Abstract
Shigella infections account for a considerable burden of acute diarrheal diseases worldwide and remain a major cause of childhood mortality in developing countries. Although, all four species of Shigella (S. dysenteriae, S. flexneri, S. boydii, and S. sonnei) cause bacillary dysentery, historically only S. dysenteriae type 1 has been recognized as carrying the genes for Shiga toxin (stx). Recent epidemiological data, however, have suggested that the emergence of stx carrying S. flexneri strains may have originated from bacteriophage-mediated inter-species horizontal gene transfer in one specific geographical area, Hispaniola. To test this hypothesis, we analyzed whole genome sequences of stx-encoding phages carried by S. flexneri strains isolated in Haiti and S. flexneri S. boydii and S. dysenteriae strains isolated from international travelers who likely acquired the infection in Haiti or the Dominican Republic. Phylogenetic analysis showed that phage sequences encoded in the Shigella strains from Hispaniola were bacteriophage φPOC-J13 and they were all closely related to a phage isolated from a USA isolate, E. coli 2009C-3133 serotype O119:H4. In addition, despite the low genetic heterogeneity of phages from different Shigella spp. circulating in the Caribbean island between 2001 and 2014, two distinct clusters emerged in Haiti and the Dominican Republic. Each cluster possibly originated from phages isolated from S. flexneri 2a, and within each cluster several instances of horizontal phage transfer from S. flexneri 2a to other species were detected. The implications of the emergence of stxproducing non-S. dysenteriae type 1 Shigella species, such as S. flexneri, spans not only the basic science behind horizontal phage spread, but also extends to medical treatment of patients infected with this pathogen.
Keywords: Shigella, Shiga toxin, Haiti, Dominican Republic, whole genome sequencing, temperate phages
1. INTRODUCTION
Bacteria of the genus Shigella are Gram negative enteric pathogens that are the causative agents of bacillary dysentery or shigellosis 1. Shigella infections account for a considerable burden of acute diarrheal diseases worldwide and are an important public health problem in developing countries where shigellosis remains a major cause of childhood mortality. Shigellosis continues to be an important public health concern even in developed countries particularly with the rising incidence of multi-antibiotic resistant strains in circulation worldwide. Shiga toxin (Stx) is a potent AB5 type cytotoxin that inhibits eukaryotic protein synthesis, eventually leading to host cell death 2. While all four species of Shigella (S. dysenteriae, S. flexneri, S. boydii, and S. sonnei) cause bacillary dysentery, historically only S. dysenteriae type 1 has been recognized as carrying the genes for Stx. The toxin genes, stx, are chromosomally encoded in S. dysenteriae3.
Strains of enterohemorrhagic Escherichia coli (EHEC) produce Stx encoded by genes that are found on a transmissible bacteriophage inserted in the bacterial chromosome 4. In the past three decades, Shiga toxin-producing E. coli (STEC) of different serotypes have emerged. Recently, isolates of non-S. dysenteriae 1 Shigella species, notably S. flexneri, S. dysenteriae 4 and S. sonnei, have also been shown to harbor a lambdoid phage that carries the Shiga toxin genes, stxAB 5–9. Epidemiological data indicated that the emergence of stx-carrying S. flexneri strains may have originated in one specific geographical area, Hispaniola. We previously characterized Shiga toxin-producing clinical isolates of non-S. dysenteriae type 1 Shigella species from Public Health Laboratories in the United States and Canada and a collection from the Institut Pasteur, Paris France. Metadata on these strains suggested a strong link of the stx-carrying phage to travelers returning to these countries from Haiti or the Dominican Republic 10,11. Clinical strains of stx-encoding S. flexneri were subsequently isolated from Haitian school children with diarrhea in Gressier, Haiti 12. These strains all carried a lambdoid phage that encoded the Shiga toxin genes.
The genus Shigella is composed of four species that are now thought to have evolved directly and independently from commensal E. coli lineages 13. Three main Shigella clusters represent the evolutionary history of each serotype. The principal step in the divergence of E. coli with Shigella spp. is the acquisition of a large virulence plasmid (pINV) by the latter. Although the genomes of Shigella and E. coli share a conserved common backbone, Shigella spp. have undergone a number of inversions and translocations. As is frequently observed with other enteric bacteria, Shigella spp. are subject to horizontal gene transfer mediated by different genetic elements, such as phages. These transmissible mobile vectors carry genetic determinants ranging from antibiotic resistance elements to metabolic pathway genes. For the Enterobacteriaceae, including STEC, the stx genes are commonly transferred via lambdoid phages 14.
The recent emergence of these Shiga toxin-producing Shigella strains can be viewed as a paradigm of rapid spread of phage-encoded toxins within a bacterial population, i.e. S. flexneri, that reside in any given habitat. The implications of the emergence of Shiga toxin producing non-S. dysenteriae type 1 Shigella spp., spans not only the basic science behind horizontal gene spread via phage but also extends to medical treatment of patients infected with this pathogen. In this study, we carried out whole genome sequence analysis of stx-encoding Shigella strains isolated from international travelers and Haitians who likely acquired the infection in Haiti or Dominican Republic, to assess phage genetic diversity and investigate the patterns of acquisition of toxin genes via phage conversion in circulating Shigella spp.
