Abstract
Atherosclerosis is a complex and chronic disease characterized by lipid deposition in the vessel wall that leads to an inflammatory and proliferative cascade involving smooth muscle, endothelial and immune cells. Despite substantial improvements in our understanding of mechanisms contributing to atherosclerosis and overall reduction in cardiovascular mortality, the absolute disease burden remains substantially high. The recent discovery of a new group of mediators known as long noncoding RNAs (lncRNAs) offers a unique opportunity for the development of novel diagnostic and therapeutic tools in atherothrombotic disease. A number of studies suggest that lncRNAs are important mediators in health and disease and rapidly accumulating evidence implicates lncRNAs in regulatory circuits controlling atherosclerosis. In this review, we outline important contributions of lncRNAs to atherosclerosis and its associated risk factors including hypercholesterolemia, diabetes, hypertension and obesity.
Keywords: Non-coding RNA; Arteriosclerosis; genetic, knockout models; genetics, bioinformatics; Atherosclerosis; Lipids and Cholesterol; Genetics; Gene expression and regulation
Condensed Abstract:
Atherosclerosis is the single most devastating cause of worldwide mortality forming the epicenter of major efforts to mitigate cardiovascular disease. Understanding how genetic sequences control function has substantially improved our understanding of common cardiovascular problems including atherosclerosis. Galvanized by evidence from the human genome project, an expanded genetic catalog has led to the discovery of new players in biology known as long noncoding RNAs (lncRNAs). In this review, we discuss the contributions of lncRNAs to atherosclerosis risk factors and key cells involved in plaque formation. The identification of lncRNAs offers newly recognized opportunities for cardiovascular risk mitigation.
Introduction
Although a precise cause for how atherosclerosis develops is not fully understood, it is well established that a number of traits increase the propensity for disease development and progression. Major modifiable risk factors for atherosclerosis development often occur in concert and include dyslipidemia, hypertension, diabetes as well as lifestyle factors such obesity and smoking. These risk factors do not equally impact disease progression, but a cornerstone of cardiovascular risk reduction includes mitigating the influence of all the above traits using lifestyle interventions or pharmacotherapy. A few of the drugs commonly used today for cardiovascular risk reduction, such as PCSK9 inhibitors, have been inspired by population genetics and complementary mechanistic studies that have dissected the relationship between gene variants and disease phenotype (1). Recently a more expanded catalog of functional genetic material has been identified with the discovery of unique set of genes referred to as long noncoding RNAs (lncRNAs). Modulation of lncRNA genes has been shown to play important roles in various stages of plaque development and may potentially offer novel diagnostic and therapeutic strategies to reduce atherosclerosis burden.
Fifty years after the term “junk DNA” became popular (2) evidence from the human genome project showed that the vast majority of human DNA bases are associated with at least one RNA transcript. Furthermore, most of these transcripts do not appear to code for functional proteins (3). These findings have helped catalyze the promulgation of the field of lncRNA biology. Today, lncRNAs are defined as noncoding transcripts >200 bp, in order to distinguish them from other types of noncoding RNA such as microRNAs. In just over a decade, there have been >10,000 studies characterizing important biological roles for these novel mediators and strongly implicating them in human disease. Although the bulk of these studies have focused on the contribution of lncRNAs in cellular development and differentiation, rapidly accumulating evidence suggests an additional contribution of lncRNAs in cardiovascular disease.
A remarkable feature of lncRNAs is that they are expressed with potent specificity. For example, roughly 80% of lncRNAs are highly tissue-specific which in theory suggests that may serve as disease markers or therapeutic targets (4). Indeed, a small number of studies have shown that lncRNAs could serve as disease markers in cancer states and cardiovascular disease (5,6). Although the precise mode of action of lncRNAs remains mysterious, diverse mechanisms have been proposed and extensively outlined elsewhere (7,8). A summary of these mechanisms is provided in Figure 1. In brief, lncRNAs can function by acting as decoys or guides to chromatin modifying and transcriptional machinery thus directly influencing gene expression. LncRNAs have also been shown to impact mRNA splicing, nuclear to cytoplasmic shuttling, and translation. Some lncRNAs may impact post-translational protein modification while others have been shown to buffer other regulatory mediators such as microRNAs. Regardless of proposed mechanism, however, functional relevance has been established for a large repertoire of lncRNAs in cardiovascular disease (9). In this review, we will highlight important contributions of lncRNAs in cardiometabolic risk factors such as dyslipidemia, hypertension, obesity and diabetes as well as lncRNAs that directly contribute to the development and propagation of atherothrombotic events (Figure 2). A summary of lncRNAs and interacting partners are provided in Table 1.
Figure 1. Long non-coding RNAs (lncRNAs) regulatory mechanisms.
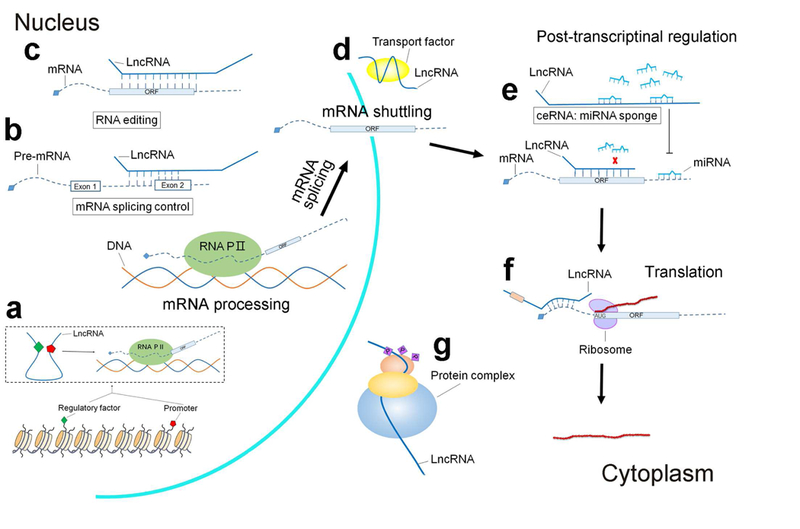
a. LncRNAs modulate chromatin architecture and enhancer-promoter looping at cognate genes to influence transcription. b. LncRNAs can regulate mRNA splicing by interfering with pre-mRNA processing. c. LncRNAs, in particular antisense lncRNAs, may direct mRNA editing through base paring interaction. d. By acting as decoys, lncRNAs influence the folding of complex three-dimensional structures affecting mRNA cytoplasmic shuttling and localization. e. LncRNAs regulate mRNA stability by acting as sponges to sequester microRNAs. f. LncRNAs may influence protein translation through interactions with binding of translation co-factors and regulators. g. LncRNAs may influence post-translational protein modification.
