Abstract
An esterase from S. mutans UA159, SMU_118c, was shown to hydrolyze methacrylate resin-based dental monomers.
Objective:
To investigate the association of SMU_118c to the whole cellular hydrolytic activity of S. mutans toward polymerized resin composites, and to examine how the bacterium adapts its hydrolytic activity in response to environmental stresses triggered by the presence of a resin composites and adhesives biodegradation by-product (BBP).
Materials and Methods:
Biofilms of S. mutans UA159 parent wild strain, SMU_118c knockout strain (ΔSMU_118c), and SMU_118c complemented strain were incubated with photo-polymerized resin composite. High performance liquid chromatography was used to quantify the amount of a universal 2,2-Bis[4-(2-hydroxy-3-methacryloxypropoxy)phenyl]propane (bisGMA)-derived BBP, bishydroxy-propoxy-phenyl-propane (bisHPPP) in the media. Fluorescence in situ hybridization (FISH) and quantitative proteomic analysis were used to measure SMU_118c gene expression and production of SMU_118c protein, respectively, from biofilms of S. mutans UA159 wild strain that were cultured with bisHPPP.
Results:
The levels of bisHPPP released from composite were similar for ΔSMU_118c and media control, and these were significantly lower compared to the parent wild-strain UA159 and complemented strain (ΔSMU_118cC) (p<0.05). Gene expression of SMU_118c and productions of SMU_118c protein were higher for bisHPPP incubated biofilms (p<0.05).
Significance:
This study suggests that SMU_118c is a dominant esterase in S. mutans and capable of catalyzing the hydrolysis of the resinous matrix of polymerized composites and adhesives. In turn, the bacterial response to BBP was to increase the expression of the esterase gene and enhance esterase production, potentially accelerating the biodegradation of the restoration, adhesive and restoration-tooth interface, ultimately contributing to premature restoration failure.
Keywords: dental restoration; dental restoration failure; recurrent caries; cariogenic bacteria; hydrolysis; degradation; 2,2-Bis[4-(2-hydroxy-3-methacryloxypropoxy)phenyl]propane; bisGMA; bishydroxy-propoxy-phenyl-propane; bisHPPP; gene response; protein regulation
Graphical Abstract
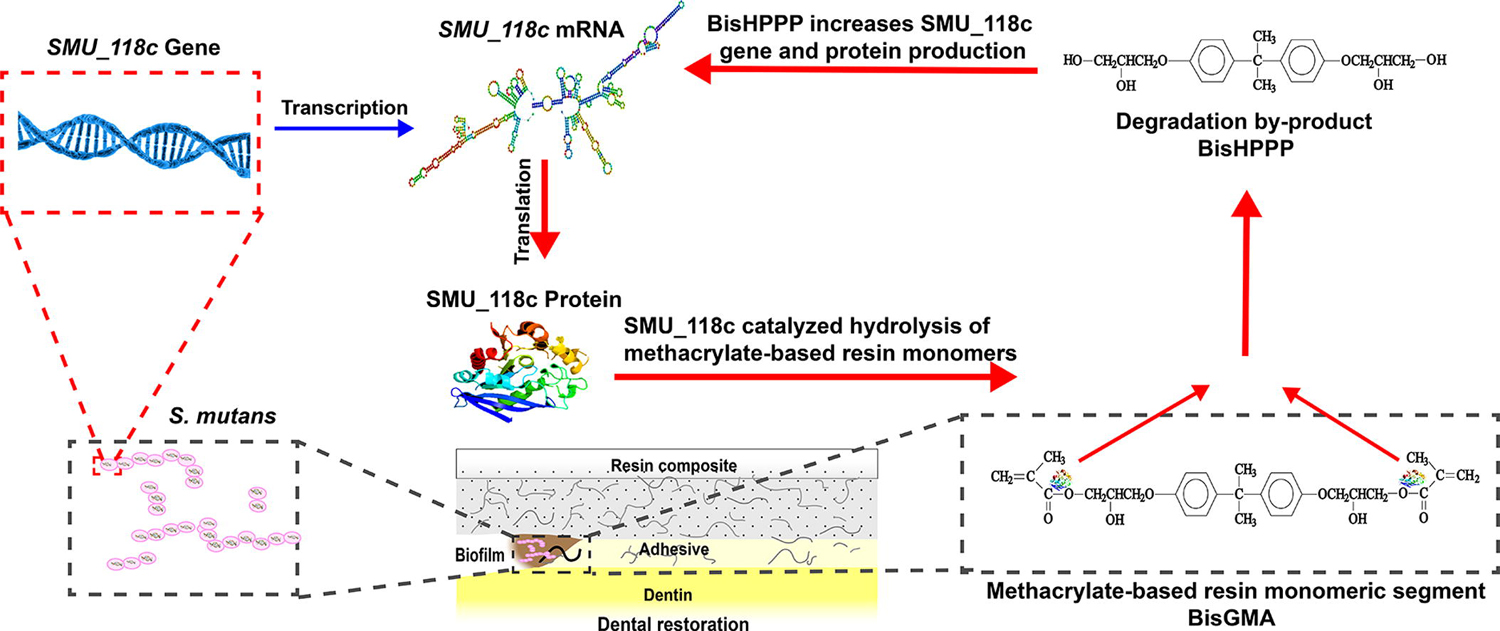
1. Introduction
Increasing concerns about possible adverse health effects of mercury, growing esthetic demands, and improved adhesive technology in dental materials have led resin composites to become the most commonly used dental restorative materials [1]. However, nearly 70% of restoration operations done by dentists are replacements of failed restorations [2–4]. The primary reason for restoration failure is recurrent or secondary caries developed along the compromised restoration-tooth interface [4–7].
Methacrylate-based resin composites and adhesives are prone to hydrolysis due to unprotected ester linkages in matrix monomers, the most abundant monomer being 2,2-Bis[4-(2-hydroxy-3-methacryloxypropoxy)phenyl]propane (bisGMA) [8]. Hydrolysis of bisGMA yields the degradation product Bishydroxy-propoxy-phenyl-propane (bisHPPP) [9, 10]. Previous studies have shown that human or model salivary esterases catalyze this hydrolysis, a process that is often referred to as biodegradation [9, 11]. This process compromises the restoration-tooth interface, allowing for cariogenic bacterial penetration into the interface and subsequent initiation and progression of secondary caries [11].
Streptococcus mutans is a major etiological agent of dental caries [12]. The main virulence factors of interest for S. mutans are its ability to form biofilm, survive in acidic environments, and produce acid by fermentation of dietary sugar. In addition to these well-understood cariogenic properties, S. mutans has been shown to contain esterase activity that degrades resin composites, which could contribute to secondary caries by compromising the restoration-tooth interface [13]. A recent study has identified and characterized an esterase from S. mutans UA159, SMU_118c, that has high hydrolytic activity toward the universal resin composites and adhesives monomer, bisGMA, with a similar activity profile to that of salivary esterases [14]. In other bacteria, esterases have been linked to virulence and pathogenesis [15, 16]. However, there are no reports on the role of bacterial esterases in the biodegradation of biomaterials, specifically dental methacrylates, as a contributing factor to bacterial virulence.
Esterase-catalyzed resin composite biodegradation enlarges the gap between restoration and tooth [11], compromising interfacial bond strength [9, 17], allowing bacterial infiltration to the gap [18, 19], and providing a unique micro-environment containing biodegradation by-products (BBPs) in close proximity to bacteria [11, 20, 21]. Bacteria have a high capacity to adapt to environmental changes as a result of their robust metabolism [22], which is controlled by a network of regulatory pathways requiring cellular programming at transcriptional and translational levels. Previous studies have reported the impact of BBPs on the growth and virulence gene expression of S. mutans [23–26]. However, these investigations relied on disruptive techniques (RNA extraction and reverse transcription polymerase chain reaction (RT-PCR)) that destroy the biofilm structure and thus do not allow for observation of spatial and temporal gene expression patterns.
On the basis of the above introduction, the first aim of this study was to investigate the association of SMU_118c to whole cellular hydrolytic activity towards polymerized resin composites, and to examine how the bacterium adapts its hydrolytic activity in response to environmental stresses triggered by the presence of a biodegradation by-product (BBP). The second aim was to establish a non-destructive methodology, based on fluorescence in situ hybridization (FISH), to analyze gene responses of the esterase gene SMU_118c (gene denoted by italic font) to BBPs in biofilms with intact and complex 3D structures. Furthermore, the effect of BBPs on SMU_118c protein production was investigated by employing label-free quantitative proteomics. The hypothesis was that SMU_118c is the major contributor to S. mutans degradative activity toward methacrylate resins, and that the bacterium responds to the presence of resin biodegradation by-products by modulating the esterase gene expression and protein synthesis.