2. MATERIALS AND METHODS
2.1. Data set of stx-encoding Shigella strains
A set of 49 clinical samples of Shigella spp. was collected between 1999 and 2014 from Haitian residents and international travelers mostly returning from the Dominican Republic and Haiti, where they likely acquired the infection 10–12,15. Epidemiological data such as isolation date and recent foreign travel destination were collected when available (Table 1). Samples were identified as Shigella “species” using conventional methods described by Nataro et al. 16 and serotyped by slide agglutination assays as described by Gray et al 6. Most of the isolates were identified as S. flexneri (40 isolates) but other Shigella subtypes such as S. sonnei (2 isolates), S. boydii (2 isolates) and S. dysenteriae (4 isolates), were also recognized (Table 1).
Table 1.
Characteristics of Shigella strains analyzed.
| Strain | Species1 | Recent foreign travel2 | Isolation Date | stx3 | φPOC-J134 |
|---|---|---|---|---|---|
| BS1041 | S. flexneri 2a | Dom. Rep. | 1999 | + | + |
| BS1022 | S. flexneri 2a | Dom. Rep. | 2004 | + | + |
| BS1023 | S. flexneri 2a | Dom. Rep. | 2005 | + | + |
| BS981 | S. flexneri 2a | Dom. Rep. | 2005 | + | + |
| BS1042 | S. flexneri 2a | Dom. Rep. | 2005 | + | + |
| BS1044 | S. flexneri 2a | Dom. Rep. | 2005 | + | + |
| BS1045 | S. flexneri 2a | Dom. Rep. | 2007 | + | + |
| BS1046 | S. flexneri 2a | Dom. Rep. | 2008 | + | + |
| BS1024 | S. flexneri 2a | French Guiana | 2005 | + | + |
| BS974 | S. flexneri 2a | Haiti | 2001 | + | + |
| BS1021 | S. flexneri 2a | Haiti | 2003 | + | + |
| BS980 | S. flexneri 2a | Haiti | 2005 | + | + |
| BS1025 | S. flexneri 2a | Haiti | 2008 | + | + |
| BS937 | S. flexneri 2a | Haiti | 2010 | + | + |
| BS968 | S. flexneri 2a | Haiti | 2010 | + | + |
| BS971 | S. flexneri 2a | Haiti | 2011 | + | + |
| BS988 | S. flexneri 2a | Haiti | 2012 | + | + |
| BS1039 | S. flexneri 2a | Haiti (r) | 2013 | + | + |
| BS1057 | S. flexneri 2a | Haiti (r) | 2013 | + | + |
| BS1059 | S. flexneri 2a | Haiti (r) | 2014 | + | + |
| BS967 | S. flexneri 2a | India | 2010 | - | - |
| BS966 | S. flexneri 2a | Mexico | 2009 | - | - |
| BS952 | S. flexneri 2a | Peru | 2005 | - | - |
| BS951 | S. flexneri 2a | NA | 2005 | + | + |
| BS956 | S. flexneri 2a | NA | 2006 | - | - |
| BS962 | S. flexneri 2a | NA | 2008 | - | - |
| BS982 | S. flexneri 2a | NA | 2008 | + | + |
| BS942 | S. flexneri 2a | NA | 2010 | + | + |
| BS970 | S. flexneri 2a | NA | 2011 | - | - |
| BS938 | S. flexneri 2a | NA | 2012 | + | + |
| BS972 | S. flexneri 2a | NA | 2012 | + | + |
| BS989 | S. flexneri 2a | NA | 2013 | + | + |
| BS1061 | S. flexneri 3a | Haiti (r) | 2014 | - | - |
| BS1038 | S. flexneri 3a | Haiti (r) | NA | - | - |
| BS1040 | S. flexneri 6 | Haiti (r) | NA | - | + |
| BS1074 | S. flexneri ? | Haiti | 2014 | + | + |
| SH199 | S. flexneri ? | Haiti | 2014 | + | + |
| SH200 | S. flexneri ? | NA | 2014 | + | + |
| BS986 | S. boydii 19 | Dom. Rep. | 2010 | + | + |
| BS984 | S. boydii 19 | Haiti | 2010 | + | + |
| BS1043 | S. flexneri Y | Haiti | 2005 | + | + |
| BS1060 | S. flexneri Y | Haiti | 2014 | + | + |
| BS978 | S. dysenteriae 1 | NA | 2004 | + | - |
| BS983 | S. dysenteriae 1 | NA | 2008 | + | - |
| BS979 | S. dysenteriae 4 | Dom. Rep. | 2005 | + | + |
| BS975 | S. dysenteriae 4 | Haiti | 2002 | + | + |
| BS1047 | S. dysenteriae 4 | Haiti | 2008 | + | + |
| BS987 | S. dysenteriae 4 | Haiti | 2010 | - | - |
| BS1058 | S. sonnei | Haiti | 2014 | - | - |
Strains positive for stx for which the full phage genome could be assembled from the bacterial genome are in bold. Strains highlighted in red were either positive for stx but the φPOC-J13 phage sequence could not be found in the bacterial genome, or negative for stx but the φPOC-J13 phage was found in the genome.