Figure 2: LncRNAs involved in mammalian atherothrombotic disease.
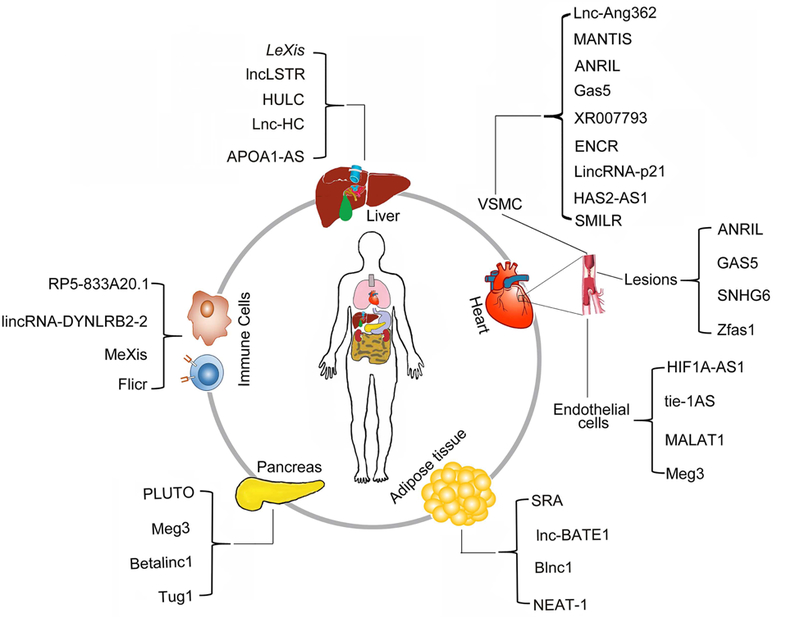
Summary of mammalian lncRNAs expressed from various tissues or cell-types involved in atherosclerosis pathways.
Table 1.
LncRNAs and their partners involved in the atherothrombotic disease
| LncRNAs/ partners | Phenotype | Putative human orthologue identified | Data on functional conservation | ref |
|---|---|---|---|---|
| lncLSTR/TDP-43 | Plasma triglyceride levels, bile acid biosynthesis | No | No | 10 |
| Lexis/Raly | Serum cholesterol levels, hepatic sterol content | Yes | No | 11, 12 |
| HULC | Lipogenesis | Yes | Yes | 13 |
| APOA1-AS | lipid transport | Yes | Yes | 14 |
| Noncoding region between Igf2 and H19 | fat deposition and obesity | 16 | ||
| LINC00237 | MOMO Syndrome | Yes | Yes | 17 |
| Noncoding RNA from chromosome 15q11-q13 |
Prader-Willi syndrome Obesity |
No | No |
17 18 |
| IPW | PWS | Yes | Yes | 19 |
| SRA/ PPARγ | adipogenesis | Yez | Yes | 22 |
| Blnc1/EBF-2 | thermogenesis | Yes | Yes | 23 |
| PLUTO/PDX1 | type 2 diabetes, glucose tolerance | Yes | Yes | 25 |
| Betalinc1 | normal islet cell formation | No | No | 27 |
| Flicr | promoting autoimmune diabetes | Yes | Yes | 28 |
| Gas5 | correlating with type 2 diabetes risk, regulating vascular remodeling in hypertensive rats | Yes | Yes | 31 |
| Lnc-Ang362 | Regulation of proliferation of VSMC | No | No | 32 |
| MANTIS/brg1 | Modulating endothelial function | Yes | Yes | 35 |
| ANRIL | Systemic hypertension and vascular aneurysms | Yes | Yes | 36–38 |
| KCNQ1OT1 | Myocardial infarction | Yes | Yes | 45 |
| RP5-833A20.1/NFIA | Function in foam-cell macrophages | Yes | Yes | 46 |
| MeXis/ Abca1 | foam cell formation and atherosclerosis | Yes | Yes | 47, 48 |
| Lnc-DC/ STAT3 | enhancing T cell activation | Yes | Yes | 49 |
| Linc-Cox2 | Regulation of inflammatory genes | No | No | 50 |
| LincRNA-EPS | Regulation of inflammatory genes | Yes | No | 51 |
| NeST | Altering chromatin methylation pattern at interferon gamma locus in T cells | Yes | Yes | 52 |
| HIF1A-AS1 | Promoting proliferation and suppressing apoptosis of HUVECs | Yes | Yes | 55 |
| tie-1AS | Differentially regulated in samples from infants and children with various vascular malformations | Yes | Yes | 56 |
| MALAT1 | Influencing proliferation of endothelial cells in vivo and neonatal retina vascularization | Yes | Yes | 59 |
| Meg3 | Controlling endothelial function | Yes | Yes | 60 |
| ENCR | Influence smooth muscle contractile machinery and inflammatory mediators | Yes | Yes | 61 |
| lincRNA-p21/hnRNP-K | Neointimal hyperplasia, coronary artery | Yes | Yes | 62, 63 |
| SMILR | In response to interleukin-1α and PDGF stimulation | Yes | Yes | 64 |
| TERMINATOR | Cardiovascular development | Yes | Yes | 65 |
| ALIEN | Cardiovascular development | Yes | Yes | 65 |
| PUNISHER | Cardiovascular development | Yes | Yes | 65 |
LncRNAs modulating atherosclerosis risk factors
Dyslipidemia
Abnormal cholesterol accumulation is a hallmark of atherosclerosis lesion development. Although the contributions of lncRNAs in sterol regulation is not well studied, multiple lines of evidence revealed important contributions of lncRNAs in sterol regulatory circuits. The vast majority of this evidence, however, comes from mechanistic animal studies. At the level of systemic lipid metabolism, Li and colleagues showed that loss of the lncRNA lncLSTR in mice markedly reduced plasma triglyceride levels (10). LncLSTR is thought to regulate expression of Cyp8b1, a rate-limiting enzyme in the bile acid biosynthesis, via interaction with the RNA-binding protein TDP-43. The change in bile acid pool alters the activity of the bile acid receptor FXR and leads to enhanced ApoC2 lipoprotein, an important component of very low-density lipoproteins and chylomicrons. Our previous work has shown that the lncRNA LeXis acts as important conduit between the nutrient-sensing nuclear receptor liver X receptor (LXR) and the master cholesterol regulator Srebp2. Under conditions where sterols accumulate in liver, LeXis is robustly induced with LXR activation, which in turn reduces de novo cholesterol synthesis by blocking the DNA binding of the transcriptional activator Raly at Srebp2-regulated genes including Hmgcr, the target of statin drugs (11). Loss of LeXis has a strong impact on serum cholesterol levels and hepatic sterol content. More recently, a gene therapy strategy utilizing LeXis was shown to reduce atherosclerosis burden in an animal model of familial hypercholesterolemia (12). Although a putative human LeXis transcript has been identified, the relevance of the LeXis pathway in humans remains undefined.
