decreased nutrient delivery to cells. Tumors with increased glycolysis gain a survival advantage in the hypoxic environment (known as the Warburg effect), and autophagy is induced in response to nutrient deprivation. As described above, chloroquine, or its derivative hydroxychloroquine, inhibits autophagy by blocking the formation of autolysosomes. While chloroquine alone is mildly effective in promoting PDAC cell death, studies indicate that it can sensitize tumors to MEK inhibitors, chemoradiotherapy, gemcitabine, and nab-paclitaxel130,131
Finally, immunotherapy is an exciting field in cancer therapeutics that has demonstrated dramatic effects in other diseases, such as melanoma. This approach employs activation of the host immune system to combat tumors by promoting tumor cell clearance via cytotoxic T-cells. The immunosuppressive environment found in pancreatic cancer prevents immunosurveillance of tumors from occurring efficiently. Thus, supercharging the immune system to overcome this immunosuppression represents an innovative therapeutic strategy. One approach is to sensitize the immune system to pancreatic cancer cells through vaccination. GVAX pancreas is created from whole tumor cells, which are genetically engineered to express granulocyte-macrophage colony-stimulating factor (GM-CSF) and then irradiated to prevent cell division. These engineered pancreatic cancer cells are capable of recruiting dendritic cells that can phagocytose the tumor cells, which are subsequently presented to T-cells to promote their activation and ability to recognize and clear tumor cells132. Along the same lines, T-cells can be modified to express chimeric antigen receptors that recognize tumor antigens. This approach has been successfully achieved in CD19-positive hematological malignancies but is still in early development for pancreatic cancer133. CD40, a cell surface protein on antigen presenting cells plays a key role in immune cell activation. Thus, CD40 agonists are being tested in combination with gemcitabine to promote accumulation of phagocytic macrophages in tumors134. PD-L1 and PD-L2 are ligands expressed by cancer cells capable of binding to the immune checkpoint receptor PD-1 on activated CD8+ T-cells, causing suppression of cytotoxic T-cell function. This impairs immunosurveillance of cancer cells. Monoclonal antibodies that target PD-L1 or PD-1, such as pembrolizumab, have been developed to promote CD8+ T-cell activation. These agents have yielded significant beneficial results in melanoma, non-small cell lung cancer, and renal cell carcinoma. As is the case with these malignancies, PD-L1 expression in pancreatic tumors is also associated with a poor prognosis88. Monoclonal antibodies against CTLA4 (e.g. ipilimumab), another T-cell immune checkpoint receptor, have also been tested in pancreatic cancer135. Unfortunately, checkpoint inhibitors have had disappointing results as single therapies in PDAC thus far. However, there is hope that they will be effective in combination with other chemotherapeutics, such as gemcitabine and nab-paclitaxel. In summary, PDAC remains one of the most deadly of all human diseases due a severe lack of effective therapeutics. Innovative therapeutic approaches for PDAC include optimizing the dosing regimens of current agents, targeting oncogene addiction, manipulating tumor metabolism, and harnessing the host immune system to fight this aggressive cancer type.
7. CONCLUSIONS
Pancreatic ductal adenocarcinoma is a highly aggressive malignancy associated with very poor clinical prognosis. Although the core genetic alterations in PDAC are well documented, their contributions to PDAC pathogenesis remain to be fully determined at the molecular level. Next generation sequencing has revealed the detailed complexity of the genomic landscape of PDAC, which is characterized by marked inter- and intratumor heterogeneity as well as a very high overall mutational burden. Gene mutations in PDAC have been shown to converge on a few critical signal transduction pathways and cellular processes including the KRAS-MAPK pathway, inflammation, and altered cellular metabolism. The complexity of PDAC pathogenesis is further illustrated by the classification of PDAC tumors into four major molecular subtypes that are distinguished by key phenotypic traits and pharmacological vulnerabilities. Detailed characterization of these subtypes could ultimately lead to the development of new precision medicines for treating PDAC. However, current therapeutic options for the disease remain limited. Understanding and attacking the complexity of PDAC pathogenesis will undoubtedly yield additional innovative therapeutic options for this aggressive and deadly disease.
