Abstract
Background
The methylotrophic yeast Pichia pastoris is a common host for the production of recombinant proteins. However, hypermannosylation hinders the use of recombinant proteins from yeast in most biopharmaceutical applications. Glyco-engineered yeast strains produce more homogeneously glycosylated proteins, but can be physiologically impaired and show tendencies for cellular agglomeration, hence are hard to cultivate. Further, comprehensive data regarding growth, physiology and recombinant protein production in the controlled environment of a bioreactor are scarce.
Results
A Man5GlcNAc2 glycosylating and a Man8–10GlcNAc2 glycosylating strain showed similar morphological traits during methanol induced shake-flask cultivations to produce the recombinant model protein HRP C1A. Both glyco-engineered strains displayed larger single and budding cells than a wild type strain as well as strong cellular agglomeration. The cores of these agglomerates appeared to be less viable. Despite agglomeration, the Man5GlcNAc2 glycosylating strain showed superior growth, physiology and HRP C1A productivity compared to the Man8–10GlcNAc2 glycosylating strain in shake-flasks and in the bioreactor. Conducting dynamic methanol pulsing revealed that HRP C1A productivity of the Man5GlcNAc2 glycosylating strain is best at a temperature of 30 °C.
Conclusion
This study provides the first comprehensive evaluation of growth, physiology and recombinant protein production of a Man5GlcNAc2 glycosylating strain in the controlled environment of a bioreactor. Furthermore, it is evident that cellular agglomeration is likely triggered by a reduced glycan length of cell surface glycans, but does not necessarily lead to lower metabolic activity and recombinant protein production. Man5GlcNAc2 glycosylated HRP C1A production is feasible, yields active protein similar to the wild type strain, but thermal stability of HRP C1A is negatively affected by reduced glycosylation.
Electronic supplementary material
The online version of this article (10.1186/s12934-018-1032-6) contains supplementary material, which is available to authorized users.
Keywords: Pichia pastoris, SuperMan5, OCH1, Bioreactor, Cellular agglomeration, Flow cytometry, Glycosylation, Horseradish peroxidase, Morphology
Background
The methylotrophic yeast Pichia pastoris, also known as Komagataella phaffii, is widely used for the production of recombinant proteins, due to its high productivity, the ability to grow on defined and inexpensive media and its capacity to perform post-translational modifications (e.g. [1]). Protein glycosylation is one of the most critical aspects in the recombinant production of proteins, especially of biopharmaceuticals, as it affects protein properties such as solubility, stability, biological activity, pharmacokinetics (e.g. [2]), clearance from the body and efficacy (e.g. [3]). In P. pastoris protein O-glycosylation still has to be fully understood, but is expected to consist of variable short, unphosphorylated/phosphorylated α-1,2- and β-1,2-mannose chains (e.g. [4]). First approaches to alter O-glycosylation in P. pastoris showed promising results [5]. Protein N-glycosylation, which is characterized by hypermannosylation, has been thoroughly investigated and is well understood (e.g. [6]). Initially, N-glycans are linked to the amido group of asparagine residues that are recognized by glycosyltransferases in the endoplasmatic reticulum (ER) at the sequence motif N-X-S/T of the proteins, where X is any amino acid but proline. After the ER, the proteins carry a Man8GlcNAc2 glycan chain, which is then subjected to hypermannosylation. The first reaction in hypermannosylation is catalyzed by an α-1,6-mannosyltransferase (Och1), which was first discovered and characterized in S. cerevisiae [7, 8]. Notably, its glycosylation activity of secreted and membrane proteins make it also a key enzyme for cell wall maintenance and integrity in yeast [9–12]. However, hypermannosylation hinders the use of recombinant proteins from yeasts in most biopharmaceutical applications, which is why numerous efforts focused on the humanization of the yeast glycosylation machinery [6, 13–17]. Although the humanization of yeast was accomplished over 10 years ago, only a few studies are known, in which biopharmaceutically relevant products with glyco-engineered strains were produced. Most of the strains have an OCH1 deficiency and retain a recombinant α-1,2-mannosidase in the ER to yield mainly Man5GlcNAc2 structures [15–23].
First studies by Vervecken et al. [16] and Jacobs et al. [15] reported higher stress sensitivity of such strains leading to reduced growth, but to homogeneously (> 90%) Man5GlcNAc2 glycosylated products. In most cases, only shake-flask experiments were performed, in which comparisons to other strains or impact of product glycosylation patterns can be biased, due to the uncontrolled behavior in terms of process control (e.g. pH, dissolved oxygen) [19, 20, 22, 23]. In literature, environmental stressors are known to affect a protein’s post-translational processing [24], which highlights the importance of analyzing a protein’s properties during the controlled production in bioreactors. To our knowledge, only a few studies exist, which analyzed the behavior of Man5GlcNAc2 glycosylating P. pastoris strains in the controlled environment of a bioreactor [18, 21, 25]. Jacobs et al. [25] were able to produce a maximum of 760 mg L−1 of a murine granulocyte-macrophage colony-stimulating factor (mGM-CSF) at high cell densities upon MeOH induction of alcohol oxidase 1 promoter (PAOX1). Although they successfully produced almost homogeneously (> 90%) Man5GlcNAc2 glycosylated mGM-CSF, a comparison of performance to a mGM-CSF expressing wild-type strain would have been interesting. Furthermore, a decrease in productivity was observed after 40 h of MeOH induction, but was not further discussed. In another study by Smith et al. [18], the authors successfully produced recombinant human mast cell chymase (rhChymase) under the control of a glyceraldehyde-3-phosphate dehydrogenase promoter (PGAP). During their glycerol fed-batch cultivation, they detected chymase-like proteolytic activity after 72 h of induction, which might have resulted from physiological stress. Recently, the cotton plant proteins GbDIR2 and GhDIR3 were produced with > 90% Man5GlcNAc2 glycosylation homogeneity, but the authors observed an increased MeOH toxicity for their glyco-engineered strain compared to conventional wild-type strains [21]. Concluding, all of these research groups experienced a decrease in Pichia’s process performance over time. We speculate that their glyco-engineered yeast strains were physiologically impaired, due to the altered glycosylation machinery. As we have shown previously for a horseradish peroxidase C1A (HRP C1A) expressing P. pastoris strain, where we knocked out OCH1 [12], an altered glycosylation machinery can have significant impacts: we found that the OCH1 knockout strain was characterized by slow growth, increased temperature sensitivity and formation of cellular agglomerates, compared to the HRP C1A expressing wild-type strain [12]. Further analysis of this cellular agglomeration showed a decrease in cell surface glycosylation, a negatively affected budding process and indicated covalently bound cells. However, the recombinant protein was still produced and carried a much more homogeneous surface glycosylation, with the majority being Man8GlcNAc2 and Man9GlcNAc2 structures [12].
In the present study, we shed more light on the physiological impairment accompanied by glyco-engineered strains, by performing a morphological investigation and analyzing the size distribution of cellular agglomerates of differently glycosylating P. pastoris strains by microscopy and flow cytometry. Furthermore, we physiologically characterized a recombinant Man5GlcNAc2 glycosylating strain (SuperMan5) in a bioreactor during un-induced phases and MeOH induced phases, produced HRP C1A as a model product and biochemically characterized it. The used strain is based on the GlycoSwitch® plasmids and is able to yield Man5GlcNAc2 glycosylated products by harboring an OCH1 disruption and an α-1,2-mannosidase from Trichoderma reesei with a C-terminal HDEL signal sequence for ER retention [15]. Due to its shortened glycan pattern, a comparable morphology to the OCH1 deficient strain was expected. We compared all results of the SuperMan5 strain to previously published data of a recombinant P. pastoris wild type (wt) (hypermannosylated product) as well as to a recombinant P. pastoris OCH1 knockout strain (∆OCH1) (Man8–10GlcNAc2 glycans; [12]).
Summarizing, in this study we compared different glyco-engineered P. pastoris strains for morphological differences and recombinant protein production. To our knowledge, we provide the first study in literature that comprehensively describes the physiology and growth behavior of a Man5GlcNAc2 glycosylating P. pastoris strain in the controlled environment of a bioreactor.
Results and discussion
Strain characterization in shake-flask screenings
Glyco-engineered strains often show a decrease in productivity over time [18, 21, 25], which might be linked to their altered glycosylation machinery and therefore a stressed metabolism. Based on our recent findings with the ∆OCH1 strain [12, 26], we found that cellular agglomeration and therefore an altered morphology, affected process performance over time. Therefore, we initially hypothesized that an altered glycosylation machinery might be the trigger for morphological deviations, due to an overall decrease in glycan length on the cell surface resulting in cellular agglomeration, as it was also shown for glyco-proteins [27]. Based on this hypothesis, we performed a shake-flask screening under inducing conditions to produce HRP C1A as recombinant model product in different P. pastoris strains and monitored cellular morphology by microscopy and flow cytometry. A hypermannosylating wt strain, the ∆OCH1 strain (Man8–10GlcNAc2 glycans) and the SuperMan5 strain (Man5GlcNAc2 glycans) were compared.