Shigella species and serotype; “?” indicates that the serotype was not determined
Dom. Rep. = Dominican Republic, “(r)” indicates strains that were isolated from Haitians residing in Haiti, i.e. not linked to foreign travel.
Presence or absence of stx genes. “+” indicate positives for stx and “-” indicates negative for stx and stx2.
Presence or absence of phage φPOC-J13 sequence assembled by Mauve
NA = data not available
2.2. DNA extraction and next generation sequencing and assembly
Shigella strains were grown in Tryptic Soy Broth (TSB) (BD Difco, Franklin Lakes, NJ) at 37°C with aeration or on TSB plates containing 1.5% agar (TSB agar) with or without 0.025% Congo red (Sigma-Aldrich, St. Louis, MO) as described by Gray et al 12. Bacterial genomic DNA was extracted from overnight cultures using the DNeasy Blood and Tissue Kit (QIAGEN, Germantown, MD) or the QIAGEN DNeasy Kit (Valencia, CA, USA). Next generation sequencing (NGS) of the whole genome of Shigella strains was performed using Illumina technology (Illumina, San Diego, CA). Briefly, DNA libraries were prepared with either the TruSeq DNA Sample Prep Kit (Illumina) or the Nextera DNA Sample Prep Kit (Illumina). Samples BS937, BS938, and BS974 were prepared for sequencing by using a Nextera XT DNA Sample Preparation Kit (Illumina). Strains were sequenced using the Illumina MiSeq Platform, generating paired-end 250 base-pair reads12. Raw reads were trimmed and assembled de novo using CLC Genomics Workbench version 7.0.4 (CLC bio, Boston, MA). Whole Shigella genome sequences have been deposited at DDBJ/ EMBL/GenBank under the accession numbers listed in Table S1.
2.3. Detection of Shiga toxin-encoding bacteriophage φPOC-J13 by in silico data analysis
The presence of the reference phage φPOC-J13 (GenBank accession no. KJ603229) from Shigella spp. contigs, obtained from de novo assembly performed previously by Gray et al 10,12, was detected using PHASTER17, a web server for the rapid identification and annotation of prophage sequences within bacterial genomes. Following PHASTER17 parameters φPOCJ13 sequence was considered present and intact if the quality score was > 90, present and questionable if the quality score was between 90 and 70, and present but incomplete if the quality score was < 70. The prediction of whether the region contains an intact or incomplete phage is called completeness of the phage and was calculated on the basis of the criteria illustrated in PHASTER (http://phaster.ca). The presence of the genes encoding Shiga toxin subunits A and B was verified by manually inspecting the Shigella spp. contigs (see section 2.4).
2.4. Extraction and alignment of the phage sequences encoded in the Shigella strains
Contigs of the Shigella spp. strains were ordered to the φPOC-J13 reference sequence (GenBank accession no. KJ603229) using MAUVE18, a system for constructing multiple genome alignments of the phage sequences in each phage positive Shigella strain18. Thirty-seven Shigella strains encoding φPOC-J13 were found (Table 1) and the complete phage sequences were manually extracted from the Shigella whole genome sequence data, using φPOC-J13 as reference. In 21 of the 36 strains carrying the stx genes, the entire stx-encoding phage was contained in one contig (Table S2). Number of contigs containing φPOC-J13 sequence for each strain is reported in Table S2. MAUVE18 was also used to obtain an alignment of 37 complete φPOC-J13 phage genomes (36 φPOC-J13 sequences plus the reference phage φPOC-J13 (accession no. KJ603229); the whole genome alignment was manually optimized using Genious version R9 (https://www.geneious.com). A second alignment of 44 sequences was obtained by adding seven additional known phage sequences, identified by Blast nucleotide search (NCBI) that shared at least 60% similarity with φPOC-J13. This allowed us to investigate the relationship of our φPOC-J13 sequences with similar phages encoded in other bacteria and the possible emergence of φPOC-J13 in the Caribbean area.
2.5. Detection of recombination within φPOC-J13 phage sequences
The presence of recombination among full phage genome sequences was assessed by inferring a Neighbor Network (NNet) and the Phi test 19 implemented in Splits Tree4 20,21 (http://www.sliptstree.org/), where p-values < 0.05 indicate statistically significant recombination signal. GARD 22, a likelihood-based recombination detection procedure that utilizes a genetic algorithm was also used to identify putative recombinant sequences and identify recombination breakpoints. Lastly, Gubbins 23, an algorithm that iteratively identifies loci containing elevated densities of base substitutions was used to generate recombinationfree alignments.