REFERENCES
- 1.Roder PV, Wu B, Liu Y, Han W. Pancreatic regulation of glucose homeostasis. Exp Mol Med. 2016;48:e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keller J, Layer P. Human pancreatic exocrine response to nutrients in health and disease. Gut. 2005;54 Suppl 6:vi1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gittes GK. Developmental biology of the pancreas: a comprehensive review. Dev Biol. 2009;326(1):4–35. [DOI] [PubMed] [Google Scholar]
- 4.Polonsky KS. The past 200 years in diabetes. N Engl J Med. 2012;367(14):1332–1340. [DOI] [PubMed] [Google Scholar]
- 5.Braganza JM, Lee SH, McCloy RF, McMahon MJ. Chronic pancreatitis. Lancet. 2011;377(9772):1184–1197. [DOI] [PubMed] [Google Scholar]
- 6.Hezel AF, Kimmelman AC, Stanger BZ, Bardeesy N, Depinho RA. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2006;20(10):1218–1249. [DOI] [PubMed] [Google Scholar]
- 7.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. [DOI] [PubMed] [Google Scholar]
- 8.Lennon AM, Wolfgang CL, Canto MI, et al. The early detection of pancreatic cancer: what will it take to diagnose and treat curable pancreatic neoplasia? Cancer Res. 2014;74(13):3381–3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garrido-Laguna I, Hidalgo M. Pancreatic cancer: from state-of-the-art treatments to promising novel therapies. Nat Rev Clin Oncol. 2015;12(6):319–334. [DOI] [PubMed] [Google Scholar]
- 10.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74(11):2913–2921. [DOI] [PubMed] [Google Scholar]
- 11.Fesinmeyer MD, Austin MA, Li CI, De Roos AJ, Bowen DJ. Differences in survival by histologic type of pancreatic cancer. Cancer Epidemiol Biomarkers Prev. 2005;14(7):1766–1773. [DOI] [PubMed] [Google Scholar]
- 12.Phan GQ, Yeo CJ, Hruban RH, Lillemoe KD, Pitt HA, Cameron JL. Surgical experience with pancreatic and peripancreatic neuroendocrine tumors: review of 125 patients. J Gastrointest Surg. 1998;2(5):472–482. [PubMed] [Google Scholar]
- 13.Kern SE, Hruban RH, Hidalgo M, Yeo CJ. An introduction to pancreatic adenocarcinoma genetics, pathology and therapy. Cancer Biol Ther. 2002;1(6):607–613. [DOI] [PubMed] [Google Scholar]
- 14.Hruban RH, Adsay NV, Albores-Saavedra J, et al. Pancreatic intraepithelial neoplasia: a new nomenclature and classification system for pancreatic duct lesions. Am J Surg Pathol. 2001;25(5):579–586. [DOI] [PubMed] [Google Scholar]
- 15.Hruban RH, Takaori K, Klimstra DS, et al. An illustrated consensus on the classification of pancreatic intraepithelial neoplasia and intraductal papillary mucinous neoplasms. Am J Surg Pathol. 2004;28(8):977–987. [DOI] [PubMed] [Google Scholar]
- 16.Sohn TA, Yeo CJ, Cameron JL, et al. Intraductal papillary mucinous neoplasms of the pancreas: an updated experience. Ann Surg. 2004;239(6):788–797; discussion 797–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones S, Zhang X, Parsons DW, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321(5897):1801–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bailey P, Chang DK, Nones K, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531(7592):47–52. [DOI] [PubMed] [Google Scholar]
- 19.Downward J Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3(1):11–22. [DOI] [PubMed] [Google Scholar]
- 20.Stephen AG, Esposito D, Bagni RK, McCormick F. Dragging ras back in the ring. Cancer Cell. 2014;25(3):272–281. [DOI] [PubMed] [Google Scholar]
- 21.Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: Mission possible? Nat Rev Drug Discov. 2014;13(11):828–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer. 2011;11(11):761–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scheffzek K, Ahmadian MR, Kabsch W, et al. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277(5324):333–338. [DOI] [PubMed] [Google Scholar]
- 24.Scheidig AJ, Burmester C, Goody RS. The pre-hydrolysis state of p21(ras) in complex with GTP: new insights into the role of water molecules in the GTP hydrolysis reaction of ras-like proteins. Structure. 1999;7(11):1311–1324. [DOI] [PubMed] [Google Scholar]