Using evidence from human hepatoma samples, a recent study showed that that the lncRNA HULC modulates the activity of the acyl-CoA synthetase subunit ACSL1, an enzyme responsible for fatty acid synthesis, through activation of the nuclear receptors PPARA and RXRA(13). The authors go on to show that HULC promotes lipogenesis in an in vivo murine model substantiating the contributions of lncRNAs in integrating metabolic signaling with cellular proliferation. Finally, lncRNAs play important roles in lipid transport. An endogenous antisense transcript, APOA1-AS, acts as a negative transcriptional regulator of APOA1 (14). Antisense oligonucleotides targeting APOA1-AS increase ApoA1 levels in primates.
Collectively these studies suggest that dysregulation in lncRNA regulatory circuits can alter sterol balance in vitro and in vivo. Despite these interesting observations, however, it should be noted that human functional conservation for the majority of these lncRNAs has not been established. In addition, it is unclear whether any of the above of the lncRNAs can be readily detected in blood or other accessible compartments. Thus, a major question moving forward is whether the lncRNAs described above have any relevance to human disease or potential diagnostic and therapeutic applications.
Obesity
Obesity, an established independent risk factor for heart disease, is a worsening health hazard in most parts of the world (6). Not surprisingly, substantial efforts have recently been devoted to dissecting pathways that influence the development of obesity, such as the role of lncRNAs in fat formation and maintenance of energy balance. Some of the earliest evidence implicating noncoding genes in metabolic phenotypes comes from studies linking the Igf2 locus with obesity (15). Epidemiological data from human blood leukocytes suggests that differences in methylation at the Igf2 gene and neighboring noncoding gene are associated with paternal obesity (15). Intriguingly, deletion of a noncoding region between Igf2 and the noncoding gene H19 in animal models was accompanied by increased fat deposition and obesity through poorly understood mechanisms (16). Similar genetic evidence suggests that the noncoding RNA LINC00237 is directly implicated in MOMO Syndrome, a rare autosomal recessive disease characterized by macrosomia, obesity, macrocephaly, and ocular abnormalities (17). Interestingly, LINC00237 is expressed in the brain; suggesting it may play a role in social disorders and feeding behavior (17). Prader-Willi syndrome (PWS) is a genetic disorder characterized by a loss of a noncoding RNA from chromosome 15q11-q13 leading to obesity from hyperphagia and intellectual disability (18). Using iPSC cells derived from PWS patients, a recent study showed that that expression of the chromatin-modifying lncRNA IPW is associated with abnormal expression of maternally expressed genes responsible for the PWS phenotype (19).
In addition to inherited obesity syndromes, systematic interrogation of lncRNAs revealed important clues to the role of these molecules in normal adipose tissue development. Work from the Rinn group showed, on a genome-wide scale, that 175 lncRNAs are specifically regulated during murine adipogenesis and that knockdown of some of these lncRNAs led to abnormal adipose tissue formation in vitro (20). A role for lncRNAs in energy homeostasis has also been corroborated in vivo. Using integrative transcriptome approaches, Yang and colleagues identified lncRNAs in diverse organs involved in energy metabolism. Using this framework, they showed that one of these lncRNAs, LncMS, acts as a suppressor of lipogenesis by modulating the activity of the master metabolic regulator srebp1c in liver (21). Finally, RNA-seq data from human tissues suggests that certain lncRNAs such as linc-DMRT2 and linc-TP53I13 may be dysregulated in human adipose tissue from obese patients compared with normal controls (22).
Since lncRNAs have been shown to directly influence transcriptional outputs, it is not surprising that a number of lncRNAs have been found to act as coactivators for important adipogenic transcription factors. For example, the SRA noncoding RNA has been shown to function as a transcriptional coactivator of PPARγ, the master regulator of adipogenesis (22). Knockout of SRA in mice is associated with decreased fat mass and enhanced insulin sensitivity.