2.6. Single nucleotide polymorphisms detection
Single nucleotide polymorphisms (SNPs) were detected in the recombinant-free whole genome alignment of the 37 φPOC-J13 phages using the software Molecular Evolutionary Genetics Analysis version 7.0. (MEGA 7.24). Gubbins 23 was utilized to produce a recombinant-free alignment filtered for polymorphic sites only, with no duplicate sequences, from the 44 phage alignment. The analysis detected 17 identical sequences within the 44 phage sequences; therefore, the final recombinant-free alignment consisted of only 27 phage sequences.
2.7. Analysis of phylogenetic signal
The presence of substitution saturation in the aligned sequences, which makes phylogeny inference unreliable, was investigated by inferring pairwise genetic distance vs. transitions and transversions plots and the Xia test 25 with the software package DAMBE6 26,27. Phylogenetic signal was also assessed on recombinant-free genome alignments by likelihood mapping analysis with TREE-PUZZLE 28 (http://www.tree-puzzle.de). In brief, a likelihood map is an equilateral triangle where each corner corresponds to one of the three possible tree topologies for a group of four sequences (a quartet) and a dot inside the map represents simultaneously the three likelihoods of the three possible trees. To evaluate the phylogenetic signal in a multiple alignment of N sequences all the possible quartets (or 10,000 randomly selected quartets if N>50) are evaluated, which results in a likelihood map where dots are distributed within different areas (regions) of the triangle. Dots in the central region and along the sides of the triangle represent phylogenetic noise (unresolved phylogenies), while dots equally distributed in the three corner regions represent tree-like signal (resolved phylogenies). Likelihood mapping with >40% phylogenetic noise are considered unreliable to infer fully resolved phylogenies.
2.8. Phylogeny inference
A Maximum Likelihood (ML) phylogenetic tree was inferred from the alignment including the sequences in this study as well as the GenBank reference sequences with the software IQ-TREE29 (http://www.iqtree.org). The best-fitting nucleotide substitution model was chosen by calculating the Bayesian Information Criterion (BIC), as implemented in IQ-TREE30. The selected model was the Kimura 2-parameter with ascertainment bias correction (K2Pu+ASC) for the 37 φPOC-J13 sequence alignment, and the transitional model and equal base frequencies with ascertainment bias correction (TIMe+ASC) for the 27 phage sequence alignment. Support for internal branches of the tree was assessed by 2000 ultrafast bootstrap (BB)31 replicates. Given the low level of genetic diversity, we also reconstructed the evolutionary history of the phages circulating in the Caribbean area by inferring the minimum spanning tree (MST) with an in-house script implemented in R using the Kruskal’s minimum spanning tree in boost (mstree.kruskal) 32. An MST is an un-directed graph that connects all the vertices (representing sequences) together, without any cycles, through edges proportional to SNPs separating any two vertices (sequences), by minimizing the possible total edge length 33. To assess compartmentalization (i.e. the existence of separate subpopulations) of isolates among Haiti and Dominican Republic geographic locations, a distance-based test was performed using the SNPs alignment and calculating four estimates of Wright’s measure of population subdivision (FST) 34–37 implemented in the software HyPHy 38. Distance matrices were calculated using the best fitting nucleotide substitution model with 1,000 bootstrapping and permutations, and statistical significance derived via a population-structure randomization test. FST values indicate evidence of weak (<0.05), moderate (0.050.15), strong (0.15–0.25) or very strong (>0.25) genetic difference between sub-populations; therefore, the higher the FST value, the higher is the level of compartmentalization39.
3. RESULTS
3.1. Detection of stx-encoding bacteriophage φPOC-J13
We identified 38 out of 49 Shigella strains as stx-positive using PHASTER14 and by searching manually the stx sequence in each of the Shigella strains as well. In 36 of these strains, stx was encoded within phages that were identified as φPOC-J13. No stx gene sequences were found in the absence of the phage with the exception of the two S. dysenteriae type 1 strains that were stx-positive despite not harboring the φPOC-J13 phage, and one S. flexneri strain (BS1040) that harbored the phage but not the stx genes (Table 1, Table S2). The absence of phage sequences in the S. dysenteriae type 1 strains was not unexpected as it is well established that these strains are not lysogens but encode the stx genes in the chromosome in the absence of intact phage sequences3. The discordance with BS1040 is possibly due to low NGS coverage. These three strains were not included in subsequent analyses. Overall, the results were in agreement with data obtained in previous studies10–12 that determined presence and insertion site of the stx-encoding bacteriophage φPOC-J13 by PCR. Phage positive strains were mostly from patients infected by S. flexneri 2a that traveled and acquired the infection in the Dominican Republic and Haiti. In each instance, the insertion site in the bacterial genomes was identified as locus S1742 or a homologous gene.