We observed that the wt and SuperMan5 strain grew similarly in terms of OD600 over the whole induction time of 71 h, but observed stagnating growth for the ∆OCH1 strain (see Additional file 1: Figure S1 for OD600 growth curve). At low cell densities (OD600 ~ 10) and after 47 h of induction, when samples for the glycosylation analysis were taken, the wt and SuperMan5 cultivations showed no significant difference in volumetric activity, however the ∆OCH1 cultivation showed a significantly lower volumetric activity (Table 1). This highlighted two aspects: first, due to the lower volumetric activity and a stagnating growth, the ∆OCH1 strain seemed to have a stressed metabolism that probably resulted in a decreased viability over time. As foam formation was more prominent for the ∆OCH1, partial cell lysis and higher abundance of host cell proteins in the cultivation broth was likely and was also observed in our previous studies [12, 26]. Second, the shake-flask screening data in Table 1 suggest that the performance of the SuperMan5 strain was not negatively affected by its altered glycosylation machinery as its performance was comparable to the wt strain.
Table 1.
Volumetric activity of HRP C1A from wt, ∆OCH1 and SuperMan5 strains from the shake-flask screening, measured in duplicates
| Volumetric activity (U mL−1) at OD600 of 10 | Volumetric activity (U mL−1) after 47 h of induction | |
|---|---|---|
| wt | 2.20 ± 0.04 | 3.20 ± 0.10 |
| ∆OCH1 | 2.02 ± 0.05 | 2.78 ± 0.08 |
| SuperMan5 | 2.13 ± 0.05 | 3.01 ± 0.09 |
In order to investigate strain morphology and previously observed agglomerate formation [12, 26], shake-flask samples were analysed via microscopy as well as flow cytometry.
Microscopy
The average cell size for P. pastoris lies between 4 and 6 μm in average cell diameter [28]. However, conventional budding cells can also encompass two or more cells. Therefore, we defined structures smaller than 15 µm as single or budding cells and structures larger than 15 µm as agglomerates [26]. In Fig. 1, microscopic pictures from shake-flask samples during MeOH induction are displayed, which enable a clear distinction between the strains. The wt strain shows typical single and budding cells. Strong agglomerate formation was seen for both glyco-engineered strains, but the SuperMan5 strain seemed to have a higher degree of agglomeration. Observed agglomerates clearly exhibited multi-budded cells and spanned up to agglomerate diameters over 25 µm. Although the microscopic analysis supported our initial hypothesis that glyco-engineered strains tend to cellular agglomeration, we aimed to analyze this phenomenon with additional methods to minimize biased results. As cellular agglomeration can be triggered by sedimentation during microscopic analysis, we also used flow cytometry to analyze cell morphology under fluid conditions, which approximated the moving environment in a bioreactor or shake-flask better than conventional microscopy.
Fig. 1.
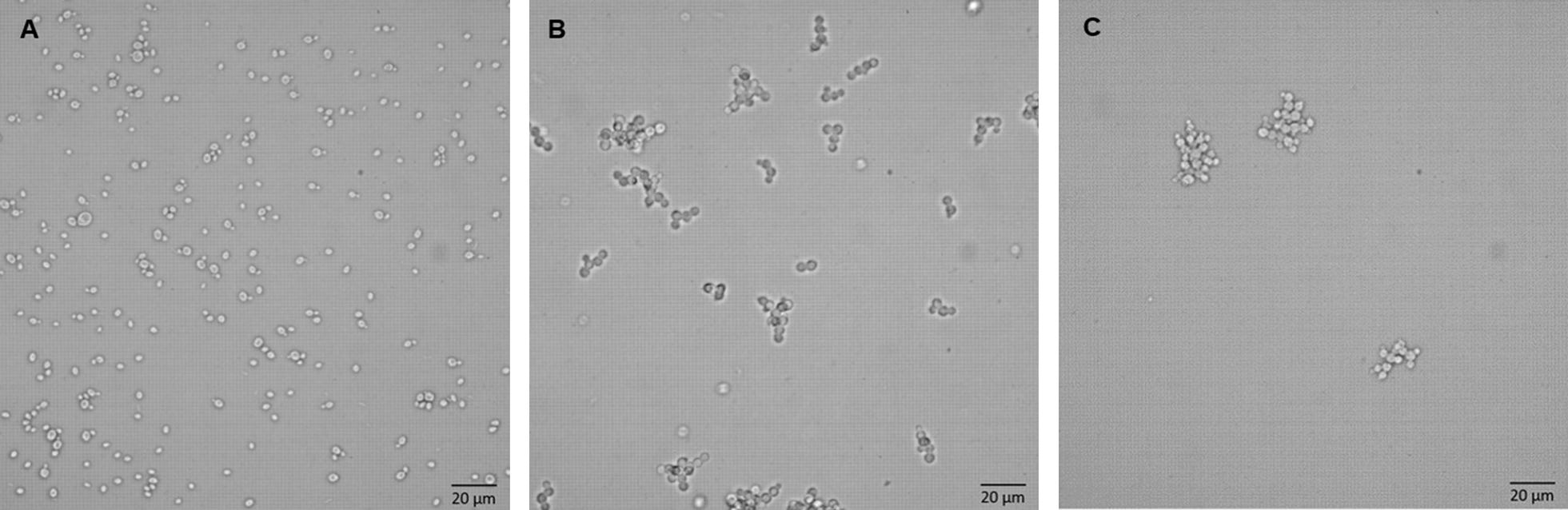
Light microscopy images taken from shake flask experiment at induction time of 11 h. Yeast cells from the different strains are shown, A wt, B ∆OCH1, C SuperMan5. The glyco-engineered strains in B, C show a distinct differentiation from the wt strain in A. Black bar signifies 20 µm
Flow cytometry
By using flow cytometry, not only sedimentation and possibly biased cellular agglomeration was minimized, but also a false-positive detection of loosely agglomerated cells could be minimized, due to the in-flow velocity of the cell suspension and the resulting force on the cells. Therefore, the detected cell agglomerates were thought to consist of cells, being attached to each other by strong non-covalent forces through their surface glycosylation or even by covalent bonds through an inefficient budding process [12]. These agglomerates were termed “cluster” in the evaluation of flow cytometry data.
Signal curve properties of various detector signals were used to differentiate morphological classes. As explained by Dubelaar and Gerritzen, forward scatter (FSC) and sideward scatter (SSC) signals represent size, shape and overall morphology of measured elements [29]. By using the flow cytometer, it was possible to distinguish between budding cells and cellular agglomerates. Furthermore, fluorescence signals derived from staining with propidium iodide (PI) and fluorescein diacetate (FDA) provided the means for biomass viability assessment [30]. Metabolic activity is shown by FDA treatment resulting in green fluorescence through esterase activity [31]. PI fluorescence is a result from DNA intercalation in cells with compromised membranes [28].
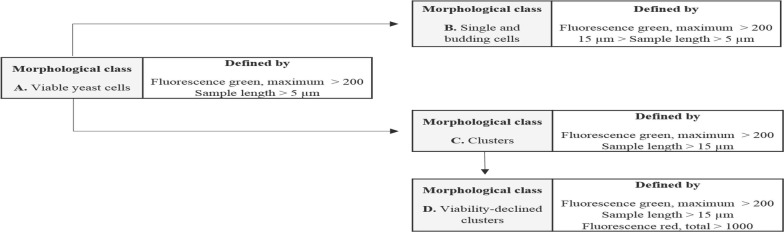
Based on initial measurements of induction medium with and without cells, a distinction of yeast cells from media background was possible and only particles above a threshold of maximum green fluorescence higher than 200, representing FDA staining, were set as viable yeast cells. In the next step, scatter plots were created and gates were set for classification. Gate setting was based on particle size in accordance with microscopic image analysis as discussed in the previous section and on our prior results with a cell agglomerate forming strain [12, 26]. The image-in-flow feature supported the visual identification of morphological classes, as single or budding cells and cluster could be distinguished. An increase in red fluorescence from PI staining indicated viability-declined agglomerates, because PI cannot cross the membrane of healthy cells (see Additional file 1: Figure S2 for comparison between viable and viability-declined clusters). Again, it was possible to set the threshold for viability-declined clusters, based on initial experiments, in which we deliberately induced cell death by heat treatment and compared the scatter plots of treated and untreated cells (data not shown). Therefore, a threshold of total red fluorescence of 1000 was set to distinguish between living and viability-declined cells. The definition of the used morphological classes for flow cytometry analysis is summarized in Fig. 2. Based on the pre-set ranges for the morphological classes, a proper distinction between different particles was possible. First, all viable yeast cells were detected, before these cells were divided into single and budding yeast cells or clusters. In a further step, these clusters were analyzed in depth, which revealed viability-declined clusters harboring a significant amount of PI permeable cells. These cells likely underwent substrate or oxygen limitation, hence the agglomerate formation led to a decrease in viability of these cells.
Fig. 2.