Materials and methods
2.1. Resin composite specimen preparation
All instrumentation was autoclaved or disinfected via 70% ethanol solution and specimen preparation was performed in a biosafety cabinet using sterile technique. Cylindrical pellets (4 mm diameter × 4 mm height) of resin composite (Z250; 3M, London, ON, Canada) were prepared using Teflon™ molds, designed to allow for an optimal curing depth of 2 mm [27]. A glass slide covered by a polyester strip provided the base for the mold and resin was inserted. The other end of the mold was then covered by another polyester strip and glass slide and the entire unit was clamped together. The specimens were photo-polymerized for 15 s from each side (Sapphire plus, DenMat, Santa Maria, CA, USA) as described previously [8]. Intensity of the curing device was verified using the built-in meter. The cured specimens were removed from the mold using a sterile plastic instrument and then post cured in a vacuum oven at 60°C for 48 hours [10, 13, 28].
2.2. Resin composite biodegradation by SMU_118c
The composite specimens (n = 3 for each incubation condition and period) were pre-incubated in phosphate-buffered saline (D-PBS, pH 7.0, 37°C) (Gibco, Grand Island, NY) for 2 days to allow for diffusion of unreacted resin monomers [13], then incubated individually in sterile vials (Chromatographic Specialties Inc., Brockville, ON, Canada) containing 1 mL of D-PBS with or without 50 U/ml of SMU_118c for up to 30 days. One unit of esterase activity was defined as the amount of enzyme producing 1 µmol p-nitrophenol from p-nitrophenyl esters per minute (ε401=1.6 × 104 M−1cm−1) [10]. 100 μL of incubation media was collected at 0, 5, 7, 17, 24 and 30 days, mixed with equal volume of pre-chilled methanol (Sigma-Aldrich, St. Louis MO, USA) to quench the hydrolytic reaction, filtered at 1000g (H-25FI, Silencer) with a 5000 Da-cut-off centrifuge filter (Ultra-0.5, 3kDa MWCO, Millipore, Bedford, MA. The processed samples were refrigerated until analysis and the original incubation solutions were replenished with the same amount of 50 U/ml SMU_118c [9, 14, 29]. A high performance liquid chromatography (HPLC) system (Waters, Mississauga, ON, Canada) equipped with photodiode detector was used for the chromatographic separation and quantification of bisHPPP and MA as described previously [9, 14, 29]. The retention time of each product was confirmed by HPLC chromatographic profile of standard monomers and mass spectrometry (MS) analysis [11, 13, 27, 28].
2.2.1. Statistics analysis
Student’s t- test was used to determine significance of differences between bisHPPP released after the two incubation conditions (D-PBS + SMU_118c or D-PBS alone) at the same time-point (p < 0.05). One-way analyses of variance (ANOVA) and Tukey’s multiple comparison tests (p < 0.05) were performed to determine the significance of differences between bisHPPP release in same conditions at different time points. Homogeneity of variance and normality were verified with Leven’s and Shapiro-Wilk tests, respectively.
2.3. Construction of S. mutans UA159 SMU_118c knockout (ΔSMU_118c) and complemented (ΔSMU_118cC) strains
The sequence of esterase gene SMU_118c was found in the S. mutans UA159 genome database as a putative esterase (http://www.genome.ou.edu/smutans.html). The SMU_118c knockout mutant (ΔSMU_118c) was constructed through PCR-ligation mutagenesis according to a previously established protocol [30–32]. The SMU_118c complemented strain (ΔSMU_118cC) was made using pIB166 plasmid that contained the S. mutans recombinant SMU_118c as described previously [33]. The primers used for mutation, confirmation, and complementation constructs are listed in Table 1 (ACGT Corporation, ON, Canada). The ΔSMU_118c deletion mutation and complementation were confirmed by PCR analysis.
Table 1:
Primers for SMU_118c gene deletion and complementation
| Primer Nucleotide sequence |
|---|
| Erm-PA a 5′ GGCGCGCCCCGGGCCCAAAA TTTGTTTGA T 3′ |
| Erm-PB b 5′ GGCCGGCCAGTCGGCAGCGACTCATAGAAT 3′ |
| SMU_118c-P1 5′ AAGAAGTCTGTTTGCTGGG 3′ |
| SMU_118c-P2 a 5′ GGCGCGCCTTGACGAATGGCTGTAGCG 3′ |
| SMU_118c-P3 b 5′ GGCCGGCCGAACCATAGAAAGTTGAGG 3′ |
| SMU_118c-P4 5′ CCCTATTAAAACAGCACCC 3′ |
| SMU_118c-Comp1 c 5′ GGATCCTTGAAATTGTTGTCCCCTCAACT3′ |
| SMU_118c-Comp2 d 5′ CTCGAGTCATTCATTACTGAGGAGGCAGG3′ |
Primers in roman (non-italic) font were used for gene deletion and primers in italics were used for gene complementation.
AscI restriction sites are in italic boldface
FseI restriction site are in boldface and underlined
BamHI restriction sites are in boldface
XhoI restriction site are in boldface and double underlined
2.4. Resin composite biodegradation by S. mutans UA159 and ΔSMU_118c strain
Cured specimens (n = 3/group) were incubated in sterile vials containing either 3 mL of 1:4 dilution brain-heart infusion (BHI) (Becton, Dickinson and Co., Sparks, MD, USA) with pH adjusted to 5.5 using lactic acid (control) (Sigma Lactic acid, Sigma-Aldrich, St. Louis MO, USA), or 1:4 dilution of BHI with overnight grown S. mutans UA159 or ΔSMU_118c (experimental groups). Incubation solutions were collected every 48 hours from each group and replaced with fresh solutions. Incubation solutions were accumulated, pooled, and biodegradation by-product bisHPPP was quantified by HPLC as previously reported [9]. The purity of the bacterial culture was assessed by gram stain at each media replacement under a light microscope (Olympus® BX 51; Olympus America Inc., NY, USA) and viability was assessed by colony forming unit (CFU) on agar plates at the time interval.
Resin composite specimens were collected from each biodegradation experimental condition (n = 3) and sonicated to remove bacteria for direct observation of the materials’ surfaces. Surface morphology of specimens incubated for 30-days was observed via scanning electron microscopy (S2500, Hitachi, Mito City, Japan) at an accelerating voltage of 10 kV as described previously [9].
2.4.1. Statistics analysis
One-way analyses of variance (ANOVA) and Tukey’s multiple comparison tests (p < 0.05) were performed to analyze bisHPPP release. Homogeneity of variance and normality were verified with Leven’s and Shapiro-Wilk tests, respectively.
2.5. Verification of the association of SMU_118c gene on resin composite biodegradation
Photo-polymerized resin composite specimens (n = 3/group) were incubated in sterile vials containing either 1 mL of Fujiwara minimum medium with pH adjusted to 5.5 using lactic acid (control) (Sigma Lactic acid, Sigma-Aldrich, St. Louis MO, USA) or medium with overnight S. mutans UA159, ΔSMU_118c or ΔSMU_118cC complemented strains (experimental groups). The purity of the bacterial cultures and viability were analysed as described above. Quantification of bisHPPP production was conducted after 8-day incubation using the HPLC protocol described above.
2.5.1. Statistics analysis
One-way analyses of variance (ANOVA) and Tukey’s multiple comparison tests (p < 0.05) were performed to analyse bisHPPP release for specimens incubated with the different strains and time points.
2.6. The effect of bisHPPP on SMU_118c gene expression by FISH
The assay was conducted based on a previously described protocol with modifications [34]. All water and buffers were treated with 0.1% vol. diethylpyrocarbonate (DEPC) (Sigma, Taufkirchen, Germany) and then autoclaved. Briefly, 18-hour biofilms of S. mutans UA 159 parent wild strain, alone or with bisHPPP in concentrations relevant to in vivo conditions (0.01, 0.1 and 1 mM) [8], or ΔSMU_118c without bisHPPP, were formed in 8-well chamber (Lab-Tek camber slide, NUNC, Naperville, IL, USA) containing 400 μ l of chemically defined Media (CDM) at 37˚C with 5% CO2. Biofilms were then fixed in 4% paraformaldehyde (pH 7.2) at 4°C for 12 hours. After fixation, all specimens were treated with freshly prepared 0.1% (vol/vol) active DEPC in D-PBS for 12 min at RT and rinsed with D-PBS and HPLC grade water for 1 min each. Specimens were then subjected to cell membrane permeabilization at 37°C for 30 min in a 200 μl solution of 10 mg/ml lysozyme (Sigma, St Louis, MO, USA) in 100 mM Tris(hydroxymethyl)aminomethane hydrochloride (Tris-HCl) buffer, pH 7.5 (Sigma, St Louis, MO, USA) supplemented with 5 mM Ethylenediaminetetraacetic acid (EDTA) (Merck, Damstadt, Germany). The biofilms were then dehydrated with a series of ethanol washes. Specimens were then incubated with green-fluorophore-labeled oligonucleotide probes (Table 2) at a concentration of 50 ng/20μl targeting the mRNA of SMU_118c in hybridization buffer (0.9 M NaCl, 20 mM (Tris-HCl) (pH 7.2), 50 % vol. formamide, and 0.01 % (w/v) sodium dodecyl sulphate (SDS). Hybridization was conducted at 38°C in a humid chamber for 12 hours. Following probe hybridization, specimens were incubated for 15 min at 42°C in wash buffer containing 20 mM Tris-HCl (pH 7.2), 5 mM EDTA, 159 mM NaCl and 0.01 % (w/v) SDS. As a control, a parallel assay was conducted to investigate the effect of bisHPPP on the expression of a housekeeping gene (16S rRNA) under same conditions except for the hybridization; red-fluorophore-labeled probes STR405 (Table 2) was hybridized to 16s rRNA at 48°C for 3 hours. All biofilms were stained with nuclear dye 4’,6’-diamidino-2-phenylindole (DAPI; blue) for further biomass analysis. Finally, the images of biofilms (n = 3 random sites/biofilm) were acquired using confocal laser scanning microscopy (CLSM; IX81 Inverted Microscope, Olympus, Tokyo, Japan) with a water immersion objective (60x/1.2 UPlanApo Water). Each CLSM image stack had a substratum coverage field area of 211 × 211 μm and z-step interval of 0.5 μm.