- 25.Witkiewicz AK, McMillan EA, Balaji U, et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun. 2015;6:6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanda M, Matthaei H, Wu J, et al. Presence of somatic mutations in most early stage pancreatic intraepithelial neoplasia. Gastroenterology. 2012;142(4):730–733 e739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aguirre AJ, Bardeesy N, Sinha M, et al. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003;17(24):3112–3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bardeesy N, Cheng KH, Berger JH, et al. Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes Dev. 2006;20(22):3130–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Collins MA, Bednar F, Zhang Y, et al. Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. J Clin Invest. 2012;122(2):639–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ying H, Kimmelman AC, Lyssiotis CA, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149(3):656–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh A, Greninger P, Rhodes D, et al. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell. 2009;15(6):489–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Collisson EA, Sadanandam A, Olson P, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. 2011;17(4):500–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lim KH, Counter CM. Reduction in the requirement of oncogenic Ras signaling to activation of PI3K/AKT pathway during tumor maintenance. Cancer Cell. 2005;8(5):381–392. [DOI] [PubMed] [Google Scholar]
- 34.Viale A, Pettazzoni P, Lyssiotis CA, et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature. 2014;514(7524):628–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kapoor A, Yao W, Ying H, et al. Yap1 activation enables bypass of oncogenic Kras addiction in pancreatic cancer. Cell. 2014;158(1):185–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shao DD, Xue W, Krall EB, et al. KRAS and YAP1 converge to regulate EMT and tumor survival. Cell. 2014;158(1):171–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muller PA, Vousden KH. p53 mutations in cancer. Nat Cell Biol. 2013;15(1):2–8. [DOI] [PubMed] [Google Scholar]
- 38.Vogelstein B, Fearon ER, Hamilton SR, et al. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319(9):525–532. [DOI] [PubMed] [Google Scholar]
- 39.Sherr CJ. The INK4a/ARF network in tumour suppression. Nat Rev Mol Cell Biol. 2001;2(10):731–737. [DOI] [PubMed] [Google Scholar]
- 40.Rozenblum E, Schutte M, Goggins M, et al. Tumor-suppressive pathways in pancreatic carcinoma. Cancer Res. 1997;57(9):1731–1734. [PubMed] [Google Scholar]
- 41.Schutte M, Hruban RH, Geradts J, et al. Abrogation of the Rb/p16 tumor-suppressive pathway in virtually all pancreatic carcinomas. Cancer Res. 1997;57(15):3126–3130. [PubMed] [Google Scholar]
- 42.Wilentz RE, Geradts J, Maynard R, et al. Inactivation of the p16 (INK4A) tumor-suppressor gene in pancreatic duct lesions: loss of intranuclear expression. Cancer Res. 1998;58(20):4740–4744. [PubMed] [Google Scholar]
- 43.Maitra A, Adsay NV, Argani P, et al. Multicomponent analysis of the pancreatic adenocarcinoma progression model using a pancreatic intraepithelial neoplasia tissue microarray. Mod Pathol. 2003;16(9):902–912. [DOI] [PubMed] [Google Scholar]
- 44.Sugimoto M, Kuo ML, Roussel MF, Sherr CJ. Nucleolar Arf tumor suppressor inhibits ribosomal RNA processing. Mol Cell. 2003;11(2):415–424. [DOI] [PubMed] [Google Scholar]
- 45.Rocha S, Campbell KJ, Perkins ND. p53- and Mdm2-independent repression of NF-kappa B transactivation by the ARF tumor suppressor. Mol Cell. 2003;12(1):15–25. [DOI] [PubMed] [Google Scholar]
- 46.Qi Y, Gregory MA, Li Z, Brousal JP, West K, Hann SR. p19ARF directly and differentially controls the functions of c-Myc independently of p53. Nature. 2004;431(7009):712–717. [DOI] [PubMed] [Google Scholar]
- 47.Paliwal S, Pande S, Kovi RC, Sharpless NE, Bardeesy N, Grossman SR. Targeting of C-terminal binding protein (CtBP) by ARF results in p53-independent apoptosis. Mol Cell Biol. 2006;26(6):2360–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barton CM, Staddon SL, Hughes CM, et al. Abnormalities of the p53 tumour suppressor gene in human pancreatic cancer. Br J Cancer. 1991;64(6):1076–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khoo KH, Verma CS, Lane DP. Drugging the p53 pathway: understanding the route to clinical efficacy. Nat Rev Drug Discov. 2014;13(3):217–236. [DOI] [PubMed] [Google Scholar]