Definition of morphological classes for flow cytometry analysis
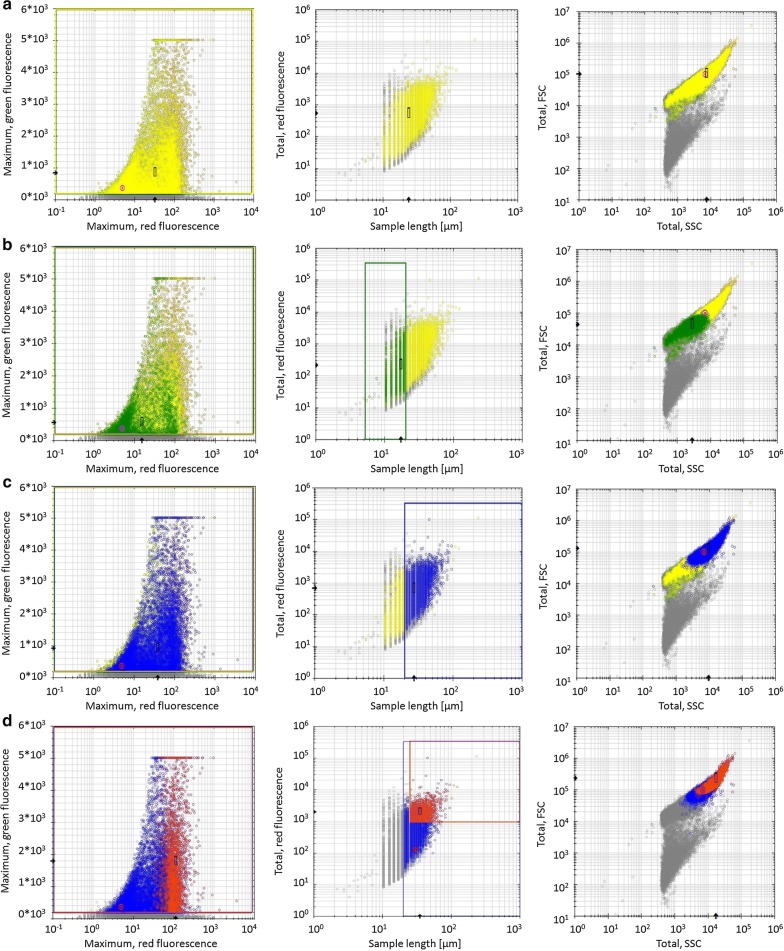
The process of morphological distinction through flow cytometry is exemplarily shown for the SuperMan5 strain from the MeOH induced shake-flask cultivation (Fig. 3). According to the classes set in Fig. 2, viable yeast cells (Fig. 3a yellow), single and budding cells (Fig. 3b green) clusters (Fig. 3c blue) as well as viability-declined clusters (Fig. 3d red) could be distinguished. Interestingly, 23 h after induction start all three strains already showed different morphological distributions for single and budding cells and for clusters (Fig. 4a). The size distribution of wt strain for single and budding yeast was visibly narrower compared to the glyco-engineered strains. This might indicate that wt cells were less stressed. In literature, it has been reported that yeast cell size can still increase, when cellular proliferation is hindered by stress, leading to larger cells [32]. Furthermore, the size distribution of ∆OCH1 clusters was narrower distributed and generally smaller in contrast to SuperMan5 clusters, hence ∆OCH1 clusters were more tightly packed. As seen in Fig. 4b, also the wt strain was found to yield a small percentage of cluster forming cells, but with a negligible amount when compared to the glyco-engineered strains. Cluster formation of the SuperMan5 strain and the ∆OCH1 strain might be triggered by the decrease in glycan length on the cell surface. It is likely that cell surface glycans help to sustain repulsive electrostatic interactions of the cells and thus prevent agglomeration, which has also been shown for glycosylated proteins in high concentrations [27]. In addition, both glyco-engineered strains showed viability-declined clusters, which correlated well to our hypothesis that cellular agglomeration can lead to limitations for inner core cells [12]. A representative signal profile of a SuperMan5 viability-declined cluster can be seen in Fig. 5a together with the corresponding image-in-flow of the measured cluster in Fig. 5b. The signal profile shows an increase in susceptibility to PI staining (see Additional file 1: Figure S2 for comparison between viable and viability-declined clusters), resulting in an increase of red fluorescence that corresponds to a decrease in viability of these cells. Interestingly, the SuperMan5 strain exhibited a stronger proportion of viability-declined clusters at the beginning of the MeOH induced shake-flask experiments (Fig. 5c). Over time, this trend diminished as the ∆OCH1 strain increased its proportion in viability-declined clusters (Fig. 5). These results together with the fact that the mean cluster size of the glyco-engineered strains remained similar over the whole induction time (Additional file 1: Figure S3) supported the morphological similarity between these two strains. However, while the overall morphology between the ∆OCH1 and SuperMan5 strain was similar, the SuperMan5 strain appeared to be fitter, due to its better HRP C1A production and no visible growth stagnation in the shake flasks. We hypothesized that the glyco-engineered strains differed in performance neither due to their altered glycosylation machinery nor due to their tendency to agglomerate. Both glyco-engineered strains strongly agglomerated and both were OCH1 deficient, which suggested another reasons for the decreased performance of the ∆OCH1 strain. Although not tested, we hypothesized that the different methods of OCH1 inactivation could be responsible for the observed difference in performance. The ∆OCH1 strain was generated by a knock-out procedure [12], but the SuperMan5 strain by an OCH1 knock-in procedure [15, 16]. Therefore, most of the OCH1 gene and promoter remained present, but inactive in the knock-in strain [16]. In literature, not only the Och1 protein, but also the OCH1 gene and its promoter region are described as important factors for cell wall integrity [10] as well as oxidative- and hypo-osmotic stress tolerance [9, 11]. For example, Li and colleagues showed that the transcription factor Skn7p, which is important in stress response pathways in S. cerevisiae, binds upstream of the OCH1 open reading frame and they further suggested that successful Skn7p binding promotes activation of other transcription factors. Therefore, it might be possible that binding/activation of Skn7p or other important factors were hindered in the OCH1 knock-out strain (∆OCH1), which diminished its stress tolerance, but future research has to elucidate this theory. Overall, the SuperMan5 strain seemed to be fit for further characterization in the bioreactor and a proper production strain for Man5GlcNAc2 glycosylated HRP C1A. Additional analysis of the glycosylation pattern of HRP C1A from the different strains also indicated that the destined glycosylation was present for each of the different HRP C1A enzymes (Additional file 1: Figure S4).
Fig. 3.
Exemplary flow cytometry scatter plots of SuperMan5 strain from shake-flask screening during induction at 23 h. From left to right: fluorescence green vs. fluorescence red, fluorescence red vs. sample length, FSC total vs. SSC total. From top to bottom: a viable yeast cells (yellow), b single and budding cells (green), c clusters (blue) and d viability-declined clusters (red). Rectangles signify chosen gates according to morphological classification
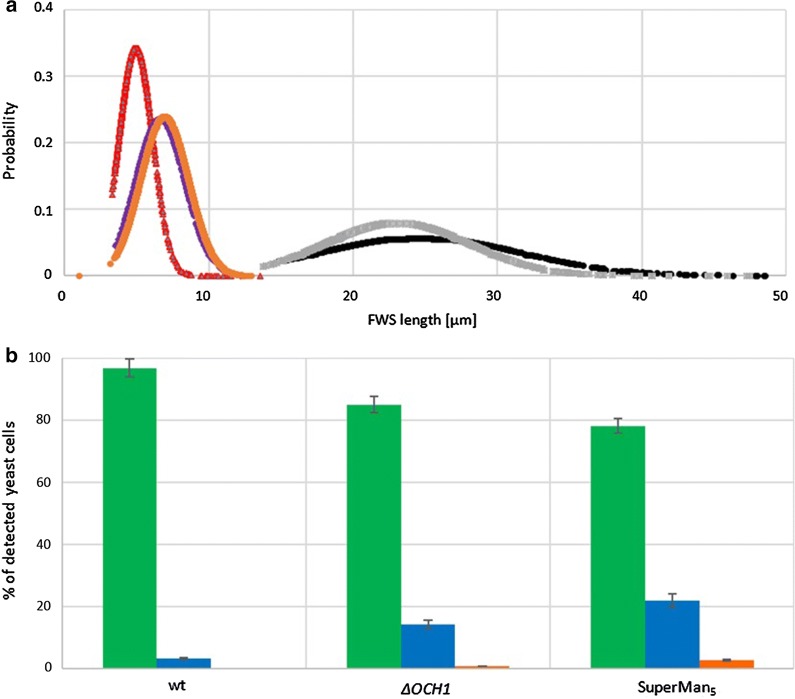
Fig. 4.
a Normal distribution of particle FSC length of single and budding cells and cluster for wt, ∆OCH1 and SuperMan5 strains at induction time of 23 h from shake-flask screening. Distribution of wt single and budding cells (red), ∆OCH1 single and budding cells (violet), SuperMan5 single and budding cells (orange), ∆OCH1 cluster distribution (grey), SuperMan5 cluster distribution (black). b Exemplary proportion of morphological classes in percentages of detected yeast cells at induction time of 23 h from shake-flask screening for wt, ΔOCH1 and SuperMan5 strain. Bar plot shows single and budding cells (green), clusters (blue), viability-declined cluster (red). Standard deviations for b were derived from multiple measurements (at least 3) of single culture shake-flask samples
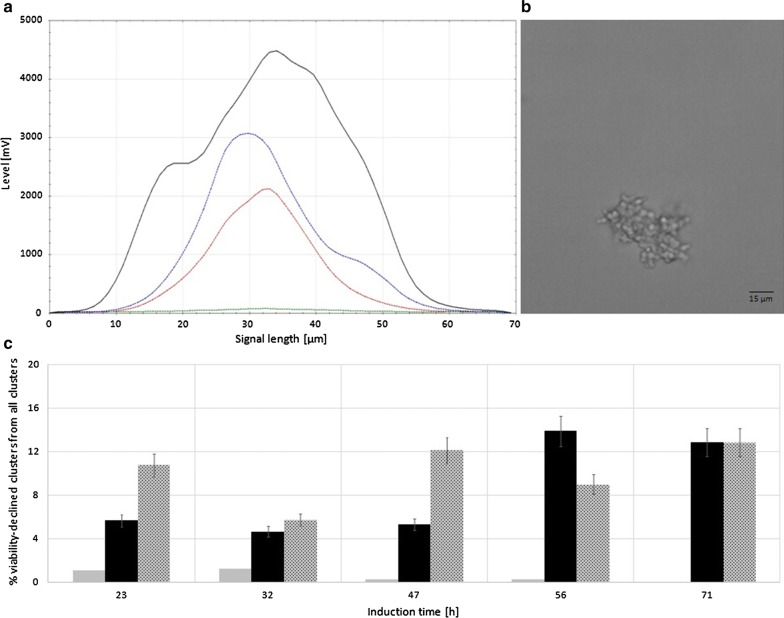
Fig. 5.