Table 2:
Probes for FISH
| UA159_SMU_118c |
|---|
| Sequence: |
| 5′-GACAAGGAGCUGGUUUUUAUCUCAACGCAACUCAGAAUCCUUGGUCGCAACAUUACCACA-3′ |
| Target: mRNA of SMU_118c in S. mutans UA 159 |
| Labeling: 5′ –fluorochrome, Alexa Fluor 488 fluorescence labeled (Ex/Em: 490/525 nm) |
| SRT 405 |
| Sequence: 5′-TAGCCGTCCCTTTCTGGT-3′ |
| Target: Streptococcus spp. 16s rRNA |
| Labeling: 5′ –fluorochrome, Alexa Fluor 594 fluorescence labeled (Ex/Em: 590/617 nm) |
2.7. Image analysis
The image stacks were converted to 3D and analyzed using a quantitative software (Imaris version 8.3, Bitplane AG, Zurich, Switzerland) to calculate the parameters of the biofilm. Biofilm formation of each group was quantified in terms of total cell number presented from DAPI staining. Gene expression level was proportional to the respective fluorescence intensity in the biofilm, allowing for quantification of the level using total voxel intensity of fluorescence from the specific probes in each biofilm, normalized by dividing by the total cell number of each biofilm. The green fluorescence from ΔSMU_118c formed biofilm was considered as baseline for the SMU_118c gene expression.
2.7.1. Statistical analysis
One-way analyses of variance (ANOVA) and Scheffe’s multiple comparison tests (p < 0.05) were performed to validate differences in gene expression between control (no bisHPPP) and experimental groups (different concentrations of bisHPPP for the wild and ΔSMU_118c strains).
2.8. Biofilm protein preparation prior to mass spectrometric analysis
18-hour biofilms of S. mutans UA 159 parent wild strain were grown at 37°C with 5% CO2 in Tryptone Glucose Yeast Extract Agar (TYEG) medium buffered at pH 5.5 with 2-(N-Morpholino)ethanesulfonic acid, 4-Morpholineethanesulfonic acid (MES) (Sigma-Aldrich, St. Louis, MO, USA), supplemented with 0.1% glucose, with no bisHPPP or with bisHPPP in concentrations relevant to in vivo conditions (0.01, 0.1 and 1 mM). Then, the biofilm cells were collected and disrupted using a homogenizer (Thermo Savant, FastPrep FP 101) at setting 6 for 45 minutes and then centrifuged at 15,700 x g for 1 minute. Supernatant was carefully removed, separated in aliquots of 50 µL, and stored at −80°C. The total protein concentration in each sample was assessed by the Micro Bicinchoninic Acid (Micro BCA) assay [35]. Equal protein amounts (20 µg) from both experimental and control groups were dried using a rotary evaporator, denatured, and reduced for 2 hours by the addition of 200 µL of 4 M urea, 10 mM dithiothreitol (DTT), and 50 mM NH4HCO3, pH 7.8. After four-fold dilution with 50 mM NH4HCO3, pH 7.8, tryptic digestion was carried out for 18 h at 37°C, following the addition of 2% (w/w) sequencing-grade trypsin (Promega, Madison, WI, USA).
2.9. Liquid Chromatography Electrospray Ionization Tandem Mass Spectrometry (LC-ESI-MS/MS) and relative proteome quantitation
Peptide separation and mass spectrometric analyses were carried out as previously described [26, 35]. Briefly, protein samples were re-suspended in 20 µL of 97.5 % H2O/2.4% acetonitrile/0.1% formic acid and subjected to liquid chromatography and electrospray ionization tandem MS (LC-ESI-MS/MS). A linear 65-min gradient ranging from 5–55% of 80% acetonitrile at a flow rate of 110 nL/min was used. The electrospray voltage and ion transfer capillary temperature was 1.8 kV and 230 °C, respectively. All samples were analyzed in triplicate. The obtained MS/MS spectra from each sample were searched against the streptococci protein database and a specific SMU_118c protein, Q8DWE3 (Swiss Prot and TrEMBL, Swiss Institute of Bioinformatics, Geneva, Switzerland, http://ca.expasy.org/sprot/), using SEQUEST algorithm in Proteome Discoverer 1.3 software (Thermo Scientific, San Jose, CA, USA).
For quantitative proteome analysis, three MS raw files from each group (control and experimental; total of 12 MS raw files) were analyzed using SIEVE software (Version 2.0 Thermo Scientific, San Jose, CA, USA) [35]. For the alignment step, a single MS raw file belonging to the control group (0 mM) was selected as the benchmark and the other files were adjusted to generate the best correlation to this reference file. After alignment, the feature detection and integration process was performed using MS-level data. For statistical analyses of protein abundance, peak integrations were summarized into protein-level annotation in SIEVE using a weighted average of intensities of LC-ESIMS/MS of a protein by run.
2.9.1. Statistical analysis
A statistical model based on an ANOVA framework with Tukey’s post hoc test was carried out. Relative abundance of SMU_118c protein from different bisHPPP concentration groups was considered significantly different from the control group (0 mM) when the values observed were >1.5 for increased and <0.5 for decreased abundance with a p-value cut-off of < 0.05 [35].
3. Results
3.1. Resin composite biodegradation by SMU_118c
A trend of increasing bisHPPP release with time throughout the incubation period was observed for both control and SMU_118c-incubated groups (p < 0.05; Fig. 1). The amount of bisHPPP released was elevated significantly in the presence of SMU_118c vs. D-PBS (control) (p < 0.05) for all incubation periods. The total amount of bisHPPP released from Z250 after 30 days of incubation with SMU_118c (8.76 ± 0.50 μg/cm 2) was 21 times higher than that released from control (0.41 ± 0.03 μg/cm2, p < 0.05).
Fig. 1.
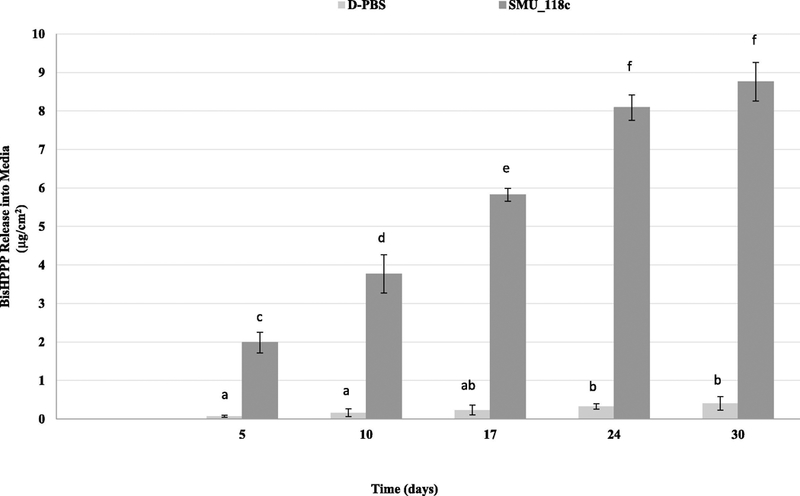
Cumulative isolated bisHPPP after incubation of resin composites in D-PBS (light gray) or with SMU_118c in D-PBS (dark gray). Data are plotted as mean ± standard error. Values with the same lower case letter denote statistically non-significant differences (p > 0.05).
3.2. Resin composite biodegradation by parent wild strain S. mutans UA159 and ΔSMU_118c strain
30-day cumulative bisHPPP release was highest for specimens incubated with the parent wild strain S. mutans UA159 (21.37 ± 1.55 μg/cm2, p < 0.05). The amount of bisHPPP released from ΔSMU_118c-incubated specimen (15.69 ± 0.27 μg/cm2) was not statistically different than that released from media control without bacteria (15.62 ± 0.23 μg/cm2, p > 0.05).