- 50.Morton JP, Timpson P, Karim SA, et al. Mutant p53 drives metastasis and overcomes growth arrest/senescence in pancreatic cancer. Proc Natl Acad Sci U S A. 2010;107(1):246–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weissmueller S, Manchado E, Saborowski M, et al. Mutant p53 drives pancreatic cancer metastasis through cell-autonomous PDGF receptor beta signaling. Cell. 2014;157(2):382–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Muller PA, Vousden KH, Norman JC. p53 and its mutants in tumor cell migration and invasion. J Cell Biol. 2011;192(2):209–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Powell E, Piwnica-Worms D, Piwnica-Worms H. Contribution of p53 to metastasis. Cancer Discov. 2014;4(4):405–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maitra A, Hruban RH. Pancreatic cancer. Annu Rev Pathol. 2008;3:157–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hezel AF, Bardeesy N. LKB1; linking cell structure and tumor suppression. Oncogene. 2008;27(55):6908–6919. [DOI] [PubMed] [Google Scholar]
- 56.Giardiello FM, Brensinger JD, Tersmette AC, et al. Very high risk of cancer in familial Peutz-Jeghers syndrome. Gastroenterology. 2000;119(6):1447–1453. [DOI] [PubMed] [Google Scholar]
- 57.Su GH, Hruban RH, Bansal RK, et al. Germline and somatic mutations of the STK11/LKB1 Peutz-Jeghers gene in pancreatic and biliary cancers. Am J Pathol. 1999;154(6):1835–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morton JP, Jamieson NB, Karim SA, et al. LKB1 haploinsufficiency cooperates with Kras to promote pancreatic cancer through suppression of p21-dependent growth arrest. Gastroenterology. 2010;139(2):586–597, 597 e581–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hahn SA, Seymour AB, Hoque AT, et al. Allelotype of pancreatic adenocarcinoma using xenograft enrichment. Cancer Res. 1995;55(20):4670–4675. [PubMed] [Google Scholar]
- 60.Hahn SA, Schutte M, Hoque AT, et al. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science. 1996;271(5247):350–353. [DOI] [PubMed] [Google Scholar]
- 61.Wilentz RE, Iacobuzio-Donahue CA, Argani P, et al. Loss of expression of Dpc4 in pancreatic intraepithelial neoplasia: evidence that DPC4 inactivation occurs late in neoplastic progression. Cancer Res. 2000;60(7):2002–2006. [PubMed] [Google Scholar]
- 62.Tascilar M, Skinner HG, Rosty C, et al. The SMAD4 protein and prognosis of pancreatic ductal adenocarcinoma. Clin Cancer Res. 2001;7(12):4115–4121. [PubMed] [Google Scholar]
- 63.David CJ, Huang YH, Chen M, et al. TGF-beta Tumor Suppression through a Lethal EMT. Cell. 2016;164(5):1015–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Padua D, Massague J. Roles of TGFbeta in metastasis. Cell Res. 2009;19(1):89–102. [DOI] [PubMed] [Google Scholar]
- 65.Fischer KR, Durrans A, Lee S, et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527(7579):472–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zheng X, Carstens JL, Kim J, et al. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527(7579):525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;29(34):4741–4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ling J, Kang Y, Zhao R, et al. KrasG12D-induced IKK2/beta/NF-kappaB activation by IL-1alpha and p62 feedforward loops is required for development of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;21(1):105–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Goggins M, Shekher M, Turnacioglu K, Yeo CJ, Hruban RH, Kern SE. Genetic alterations of the transforming growth factor beta receptor genes in pancreatic and biliary adenocarcinomas. Cancer Res. 1998;58(23):5329–5332. [PubMed] [Google Scholar]
- 70.Blasco MA, Lee HW, Hande MP, et al. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91(1):25–34. [DOI] [PubMed] [Google Scholar]
- 71.Lee HW, Blasco MA, Gottlieb GJ, Horner JW 2nd, Greider CW, DePinho RA. Essential role of mouse telomerase in highly proliferative organs. Nature. 1998;392(6676):569–574. [DOI] [PubMed] [Google Scholar]
- 72.Artandi SE, Chang S, Lee SL, et al. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature. 2000;406(6796):641–645. [DOI] [PubMed] [Google Scholar]