3.2. Phylogenetic inference of φPOC-J13 phage sequences
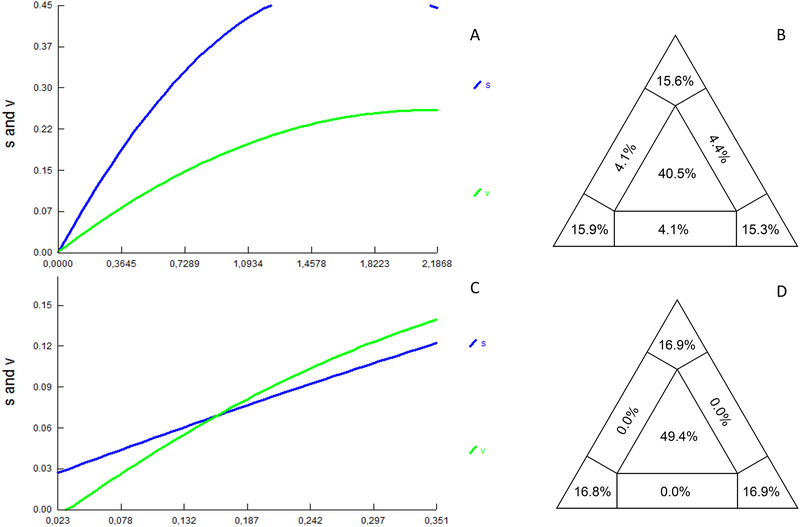
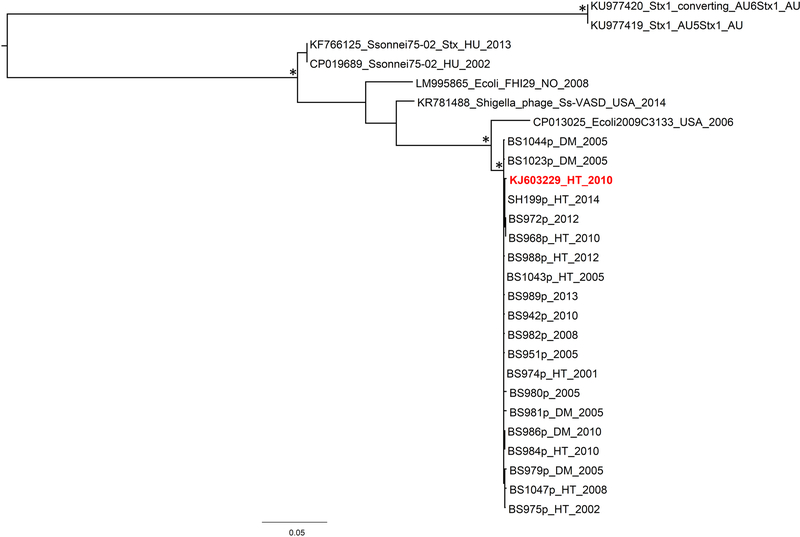
The phage φPOC-J13 sequences extracted from all stx-positive bacterial strains and the φPOC-J13 reference sequence from BS93711, a clinical isolate of stx-producing S. flexneri with an epidemiological link to travel to Hispaniola, were used to build a φPOC-J13 full genome phage alignment (37 strains) of 62,741 bp. To investigate the relationship of the Shigella phages circulating in Hispaniola with evolutionarily related phages from other species in different areas of the world, we data mined available sequences in GenBank by Blastn and found seven additional phage sequences with at least 60% similarity to the φPOCJ13 that were added to our SNPs alignment. None of these phages was from S. dysenteriae type 1; three were carried by S. sonnei isolates and identified as Shigella phage 75/02 (GenBank accession no. KF766125 and CP019689) and as Shigella phage Ss-VASD (GenBank accession no. KR781488); the other four were E. coli phages (GenBank accession no. CP013025 and LM995865) from STEC 2009C-3133 and FHI29, respectively, and from Australian E. coli O157 isolates (GenBank accession no. KU977420 and KU977419). After removing 17 duplicate sequences (several sequences from Hispaniola were identical, see Supplementary Results for details), the final alignment of 44 full genome phages showed highly significant statistical signal (p < 10−99) for recombination using split decomposition network analysis 20,21 and the Phi test19, which was also confirmed by GARD22 (see section 2.5). Specific recombination breakpoints were inferred with Gubbins 23, and average recombination to mutation (r/m), the ratio of SNPs imported through recombination to those presented through mutation, was 5,56. Recombination analysis showed higher level of recombination for the reference sequences from GenBank, which was expected given the high similarity of the φPOC-J13 phages (Figure S1). After obtaining these results a recombinant-free alignment of 711 SNPs was finally assembled to carry out the subsequent analysis. Pairwise transition/transversion vs. genetic diversity plots show the presence of transversion saturation for genetic distances > 1.5 (Figure 1A), thus indicating the presence of moderate phylogenetic noise in the data set, as also confirmed by 40.5% of the dots in the center area of the likelihood map (Figure 1B). The Xia test, however, indicated the presence of sufficient signal to infer the phylogeny (p < 0.0001). Therefore, a ML tree was inferred to investigate further the evolutionary relationships among phage sequences among different Shigella spp. (Figure 2). Phages circulating in Hispaniola clustered within a highly supported monophyletic clade (Figure 2), which in turn clustered, with high support, with a phage isolated from E. coli 2009C-3133 serotype O119:H440, infecting a patient in the United States. Although the ML tree was unrooted and a molecular clock could not be calibrated due to insufficient clock-like signal (data not shown), the divergent phage sequences from GenBank were obviously a natural outgroup from the Hispaniola strains that could be used to root the tree. The evolutionary direction of the rooted tree suggests a horizontal transfer from E. coli to S. flexneri 2a at the origin of the phages circulating in Haiti and the Dominican Republic.