SEM micrographs (Fig. 2) demonstrated that the surfaces of specimens incubated with the parent wild strain S. mutans UA159 for 30 days appear roughest, while the surface of ΔSMU_118c-incubated specimen after 30 days were similar to media-incubated and non-incubated specimens. The incubation media alone did not have a significant effect on surface roughness vs. non-incubated specimens.
Fig. 2.
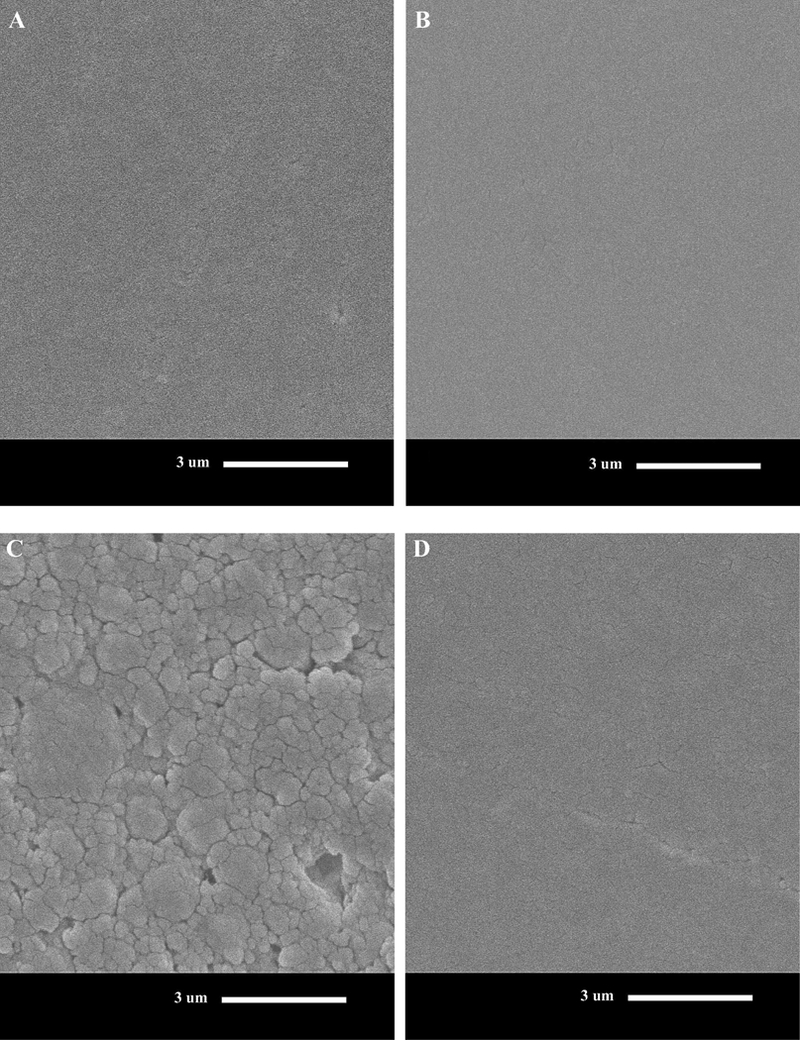
Scanning electron micrographs of resin composite at day 0 (A), or following 30-day incubation in BHI with no bacteria (pH 5.5) (B), S. mutans UA159 (C) and ΔSMU_118c (D) SMU_118c (D) in BHI (104 X original magnification). Scale bar applies to all figures and represents 3 lm. Note the rougher surfaces for S. mutans UA159 (parent wild strain) vs. ΔSMU_118c and BHI media control.
3.3. Verification of the association of SMU_118c to the degradative activity of the whole bacterial cell towards resin composite
Results for bisHPPP release are provided as a percentage of the amount of bisHPPP isolated from the resin composites incubated with S. mutans UA159 parent wild strain (Fig. 3). Relative bisHPPP amounts were lowest for specimens incubated with ΔSMU_118c (52.9% ± 8.9%, p < 0.05), while the S. mutans UA159 and ΔSMU_118c complemented strain (ΔSMU_118cC) incubated groups had similar bisHPPP release (100.0% ± 7.4% vs. 103.8% ± 6.1%, p > 0.05).
Fig. 3.
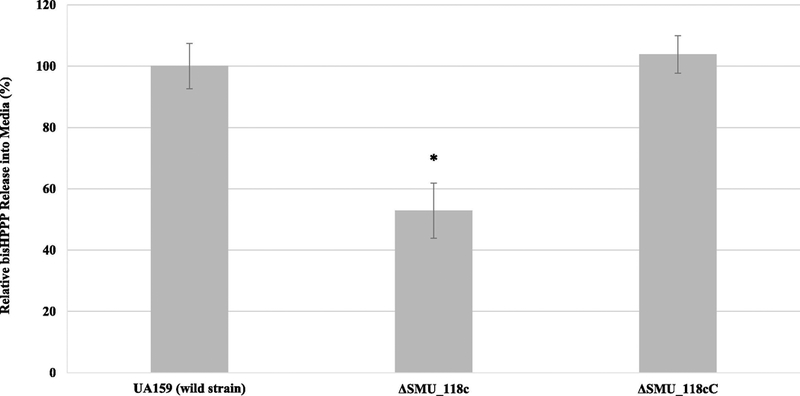
BisHPPP isolated from the media after incubation of resin composites with parent wild strain S. mutans UA 159, ΔSMU_118c or ΔSMU_118cC. Data were normalized to media background and plotted as mean ± standard error. * represents significant differences between groups with different bisHPPP concentration (p < 0.05).
3.4. The effect of bisHPPP on SMU_118c gene expression
To investigate the effect of bisHPPP on SMU_118c gene expression, the biofilm incorporated both DAPI and UA159_SMU_118 probes presented as blue and green colors, respectively. The specificity of the probe targeting mRNA of SMU_118c was demonstrated by comparing green fluorescence signal from biofilm formed by ΔSMU_118c to that of S. mutans UA159 (Fig. 3). For the control groups, biofilms incorporated DAPI and STR405 probes presented in blue and red florescence range, respectively (data not shown). A reduced biofilm mass was observed at 1 mM bisHPPP (Fig. 3A; p < 0.05). There were no differences in biofilm formation of ΔSMU_118c and the parent wild strain, indicating that the mutation had no effect on bacterial viability and the ability to form biofilm (Fig. 3A; p > 0.05). FISH images showed enhanced fluorescence intensity in biofilms with increased bisHPPP concentration from 0.01 mM to 1 mM, indicating possible up-regulation of SMU_118c (Fig. 4A). Due to the non-specific binding and auto-fluorescence of biofilm and/or culture media, fluorescence detected in ΔSMU_118c biofilm was used as baseline (background) and was subtracted from all groups for the quantification of SMU_118c gene expression. Quantified results were normalized by total cell number in each biofilm (Fig. 4B). Up-regulation of SMU_118c was concentration dependent, with a significant increase in biofilms incubated with 1 mM of bisHPPP (p < 0.05). In all strains, the expression of the housekeeping gene 16S rRNA did not change in response to bisHPPP (data not shown; p > 0.05).
Fig. 4.
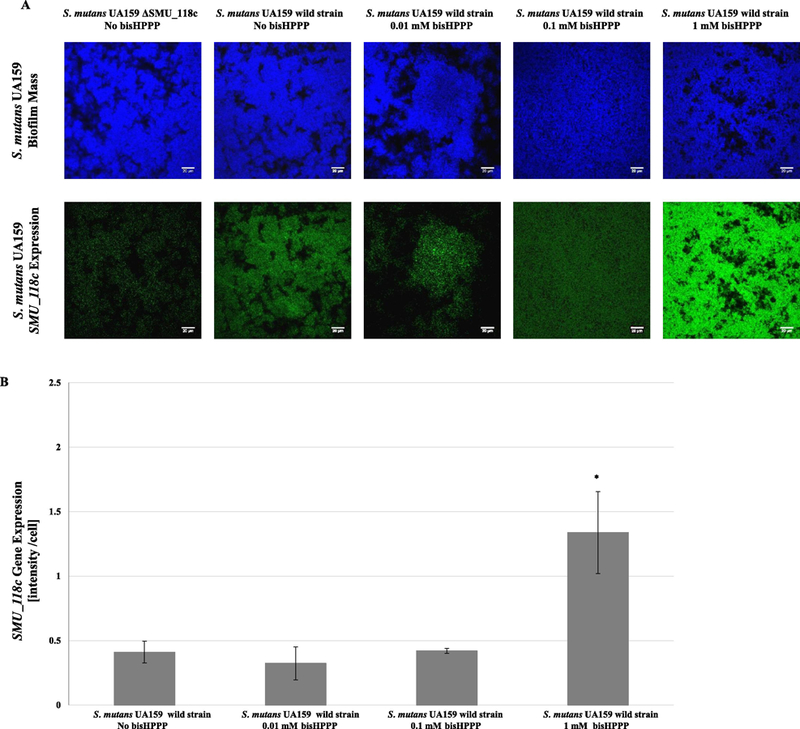
A) CLSM images of the SMU_118c gene expression in S. mutans biofilm formed with different concentrations of bisHPPP (A) andΔSMU_118c gene expression quantification analysis (B). Gene expression level is proportional to the fluorescence intensity in the biofilm. Blue signal by 4׳ 6-diamidino-2-phenylindole (DAPI) staining indicates biofilm biomass. Green signal from the UA159_SMU_118c probe represents levels of mRNA of SMU_118c expression. Increasing concentrations of bisHPPP resulted in an increase in the expression of SMU_118c gene in the parent wild strain of S. mutans UA159. Negative control (ΔSMU_118c) confirms specificity of the probe. Gene expression is normalized to number of cells in biofilm. For ΔSMU_118c gene expression, data were normalized to ΔSMU_118c groups (background). * represents significant differences between groups with different bisHPPP concentration (p < 0.05). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.).