a Exemplary signal profile from flow cytometer of SuperMan5 cluster after 23 h induction time in shake-flasks. FSC (black line -), SSC (blue line –), green (green line –) and red (red line –) fluorescence signals. b The corresponding image-in-flow picture of the cellular cluster, black bar represents 15 µm. c Percentages of viability-declined clusters from all detected clusters over induction time from shake-flask screening: wt (grey bars), ∆OCH1 (black bars), SuperMan5 (dotted grey bars). Standard deviations for c were derived from multiple measurements (at least 3) of single culture shake-flask samples
Summarizing, we compared the HRP C1A production in three different P. pastoris strains that yielded differently glycosylated products and analysed strain morphological properties by microscopy and flow cytometry. The ∆OCH1 and SuperMan5 strain showed similar morphological traits. Both glyco-engineered strains had larger single and budding cells than the wt strain and showed strong cellular agglomeration, as we had described for the ∆OCH1 strain before [12]. This phenomenon of agglomeration is believed to be triggered by the shorter glycan structures on the cell surface and a disturbed budding process. We also found that inner core cells of these agglomerates appeared to be less viable, due to possible limitations, hence one should consider this fact when working with other glyco-engineered strains. Additionally, our results suggested that the diminished performance of the ∆OCH1 strain did not result from the altered glycosylation machinery, but rather from different phenomena, likely including hindered stress tolerance pathways. Nevertheless, a high volumetric activity of HRP C1A was found for the SuperMan5 strain, which led to further strain characterization in the bioreactor.
Physiological strain characterization in the bioreactor
Based on the prior shake-flask screening, the SuperMan5 strain was seen as a suitable strain for the production of Man5GlcNAc2 glycosylated HRP C1A in the controlled environment of a bioreactor. First, we characterized the SuperMan5 strain with our published method of conducting dynamic experiments with pulse-wise feeding during batch cultivations in the bioreactor [12, 33–36] and to our knowledge, this study represents the first comprehensive analysis of the temperature dependent physiology and growth behavior of a Man5GlcNAc2 glyco-engineered P. pastoris strain. The strain specific physiological parameters of SuperMan5 for the batch phase and the MeOH induction phase at different temperatures are summarized in Table 2.
Table 2.
Strain specific parameters of SuperMan5
| SuperMan5 | |
|---|---|
| max. µGly (h−1) | 0.28 ± 0.03 |
| YX/Gly (Cmol Cmol−1) | 0.55 ± 0.03 |
| (Cmol Cmol−1) | 0.47 ± 0.02 |
| Δtimeadapt (h) | 4.85 |
| 15 °C | 20 °C | 25 °C | 30 °C | |
|---|---|---|---|---|
| qMeOH (mmol g−1 h−1) | 0.76 ± 0.12 | 1.17 ± 0.05 | 1.41 ± 0.18 | 1.52 ± 0.11 |
| YX/MeOH (Cmol Cmol−1) | 0.037 ± 0.001 | 0.024 ± 0.002 | 0.022 ± 0.006 | 0.033 ± 0.001 |
| (Cmol Cmol−1) | 0.924 ± 0.001 | 0.923 ± 0.048 | 0.961 ± 0.046 | 0.972 ± 0.033 |
| C-balance | 0.961 ± 0.003 | 0.947 ± 0.071 | 0.983 ± 0.074 | 1.005 ± 0.048 |
| qp (U g−1 h−1) | 0.62 ± 0.06 | 0.94 ± 0.07 | 1.01 ± 0.06 | 1.05 ± 0.05 |
The batch phase on glycerol and the adaptation to methanol were performed at 30 °C. Subsequent methanol pulses were added at different temperatures. Parameters are defined in the footnote
max. µGly maximum specific growth rate on glycerol, qGly specific uptake rate of glycerol during the batch, YX/Gly biomass yield on glycerol, CO2 yield on glycerol, Δtimeadapt time from first addition of methanol to a maximum in offgas activity, qMeOH average specific uptake rate of methanol during consecutive methanol pulses, qP specific product formation rate, YX/MeOH biomass yield on methanol, CO2 yield on methanol, C-balance sum of biomass and CO2 yields
As shown in Table 2, the specific uptake rate for MeOH (qMeOH) increased with increasing temperature until 25 °C. The specific productivity (qp) showed only a significant difference at 15 °C. At 25 °C and 30 °C, the only significant difference was found in the biomass yield on MeOH. To assess the physiology of the SuperMan5 strain in a more comprehensive way, we compared its strain specific physiological parameters at 30 °C to our published results for a HRP C1A producing wt strain and ∆OCH1 strain under the same conditions (Table 3).
Table 3.
Strain specific physiological parameters of the different glyco-engineered P. pastoris MutS strains expressing HRP C1A as model enzyme at 30 °C
| wt [12] | ∆OCH1 [12] | SuperMan5 | |
|---|---|---|---|
| max. µGly (h−1) | 0.31 | 0.20 | 0.28 ± 0.03 |
| YX/Gly (Cmol Cmol−1) | 0.41 | 0.54 | 0.55 ± 0.03 |
| (Cmol Cmol−1) | 0.64 | 0.44 | 0.47 ± 0.02 |
| Δtimeadapt (h) | 19.9 | 8.90 | 4.85 |
| qMeOH (mmol g−1 h−1) | 0.70 | 0.43 | 1.52 ± 0.11 |
| YX/MeOH (Cmol Cmol−1) | 0.071 | 0.046 | 0.033 ± 0.001 |
| (Cmol Cmol−1) | 1.02 | Constantly decreasing | 0.97 ± 0.03 |
| C-balance | 1.09 | Constantly decreasing | 1.01 ± 0.05 |
| qp (U g−1 h−1) | 0.77 | 0.50 | 1.05 ± 0.05 |
All results were gathered from a previous batch phase on glycerol, a subsequent MeOH adaptation and dynamic pulsing experiment with MeOH. Values for wt and ∆OCH1 strain were taken from [12]
As shown in Table 3, the HRP C1A expressing P. pastoris strains showed differences in physiological parameters during batch on glycerol, hence their growth behaviour was already distinguishable. The SuperMan5 strain showed a similar maximum specific growth rate (µmax) to the wt strain, but the ∆OCH1 strain clearly exhibited slower growth. The above discussed shake-flask screening showed that both glyco-engineered strains have similar morphology and thus, cellular agglomeration does not seem to trigger the slow growth of the ∆OCH1 strain, but rather hindered stress tolerance pathways. More interestingly, the glyco-engineered strains had a better conversion of substrate to biomass, as seen by a higher biomass yield and produced less CO2 per substrate compared to the wt strain. We hypothesized that this might be connected to the altered glycosylation machinery in both strains that led to smaller glycans. Smaller glycans also mean that less carbon is used to build sugar molecules, hence the carbon can be re-routed for biomass conversion.
All three strains behaved very differently during adaptation to MeOH and induction with MeOH. The decrease in glycan length correlated with the adaptation time (Δtimeadapt) for MeOH, but the cause of this faster adaptation has to be elucidated yet. As also seen during the shake-flask screening, the SuperMan5 strain was superior during induction by yielding the best results for HRP C1A productivity and the highest qMeOH. Whereas ∆OCH1 lost metabolic activity over time, shown in a constantly decreasing [12], the wt and SuperMan5 strain were easily cultivated at 30 °C for a prolonged induction time. Closing C-balances for wt and SuperMan5 underlined the validity of the calculated strain specific physiological data.
Summarizing, it was possible to characterize the Man5GlcNAc2 glycosylating SuperMan5 strain in the controlled environment of a bioreactor during un-induced batch on glycerol and induced batches on MeOH. The use of dynamic substrate pulsing made it possible to characterize the SuperMan5 strain at 30 °C, 25 °C, 20 °C and 15 °C during induction with MeOH in only one experiment. HRP C1A productivity was similar between 20 and 30 °C. Furthermore, a comparison between the SuperMan5, ∆OCH1 and wt strain was possible at 30 °C to yield comprehensive data on the impact of glyco-engineering on growth behaviour and HRP C1A productivity. The SuperMan5 strain exhibited similar µmax as the wt strain during batch, but the glyco-engineered strains seemed to have a more efficient substrate to biomass conversion. We hypothesized that this resulted from decreased carbon demand for the glycosylation machinery. The decrease in glycan length of protein and cell surface glycosylation of the SuperMan5 strain did not lead to a lower metabolic activity, growth or protein productivity compared to the wt strain.
Enzyme characterization
Biochemical enzyme characterization
To check whether the kinetic constants and the stability of the recombinantly produced model protein were affected by the altered glycosylation pattern, we characterized the twofold concentrated and diafiltrated HRP C1A from the SuperMan5 bioreactor cultivation.