Figure 1. Transition and transversion versus divergence plots and likelihood mapping analysis for the POC-J13 phage data sets.
Pairwise transition and transversion versus divergence plots and likelihood mapping analyses are shown in the left and right panels, respectively. (A and B) Analysis of the 44 sequence alignment including the 36 phage sequences described in the paper (Table 1 and S1), the φPOC-J13 reference sequence (accession no. KJ603229), and seven additional divergent phage strains from GenBank. (C and D) Analysis of the alignment including only the 36 phage sequences described in the paper.
Figure 2. Maximum Likelihood tree inferred on the alignment of unique phage sequences from Hispaniola plus reference sequences from GenBank.
The reference sequence of Shigella phage φPOCJ13 (GenBank accession no. KJ603229) is reported in red. BS984 and BS986 are phages extracted from S. boydii 19 strains, while strains BS975, BS979 and BS1047 are extracted from S. dysenteriae 4 strains. All the other phages are encoded in S. flexneri strains. The country of origin and time of isolation (when known) are indicated in the label of each strain by a two/three letter code (AU = Australia, DM = Dominican Republic, HT= Haiti, HU = Hungary, NO = Norway, USA = United States) followed by the year. The tree was rooted using the divergent Australian phage sequences from E. coli as an outgroup. Branch lengths are drawn to scale proportional to SNPs distance per SNP site according to the scale bar at the bottom of the tree. Ultrafast bootstrap support >90% for internal branches of the tree is indicated by an asterisk.
3.3. Cluster analysis of φPOC-J13 phages circulating in Hispaniola
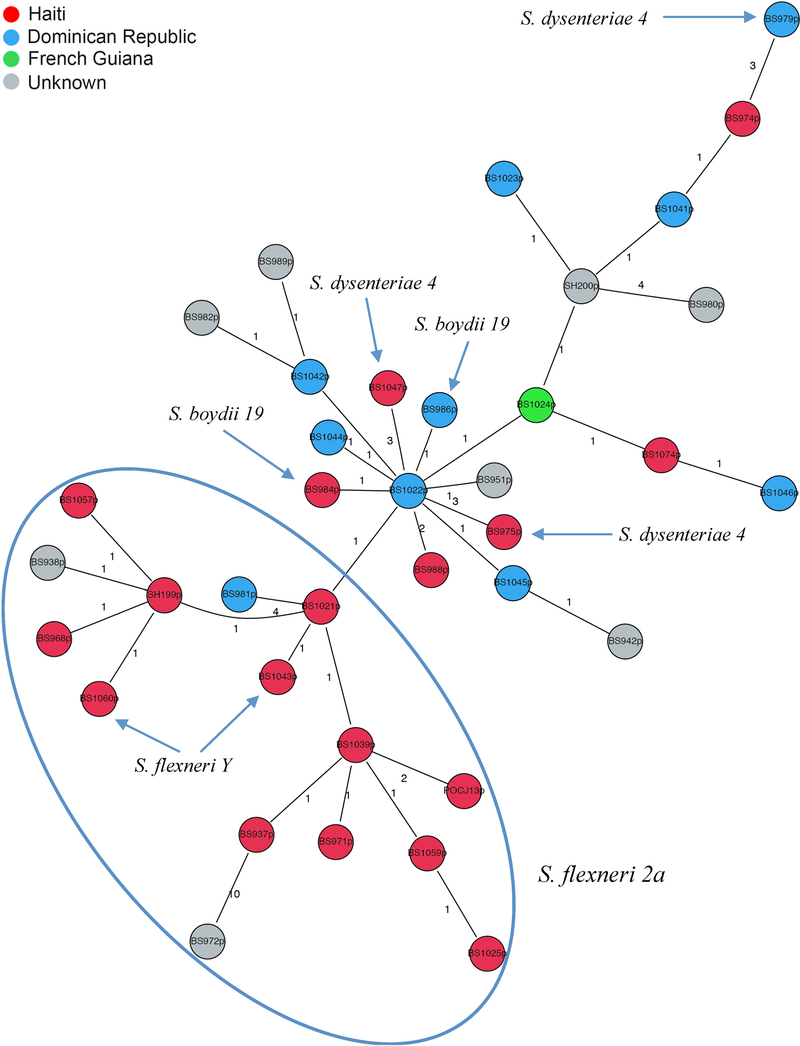
Overall, φPOC-J13 sequences in our data set (Table 1) were highly conserved, with several identical sequences (see Supplementary Material), and 33 SNP sites, only eight of which were parsimony informative. Among these sequences, no evidence for recombination was found based on the split decomposition algorithm41 and the Phi Test20 of recombination (p value = 1.0). Also, as expected given such a low level of genetic heterogeneity, no substitution saturation was detected by the Xia test (p <0.0001), or the transition/transversion vs. genetic diversity plot (Figure 1C). Likelihood mapping analysis, on the other hand, showed very low phylogenetic signal in the data (49.4% unresolved quartets, see Figure 1D), making the data set unsuited for robust phylogeny inference, in agreement with the small number of parsimony informative sites. The lack of diversity is a strong indication of a single clonal introduction of φPOC-J13 phage in the Shigella strains circulating in Haiti and Dominican Republic, likely followed by clonal expansion. To investigate this scenario, we further examined the relationship among phage sequences from Hispaniola by calculating a minimum spanning tree (MST). MST is a clustering method that allows exploration of potential relationships among closely related sequences, under the assumption that, in an outbreak, a chain of transmission can be represented by a graph connecting all strains with the minimum genetic distance among them 42. A main “Haitian” cluster is evident in the MST (Figure 3). The “Haitian” cluster includes 12 out of the 17 Haitian sequences (70% of the sample), as well as one sequence from the Dominican Republic and two of unknown origin, all isolated from S. flexneri 2a except two from S. flexneri Y, possibly emerging from a Haitian sequence (central node). The remaining sequences cluster together in a mixed Dominican/Haitian cluster, which includes phage sequences obtained from S. flexneri 2a, as well as two from S. boydii 19 and three from S. dysenteriae 4. Haitian and non-Haitian sequences from different Shigella spp. are intermixed, although the central node from which most of the other sequences emerge is Dominican sequence BS1022p (Figure 3), suggesting