3.5. The effect of bisHPPP on SMU_118c protein synthesis
SMU_118c from all four different groups (S. mutans UA159+ 0.0 mM, 0.01 mM, 0.1 mM and 1 mM bisHPPP) showed a consistent elution of protein/peptides. Compared to the control group (0.0 mM bisHPPP), increased levels of SMU_118c protein production were observed in two experimental groups ranging from 2.21 (0.1 mM bisHPPP; p < 0.05) to 2.32 (0.01 mM bisHPPP; p < 0.05) (Table 3). S. mutans UA159 incubated with 1 mM bisHPPP show statistical but not physiologically significant changes (Table 3).
Table 3:
SMU_118c protein (accession number Q8DWE3) abundance ratios from bisHPPP-incubated biofilms
| BBP | Gene name |
Protein function |
Ratio 1mM/0mM |
P | Ratio 0.1mM/0mM |
P | Ratio 0.01mM/0mM |
P |
|---|---|---|---|---|---|---|---|---|
| bisHPPP | SMU_118c |
putative esterase |
0.89 | 0.00 | 2.21 | 0.30 | 2.32 | 0.04 |
4. Discussion
Resin composites are the most popular dental restorative materials [36], and their physical and mechanical properties have been well studied. Recent studies have reported that the biostability of resin composites can affect their biocompatibility, since the release of biodegradation by-products affects gene expression of cariogenic bacteria [25, 26]. Esterase activities in human saliva degrade the bulk of the materials and the restoration-tooth interface [9–11, 17] and the cariogenic bacterium S. mutans UA159 was found to hydrolyze resin composites and adhesives [13]. A specific esterase, SMU_118c from this cariogenic bacterium was suggested as a contributor to hydrolysis of dental resins by this species [14]. The current study adds to the knowledge of the material’s biodegradation by exploring the contribution of this specific esterase from S. mutans to the whole bacterial cell biodegradative activity of resinous matrix in resin composites and the reciprocal effect of the biodegradation by-product on the bacteria at both gene and protein levels. These findings contribute to our understanding of the interaction between materials, host, and bacteria and could provide guidance to materials manufacturers.
In this study, the commercial resin composite Z250 was chosen as a substrate for biodegradation investigation because it contains the most abundant matrix monomer used in commercial resin-based materials and adhesives, bisGMA [37, 38]. BisHPPP, a bisGMA-derived degradation by-product, was chosen as an indicator of esterase-catalyzed biodegradation due to its very limited diffusion through the resin matrix and hydrophobic nature that significantly limits its solubility in aqueous medium of its precursor, bisGMA after polymerization [28]. In addition to esterase-mediated hydrolysis, hydrogen ions produced by oral bacteria can catalyze the hydrolysis of the ester bonds present in the polymer matrix [39–41]. To prevent the latter effect on resin biodegradation, all culture media controls were adjusted to pH 5.5 by lactic acid, which is the primary organic acid produced by S. mutans.
4.1. The role of SMU_118c in S. mutans-mediated resin composites biodegradation
In our previous work (Huang et al.) [14]) we reported that the esterase SMU_118c has a similar enzymatic profile to that of human saliva [10] and S. mutans whole bacterial cell [13] towards ester substrates, indicating its potential to degrade resin-based materials and thereby promote secondary caries. By knocking out and then restoring SMU_118c, the current study confirmed that SMU_118c is able to continuously degrade cured composite materials and that it is associated with the whole bacterial esterase activity of S. mutans. Compared to the parent wild strain, ΔSMU_118c-mediated biodegradation was reduced to a level similar to the media control and the complemented strain recovered whole bacterial hydrolytic activity, demonstrating that SMU_118c is the dominant esterase in S. mutans responsible for esterase-mediated hydrolysis of dental resin composites.
SEM micrographs showed that the surfaces of pre-incubated specimens were smooth and covered by resin, which excludes any effect of the filler on the availability of resin surface for biodegradation. After a 30-day incubation, SEM micrographs demonstrated rougher surfaces for specimens incubated with S. mutans vs. media control and ΔSMU_118c. The observation for the parent wild strain was consistent with previous studies [13, 42, 43] and corroborated the HPLC analysis of biodegradation. However, inconsistent results between HPLC and SEM analysis were observed from the media control groups where bisHPPP release from media-incubated specimens was detected by HPLC but the SEM images showed that the surfaces of specimens incubated with media were similar to non-incubated specimens. This could be explained by the different sensitivity and resolution of the two methods to analysing the degree of biodegradation. Unlike the quantitative HPLC analysis, SEM cannot detect minor changes and provides descriptive (qualitative) results of only the surface. In addition, the apparent discrepancy could be due to production of bisHPPP from unreacted bisGMA monomers that diffused from the bulk of the specimens rather than the surface of the sample. The latter process does not result in surface changes that can be observed by SEM, but produces sufficient amounts of the degradation by-product bisHPPP to be detected by HPLC.
Another possible explanation to the apparent differences between the SEM and HPLC biodegradation analyses is the incubation media used to grow bacteria. BHI media, required for longer bacterial growth periods, has esterase-like activity [13]. To reduce its effect on the surface biodegradation of the composite, which would potentially mask the true bacterial biodegradation, a 2.5 times diluted BHI media was used for the SEM study vs. the media used for the analyses by HPLC (it was not critical for the latter analysis, since all results were normalized or related to the BHI media alone). Therefore, it was not surprising that reduced degradative activity with diluted BHI was observed via SEM analysis. Although the exact composition of BHI medium and the identity of the specific degradative activity from this medium are not fully defined, use of this medium demonstrated the importance of the incubation condition and the steps that needed to be taken when analyzing the impact of different cell cultures on material biostability. The findings further demonstrate the significance of the interaction between bacteria, host, and material, and the important of using in vitro conditions that mimic these interactions. It also highlights the limitation of surface observation vs. quantification of degradation products when assessing degradation of resin composite dental biomaterials.
4.2. The effect of bisHPPP on SMU_118c gene expression and SMU_118c protein synthesis
Previous studies confirmed that in addition to temperature, pH, and chemical agents, BBPs induced adaptation mechanisms in S. mutans by stimulating bacterial growth and up-regulating key virulence genes [24–26]. The effect of bisHPPP on the expression of bacterial virulence genes was reported to be concentration dependent and only in biofilms growth mode at cariogenic pH, regarded as optimum conditions for the development of dental caries [26]. Hence, in this study, experiments were conducted only under conditions deemed optimum for the development of dental caries.
To analyze the effect of bisHPPP on SMU_118c gene expression, clinically relevant bisHPPP concentrations were tested [8]. However, while concentrations up to 0.1 mM bisHPPP are considered clinically relevant, these values are based on analysis of bisHPPP content in saliva after restoration [8]. Considering the confined local space in the restoration-tooth interface, even higher concentrations of bisHPPP (1 mM) could be expected due to accumulation of bisHPPP within the restoration-tooth interface [11, 26, 44]. In addition, considering that all measurements were conducted in situ with intact biofilms without planktonic counterparts, higher concentrations (1 mM) of bisHPPP were employed to test its adverse effects on biofilm biomass, which could affect gene expression and protein levels.
In situ hybridization of mRNA sequences is a popular technique for studying gene expression in eukaryotic cells and tissues [45]. In microorganisms, FISH with rRNA-targeted probes is routinely used for bacterial identification [46]. With the development of new probe designs and detection techniques, FISH is able to detect bacterial responses to environmental changes at the transcription level. Compared to the more commonly used qRT-PCR method for investigating gene expression, the design and labeling of specific probes in FISH is challenging, but the results are more informative due to the undisrupted biofilm structure that allows for spatial analysis and more complex biological growth conditions.