Overall a preponderance of evidence supports the notion that lncRNAs are differentially regulated during obesity and that perturbations of lncRNAs may be causally linked to human adiposity. More recently, an area that has attracted growing attention is the contribution of lncRNAs to brown adipose tissue function, a critical determinant of energy expenditure. Recent evidence suggests that the conserved lncRNA Blnc1 enhances the activity of the browning adipose transcription factor EBF-2 to stimulate thermogenesis (23). Given the substantial interest in developing therapies that enhance thermogenesis as means of treating obesity, continued interrogation of lncRNAs in this setting may lead to fruitful results.
Diabetes
The first evidence suggesting that non-coding transcripts play important metabolic roles came from systematic interrogation of human pancreatic islet and β cells (24). This work catalogs over 1000 dynamically regulated lncRNAs in type I diabetes and reinforces the tissue-specific expression of some of these lncRNAs. In addition, a number of lncRNAs have been shown to be important for human islet cell function by acting in concert with pancreatic lineage specific transcription factors. For example, the lncRNA PLUTO influences the β cell transcription factor PDX1 by regulating chromatin architecture. PLUTO levels are downregulated in islets from donors with type 2 diabetes or impaired glucose tolerance (25). Similar unbiased high throughput studies analyzing human pancreatic islets revealed that certain lncRNAs are highly enriched when comparing patients with hyperglycemia to normal glucose controls (26). In addition, mechanistic evidence has linked dysregulation of lncRNAs with serum glucose levels. For example, the conserved lncRNA Betalinc1 is involved in normal islet cell formation knockout of this lncRNA in a murine model resulted in glucose intolerance through regulation of β-cell rich genes (27). Given the strong influence of autoimmunity in type I diabetes, it is not surprising that lncRNAs have been implicated in organ-specific immune activation. Flicr (Foxp3 long intergenic noncoding RNA) is a cis-acting noncoding RNA in human and mouse regulatory T cells which acts as negative regulator of Foxp3 expression by modifying chromatin accessibility (28). Flicr promotes autoimmune diabetes whereas its loss of function is associated with improved glucose control in mice. Taken together, the above studies strongly suggest that lncRNAs directly contribute to the regulation of systemic glucose homeostasis by modulating islet cell function as well as priming immune pathways and impacting disease progression.
Unbiased population genetic studies found a strong association between SNPs at intergenic noncoding regions or intronic transcripts with diabetes risk. For example, a SNP in the first intron of ABO at the putative promoter of an antisense lncRNA was significantly associated with elevated fasting glucose levels (29). Mechanistically, how this noncoding region may be conferring disease risk or whether biological effects could be attributed to other genes in the vicinity remains largely unexplored. A number of clinical studies investigating the potential of lncRNAs to serve as diabetes clinical disease markers have shown promising initial results (30). For example, profiling of human serum samples revealed that the lncRNA Gas5 strongly correlates with type 2 diabetes risk (31). However, more robust clinical evidence systematically validating these results is required. In summary, the weight of evidence suggests that lncRNAs play important roles in human diabetes development but additional insight into the pathophysiology of these lncRNAs remains lacking.
Hypertension
A limited number of studies have also demonstrated a role for lncRNAs in hypertension. It is well established that dysregulation in the renin angiotensin system is causally associated with hypertension and heart disease risk. Using unbiased transcriptional profiling coupled with epigenetic interrogation, one study identified differentially regulated lncRNAs in vascular smooth muscle cells in response to angiotensin II (32). Loss of one these lncRNAs, Lnc-Ang362, resulted in reduced proliferation of VSMC thereby corroborating an important role for lncRNAs in molecular mechanisms driving the development of hypertension. Similarly, a number of studies profiling lncRNAs in rat models of hypertension have observed unique transcriptional signatures (33,34). Recently, an elegant report implicated the nuclear lncRNA MANTIS in modulating human endothelial function through direct interactions with brg1, an important component of chromatin modifying machinery (35). Finally, GWAS has provided further evidence linking noncoding regions with hypertension. Variants at the lncRNA ANRIL locus have been strongly associated with systemic hypertension and vascular aneurysms (36–38) . Although ANRIL is a commonly studied lncRNA, its precise role in impacting a number of cardiovascular traits including hypertension remains poorly understood. Remarkably, very few studies have investigated the contributions of lncRNAs to systemic blood pressure in humans.
LncRNAs inside the lesion
Genome wide association studies have shed important light on disease regulatory circuits controlling heart disease. An astonishing finding validated by the multiple groups is that the strongest GWAS hit associated with risk of heart disease in humans by far is the noncoding RNA ANRIL located at chromosome 9p21 region (39,40). The function of this lncRNA has been completely puzzling but a number of studies suggest an important role in cellular proliferative responses within lesions (41). This is particularly timely since growing evidence suggests that regulation of smooth muscle or macrophage proliferation has a profound impact on lesion progression (42,43). A number of studies directly measuring lncRNAs in human atherosclerosis lesions including carotid plaque have found a number of lncRNAs, including GAS5, SNHG6 and Zfas1, to be dramatically increased in this context (44). More recently, several lncRNAs, such as ANRIL and KCNQ1OT1, have been measured in blood from patients with myocardial infarction and their levels have been found to predict left ventricular dysfunction in multivariable analysis models (45). Given that atherosclerosis development involves an orchestrated interplay between endothelial, immune and smooth muscle cells, we review the contributions of lncRNAs in each of these cells below.
Immune Cells
A number of studies have shown important contributions of lncRNAs to sterol regulation in macrophages, key drivers of atherogenesis and integrators of metabolic and inflammatory signaling. The lncRNA RP5–833A20.1 is an intronic lncRNA that regulates the transcription factor NFIA in human foam-cell macrophages by modulating microRNA hsa-miR-382–5p (46). Recent work identified the lncRNA MeXis as an important contributor to cellular responses to cholesterol overload in macrophages (47). MeXis influences chromatin architecture at the locus of critical cholesterol efflux regulator Abca1 to enhance Abca1 expression. This effect requires the transcriptional coactivator DDX17. Loss of MeXis from immune cells using a genetic deletion approach markedly enhances foam cell formation and accelerates atherosclerosis in a murine model. In addition, perturbing a human orthologue of MeXis influences Abca1 levels and function in human macrophages. A SNP overlapping the human MeXis orthologue is associated with coronary artery disease risk, hinting at functional conservation of this lncRNA. In addition, deep sequencing of human-derived macrophage transcriptomes corroborates dynamic regulation of macrophage lncRNAs in response to exogenous stimuli and loss of one these lncRNAs influenced IFN-γ signaling (48).