that the clade may have originated in the Dominican and subsequently spread. Genetic compartmentalization of phages between the two main clusters in the MST was supported by FST values > 0.25 34–37. The result suggests two independent introductions of the φPOC-J13 phage, one in Haiti and the other one in the Dominican Republic, followed by clonal expansion. In addition, although most of the phages in our study were isolated from S. flexneri 2a, the phages from other Shigella spp. consistently appear to be intermixed with phages from S. flexneri 2a (Figure 3). Phage sequences from non-S. flexneri 2a are always in terminal nodes connected to internal nodes of S. flexneri 2a sequences. Therefore, although an MST is an undirected graph, its general topology suggests several instances of horizontal phage transmission from S. flexneri 2a to S. flexneri Y, S. boydii, and S. dysenteriae 4. Despite these findings, given low genetic heterogeneity of phages and the small sampling size of the strains analyzed, a common origin for the stx-encoding phages cannot be excluded. Further studies with additional samples could help in reconstructing the precise picture of the evolutionary pattern of these strains.
Figure 3. Minimum spanning tree (MST) of φPOC-J13 phage sequences.
The MST was inferred from the alignment including 36 full genome φPOC-J13 sequences isolated from stx-encoding S. flexneri 2a strains (except for a few phages isolated from other Shigella spp., indicated by arrows), plus the φPOC-J13 reference sequence (GenBank accession no. KJ603229). Each circle represents a phage sequence colored by country of origin according to the legend in the figure. Unknown country of origin indicates that the infected patient did not have a known travel history available. The last two digits of each strain label indicate the time of sampling (2001–2014). The number along an edge is the number of SNPs separating two connected sequences. The clade including most of the Haitian strains is indicated with an ellipse.
4. DISCUSSION
Although the role of the Shiga toxin in Shigella pathogenesis has not been fully elucidated, it is responsible for the production of hemolytic uremic syndrome (HUS), a sequela of bacillary dysentery (shigellosis) in infected individuals 43,44. Recent reports have shown that other Shigella strains in addition to S. dysenteriae type 1, carry the stx genes and, notably, are encoded in a lambdoid type phage 6–11. We sequenced the complete genome of φPOC-J13, the reference lambdoid phage carrying the stx genes in S. flexneri 11. Using the φPOC-J13 sequence as a reference point, we studied the evolutionary relationship among phages carrying the stx genes from the other Shigella spp. strains in our collection. Phylogenetic analysis showed that phage sequences from Hispaniola were closely related to a phage isolated from an E. coli strain 2009C-3133 serotype O119:H4 (GenBank accession no. CP013025), isolated from a patient in New York in 2009. In addition, despite the low genetic heterogeneity of phages from different Shigella spp. circulating on the Caribbean island between 2001 and 2014, two distinct clusters seemed to have emerged in Haiti and the Dominican Republic. Each cluster possibly originated from phages isolated from S. flexneri 2a, and within each cluster several instances of horizontal phage transfer from S. flexneri 2a to other Shigella spp. were detected. Given the small sampling size of the strains analyzed, as well as the oversampling of S. flexneri 2a strains, a common source for stx-encoding phages cannot be excluded, but the precise picture of the evolutionary pattern of these phages may be improved with additional samples. Nevertheless, the results suggest that phage-mediated horizontal transfer of the stx genes from S. flexneri 2a to other S. flexneri serotypes or Shigella spp. played a major role in the emergence of toxigenic non-S. dysenteriae type 1 strains in Hispaniola.