The inhibitory effect of bisHPPP on S. mutans UA159 biofilm formation with respect to reduced total cell numbers was observed at 1 mM, a much higher concentration than what has previously been reported for the S. mutans NG8 strain (0.1 and 0.25 mM) [47]. However, previous studies reported these two concentrations based on the growth rates of planktonic cells, which are more vulnerable to extracellular agents [48]. Based on FISH images, more mRNA copies of SMU_118c, presented as higher intensity of fluorescence from mRNA probes, were produced in biofilms with 1 mM of bisHPPP. Further quantification of gene expression confirmed this observation. This concentration-dependent up-regulation of SMU_118c may be explained by the accumulative effect of bisHPPP on the bacterial growth environment. Since bisHPPP is hydrophobic and weakly charged in an acidic environment, its accumulation around bacterial cells at high concentration could change the cell’s microenvironment, leading to pH fluctuations, fluid flow interruption, and nutrient blockage [49–51]. Previous studies reported that S. mutans could sense these changes and regulate gene expression for adaptation via two-component signal transduction systems, which are comprised of surface sensors and response regulators [33, 52, 53]. In addition, the direct interaction between bisHPPP and bacteria might create a new bacterial/environmental interface inducing auto-phosphorylation of sensor kinases in the bacterial membrane, which could activate SMU_118c expression by the intracellular response regulator [54].
The bisHPPP concentration that triggered an increase in gene expression was higher in this study than has previously been reported [26]. This could be explained by different biofilm biological conditions employed in each study. In this study, chemically defined media with limited nutrients was employed for FISH analysis to form a thin biofilm, necessary to facilitate cell permeabilization and probe penetration, and to reduce unnecessary interactions between compounds in the media and FISH working solutions. As reported previously, nutrients can influence bacterial physiochemical properties in biofilm development [55–57], which may explain the differential gene regulation associated with different culture media used for FISH and qRT-PCR. Different pH conditions may also contribute to differential gene responses. Previous studies investigated gene responses in biofilm with controlled pH (5.5) by adding organic acid. In the current study, the acidic environment, which had a similar measured endpoint pH (5.2), was created by S. mutans metabolism with natural pH fluctuation during biofilm formation. Changes in pH can induce a bacterial acid tolerance response associated with changes in expression of over 30 proteins [58, 59], which may affect SMU_118c gene expression. The differences between in situ detection vs. qRT-PCR analysis highlight the importance of using relevant in vitro models to reflect in vivo environments and to understand the precise effects of biomaterials in a biological environment.
Bacterial biological characteristics and activities are directly controlled by the expression of various functional proteins. Gene responses to environmental fluctuations must be reflected at the protein level to carry out bacterial adaptation to a wide range of conditions [60]. In the current study, bisHPPP effect on the hydrolytic activity of S. mutans was explored at the protein level since this protein directly contributes to resinous restoration biodegradation. Proteomic analysis demonstrated significant increase of SMU_118c production in groups with low concentrations of bisHPPP (0.01 and 0.1 mM). The greatest increase was found in S. mutans biofilm incubated with 0.01 mM bisHPPP, at 2.32-fold increase. In contrast, a statistically significant but not physiological meaningful decrease of 0.89-fold in SMU_118c protein production was found in the group exposed to 1 mM bisHPPP, while the SMU_118c gene expression was 3.2-fold up-regulated. These conflicting results are not surprising as previous studies have highlighted the weak correlation between mRNA levels and protein abundance, with greater variations observed for the latter [61–64]. In bacteria, mRNA concentration alone can explain less than 50% of total variation in protein quantity [61, 63, 65] due to post translational regulation by which bacteria are able to fine-tune their protein expression levels [66]. Bacteria regulate translation as a stress response mechanism [62, 66, 67]. For instance, Escherichia coli can rearrange mRNA structures to facilitate protein synthesis in response to environmental changes such as pH and temperature [68–72]. In addition, Bacillis subtilis and E. coli can conduct translational selectivity by altering and modifying rRNA when encountering extracellular stresses.
The exact reason for the lowest bisHPPP concentration inducing the greatest SMU_118c protein production by the bacteria is not completely clear. However, the bacteria have evolved global protein-mediated translational regulation mechanisms, which allow it to modulate translation, leading to a large variation in cellular mRNA levels in response to environmental stressors [66, 73, 74]. Therefore, it can be hypothesized that even without up-regulation of the SMU_118c gene, the bacteria could have still promoted SMU_118c synthesis in response to bisHPPP by enhancing the mRNA stability and/or increasing translational efficiency/selectivity [66]. It may also be that S. mutans survival was not threatened by the lowest concentration of bisHPPP, so SMU_118c protein synthesis was selected and promoted over other critical proteins associated with bacterial survival under harsh conditions. As bisHPPP concentrations increased to a toxic level, the bacteria selectively reduced the translation of SMU_118c or enhanced the biodegradation of SMU_118c to facilitate expression of other critical proteins, which is in agreement with the observation of reduced protein production and biofilm biomass formation at 1 mM. The findings indicate that mRNA levels do not determine total protein expression levels and that the overall regulation is highly dependent on bacterial stresses, such as the possible toxicity of high concentrations of bisHPPP on biofilm formation.
In summary, unlike the limited effect of bisHPPP on SMU_118c up-regulation that required high concentrations of bisHPPP (1 mM), SMU_118c protein synthesis can be significantly enhanced by bisHPPP at a wider range of concentrations (0.01 – 0.1 mM). With constant bisHPPP release due to biodegradation of resin composites by both salivary and bacterial esterases, SMU_118c production is likely to be continuously accelerated by the release of this BBP, further increasing the biodegradation of the restoration and restoration-tooth interface and potentially promoting the premature failure of restorations.
5. Conclusion
This study presents a significant finding regarding S. mutans virulence factors and the prevalence of secondary caries due to the vulnerability of current restorative materials to biodegradation. SMU_118c is the dominant esterase in S. mutans whole bacterial cells, and could act synergistically with salivary esterase to degrade resin composite and release BBPs such as bisHPPP. BisHPPP in turn increases SMU_118c production, resulting in elevated hydrolytic activity of S. mutans. Therefore, the hypothesis that SMU_118c is the major contributor to S. mutans degradative activity toward methacrylate resins, and that the bacterium responds to the presence of resin biodegradation by-product by modulating the esterase gene expression and protein synthesis, is accepted. The reciprocal effect of BBPs on the bacteria creates a positive feedback loop that accelerates resin composites degradation, potentially resulting in an increased rate of secondary caries and more frequent replacement of resin-based restorations. Future studies should further examine the mechanisms of interaction between bacteria and dental materials to better understand the development of secondary caries in vivo and improve the design of dental materials.
Statement of Significance.
We recently reported (Huang et al., 2018) on the isolation and initial characterization of a specific esterase (SMU_118c) from S. mutans that show degradative activity toward the hydrolysis of dental monomers. The current study further characterize this enzyme and shows that SMU_118c is a dominant degradative esterase activity in the whole cell cariogenic bacterium S. mutans and is capable of catalyzing the hydrolysis of the resinous matrix of polymerized composites and adhesives. In turn, the bacterial response to biodegradation by-products from composites and adhesives was to increase the expression of the esterase gene and enhance esterase production, accelerating the biodegradation of the restoration, adhesive and the restoration-tooth interface, potentially contributing to the pathogenesis of recurrent caries around resin composite restorations.
Acknowledgments
The authors wish to thank Dr. A. Savchenko for his assistance in protein synthesis and purification. This investigation was funded by National Institute of Dental & Craniofacial Research R01DE021385, Canadian Institute of Health Research MOP115113, Canada Foundation for Innovation John R. Evans Leaders Fund (CFI_JELF) [project #35378], and Ministry of Research and Innovation (MRI), Ontario Research Fund (ORF) [ORF-35378].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
This is an independent study free of conflict of interest.