We compared published results for substrate affinity and thermal stability of HRP C1A produced in the wt and ∆OCH1 strain from our recent study (Table 4; [12]). The affinity towards the substrate ABTS was not significantly affected by altered protein glycosylation. However, the thermal stability of HRP C1A decreased clearly together with the amount of sugars attached to the protein surface. This phenomenon has already been described in literature for glycosylated proteins before [37–39].
Table 4.
Comparison of KM_ABTS and thermal stability at 60 °C between recombinant HRP C1A from wt, ∆OCH1 and SuperMan5
Values for wt and ∆OCH1 produced HRP C1A were taken from [12]
Summarizing, we analyzed enzyme kinetics and thermal stability of HRP C1A from the SuperMan5 strain. Although enzyme affinity towards ABTS was comparable to wt and ∆OCH1 products, the thermal stability was diminished 3-fold compared to the HRP C1A from the wt strain, hence proofing that protein stability is strongly affected by glycosylation.
Conclusion
In this study, we report the first comprehensive evaluation of growth, physiology and recombinant protein productivity of a Man5GlcNAc2 glycosylating P. pastoris strain (SuperMan5) in the controlled environment of a bioreactor. The strain showed superior growth, physiology and HRP C1A productivity compared to a Man8–10GlcNAc2 glycosylating ∆OCH1 strain.
Additionally, we shed more light on the often seen impairment in process performance of glyco-engineered strains in a detailed morphological study: Flow cytometry and microscopic analysis revealed the formation of cellular agglomerates with impaired viability of inner core cells. Although agglomeration was prominent in both glyco-engineered strains, our results suggest that a decreased process performance might not necessarily derive from the altered glycosylation machinery. It can be rather attributed to an additional metabolic burden, like hindered stress tolerance pathways, which can make strains more sensitive to environmental stressors.
Methods
Chemicals
Enzymes, deoxynucleotide triphosphates and Phusion™ high-fidelity DNA-polymerase was obtained from ThermoFisher Scientific (Vienna, Austria). 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) diammonium salt (ABTS) and hemin were purchased from Sigma-Aldrich (Vienna, Austria). Difco™ yeast nitrogen base w/o amino acids (YNB), Difco™ yeast nitrogen base w/o amino acids and ammonia sulfate (YNB2), Bacto™ tryptone and Bacto™ yeast extract were purchased from Becton–Dickinson (Vienna, Austria). Zeocin™ was purchased from InvivoGen (Toulouse, France) via Eubio (Vienna, Austria).
Microorganisms
For this study, a HRP C1A gene, codon-optimized for P. pastoris, was ordered from GenScript (Nanjing, China) and cloned into a pPICZαC vector, providing a Zeocin™ (Zeo) resistance gene as well as an α-prepro mating signal sequence from Saccharomyces cerevisiae for product secretion, using standard methods. Correct integration was verified by sequencing. The pPICZαC vector was successfully integrated in a P. pastoris GS115 strain (HIS+, pep4Δ, aox1∆), kindly provided by Biogrammatics, Inc. (California, United States) and should yield mainly Man5GlcNAc2 glycosylated HRP C1A after transformation (SuperMan5) [15]. The Och1 deficiency of the SuperMan5 strain is based on the disruption, but not deletion of the OCH1 gene. The strain CBS 7435 (identical to NRRL Y-11430 or ATCC 76273) was used as a benchmark wild type strain (wt), which yields natively hypermannosylated HRP C1A [12]. As described in our previous study, we used a genetically engineered wt strain harboring a knock-out deletion of the OCH1 gene (∆OCH1) to avoid hypermannosylation that yielded mainly Man8–10GlcNAc2 glycosylated HRP C1A upon transformation [12]. Both, the wt and ∆OCH1 strain contained a pPpT4_S, which contained the codon-optimized HRP C1A gene under equal conditions [12, 40]. Therefore, all resulting strains had a MutS phenotype, expressed and secreted HRP C1A upon induction of the AOX1 promoter with MeOH.
Culture media
The growth medium [buffered medium with glycerol for yeast (BMGY)] for the shake-flask screenings contained: 10 g L−1 yeast extract, 20 g L−1 peptone, 13.4 g L−1 YNB2, 4 mg L−1 d(+)-biotin, 10 g L−1 glycerol and 100 mL of a 1 M potassium phosphate buffer pH 6.0. The induction medium [buffered medium with MeOH for yeast (BMMY)] for the shake-flask screenings contained: 10 g L−1 yeast extract, 20 g L−1 peptone, 13.4 g L−1 YNB2, 4 mg L−1 d(+)-biotin, 5 g L−1 MeOH and 100 mL of a 1 M potassium phosphate buffer pH 6.0. The preculture medium for the bioreactor cultivations [yeast nitrogen base medium (YNBM)] contained: 20 g L−1 α-d(+)-glucose monohydrate, 3.4 g L−1 YNB2, 10 g L−1 (NH4)2SO4, 0.4 g L−1 d(+)-biotin, 0.1 M potassium phosphate buffer, pH 6.0. Trace element solution (PTM1) for the bioreactor cultivation contained: 6 g L−1 CuSO4·5H2O, 0.08 g L−1 NaCI, 3 g L−1 MnSO4·H2O, 0.2 g L−1 Na2MoO4·2H2O, 0.02 g L−1 H3BO3, 0.5 g L−1 CoCl2, 20 g L−1 ZnCl2, 65 g L−1 FeSO4·7H2O, 0.2 g L−1 d(+)-biotin, 5 mL L−1 95–98% H2SO4. Basal salt medium (BSM) for the bioreactor cultivations contained: 60 g L−1 glycerol, 1.17 g L−1 CaSO4·2H2O, 18.2 g L−1 K2SO4, 14.9 g L−1 MgSO4·7H2O, 4.13 g L−1 KOH, 26.7 mL L−1 85% (v/v) o-phosphoric acid, 0.2 mL L−1 Antifoam Struktol J650, 4.35 mL L−1 PTM1, NH4OH as N-source. pH was maintained by using 12.5% NH3,aq. Throughout all shake-flask cultivations, Zeocin™ was used in a concentration of 50 µg mL−1.
Strain selection
After transformation, 10 Zeo resistant clones were picked and grown overnight in 10 mL BMGY-Zeo medium in 100 mL baffled shake-flasks at 230 rpm and 30 °C. Then, cells were harvested by centrifugation (1800×g, 4 °C, 10 min) and re-suspended in BMMY-Zeo for adaptation of the cells to MeOH. Again, cells were grown at 230 rpm and 30 °C. Recombinant protein production was induced by adding 1.5% (v/v) pulses of pure MeOH supplemented with 12 mL PTM1/L MeOH to the culture each day, for 5 days. Each day, a sample was taken and analyzed for OD600, total protein content in the cell-free cultivation broth (Bradford assay) as well as the presence of recombinant HRP C1A by SDS-PAGE. A recombinant Pichia pastoris strain carrying the empty pPICZαC vector was included as negative control in all experiments.
Analysis of strain morphology and glycosylation
To understand a possible impact of the genotype and phenotype on overall strain physiology and productivity, an initial shake-flask screening, including morphological analysis was performed. The strain morphology was analyzed under inducing conditions for the wt, ∆OCH1 and SuperMan5 strain. In parallel, growth and product formation were monitored to assure product presence for later glycosylation pattern analysis.
Shake-flask screening
A fresh cryo tube (− 80 °C) was thawed for each HRP C1A containing strain, added to 200 mL BMGY-Zeo medium in a 1000 mL shake-flask and incubated at 28 °C and 230 rpm overnight. The next day, 50 mL of each culture were transferred to 450 mL BMMY-Zeo, including also 10 µM Hemin (Heme) to ease HRP C1A induction [41]. Induced cultures were grown in 2.5 L baffled flasks and a working volume of 500 mL. For comparability, HRP C1A induction was performed at 28 °C for all 3 strains. To assure complete depletion of the initial C-source (glycerol) and accurate adaptation to the inducing C-source in the shake-flasks (MeOH), cells were grown for 23 h in BMMY-Zeo-Heme before the first MeOH pulse was given. MeOH pulses were given each day as 1% (v/v) with PTM1 (12 mL L−1 MeOH). Sampling of the cultures was done approximately every 12 h. After 47 h of induction, 100 mL of each culture were harvested, centrifuged (4000×g, 10 min, 4 °C), the cell free supernatant was concentrated 20× with a 10 kDa centrifugal filter membrane (Amicon®Ultra—15) tube (Merck Millipore Ltd., Carrigtwohill, IRL) and stored at − 20 °C for further analysis. Enzyme activity and total protein content of the concentrates were measured and aliquots of the concentrates were used for identification of the respective HRP C1A glycosylation pattern of each strain. However, the total induction time of the shake-flask cultures was 71 h to further monitor growth and the morphological behavior of the different strains.
Microscopy
Twenty microlitres of the cultivation broth were pipetted onto a standard glass slide (25 × 75 mm) and then covered with an extra-large cover slide (24 × 60 mm). Images were recorded at a 40× magnification with a five megapixel microscopy CCD colour camera (Olympus, Austria). These images were used as a rough estimation of cell agglomerate formation and agglomerate diameter.
Flow cytometry
Samples of the shake-flask screening were diluted in phosphate buffered saline (PBS) (2.65 g L−1 CaCl2 solution, 0.2 g L−1 KCl, 0.2 g L−1 KH2PO4, 0.1 g L−1 MgCl·6 H2O, 8 g L−1 NaCl and 0.764 g L−1 Na2HPO4·2H2O at pH 6.5) to an OD600 of 1. Then, 0.5 µL of 20 mM propidium iodide stock in dimethyl sulfoxide (both from Sigma Aldrich, St. Louis, United States) and 5 µL of 12 mM fluorescein diacetate (Sigma Aldrich, St. Louis, United States) stock in acetone were added to 0.5 mL of the cell suspension. After 10 min incubation in the dark at room temperature, the sample was further diluted (1:10 in PBS) for flow cytometric analysis.