A number of lncRNAs have been shown to play important roles in other immune cell subtypes thought to be highly relevant to lesion development; although their effects on atherosclerosis have not been directly tested. Lnc-DC is a noncoding transcript expressed with potent specificity in human dendritic cells. This STAT3 binding noncoding RNA promotes STAT3 phosphorylation and enhances T cell activation (49). LincCox2 and LincRNA EPS are two conserved lncRNAs found to be induced by TLR ligands in BMDMs leading to activation or repression of a subset of inflammatory genes in vivo(50) (51). Similarly, the murine lncRNA NeST alters chromatin methylation patterns at the interferon gamma locus in T cells (52).
Since most of the lncRNAs described here have been shown to modulate inflammatory signaling in vivo, it would be intriguing to directly test their contribution to lesion development since this has only been tested in a few studies. Furthermore, a number of lncRNAs described in this section have been shown to potently modulate IL-1B, a key target of anti-inflammatory therapy shown to reduce cardiovascular disease burden in the CANTOS trial (53). Given the growing interest in developing novel strategies that target lesions directly, interrogation of lncRNAs may be one promising approach. However, it cannot be overstated that future studies need to directly delineate the role of lncRNAs in human lesions and immune-subsets from disease patients and matching controls.
Endothelial Cells
Dysfunction of endothelial lining of blood vessels is a key inciting event in atherosclerosis initiation. Multiple lines of evidence corroborate important roles for lncRNAs in endothelial cell function. Interrogation of human umbilical vein endothelial cells (HUVECs) responses to oxidized LDL loading revealed dysregulation of hundreds of lncRNAs (54). More recently, a number of studies have shown that commonly used cardiovascular drugs may function through modulation of lncRNA activity. For example, clopidogrel reduces cell death and promotes proliferation of human vascular endothelial cells through the expression of LncRNA HIF 1 alpha-antisense RNA 1 (HIF1A-AS1) (55). In fact, knockdown of HIF1A-AS1 is sufficient to promote proliferation and suppress apoptosis of HUVECs. Another antisense transcript to the tyrosine kinase Tie1 (tie-1AS) has been shown to modulate tie1 protein activity and is differentially regulated in samples from infants and children with various vascular malformations (56).
More recently, various lncRNA regulatory circuits have been shown to impact autophagy, proliferation and inflammation in vascular endothelial cells (57,58). MALAT1 is a notable conserved lncRNA shown to influence proliferation of endothelial cells in vivo and neonatal retina vascularization (59). An interesting report suggests that the lncRNA Meg3 is also important in controlling endothelial function (60). Inhibition of Meg3 in HUVECs prevented aging-mediated inhibition of angiogenic sprouting, and in vivo silencing led to improved blood flow in an ischemic hindlimb mouse model. It is tempting to speculate that inhibition of Meg3 may represent a therapeutic strategy for mitigating age-related endothelial dysfunction. Overall, the rapidly growing evidence presented here highlights the important contributions of lncRNAs to endothelial cell function, particularly in human endothelial cell models. However, the relevance of these lncRNAs to whole organism development and function is less well understood.
Vascular Smooth Muscle Cells
A number of studies have cataloged lncRNAs in human vascular smooth muscle cells. Bell and colleagues identified a number of lncRNAs in human coronary artery smooth muscle cells including ENCR (Smooth muscle and Endothelial cell enriched migration/differentiation-associated long Non-Coding RNA), which is thought to influence smooth muscle contractile machinery and inflammatory mediators (61). Lincp21 a well-characterized lncRNA that is conserved between mice and humans and was originally identified as a noncoding transcript regulated by p53 and required for p53 mediated transcriptional repression by interacting with hnRNP-K (62). Recent evidence suggests that inhibition of lincRNA-p21 results in neointimal hyperplasia in a mouse carotid disease model and that lincRNA-p21 expression is decreased in patients with coronary artery disease (63). Lately, the novel lncRNA SMILR was shown to be induced in proliferating smooth muscle cells in response to interleukin-1α and PDGF stimulation (64). Remarkably, SMILR expression was more enhanced in serum samples from patients with unstable atherosclerotic plaques. Considerable interest lately has also been devoted to understanding cardiovascular progenitor to disease transitions. A recent study identified 3 novel lncRNAs, TERMINATOR, ALIEN, and PUNISHER, in cardiovascular development (65). Given the substantial recent interest in linking proliferative pathways and plaque development, lncRNAs such lincRNA-p21 and SMILR may be viable targets for disease modulation. In addition, it is also plausible that lineage tracing of lncRNA expression patterns can provide clues to the origin and fates of smooth muscle cells within lesions a topic of much debate in scientific circuits (42).
Conclusions and Future Perspective
The study of lncRNAs has undoubtedly changed the way we think about health and disease, yet a number of important questions remain unresolved. The expression level of many lncRNAs remains quite low, which brings some uncertainty to the reliability and reproducibility of large-scale lncRNA interrogations. Protein-coding transcripts are transported to the cytoplasm and bound by ribosomes whereas most non-coding transcripts are retained in the nucleus? Understanding the molecular basis of why seemingly structurally similar transcripts are destined for different cellular fates remains unresolved. A few studies reported that lncRNAs can be detected in exosomes, extracellular transport vesicles that carry biologic mediators, but the triaging of some lncRNAs to the circulation remains poorly understood (66,67). More importantly, de novo discovery of lncRNAs is costly and often requires extensive sequencing with no established standards regarding data analysis methodologies. Another major challenge in studying lncRNAs is the lack of predictive function based on sequence or structural features. One can predict the function of a protein-coding gene based on known homology domains and the function of a micoRNA gene based on bioinformatic target prediction sites, but the primary sequence of a lncRNA offers almost no clues into the function of the transcript.