- 73.O’Hagan RC, Chang S, Maser RS, et al. Telomere dysfunction provokes regional amplification and deletion in cancer genomes. Cancer Cell. 2002;2(2):149–155. [DOI] [PubMed] [Google Scholar]
- 74.van Heek NT, Meeker AK, Kern SE, et al. Telomere shortening is nearly universal in pancreatic intraepithelial neoplasia. Am J Pathol. 2002;161(5):1541–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Greenberg RA, Chin L, Femino A, et al. Short dysfunctional telomeres impair tumorigenesis in the INK4a(delta2/3) cancer-prone mouse. Cell. 1999;97(4):515–525. [DOI] [PubMed] [Google Scholar]
- 76.Chin L, Artandi SE, Shen Q, et al. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell. 1999;97(4):527–538. [DOI] [PubMed] [Google Scholar]
- 77.Sato N, Mizumoto K, Nagai E, Tanaka M. Telomerase as a new target for pancreatic cancer treatment. J Hepatobiliary Pancreat Surg. 2002;9(3):322–327. [DOI] [PubMed] [Google Scholar]
- 78.Thiery JP. Epithelial-mesenchymal transitions in development and pathologies. Curr Opin Cell Biol. 2003;15(6):740–746. [DOI] [PubMed] [Google Scholar]
- 79.Wu Q, Miele L, Sarkar FH, Wang Z. The Role of EMT in Pancreatic Cancer Progression. Pancreat Disord Ther. 2012;2(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dvorak HF. Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. N Engl J Med. 1986;315(26):1650–1659. [DOI] [PubMed] [Google Scholar]
- 81.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2(6):442–454. [DOI] [PubMed] [Google Scholar]
- 82.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7(6):415–428. [DOI] [PubMed] [Google Scholar]
- 83.Gal A, Sjoblom T, Fedorova L, Imreh S, Beug H, Moustakas A. Sustained TGF beta exposure suppresses Smad and non-Smad signalling in mammary epithelial cells, leading to EMT and inhibition of growth arrest and apoptosis. Oncogene. 2008;27(9):1218–1230. [DOI] [PubMed] [Google Scholar]
- 84.Shah AN, Summy JM, Zhang J, Park SI, Parikh NU, Gallick GE. Development and characterization of gemcitabine-resistant pancreatic tumor cells. Ann Surg Oncol. 2007;14(12):3629–3637. [DOI] [PubMed] [Google Scholar]
- 85.Arumugam T, Ramachandran V, Fournier KF, et al. Epithelial to mesenchymal transition contributes to drug resistance in pancreatic cancer. Cancer Res. 2009;69(14):5820–5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li Y, VandenBoom TG 2nd, Kong D, et al. Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer Res. 2009;69(16):6704–6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Du Z, Qin R, Wei C, et al. Pancreatic cancer cells resistant to chemoradiotherapy rich in “stem-cell-like” tumor cells. Dig Dis Sci. 2011;56(3):741–750. [DOI] [PubMed] [Google Scholar]
- 88.Nomi T, Sho M, Akahori T, et al. Clinical significance and therapeutic potential of the programmed death-1 ligand/programmed death-1 pathway in human pancreatic cancer. Clin Cancer Res. 2007;13(7):2151–2157. [DOI] [PubMed] [Google Scholar]
- 89.Troiani T, Martinelli E, Capasso A, et al. Targeting EGFR in pancreatic cancer treatment. Curr Drug Targets. 2012;13(6):802–810. [DOI] [PubMed] [Google Scholar]
- 90.Pao W, Chmielecki J. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat Rev Cancer. 2010;10(11):760–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Grabocka E, Pylayeva-Gupta Y, Jones MJ, et al. Wild-type H- and N-Ras promote mutant K-Ras-driven tumorigenesis by modulating the DNA damage response. Cancer Cell. 2014;25(2):243–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Eser S, Schnieke A, Schneider G, Saur D. Oncogenic KRAS signalling in pancreatic cancer. Br J Cancer. 2014;111(5):817–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Asano T, Yao Y, Zhu J, Li D, Abbruzzese JL, Reddy SA. The PI 3-kinase/Akt signaling pathway is activated due to aberrant Pten expression and targets transcription factors NF-kappaB and c-Myc in pancreatic cancer cells. Oncogene. 2004;23(53):8571–8580. [DOI] [PubMed] [Google Scholar]
- 94.Baer R, Cintas C, Therville N, Guillermet-Guibert J. Implication of PI3K/Akt pathway in pancreatic cancer: When PI3K isoforms matter? Adv Biol Regul. 2015;59:19–35. [DOI] [PubMed] [Google Scholar]