A CytoSense flow cytometer (CytoBuoy, Woerden, Netherlands) with two forward scatter (FSC), one sideward scatter (SSC) and two fluorescence channels (green, red) was used for single cell analysis. The implemented laser had a wavelength of 488 nm. The configuration of the emission wavelength filter set was 515–562 ± 5 nm for the green fluorescence channel (used for fluorescein diacetate) and 605–720 ± 5 nm for the red fluorescence channel (used for propidium iodide). The flow cytometer was equipped with a PixeLINK PL-B741 1.3 MP monochrome camera for image-in-flow acquisition, which made real-time imaging of cell agglomerates possible. For data evaluation, the software CytoClus3 (CytoBuoy, Woerden, Netherlands) was used.
The CytoSense flow cytometer provides multiple spatially resolved data points per channel per particle. This signal is achieved for both scatter channels as well as for green and red fluorescence channels [29], which is the basis for multiple curve parameters. Except for length parameters in µm, all parameters are in arbitrary units, as the user can set the sensitivity of the detector. The following parameters were used for the distinction of morphological classes: maximum (maximum of signal curve), total (area under curve), length (length of the signal) and sample length (length of signal above trigger level). Furthermore, image-in-flow feature enabled visual identification of yeast agglomerates, termed clusters. It should be noted that while FSC signals are closely linked to particle size (sample length), FSC length signals do not always correspond entirely to sample length due to overlays of other signals, which was seen during calibration with defined beads.
Glycosylation analysis
A glycopeptide analysis using an LC–ESI–MS system was performed, as we have reported before [12, 42]. The concentrated samples of the shake-flask screening were digested in solution. The proteins were S-alkylated with iodoacetamide and digested with trypsin (Promega, Madison, United States). The peptide mixtures were loaded on a BioBasic C18 column (BioBasic-18, 150 × 0.32 mm, 5 µm; ThermoFisher Scientific, Vienna, Austria) using 80 mM ammonium formiate buffer as the aqueous solvent. A gradient from 5% B (B: 80% acetonitrile) to 40% B in 45 min was applied, followed by a 15 min gradient from 40% B to 90% B that facilitates elution of large peptides, at a flow rate of 6 µL min−1. Detection was performed with a QTOF MS (Bruker maXis 4G) equipped with the standard ESI source in positive ion, DDA mode (= switching to MSMS mode for eluting peaks). MS-scans were recorded (range 150–2200 Da) and the 3 highest peaks were selected for fragmentation. Instrument calibration was performed using an ESI calibration mixture (Agilent, Santa Clara, United States). The nine possible glycopeptides were identified as sets of peaks consisting of the peptide moiety and the attached N-glycan varying in the number of HexNAc, hexose and phosphate residues. The theoretical masses of these glycopeptides were determined with a spreadsheet using the monoisotopic masses for amino acids and monosaccharides. Manual glycopeptide searches were made using DataAnalysis 4.0 (Bruker, Billerica, United States).
Bioreactor cultivations
After conducting the shake-flask screening, we characterized the recombinant SuperMan5 strain in terms of physiology, biomass growth and productivity using a dynamic strategy of conducting MeOH pulses during batch cultivations in the controlled environment of a bioreactor, which we have described several times before [12, 33–36]. This cultivation was used for subsequent purification to perform product kinetics and thermal stability analysis.
Preculture
Frozen stocks (− 80 °C) from working cell banks were incubated in 100 mL of YNBM-Zeo in 1 L shake-flasks at 30 °C and 230 rpm for 24 h. The preculture was transferred aseptically to the respective culture vessel. The inoculation volume was 10% of the final starting volume.
Cultivation
Batch cultivation was carried out in a 5 L working volume Labfors glass bioreactor (Infors, Bottmingen, Switzerland). BSM was sterilized in the bioreactor and pH was adjusted to pH 5.0 by using 12.5% NH3,aq after autoclavation. Sterile filtered PTM1 was transferred to the reactor aseptically. pH and dissolved oxygen probes were calibrated prior to cultivation start. Dissolved oxygen (dO2) was measured with a sterilizable polarographic dissolved oxygen electrode (Mettler Toledo, Vienna, Austria) and maintained above 20% throughout the cultivation. The pH was measured with a sterilizable electrode (Mettler Toledo, Vienna, Austria) and maintained constant at pH 5.0 with a step controller using 12.5% NH3,aq. Base consumption was determined gravimetrically. Agitation was fixed to 1495 rpm. The culture was aerated with 2.0 vvm dried air and offgas of the culture was measured by using an infrared cell for CO2 and a paramagnetic cell for O2 concentration (Servomax, Egg, Switzerland). Temperature, pH, dO2, agitation as well as CO2 and O2 in the offgas were measured on-line and logged in a process information management system (PIMS Lucullus; Applikon Biotechnology, Delft, Netherlands).
The end of the initial batch phase at 30 °C and therefore complete glycerol consumption was indicated by an increase in dO2, a drop in offgas CO2 and an increase in offgas O2. The first MeOH pulse (adaptation pulse) of a final concentration of 0.5% (v/v) was conducted with MeOH supplemented with 12 mL PTM1 per 1 L of added MeOH (MeOH/PTM1 pulse). Subsequently, at least two MeOH/PTM1 pulses were given to 1% (v/v) at 30 °C, then 25 °C, 20 °C and finally 15 °C. For each pulse, at least two samples were taken to determine the concentrations of MeOH and product as well as dry cell weight (DCW) and OD600 to calculate the strain specific physiological parameters. Induction was carried out in the presence of 1 mM hemin, which was added prior to the adaptation pulse [43].
Sample analysis
Analysis of growth- and expression-parameters
Dry cell weight (DCW) was determined by centrifugation of 5 mL culture broth (4000×g, 4 °C, 10 min), washing the pellet twice with 5 mL water and subsequent drying for 72 h at 105 °C. Determination was performed in triplicates. OD600 of the culture broth was measured in duplicates using a spectrophotometer (Genesys 20; ThermoFisher Scientific, Vienna, Austria). The activity of HRP C1A in the cell free supernatant was determined with a CuBiAn XC enzymatic robot (Optocell, Bielefeld, Germany) in duplicates. Cell free samples (60 µL) were added to 840 µL of 1 mM ABTS in 50 mM potassium phosphate buffer, pH 6.5. The reaction mixture was incubated for 5 min at 37 °C and was started by the addition of 100 µL of 0.078% H2O2. Changes of absorbance at 420 nm were measured for 180 s and rates were calculated. Calibration was done by using commercially available horseradish peroxidase (Type VI-A, P6782; Sigma-Aldrich, Vienna, Austria,) as standard at six different concentrations (0.02; 0.05; 0.1; 0.25; 0.5 and 1.0 U mL−1). Protein concentration of cell free supernatant was determined at 595 nm using the Bradford Protein Assay Kit (Bio-Rad Laboratories GmbH, Vienna, Austria) with bovine serum albumin (protein standard; micro standard, liquid; P0914; Sigma Aldrich, Vienna, Austria) as standard.
Substrate concentrations
Concentration of glycerol and MeOH was determined in cell free samples of the bioreactor cultivation by HPLC (Agilent Technologies, Santa Clara, United States) equipped with a Supelco guard column, a Supelco gel C-610H ion-exchange column (Sigma-Aldrich, Vienna, Austria) and a refractive index detector (Agilent Technologies, Santa Clara, United States). The mobile phase was 0.1% H3PO4 with a constant flow rate of 0.5 mL min−1 and the system was run isocratically. Calibration was done by measuring standard points in the range of 0.1–10 g L−1 glycerol and MeOH.
Data analysis
Strain characteristic parameters of the bioreactor cultivation were determined at a carbon dioxide evolution rate (CER) above 2.5 mmol g−1 h−1 during each MeOH pulse. Along the observed standard deviation for the single measurement, the error was propagated to the specific rates (qs and qp) as well as to the yield coefficients. The error of determination of the specific rates and the yields was therefore set to 10% and 5%, respectively for single measurement-derived values like seen in the batch phase [34]. For the pulse experiments, the average value and the standard deviation were used as two pulses were given for each temperature.
Enzyme characterization
Kinetic constants
A cell free bioreactor supernatant with HRP C1A from the SuperMan5 strain was twofold concentrated and diafiltrated with buffer (500 mM NaCl, 20 mM NaOAc, pH 6.0) [44, 45]. Protein concentration of the HRP C1A preparation was determined at 595 nm using the Bradford Protein Assay Kit (Bio-Rad Laboratories GmbH, Austria) with bovine serum albumin as standard. The kinetic constants for ABTS and H2O2 were determined. The reaction was started by adding 10 μL enzyme solution (1.0 mg mL−1) to 990 μL reaction buffer containing either ABTS in varying concentrations (0.01–5 mM) and 1 mM H2O2 or H2O2 in varying concentrations (0.001–0.5 mM) and 5 mM ABTS in 50 mM potassium phosphate buffer at pH 6.5. The change in absorbance at 420 nm was recorded in a spectrophotometer UV-1601 (Shimadzu, Japan) at 30 °C. Absorption curves were recorded with a software program (UVPC Optional Kinetics; Shimadzu, Japan). Measurements were performed in triplicates.