Perhaps the most important outstanding question is how do we convert these recent discoveries in lncRNA biology from being merely interesting observations into viable therapeutic and diagnostic options. The fact that the vast majority of noncoding transcripts are not conserved remains a challenge; hence functional relevance of many identified lncRNAs to human disease is often questioned. This underscores the importance of a “patient-centric” discovery approach. Finally, the potent tissue-specific expression pattern of lncRNAs is a double-edge sword. On the one hand, it implies that one must strive to find the right context, a key to accurate cataloging of lncRNAs. On the flipside, however, this specificity imparts exciting potential gains even with lack of mechanistic detail. Fifty-five years since the identification of troponin, perhaps the ultimate biomarker in cardiovascular disease, we are still searching for the next big one. It is tempting to speculate that more rigorous studies of lncRNAs may offer breakthroughs on that front.
Central Illustration: Long non-coding RNAs in Atherosclerosis.
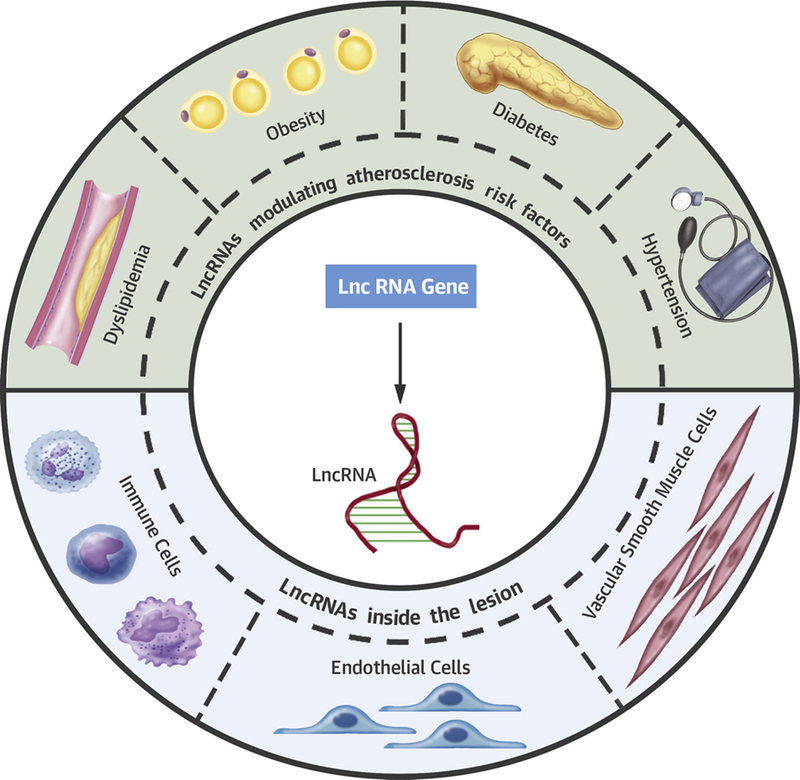
Long non-coding RNAs are actively transcribed genes that do not “appear” to code for proteins. Emerging evidence suggests that lncRNAs serve as key regulators of atherosclerosis and related risk factors.
Acknowledgments:
We thank our colleagues within the Atherosclerosis Research Unit at UCLA for critical review of the manuscript. We apologize to many of our colleagues whose work could not be included due to space constraints.
Sources of Funding: This work was supported by grants HL128822, HL139549., & DK118086
Abbreviations and Acronyms
- LncRNA
Long noncoding RNA
- hnRNP
Heterogeneous nuclear ribonucleoproteins
- SREBP
Sterol regulatory element-binding protein
- PPARA
Peroxisome proliferator-activated receptor alpha
- RXRA
Retinoid X receptor alpha
- APOA1
Apolipoprotein A1
- BMDM
Bone marrow derived macrophages
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: None
References
- 1.Horton JD, Cohen JC, Hobbs HH. Molecular biology of PCSK9: its role in LDL metabolism. Trends biochemical sciences 2007;32:71–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ehret CF, De Haller G. Origin, Development and Maturation of Organelles and Organelle Systems of the Cell Surface in Paramecium. J ultrastructure research 1963;23:SUPPL6:1–42. [DOI] [PubMed] [Google Scholar]
- 3.Consortium EP, Birney E, Stamatoyannopoulos JA et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 2007;447:799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cabili MN, Trapnell C, Goff L et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes & development 2011;25:1915–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dai M, Chen X, Mo S et al. Meta-signature LncRNAs serve as novel biomarkers for colorectal cancer: integrated bioinformatics analysis, experimental validation and diagnostic evaluation. Scientific reports 2017;7:46572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collaborators GBDO, Afshin A, Forouzanfar MH et al. Health Effects of Overweight and Obesity in 195 Countries over 25 Years. N Engl J med 2017;377:13–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics 2013;193:651–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annual rev biochemistry 2012;81:145–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devaux Y, Zangrando J, Schroen B et al. Long noncoding RNAs in cardiac development and ageing. Nature reviews Cardiology 2015;12:415–25. [DOI] [PubMed] [Google Scholar]
- 10.Li P, Ruan X, Yang L et al. A liver-enriched long non-coding RNA, lncLSTR, regulates systemic lipid metabolism in mice. Cell metabolism 2015;21:455–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sallam T, Jones MC, Gilliland T et al. Feedback modulation of cholesterol metabolism by the lipid-responsive non-coding RNA LeXis. Nature 2016;534:124–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tontonoz P, Wu X, Jones M, Zhang Z, Salisbury D, Sallam T. Long Noncoding RNA Facilitated Gene Therapy Reduces Atherosclerosis in a Murine Model of Familial Hypercholesterolemia. Circulation 2017;136:776–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui M, Xiao Z, Wang Y et al. Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Cancer research 2015;75:846–57. [DOI] [PubMed] [Google Scholar]
- 14.Halley P, Kadakkuzha BM, Faghihi MA et al. Regulation of the apolipoprotein gene cluster by a long noncoding RNA. Cell reports 2014;6:222–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soubry A, Schildkraut JM, Murtha A et al. Paternal obesity is associated with IGF2 hypomethylation in newborns: results from a Newborn Epigenetics Study (NEST) cohort. BMC med 2013;11:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones BK, Levorse J, Tilghman SM. Deletion of a nuclease-sensitive region between the Igf2 and H19 genes leads to Igf2 misregulation and increased adiposity. Human molecular genetics 2001;10:807–14. [DOI] [PubMed] [Google Scholar]
- 17.Vu PY, Toutain J, Cappellen D et al. A homozygous balanced reciprocal translocation suggests LINC00237 as a candidate gene for MOMO (macrosomia, obesity, macrocephaly, and ocular abnormalities) syndrome. Am j medical genetics Part A 2012;158A:2849–56. [DOI] [PubMed] [Google Scholar]