- 95.Soler A, Figueiredo AM, Castel P, et al. Therapeutic benefit of selective inhibition of p110alpha PI3-kinase in pancreatic neuroendocrine tumors. Clin Cancer Res. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhao B, Li L, Lei Q, Guan KL. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 2010;24(9):862–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer. 2013;13(4):246–257. [DOI] [PubMed] [Google Scholar]
- 98.Moroishi T, Hansen CG, Guan KL. The emerging roles of YAP and TAZ in cancer. Nat Rev Cancer. 2015;15(2):73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Meng Z, Moroishi T, Guan KL. Mechanisms of Hippo pathway regulation. Genes Dev. 2016;30(1):1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Morvaridi S, Dhall D, Greene MI, Pandol SJ, Wang Q. Role of YAP and TAZ in pancreatic ductal adenocarcinoma and in stellate cells associated with cancer and chronic pancreatitis. Sci Rep. 2015;5:16759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dong J, Feldmann G, Huang J, et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130(6):1120–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang W, Nandakumar N, Shi Y, et al. Downstream of mutant KRAS, the transcription regulator YAP is essential for neoplastic progression to pancreatic ductal adenocarcinoma. Sci Signal. 2014;7(324):ra42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Overholtzer M, Zhang J, Smolen GA, et al. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc Natl Acad Sci U S A. 2006;103(33):12405–12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.McPherson JP, Tamblyn L, Elia A, et al. Lats2/Kpm is required for embryonic development, proliferation control and genomic integrity. EMBO J. 2004;23(18):3677–3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Morin-Kensicki EM, Boone BN, Howell M, et al. Defects in yolk sac vasculogenesis, chorioallantoic fusion, and embryonic axis elongation in mice with targeted disruption of Yap65. Mol Cell Biol. 2006;26(1):77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lee JH, Kim TS, Yang TH, et al. A crucial role of WW45 in developing epithelial tissues in the mouse. EMBO J. 2008;27(8):1231–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhou D, Conrad C, Xia F, et al. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell. 2009;16(5):425–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nishio M, Hamada K, Kawahara K, et al. Cancer susceptibility and embryonic lethality in Mob1a/1b double-mutant mice. J Clin Invest. 2012;122(12):4505–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wierzbicki PM, Adrych K, Kartanowicz D, et al. Underexpression of LATS1 TSG in colorectal cancer is associated with promoter hypermethylation. World J Gastroenterol. 2013;19(27):4363–4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Takahashi Y, Miyoshi Y, Takahata C, et al. Down-regulation of LATS1 and LATS2 mRNA expression by promoter hypermethylation and its association with biologically aggressive phenotype in human breast cancers. Clin Cancer Res. 2005;11(4):1380–1385. [DOI] [PubMed] [Google Scholar]
- 111.Johnson R, Halder G. The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. Nat Rev Drug Discov. 2014;13(1):63–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gao T, Zhou D, Yang C, et al. Hippo signaling regulates differentiation and maintenance in the exocrine pancreas. Gastroenterology. 2013;144(7):1543–1553, 1553 e1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.George NM, Day CE, Boerner BP, Johnson RL, Sarvetnick NE. Hippo signaling regulates pancreas development through inactivation of Yap. Mol Cell Biol. 2012;32(24):5116–5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Guerra C, Schuhmacher AJ, Canamero M, et al. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell. 2007;11(3):291–302. [DOI] [PubMed] [Google Scholar]
- 115.Gukovsky I, Li N, Todoric J, Gukovskaya A, Karin M. Inflammation, autophagy, and obesity: common features in the pathogenesis of pancreatitis and pancreatic cancer. Gastroenterology. 2013;144(6):1199–1209 e1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pylayeva-Gupta Y, Lee KE, Hajdu CH, Miller G, Bar-Sagi D. Oncogenic Kras-induced GM-CSF production promotes the development of pancreatic neoplasia. Cancer Cell. 2012;21(6):836–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bayne LJ, Beatty GL, Jhala N, et al. Tumor-derived granulocyte-macrophage colony-stimulating factor regulates myeloid inflammation and T cell immunity in pancreatic cancer. Cancer Cell. 2012;21(6):822–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Iacobuzio-Donahue CA, Herman JM. Autophagy, p53, and pancreatic cancer. N Engl J Med. 2014;370(14):1352–1353. [DOI] [PubMed] [Google Scholar]