Thermal stability
The purified enzyme solution was incubated at 60 °C. At different time points, aliquots were withdrawn, the solutions were immediately cooled and centrifuged (20,000×g, 15 min) to pellet precipitated proteins and the remaining catalytic activity in the supernatants was measured [46].
Additional file
Additional file 1: Figure S1. OD600 over induction time in shake-flask experiment, black-filled circle = wt, unfilled white circle = SuperMan5, grey triangle = ∆OCH1. Figure S2. Comparison of signal profiles from flow cytometer. FSC (black line -), SSC (blue line --), green (green line --) and red (red line --) fluorescence signals of SuperMan5 clusters after 23 h induction time in shake-flasks. In viability-declined cluster (A) a clear increase in red fluorescence is visible from PI staining in contrast to the viable cluster (B). Figure S3. Mean cluster size of ∆OCH1 (black bars) and SuperMan5 (dotted grey bars) over induction time in shake-flask cultivation. Standard deviations are derived from multiple measurements (at least 3) of single culture shake-flask samples. Figure S4. ESI–MS spectra of HRP glycopeptides at n-glycosylation site 7. The triply (or doubly) charged site 7 N-glycopeptide GLIQSDQELFSSPNATDTIPLVR of natural HRP and HRP produced in a ∆OCH1, in SuperMan5 and in wt P. pastoris are shown. The wt_HRP exhibits also phosphorylated versions (P) of the oligomannosidic glycans.
Authors’ contributions
OSP, APE and LVE planned the experiments. APE, LVE and VRA performed the experiments and analyzed the data. LVE analyzed the flow cytometry data. CGG and FAL conducted and analyzed the ESI–MS measurements. CHE gave valuable scientific input. APE, LVE and OSP wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We especially thank Prof. Anton Glieder, TU Graz, for fruitful collaboration in former project P24861-B19, which represents the starting point for this contribution by delivering all the (published) reference data of the wildtype and the OCH1 k.o. strain. Furthermore, we thank Biogrammatics Inc. for kindly providing the SM5 strain for our research purposes. Finally, we thank the Austrian Ministry of Education, Science and Research in course of the Christian Doppler Laboratory for Mechanistic and Physiological Methods for Improved Bioprocesses for financial support.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The dataset(s) supporting the conclusions of this article are all included within the article. The datasets used and/or analyzed during the current study, if not shown in the text or additional files, are available from the corresponding author on reasonable request.
Consent of publication
Not applicable.
Ethics approval and consent to participate
Not applicable. The manuscript does not contain data collected from humans or animals.
Funding
The authors are very grateful to the Austrian Science Fund (FWF): project P30872-B26.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- FSC
flow cytometry forward scatter signal
- SSC
flow cytometry sideward scatter signal
- max. µGly
maximum specific growth rate on glycerol (h−1)
- YX/Gly
biomass yield on glycerol (Cmol Cmol−1)
CO2 yield on glycerol (Cmol Cmol−1)
- Δtimeadapt
time from first MeOH addition to maximum offgas activity (h)
- qMeOH
average specific uptake rate of MeOH during MeOH pulses (mmol g−1 h−1)
- qP
specific product formation rate (U g−1 h−1)
- YX/MeOH
biomass yield on MeOH (Cmol Cmol−1)
CO2 yield on MeOH (Cmol Cmol−1)
- C-balance
sum of biomass and CO2 yields
Contributor Information
Alexander Pekarsky, Email: alexander.pekarsky@tuwien.ac.at.
Lukas Veiter, Email: lukas.veiter@tuwien.ac.at.
Vignesh Rajamanickam, Email: vignesh.rajamanickam@tuwien.ac.at.
Christoph Herwig, Email: christoph.herwig@tuwien.ac.at.
Clemens Grünwald-Gruber, Email: clemens.gruber@boku.ac.at.
Friedrich Altmann, Email: friedrich.altmann@boku.ac.at.
Oliver Spadiut, Phone: +43 1 58801 166473, Email: oliver.spadiut@tuwien.ac.at.
References
- 1.Cregg JM, Cereghino JL, Shi JY, Higgins DR. Recombinant protein expression in Pichia pastoris. Mol Biotechnol. 2000;16(1):23–52. doi: 10.1385/MB:16:1:23. [DOI] [PubMed] [Google Scholar]
- 2.Cregg JM, Tolstorukov I, Kusari A, Sunga J, Madden K, Chappell T. Expression in the yeast Pichia pastoris. Methods Enzymol. 2009;463:169–189. doi: 10.1016/S0076-6879(09)63013-5. [DOI] [PubMed] [Google Scholar]
- 3.Parekh R. Automation of glycosylation analysis: a way forward for recombinant therapeutics. Glycoconj J. 1991;8(2):63–65. doi: 10.1007/BF00731011. [DOI] [PubMed] [Google Scholar]
- 4.Lommel M, Strahl S. Protein O-mannosylation: conserved from bacteria to humans. Glycobiology. 2009;19(8):816–828. doi: 10.1093/glycob/cwp066. [DOI] [PubMed] [Google Scholar]
- 5.Hamilton SR, Cook WJ, Gomathinayagam S, Burnina I, Bukowski J, Hopkins D, et al. Production of sialylated O-linked glycans in Pichia pastoris. Glycobiology. 2013;23(10):1192–1203. doi: 10.1093/glycob/cwt056. [DOI] [PubMed] [Google Scholar]
- 6.Hamilton SR, Gerngross TU. Glycosylation engineering in yeast: the advent of fully humanized yeast. Curr Opin Biotechnol. 2007;18(5):387–392. doi: 10.1016/j.copbio.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 7.Nakayama K, Nagasu T, Shimma Y, Kuromitsu J, Jigami Y. OCH1 encodes a novel membrane bound mannosyltransferase: outer chain elongation of asparagine-linked oligosaccharides. EMBO J. 1992;11(7):2511–2519. doi: 10.1002/j.1460-2075.1992.tb05316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagasu T, Shimma Y, Nakanishi Y, Kuromitsu J, Iwama K, Nakayama K, et al. Isolation of new temperature-sensitive mutants of Saccharomyces cerevisiae deficient in mannose outer chain elongation. Yeast. 1992;8(7):535–547. doi: 10.1002/yea.320080705. [DOI] [PubMed] [Google Scholar]
- 9.Lee J, Godon C, Lagniel G, Spector S, Garin J, Labarre J, et al. Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J Biol Chem. 1999;274(33):16040–16046. doi: 10.1074/jbc.274.23.16040. [DOI] [PubMed] [Google Scholar]
- 10.Tang H, Wang S, Wang J, Song M, Xu M, Zhang M, et al. N-hypermannose glycosylation disruption enhances recombinant protein production by regulating secretory pathway and cell wall integrity in Saccharomyces cerevisiae. Sci Rep. 2016;6:25654. doi: 10.1038/srep25654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li S, Dean S, Li Z, Horecka J, Deschenes RJ, Fassler JS. The eukaryotic two-component histidine kinase Sln1p regulates OCH1 via the transcription factor, Skn7p. Mol Biol Cell. 2002;13(2):412–424. doi: 10.1091/mbc.01-09-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krainer FW, Gmeiner C, Neutsch L, Windwarder M, Pletzenauer R, Herwig C, et al. Knockout of an endogenous mannosyltransferase increases the homogeneity of glycoproteins produced in Pichia pastoris. Sci Rep. 2013;3:3279. doi: 10.1038/srep03279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamilton SR, Davidson RC, Sethuraman N, Nett JH, Jiang Y, Rios S, et al. Humanization of yeast to produce complex terminally sialylated glycoproteins. Science. 2006;313(5792):1441–1443. doi: 10.1126/science.1130256. [DOI] [PubMed] [Google Scholar]
- 14.Nett JH, Stadheim TA, Li H, Bobrowicz P, Hamilton SR, Davidson RC, et al. A combinatorial genetic library approach to target heterologous glycosylation enzymes to the endoplasmic reticulum or the Golgi apparatus of Pichia pastoris. Yeast. 2011;28(3):237–252. doi: 10.1002/yea.1835. [DOI] [PubMed] [Google Scholar]
- 15.Jacobs PP, Geysens S, Vervecken W, Contreras R, Callewaert N. Engineering complex-type N-glycosylation in Pichia pastoris using GlycoSwitch technology. Nat Protoc. 2009;4(1):58–70. doi: 10.1038/nprot.2008.213. [DOI] [PubMed] [Google Scholar]
- 16.Vervecken W, Kaigorodov V, Callewaert N, Geysens S, De Vusser K, Contreras R. In vivo synthesis of mammalian-like, hybrid-type N-glycans in Pichia pastoris. Appl Environ Microbiol. 2004;70(5):2639–2646. doi: 10.1128/AEM.70.5.2639-2646.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi BK, Bobrowicz P, Davidson RC, Hamilton SR, Kung DH, Li HJ, et al. Use of combinatorial genetic libraries to humanize N-linked glycosylation in the yeast Pichia pastoris. PNAS. 2003;100(9):5022–5027. doi: 10.1073/pnas.0931263100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith ET, Perry ET, Sears MB, Johnson DA. Expression of recombinant human mast cell chymase with Asn-linked glycans in glycoengineered Pichia pastoris. Protein Expr Purif. 2014;102:69–75. doi: 10.1016/j.pep.2014.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Meyer T, Laukens B, Nolf J, Van Lerberge E, De Rycke R, De Beuckelaer A, et al. Comparison of VHH-Fc antibody production in Arabidopsis thaliana, Nicotiana benthamiana and Pichia pastoris. Plant Biotechnol J. 2015;13(7):938–947. doi: 10.1111/pbi.12330. [DOI] [PubMed] [Google Scholar]