- 18.Powell WT, Coulson RL, Crary FK et al. A Prader-Willi locus lncRNA cloud modulates diurnal genes and energy expenditure. Human molecular genetics 2013;22:4318–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stelzer Y, Sagi I, Yanuka O, Eiges R, Benvenisty N. The noncoding RNA IPW regulates the imprinted DLK1-DIO3 locus in an induced pluripotent stem cell model of Prader-Willi syndrome. Nature genetics 2014;46:551–7. [DOI] [PubMed] [Google Scholar]
- 20.Sun L, Goff LA, Trapnell C et al. Long noncoding RNAs regulate adipogenesis. Proceed National Acad Sciences USA 2013;110:3387–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang L, Li P, Yang W et al. Integrative Transcriptome Analyses of Metabolic Responses in Mice Define Pivotal LncRNA Metabolic Regulators. Cell metabolism 2016;24:627–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu S, Sheng L, Miao H et al. SRA gene knockout protects against diet-induced obesity and improves glucose tolerance. J biological chemistry 2014;289:13000–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao XY, Li S, Wang GX, Yu Q, Lin JD. A long noncoding RNA transcriptional regulatory circuit drives thermogenic adipocyte differentiation. Molecular cell 2014;55:372–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moran I, Akerman I, van de Bunt M et al. Human beta cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell metabolism 2012;16:435–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akerman I, Tu Z, Beucher A et al. Human Pancreatic beta Cell lncRNAs Control Cell-Specific Regulatory Networks. Cell metabolism 2017;25:400–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fadista J, Vikman P, Laakso EO et al. Global genomic and transcriptomic analysis of human pancreatic islets reveals novel genes influencing glucose metabolism. Proceed National Acad Sciences USA 2014;111:13924–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arnes L, Akerman I, Balderes DA, Ferrer J, Sussel L. betalinc1 encodes a long noncoding RNA that regulates islet beta-cell formation and function. Genes & development 2016;30:502–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zemmour D, Pratama A, Loughhead SM, Mathis D, Benoist C. Flicr, a long noncoding RNA, modulates Foxp3 expression and autoimmunity. Proceed National Acad Sciences USA 2017;114:E3472–E3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wessel J, Chu AY, Willems SM et al. Low-frequency and rare exome chip variants associate with fasting glucose and type 2 diabetes susceptibility. Nature communications 2015;6:5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X, Zhao Z, Gao C et al. The Diagnostic Value of Whole Blood lncRNA ENST00000550337.1 for Pre-Diabetes and Type 2 Diabetes Mellitus. Experimental and clinical endocrinology & diabetes 2017. [DOI] [PubMed]
- 31.Carter G, Miladinovic B, Patel AA, Deland L, Mastorides S, Patel NA. Circulating long noncoding RNA GAS5 levels are correlated to prevalence of type 2 diabetes mellitus. BBA clinical 2015;4:102–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leung A, Trac C, Jin W et al. Novel long noncoding RNAs are regulated by angiotensin II in vascular smooth muscle cells. Circulation research 2013;113:266–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang YN, Shan K, Yao MD et al. Long Noncoding RNA-GAS5: A Novel Regulator of Hypertension-Induced Vascular Remodeling. Hypertension 2016;68:736–48. [DOI] [PubMed] [Google Scholar]
- 34.Yao QP, Xie ZW, Wang KX et al. Profiles of long noncoding RNAs in hypertensive rats: long noncoding RNA XR007793 regulates cyclic strain-induced proliferation and migration of vascular smooth muscle cells. J hypertension 2017;35:1195–1203. [DOI] [PubMed] [Google Scholar]
- 35.Leisegang MS, Fork C, Josipovic I et al. Long Noncoding RNA MANTIS Facilitates Endothelial Angiogenic Function. Circulation 2017. [DOI] [PMC free article] [PubMed]
- 36.Bayoglu B, Yuksel H, Cakmak HA, Dirican A, Cengiz M. Polymorphisms in the long non-coding RNA CDKN2B-AS1 may contribute to higher systolic blood pressure levels in hypertensive patients. Clinical biochemistry 2016;49:821–7. [DOI] [PubMed] [Google Scholar]
- 37.Foroud T, Koller DL, Lai D et al. Genome-wide association study of intracranial aneurysms confirms role of Anril and SOX17 in disease risk. Stroke 2012;43:2846–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang W, Chen Y, Liu P et al. Variants on chromosome 9p21.3 correlated with ANRIL expression contribute to stroke risk and recurrence in a large prospective stroke population. Stroke 2012;43:14–21. [DOI] [PubMed] [Google Scholar]
- 39.Congrains A, Kamide K, Oguro R et al. Genetic variants at the 9p21 locus contribute to atherosclerosis through modulation of ANRIL and CDKN2A/B. Atherosclerosis 2012;220:449–55. [DOI] [PubMed] [Google Scholar]
- 40.Motterle A, Pu X, Wood H et al. Functional analyses of coronary artery disease associated variation on chromosome 9p21 in vascular smooth muscle cells. Human molecular genetics 2012;21:4021–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holdt LM, Hoffmann S, Sass K et al. Alu elements in ANRIL non-coding RNA at chromosome 9p21 modulate atherogenic cell functions through trans-regulation of gene networks. PLoS genetics 2013;9:e1003588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bennett MR, Sinha S, Owens GK. Vascular Smooth Muscle Cells in Atherosclerosis. Circulation research 2016;118:692–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robbins CS, Hilgendorf I, Weber GF et al. Local proliferation dominates lesional macrophage accumulation in atherosclerosis. Nature medicine 2013;19:1166–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cui X, You L, Li Y et al. A transcribed ultraconserved noncoding RNA, uc.417, serves as a negative regulator of brown adipose tissue thermogenesis. FASEB J 2016;30:4301–4312. [DOI] [PubMed] [Google Scholar]