- 119.Kimmelman AC. The dynamic nature of autophagy in cancer. Genes Dev. 2011;25(19):1999–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Perera RM, Stoykova S, Nicolay BN, et al. Transcriptional control of autophagylysosome function drives pancreatic cancer metabolism. Nature. 2015;524(7565):361–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hashimoto D, Blauer M, Hirota M, Ikonen NH, Sand J, Laukkarinen J. Autophagy is needed for the growth of pancreatic adenocarcinoma and has a cytoprotective effect against anticancer drugs. Eur J Cancer. 2014;50(7):1382–1390. [DOI] [PubMed] [Google Scholar]
- 122.Antonucci L, Fagman JB, Kim JY, et al. Basal autophagy maintains pancreatic acinar cell homeostasis and protein synthesis and prevents ER stress. Proc Natl Acad Sci U S A. 2015;112(45):E6166–6174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rosenfeldt MT, O’Prey J, Morton JP, et al. p53 status determines the role of autophagy in pancreatic tumour development. Nature. 2013;504(7479):296–300. [DOI] [PubMed] [Google Scholar]
- 124.Rhim AD, Mirek ET, Aiello NM, et al. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148(1–2):349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Provenzano PP, Cuevas C, Chang AE, Goel VK, Von Hoff DD, Hingorani SR. Enzymatic targeting of the stroma ablates physical barriers to treatment of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;21(3):418–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sun JD, Liu Q, Wang J, et al. Selective tumor hypoxia targeting by hypoxia-activated prodrug TH-302 inhibits tumor growth in preclinical models of cancer. Clin Cancer Res. 2012;18(3):758–770. [DOI] [PubMed] [Google Scholar]
- 127.Borad MJ, Reddy SG, Bahary N, et al. Randomized Phase II Trial of Gemcitabine Plus TH-302 Versus Gemcitabine in Patients With Advanced Pancreatic Cancer. J Clin Oncol. 2015;33(13):1475–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Drummond DC, Noble CO, Guo Z, Hong K, Park JW, Kirpotin DB. Development of a highly active nanoliposomal irinotecan using a novel intraliposomal stabilization strategy. Cancer Res. 2006;66(6):3271–3277. [DOI] [PubMed] [Google Scholar]
- 129.Singh A, Settleman J. Oncogenic K-ras “addiction” and synthetic lethality. Cell Cycle. 2009;8(17):2676–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wolpin BM, Rubinson DA, Wang X, et al. Phase II and pharmacodynamic study of autophagy inhibition using hydroxychloroquine in patients with metastatic pancreatic adenocarcinoma. Oncologist. 2014;19(6):637–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chen Z, Cheng K, Walton Z, et al. A murine lung cancer co-clinical trial identifies genetic modifiers of therapeutic response. Nature. 2012;483(7391):613–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Laheru D, Lutz E, Burke J, et al. Allogeneic granulocyte macrophage colony-stimulating factor-secreting tumor immunotherapy alone or in sequence with cyclophosphamide for metastatic pancreatic cancer: a pilot study of safety, feasibility, and immune activation. Clin Cancer Res. 2008;14(5):1455–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Beatty GL, Haas AR, Maus MV, et al. Mesothelin-specific chimeric antigen receptor mRNA-engineered T cells induce anti-tumor activity in solid malignancies. Cancer Immunol Res. 2014;2(2):112–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Beatty GL, Chiorean EG, Fishman MP, et al. CD40 agonists alter tumor stroma and show efficacy against pancreatic carcinoma in mice and humans. Science. 2011;331(6024):1612–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ozdemir BC, Pentcheva-Hoang T, Carstens JL, et al. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell. 2014;25(6):719–734. [DOI] [PMC free article] [PubMed] [Google Scholar]