4.7 References
- [1].Wakefield CW, Kofford KR, Advances in restorative materials, Dental Clinics of North America 45(1) (2001) 7. [PubMed] [Google Scholar]
- [2].Murray PE, Windsor LJ, Smyth TW, Hafez AA, Cox CF, Analysis of pulpal reactions to restorative procedures, materials, pulp capping, and future therapies, Critical Reviews in Oral Biology & Medicine 13(6) (2002) 509. [DOI] [PubMed] [Google Scholar]
- [3].Simecek JW, Diefenderfer KE, Cohen ME, An evaluation of replacement rates for posterior resin-based composite and amalgam restorations in US Navy and Marine Corps recruits, The Journal of the American Dental Association 140(2) (2009) 200. [DOI] [PubMed] [Google Scholar]
- [4].Brunthaler A, König F, Lucas T, Sperr W, Schedle A, Longevity of direct resin composite restorations in posterior teeth: a review, Clinical oral investigations 7(2) (2003) 63–70. [DOI] [PubMed] [Google Scholar]
- [5].Ferracane JL, Resin composite--state of the art, Dent Mater 27(1) (2011) 29–38. [DOI] [PubMed] [Google Scholar]
- [6].Opdam NJ, Bronkhorst EM, Roeters JM, Loomans BA, A retrospective clinical study on longevity of posterior composite and amalgam restorations, Dental materials 23(1) (2007) 2–8. [DOI] [PubMed] [Google Scholar]
- [7].Bernardo M, Luis H, Martin MD, Leroux BG, Rue T, Leitão J, DeRouen TA, Survival and reasons for failure of amalgam versus composite posterior restorations placed in a randomized clinical trial, The Journal of the American Dental Association 138(6) (2007) 775–783. [DOI] [PubMed] [Google Scholar]
- [8].Jaffer F, Finer Y, Santerre JP, Interactions between resin monomers and commercial composite resins with human saliva derived esterases, Biomaterials 23(7) (2002) 1707–19. [DOI] [PubMed] [Google Scholar]
- [9].Shokati B, Tam LE, Santerre JP, Finer Y, Effect of salivary esterase on the integrity and fracture toughness of the dentin resin interface, Journal of Biomedical Materials Research Part B: Applied Biomaterials 94(1) (2010) 230–237. [DOI] [PubMed] [Google Scholar]
- [10].Finer Y, Santerre JP, Salivary esterase activity and its association with the biodegradation of dental composites, Journal of dental research 83(1) (2004) 22–6. [DOI] [PubMed] [Google Scholar]
- [11].Kermanshahi S, Santerre JP, Cvitkovitch DG, Finer Y, Biodegradation of resin-dentin interfaces increases bacterial microleakage, Journal of dental research 89(9) (2010) 996–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hamada S, Koga T, Ooshima T, Virulence factors of Streptococcus mutans and dental caries prevention, Journal of dental research 63(3) (1984) 407–411. [DOI] [PubMed] [Google Scholar]
- [13].Bourbia M, Ma D, Cvitkovitch DG, Santerre JP, Finer Y, Cariogenic bacteria degrade dental resin composites and adhesives, Journal of dental research 92(11) (2013) 989–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Huang B, Siqueira WL, Cvitkovitch DG, Finer Y, Esterase from a cariogenic bacterium hydrolyzes dental resins, Acta Biomater 71 (2018) 330–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lun S, Bishai WR, Characterization of a novel cell wall-anchored protein with carboxylesterase activity required for virulence in Mycobacterium tuberculosis, Journal of Biological Chemistry 282(25) (2007) 18348–18356. [DOI] [PubMed] [Google Scholar]
- [16].Wall T, Båth K, Britton RA, Jonsson H, Versalovic J, Roos S, The early response to acid shock in Lactobacillus reuteri involves the ClpL chaperone and a putative cell wall-altering esterase, Applied and environmental microbiology 73(12) (2007) 3924–3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Serkies KB, Garcha R, Tam LE, De Souza GM, Finer Y, Matrix metalloproteinase inhibitor modulates esterase-catalyzed degradation of resin–dentin interfaces, Dental Materials 32(12) (2016) 1513–1523. [DOI] [PubMed] [Google Scholar]
- [18].Matharu S, Spratt D, Pratten J, Ng Y, A new in vitro model for the study of microbial microleakage around dental restorations: A preliminary qualitative evaluation, International Endodontic Journal 34(7) (2001) 547–553. [DOI] [PubMed] [Google Scholar]
- [19].Zivkovi’c S, Bojovi’c S, Pavlica D, Bacterial penetration of restored cavities, Oral Surgery, Oral Medicine, Oral Pathology, Oral Radiology & Endodontics 91(3) (2001) 353–358. [DOI] [PubMed] [Google Scholar]
- [20].Delaviz Y, Finer Y, Santerre JP, Biodegradation of resin composites and adhesives by oral bacteria and saliva: a rationale for new material designs that consider the clinical environment and treatment challenges, Dental materials : official publication of the Academy of Dental Materials 30(1) (2014) 16–32. [DOI] [PubMed] [Google Scholar]
- [21].Bourbia M, Finer Y, Biochemical Stability and Interactions of Dental Resin Composites and Adhesives with Host and Bacteria in the Oral Cavity: A Review, Journal 84 (2018) i1. [PubMed] [Google Scholar]
- [22].Ishii N, Nakahigashi K, Baba T, Robert M, Soga T, Kanai A, Hirasawa T, Naba M, Hirai K, Hoque A, Ho PY, Kakazu Y, Sugawara K, Igarashi S, Harada S, Masuda T, Sugiyama N, Togashi T, Hasegawa M, Takai Y, Yugi K, Arakawa K, Iwata N, Toya Y, Nakayama Y, Nishioka T, Shimizu K, Mori H, Tomita M, Multiple high-throughput analyses monitor the response of E. coli to perturbations, Science 316(5824) (2007) 593–7. [DOI] [PubMed] [Google Scholar]
- [23].Khalichi P, Cvitkovitch DG, Santerre JP, Effect of composite resin biodegradation products on oral streptococcal growth, Biomaterials 25(24) (2004) 5467–72. [DOI] [PubMed] [Google Scholar]
- [24].Khalichi P, Singh J, Cvitkovitch DG, Santerre JP, The influence of triethylene glycol derived from dental composite resins on the regulation of Streptococcus mutans gene expression, Biomaterials 30(4) (2009) 452–9. [DOI] [PubMed] [Google Scholar]
- [25].Sadeghinejad L, Cvitkovitch DG, Siqueira WL, Santerre JP, Finer Y, Triethylene Glycol Up-Regulates Virulence-Associated Genes and Proteins in Streptococcus mutans, PloS one 11(11) (2016) e0165760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sadeghinejad L, Cvitkovitch DG, Siqueira WL, Merritt J, Santerre JP, Finer Y, Mechanistic, genomic and proteomic study on the effects of BisGMA-derived biodegradation product on cariogenic bacteria, Dental materials : official publication of the Academy of Dental Materials 33(2) (2017) 175–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Caughman WF, Rueggeberg FA, Curtis JW Jr., Clinical guidelines for photocuring restorative resins, Journal of the American Dental Association 126(9) (1995) 1280-2, 1284, 1286. [DOI] [PubMed] [Google Scholar]
- [28].Finer Y, Santerre JP, Influence of silanated filler content on the biodegradation of bisGMA/TEGDMA dental composite resins, J Biomed Mater Res A 81(1) (2007) 75–84. [DOI] [PubMed] [Google Scholar]
- [29].Huang B, Cvitkovitch DG, Santerre JP, Finer Y, Biodegradation of resin-dentin interfaces is dependent on the restorative material, mode of adhesion, esterase or MMP inhibition, Dental Materials In Press (2018). [DOI] [PubMed]
- [30].Lau PC, Sung CK, Lee JH, Morrison DA, Cvitkovitch DG, PCR ligation mutagenesis in transformable streptococci: application and efficiency, J Microbiol Methods 49(2) (2002) 193– 205. [DOI] [PubMed] [Google Scholar]
- [31].Li YH, Lau PC, Lee JH, Ellen RP, Cvitkovitch DG, Natural genetic transformation of Streptococcus mutans growing in biofilms, Journal of bacteriology 183(3) (2001) 897–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Senadheera MD, Guggenheim B, Spatafora GA, Huang YC, Choi J, Hung DC, Treglown JS, Goodman SD, Ellen RP, Cvitkovitch DG, A VicRK signal transduction system in Streptococcus mutans affects gtfBCD, gbpB, and ftf expression, biofilm formation, and genetic competence development, Journal of bacteriology 187(12) (2005) 4064–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Li YH, Tang N, Aspiras MB, Lau PC, Lee JH, Ellen RP, Cvitkovitch DG, A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation, Journal of bacteriology 184(10) (2002) 2699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Fröjd V, Chávez de Paz L, Andersson M, Wennerberg A, Davies JR, Svensäter G, In situ analysis of multispecies biofilm formation on customized titanium surfaces, Molecular Oral Microbiology 26(4) 241–252. [DOI] [PubMed] [Google Scholar]
- [35].Siqueira WL, Bakkal M, Xiao Y, Sutton JN, Mendes FM, Quantitative proteomic analysis of the effect of fluoride on the acquired enamel pellicle, PLoS One 7(8) (2012) e42204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Sunnegårdh-Grönberg K, van Dijken JW, Funegård U, Lindberg A, Nilsson M, Selection of dental materials and longevity of replaced restorations in Public Dental Health clinics in northern Sweden, Journal of dentistry 37(9) (2009) 673–678. [DOI] [PubMed] [Google Scholar]