- 20.Baeshen MN, Bouback TA, Alzubaidi MA, Bora RS, Alotaibi MA, Alabbas OT, et al. Expression and purification of C-peptide containing insulin using Pichia pastoris expression system. Biomed Res Int. 2016;2016:3423685. doi: 10.1155/2016/3423685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Effenberger I, Harport M, Pfannstiel J, Klaiber I, Schaller A. Expression in Pichia pastoris and characterization of two novel dirigent proteins for atropselective formation of gossypol. Appl Microbiol Biotechnol. 2017;101(5):2021–2032. doi: 10.1007/s00253-016-7997-3. [DOI] [PubMed] [Google Scholar]
- 22.Maciola AK, Pietrzak MA, Kosson P, Czarnocki-Cieciura M, Smietanka K, Minta Z, et al. The length of N-glycans of recombinant H5N1 hemagglutinin influences the oligomerization and immunogenicity of vaccine antigen. Front Immunol. 2017;8:444. doi: 10.3389/fimmu.2017.00444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu CP, Tsai TI, Cheng T, Shivatare VS, Wu CY, Wu CY, et al. Glycoengineering of antibody (herceptin) through yeast expression and in vitro enzymatic glycosylation. Proc Natl Acad Sci USA. 2018;115(4):720–725. doi: 10.1073/pnas.1718172115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Puxbaum V, Mattanovich D, Gasser B. Quo vadis? The challenges of recombinant protein folding and secretion in Pichia pastoris. Appl Microbiol Biotechnol. 2015;99(7):2925–2938. doi: 10.1007/s00253-015-6470-z. [DOI] [PubMed] [Google Scholar]
- 25.Jacobs PP, Inan M, Festjens N, Haustraete J, Van Hecke A, Contreras R, et al. Fed-batch fermentation of GM-CSF-producing glycoengineered Pichia pastoris under controlled specific growth rate. Microb Cell Fact. 2010;9:93. doi: 10.1186/1475-2859-9-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gmeiner C, Saadati A, Maresch D, Krasteva S, Frank M, Altmann F, et al. Development of a fed-batch process for a recombinant Pichia pastoris Deltaoch1 strain expressing a plant peroxidase. Microb Cell Fact. 2015;14:1. doi: 10.1186/s12934-014-0183-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Høiberg-Nielsen R, Westh P, Skov LK, Arleth L. Interrelationship of steric stabilization and self-crowding of a glycosylated protein. Biophys J. 2009;97(5):1445–1453. doi: 10.1016/j.bpj.2009.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alain R, Yves C, Ludmila O, Francois K. Three-dimensional structure of tubular networks, presumably Golgi in nature, in various yeast strains: a comparative study. Anat Rec. 1995;243(3):283–293. doi: 10.1002/ar.1092430302. [DOI] [PubMed] [Google Scholar]
- 29.Dubelaar GBJ, Gerritzen PL. CytoBuoy: a step forward towards using flow cytometry in operational oceanography. Scientia Marina. 2000;64(2):255–265. doi: 10.3989/scimar.2000.64n2255. [DOI] [Google Scholar]
- 30.Ehgartner D, Herwig C, Neutsch L. At-line determination of spore inoculum quality in Penicillium chrysogenum bioprocesses. Appl Microbiol Biotechnol. 2016;100(12):5363–5373. doi: 10.1007/s00253-016-7319-9. [DOI] [PubMed] [Google Scholar]
- 31.Söderström BE. Vital staining of fungi in pure cultures and in soil with fluorescein diacetate. Soil Biol Biochem. 1977;9:59–63. doi: 10.1016/0038-0717(77)90061-X. [DOI] [Google Scholar]
- 32.Tripathi K, Matmati N, Zheng WJ, Hannun YA, Mohanty BK. Cellular morphogenesis under stress is influenced by the sphingolipid pathway gene ISC1 and DNA integrity checkpoint genes in Saccharomyces cerevisiae. Genetics. 2011;189(2):533–547. doi: 10.1534/genetics.111.132092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dietzsch C, Spadiut O, Herwig C. A fast approach to determine a fed batch feeding profile for recombinant Pichia pastoris strains. Microb Cell Fact. 2011;10:85. doi: 10.1186/1475-2859-10-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dietzsch C, Spadiut O, Herwig C. A dynamic method based on the specific substrate uptake rate to set up a feeding strategy for Pichia pastoris. Microb Cell Fact. 2011;10:14. doi: 10.1186/1475-2859-10-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Capone S, Horvat J, Herwig C, Spadiut O. Development of a mixed feed strategy for a recombinant Pichia pastoris strain producing with a de-repression promoter. Microb Cell Fact. 2015;14:101. doi: 10.1186/s12934-015-0292-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krainer FW, Dietzsch C, Hajek T, Herwig C, Spadiut O, Glieder A. Recombinant protein expression in Pichia pastoris strains with an engineered methanol utilization pathway. Microb Cell Fact. 2012;11:22. doi: 10.1186/1475-2859-11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tams JW, Welinder KG. Glycosylation and thermodynamic versus kinetic stability of horseradish peroxidase. FEBS Lett. 1998;421(3):234–236. doi: 10.1016/S0014-5793(97)01573-1. [DOI] [PubMed] [Google Scholar]
- 38.Zou S, Xie L, Liu Y, Kaleem I, Zhang G, Li C. N-linked glycosylation influences on the catalytic and biochemical properties of Penicillium purpurogenum beta-d-glucuronidase. J Biotechnol. 2012;157(3):399–404. doi: 10.1016/j.jbiotec.2011.12.017. [DOI] [PubMed] [Google Scholar]
- 39.Wang Z, Guo C, Liu L, Huang H. Effects of N-glycosylation on the biochemical properties of recombinant bEKL expressed in Pichia pastoris. Enzyme Microb Technol. 2018;114:40–47. doi: 10.1016/j.enzmictec.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 40.Naatsaari L, Mistlberger B, Ruth C, Hajek T, Hartner FS, Glieder A. Deletion of the Pichia pastoris KU70 homologue facilitates platform strain generation for gene expression and synthetic biology. PLoS ONE. 2012;7(6):e39720. doi: 10.1371/journal.pone.0039720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gmeiner C, Spadiut O. Effects of different media supplements on the production of an active recombinant plant peroxidase in a Pichia pastoris ΔOCH1 strain. Bioengineered. 2015;6(3):175–178. doi: 10.1080/21655979.2015.1036208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gruber C, Altmann F. Glyco-engineering methods and protocols. New York: Humana Press; 2015. Site-specific glycosylation profiling using liquid chromatography–tandem mass spectrometry (LC–MS) pp. 407–415. [DOI] [PubMed] [Google Scholar]
- 43.Krainer FW, Capone S, Jager M, Vogl T, Gerstmann M, Glieder A, et al. Optimizing cofactor availability for the production of recombinant heme peroxidase in Pichia pastoris. Microb Cell Fact. 2015;14(1):4. doi: 10.1186/s12934-014-0187-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krainer FW, Pletzenauer R, Rossetti L, Herwig C, Glieder A, Spadiut O. Purification and basic biochemical characterization of 19 recombinant plant peroxidase isoenzymes produced in Pichia pastoris. Protein Expr Purif. 2014;95:104–112. doi: 10.1016/j.pep.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spadiut O, Rossetti L, Dietzsch C, Herwig C. Purification of a recombinant plant peroxidase produced in Pichia pastoris by a simple 2-step strategy. Protein Expr Purif. 2012;86(2):89–97. doi: 10.1016/j.pep.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 46.Spadiut O, Leitner C, Salaheddin C, Varga B, Vertessy BG, Tan TC, et al. Improving thermostability and catalytic activity of pyranose 2-oxidase from Trametes multicolor by rational and semi-rational design. FEBS J. 2009;276(3):776–792. doi: 10.1111/j.1742-4658.2008.06823.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. OD600 over induction time in shake-flask experiment, black-filled circle = wt, unfilled white circle = SuperMan5, grey triangle = ∆OCH1. Figure S2. Comparison of signal profiles from flow cytometer. FSC (black line -), SSC (blue line --), green (green line --) and red (red line --) fluorescence signals of SuperMan5 clusters after 23 h induction time in shake-flasks. In viability-declined cluster (A) a clear increase in red fluorescence is visible from PI staining in contrast to the viable cluster (B). Figure S3. Mean cluster size of ∆OCH1 (black bars) and SuperMan5 (dotted grey bars) over induction time in shake-flask cultivation. Standard deviations are derived from multiple measurements (at least 3) of single culture shake-flask samples. Figure S4. ESI–MS spectra of HRP glycopeptides at n-glycosylation site 7. The triply (or doubly) charged site 7 N-glycopeptide GLIQSDQELFSSPNATDTIPLVR of natural HRP and HRP produced in a ∆OCH1, in SuperMan5 and in wt P. pastoris are shown. The wt_HRP exhibits also phosphorylated versions (P) of the oligomannosidic glycans.
Data Availability Statement
The dataset(s) supporting the conclusions of this article are all included within the article. The datasets used and/or analyzed during the current study, if not shown in the text or additional files, are available from the corresponding author on reasonable request.