- 45.Vausort M, Wagner DR, Devaux Y. Long noncoding RNAs in patients with acute myocardial infarction. Circulation research 2014;115:668–77. [DOI] [PubMed] [Google Scholar]
- 46.Hu YW, Zhao JY, Li SF et al. RP5–833A20.1/miR-382–5p/NFIA-dependent signal transduction pathway contributes to the regulation of cholesterol homeostasis and inflammatory reaction. Arteriosclerosis, thrombosis, vascular bioll 2015;35:87–101. [DOI] [PubMed] [Google Scholar]
- 47.Sallam T, Jones M, Thomas BJ et al. Transcriptional regulation of macrophage cholesterol efflux and atherogenesis by a long noncoding RNA. Nature medicine 2018;24:304–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang H, Xue C, Wang Y et al. Deep RNA Sequencing Uncovers a Repertoire of Human Macrophage Long Intergenic Noncoding RNAs Modulated by Macrophage Activation and Associated With Cardiometabolic Diseases. J Am Heart Assoc 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang P, Xue Y, Han Y et al. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science 2014;344:310–3. [DOI] [PubMed] [Google Scholar]
- 50.Carpenter S, Aiello D, Atianand MK et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science 2013;341:789–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Atianand MK, Hu W, Satpathy AT et al. A Long Noncoding RNA lincRNA-EPS Acts as a Transcriptional Brake to Restrain Inflammation. Cell 2016;165:1672–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gomez JA, Wapinski OL, Yang YW et al. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-gamma locus. Cell 2013;152:743–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ridker PM, Everett BM, Thuren T et al. Antiinflammatory Therapy with Canakinumab for Atherosclerotic Disease. The New England journal of medicine 2017;377:1119–1131. [DOI] [PubMed] [Google Scholar]
- 54.Singh KK, Matkar PN, Pan Y et al. Endothelial long non-coding RNAs regulated by oxidized LDL. Molecular cellular biochemistry 2017. [DOI] [PubMed]
- 55.Wang J, Chen L, Li H et al. Clopidogrel reduces apoptosis and promotes proliferation of human vascular endothelial cells induced by palmitic acid via suppression of the long non-coding RNA HIF1A-AS1 in vitro. Molecular cellular biochemistry 2015;404:203–10. [DOI] [PubMed] [Google Scholar]
- 56.Li K, Blum Y, Verma A et al. A noncoding antisense RNA in tie-1 locus regulates tie-1 function in vivo. Blood 2010;115:133–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fiedler J, Breckwoldt K, Remmele CW et al. Development of Long Noncoding RNA-Based Strategies to Modulate Tissue Vascularization. J Am Coll Cardiol 2015;66:2005–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Voellenkle C, Garcia-Manteiga JM, Pedrotti S et al. Implication of Long noncoding RNAs in the endothelial cell response to hypoxia revealed by RNA-sequencing. Scientific reports 2016;6:24141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Michalik KM, You X, Manavski Y et al. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circulation research 2014;114:1389–97. [DOI] [PubMed] [Google Scholar]
- 60.Boon RA, Hofmann P, Michalik KM et al. Long Noncoding RNA Meg3 Controls Endothelial Cell Aging and Function: Implications for Regenerative Angiogenesis. J Am Coll Cardiol 2016;68:2589–2591. [DOI] [PubMed] [Google Scholar]
- 61.Bell RD, Long X, Lin M et al. Identification and initial functional characterization of a human vascular cell-enriched long noncoding RNA. Arteriosclerosis, thrombosis, and vascular biology 2014;34:1249–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huarte M, Guttman M, Feldser D et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 2010;142:409–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu G, Cai J, Han Y et al. LincRNA-p21 regulates neointima formation, vascular smooth muscle cell proliferation, apoptosis, and atherosclerosis by enhancing p53 activity. Circulation 2014;130:1452–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ballantyne MD, Pinel K, Dakin R et al. Smooth Muscle Enriched Long Noncoding RNA (SMILR) Regulates Cell Proliferation. Circulation 2016;133:2050–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kurian L, Aguirre A, Sancho-Martinez I et al. Identification of novel long noncoding RNAs underlying vertebrate cardiovascular development. Circulation 2015;131:1278–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Isin M, Uysaler E, Ozgur E et al. Exosomal lncRNA-p21 levels may help to distinguish prostate cancer from benign disease. Frontiers in genetics 2015;6:168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ruan Y, Lin N, Ma Q et al. Circulating LncRNAs Analysis in Patients with Type 2 Diabetes Reveals Novel Genes Influencing Glucose Metabolism and Islet beta-Cell Function. Cellular physiology and biochemistry 2018;46:335–350. [DOI] [PubMed] [Google Scholar]