- [37].Zimmerli B, Strub M, Jeger F, Stadler O, Lussi A, Composite materials: composition, properties and clinical applications. A literature review, Schweiz Monatsschr Zahnmed 120(11) (2010) 972–86. [PubMed] [Google Scholar]
- [38].Peutzfeldt A, Resin composites in dentistry: the monomer systems, European journal of oral sciences 105(2) (1997) 97–116. [DOI] [PubMed] [Google Scholar]
- [39].Moszner N, Salz U, Zimmermann J, Chemical aspects of self-etching enamel–dentin adhesives: a systematic review, Dental Materials 21(10) (2005) 895–910. [DOI] [PubMed] [Google Scholar]
- [40].Borges MA, Matos IC, Mendes LC, Gomes AS, Miranda MS, Degradation of polymeric restorative materials subjected to a high caries challenge, dental materials 27(3) (2011) 244–252. [DOI] [PubMed] [Google Scholar]
- [41].Silva EM, Almeida GS, Poskus LT, Guimarães JGA, Influence of organic acids present in the oral biofilm on the microtensile bond strength of adhesive systems to human dentin, Journal of Biomedical Materials Research Part B: Applied Biomaterials 100(3) (2012) 735–741. [DOI] [PubMed] [Google Scholar]
- [42].Fúcio SB, Carvalho FG, Sobrinho LC, Sinhoreti MA, Puppin-Rontani RM, The influence of 30-day-old Streptococcus mutans biofilm on the surface of esthetic restorative materials—An in vitro study, Journal of dentistry 36(10) (2008) 833–839. [DOI] [PubMed] [Google Scholar]
- [43].Gregson KS, Shih H, Gregory RL, The impact of three strains of oral bacteria on the surface and mechanical properties of a dental resin material, Clinical oral investigations 16(4) (2012) 1095–1103. [DOI] [PubMed] [Google Scholar]
- [44].Huang B, Cvitkovitch DG, Santerre JP, Finer Y, Biodegradation of resin-dentin interfaces is dependent on the restorative material, mode of adhesion, esterase or MMP inhibition, Dental materials : official publication of the Academy of Dental Materials 34(9) (2018) 1253–1262. [DOI] [PubMed] [Google Scholar]
- [45].Wendeberg A, Zielinski FU, Borowski C, Dubilier N, Expression patterns of mRNAs for methanotrophy and thiotrophy in symbionts of the hydrothermal vent mussel Bathymodiolus puteoserpentis, ISME J 6(1) (2012) 104–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Al-Ahmad A, Wunder A, Auschill TM, Follo M, Braun G, Hellwig E, Arweiler NB, The in vivo dynamics of Streptococcus spp., Actinomyces naeslundii, Fusobacterium nucleatum and Veillonella spp. in dental plaque biofilm as analysed by five-colour multiplex fluorescence in situ hybridization, J Med Microbiol 56(Pt 5) (2007) 681–7. [DOI] [PubMed] [Google Scholar]
- [47].Singh J, Khalichi P, Cvitkovitch DG, Santerre JP, Composite resin degradation products from BisGMA monomer modulate the expression of genes associated with biofilm formation and other virulence factors in Streptococcus mutans, J Biomed Mater Res A 88(2) (2009) 551–60. [DOI] [PubMed] [Google Scholar]
- [48].Jefferson KK, What drives bacteria to produce a biofilm?, FEMS microbiology letters 236(2) (2004) 163–173. [DOI] [PubMed] [Google Scholar]
- [49].Rohde KH, Abramovitch RB, Russell DG, Mycobacterium tuberculosis invasion of macrophages: linking bacterial gene expression to environmental cues, Cell host & microbe 2(5) (2007) 352–364. [DOI] [PubMed] [Google Scholar]
- [50].Olson ER, Influence of pH on bacterial gene expression, Molecular microbiology 8(1) (1993) 5–14. [DOI] [PubMed] [Google Scholar]
- [51].Fux C, Costerton J, Stewart P, Stoodley P, Survival strategies of infectious biofilms, Trends in microbiology 13(1) (2005) 34–40. [DOI] [PubMed] [Google Scholar]
- [52].Hudson MC, Curtiss R, Regulation of expression of Streptococcus mutans genes important to virulence, Infection and immunity 58(2) (1990) 464–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Burne RA, Chen Y-YM, Penders JE, Analysis of gene expression in Streptococcus mutans in biofilms in vitro, Advances in dental research 11(1) (1997) 100–109. [DOI] [PubMed] [Google Scholar]
- [54].Stock J, Da Re S, Signal transduction: response regulators on and off, Curr Biol 10(11) (2000) R420–4. [DOI] [PubMed] [Google Scholar]
- [55].Bowden GH, Li YH, Nutritional influences on biofilm development, Adv Dent Res 11(1) (1997) 81–99. [DOI] [PubMed] [Google Scholar]
- [56].Sauer K, Cullen M, Rickard A, Zeef L, Davies D, Gilbert P, Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm, Journal of bacteriology 186(21) (2004) 7312–7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Moller S, Korber DR, Wolfaardt GM, Molin S, Caldwell DE, Impact of nutrient composition on a degradative biofilm community, Appl Environ Microbiol 63(6) (1997) 2432–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Hamilton I, Svensäter G, Acid‐regulated proteins induced by Streptococcus mutans and other oral bacteria during acid shock, Oral microbiology and immunology 13(5) (1998) 292–300. [DOI] [PubMed] [Google Scholar]
- [59].Wilkins JC, Homer KA, Beighton D, Analysis of Streptococcus mutans proteins modulated by culture under acidic conditions, Applied and environmental microbiology 68(5) (2002) 2382–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Chubukov V, Gerosa L, Kochanowski K, Sauer U, Coordination of microbial metabolism, Nature Reviews Microbiology 12(5) (2014) 327–340. [DOI] [PubMed] [Google Scholar]
- [61].Lu P, Vogel C, Wang R, Yao X, Marcotte EM, Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation, Nature biotechnology 25(1) (2007) 117–124. [DOI] [PubMed] [Google Scholar]
- [62].Nie L, Wu G, Zhang W, Correlation of mRNA expression and protein abundance affected by multiple sequence features related to translational efficiency in Desulfovibrio vulgaris: a quantitative analysis, Genetics 174(4) (2006) 2229–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Dressaire C, Laurent B, Loubière P, Besse P, Cocaign-Bousquet M, Linear covariance models to examine the determinants of protein levels in Lactococcus lactis, Mol Biosyst 6(7) (2010) 1255–64. [DOI] [PubMed] [Google Scholar]
- [64].Nie L, Wu G, Zhang W, Correlation between mRNA and protein abundance in Desulfovibrio vulgaris: a multiple regression to identify sources of variations, Biochem Biophys Res Commun 339(2) (2006) 603–10. [DOI] [PubMed] [Google Scholar]
- [65].Corbin RW, Paliy O, Yang F, Shabanowitz J, Platt M, Lyons CE, Root K, McAuliffe J, Jordan MI, Kustu S, Toward a protein profile of Escherichia coli: comparison to its transcription profile, Proceedings of the National Academy of Sciences 100(16) (2003) 9232– 9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Picard F, Milhem H, Loubière P, Laurent B, Cocaign-Bousquet M, Girbal L, Bacterial translational regulations: high diversity between all mRNAs and major role in gene expression, BMC genomics 13(1) (2012) 528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].de Sousa Abreu R, Penalva LO, Marcotte EM, Vogel C, Global signatures of protein and mRNA expression levels, Molecular BioSystems 5(12) (2009) 1512–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Jiang W, Hou Y, Inouye M, CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone, Journal of Biological Chemistry 272(1) (1997) 196–202. [DOI] [PubMed] [Google Scholar]
- [69].Giuliodori AM, Di Pietro F, Marzi S, Masquida B, Wagner R, Romby P, Gualerzi CO, Pon CL, The cspA mRNA is a thermosensor that modulates translation of the cold-shock protein CspA, Molecular cell 37(1) (2010) 21–33. [DOI] [PubMed] [Google Scholar]
- [70].Nechooshtan G, Elgrably-Weiss M, Sheaffer A, Westhof E, Altuvia S, A pH-responsive riboregulator, Genes & development 23(22) (2009) 2650–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Weixlbaumer A, Leon K, Landick R, Darst SA, Structural basis of transcriptional pausing in bacteria, Cell 152(3) (2013) 431–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Bingham RJ, Hall KS, Slonczewski JL, Alkaline induction of a novel gene locus, alx, in Escherichia coli, Journal of bacteriology 172(4) (1990) 2184–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Duval M, Simonetti A, Caldelari I, Marzi S, Multiple ways to regulate translation initiation in bacteria: Mechanisms, regulatory circuits, dynamics, Biochimie (2015). [DOI] [PubMed] [Google Scholar]
- [74].Romeo T, Vakulskas CA, Babitzke P, Post‐transcriptional regulation on a global scale: form and function of Csr/Rsm systems, Environmental microbiology 15(2) (2013) 313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]