Our results suggest that such an ancestral horizontal gene transfer event has been followed in recent years by additional events among different Shigella spp., which are likely at the origin of the emergence of pathogenic S. flexneri strains in the Caribbean. In particular, the central role played by S. flexneri 2a strains in the emergence of stx-producing non-S. dysenteriae type 1 Shigella spp., may have significant implications to understand not only the basic science behind the patterns of acquisition of toxin genes via phage conversion, but also extends to medical treatment of patients infected with this pathogen.
Moreover, even if multiple studies have investigated the ratio of recombination to mutation, which is an important parameter to evaluate the contribution of vertical and horizontal processes to genome evolution in bacterial populations, few estimates have been reported for phages45. Therefore our results could help future research on the evolution of phages encoded by stx-producing non-S. dysenteriae type 1 Shigella spp.
A major factor in the global rise of multiple drug resistant bacterial pathogens, including Shigella spp., is the dissemination of antibiotic resistance genes by transfer systems such as plasmids and phages. The critical role that horizontal gene transfer via lysogenic phages plays in bacterial evolution is well defined. In a similar fashion, the contribution of international travel to the finding of lambdoid phages carrying stx genes parallels the global spread of antibiotic resistance genes. Moreover, we note the potential risk to patients with shigellosis with regard to medical treatment with antibiotics. In addition to selection of the appropriate antibiotic regimen, clinicians should be aware of the possibility of phage induction (and increased toxin production) by such treatment and the increased risk of HUS in these patients. Thus, these strains of stx-producing strains of Shigella represent another example of the complex issues now facing the scientific and medical communities as they address rising multiple antibiotic resistance and the role of international travel in the spread of emerging bacterial pathogens.
5. CONCLUSIONS
New strains of non-S. dysenteriae type 1 Shigella spp. that carry the Shiga toxin (stx) genes on a bacteriophage are emerging. There is strong evidence to support their emergence as two distinct clusters originating in Haiti and the Dominican Republic. Each cluster of strains acquired the stx genes via horizontal gene transfer mediated by phage φPOC-J13.
International travel serves a vehicle for global spread of these emerging pathogens. Caution should be taken when antibiotic treatment of patients infected with these strains is considered. Such treatment may induce the phage lytic cycle and thus higher expression of the toxin genes, increasing the risk of hemolytic uremic syndrome.
Supplementary Material
HIGHLIGHTS.
Phylogenetic analyses show that phage sequences from Hispaniola are closely related to a phage isolated from an E. coli strain 2009C-3133, serotype O119:H4.
The low level of genetic heterogeneity of the stx-encoding phages carried by S. flexneri is a strong indication of a single clonal introduction of φPOC-J13 phage in the Shigella strains circulating in Haiti and the Dominican Republic, likely followed by clonal expansion.
Two distinct clusters emerged in Haiti and in the Dominican Republic. Each cluster possibly originated from phages isolated from S. flexneri 2a, and within each cluster several instances of horizontal phage transfer from S. flexneri 2a to other species were detected.
Phage-mediated horizontal transfer of the stx genes from S. flexneri 2a to other S. flexneri serotypes or Shigella spp. could play a major role in the emergence of toxigenic non-S. dysenteriae type 1 strains in Hispaniola.
Acknowledgements
We would like to thank Jayanthi Gangiredla for her efforts in depositing the sequences in GenBank.
Funding
This work was supported by the National Institute of Allergy and Infectious Diseases to ATM [grant number R01 AI024656-23].
Footnotes
Declaration of interest: None
Disclaimer for Dr. Lampel
The views expressed in this article are those of the author (KAL) and do not necessarily reflect the official policy of the Department of Health and Human Services, the U.S. Food and Drug Administration (FDA), or the U.S. Government. Reference to any commercial materials, equipment, or process does not in any way constitute approval, endorsement, or recommendation by the FDA.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFRENCES
- 1.Mandell GL, Bennett JE, & Dolin R Mandell, Douglas, and Bennett’s principles and practice of infectious diseases. 7th ed. edn, (Churchill Livingstone/Elsevier, 2010). [Google Scholar]
- 2.Melton-Celsa AR Shiga Toxin (Stx) Classification, Structure, and Function. Microbiol Spectr 2, EHEC-0024–2013 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McDonough MA & Butterton JR Spontaneous tandem amplification and deletion of the shiga toxin operon in Shigella dysenteriae 1. Mol Microbiol 34, 1058–1069 (1999). [DOI] [PubMed] [Google Scholar]
- 4.Strockbine NA et al. Two toxin-converting phages from Escherichia coli O157:H7 strain 933 encode antigenically distinct toxins with similar biologic activities. Infect Immun 53, 135–140 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beutin L, Strauch E & Fischer I Isolation of Shigella sonnei lysogenic for a bacteriophage encoding gene for production of Shiga toxin. Lancet 353, 1498, doi:10.1016/S0140-6736(99)00961-7 (1999). [DOI] [PubMed] [Google Scholar]
- 6.Gupta SK et al. Emergence of Shiga toxin 1 genes within Shigella dysenteriae type 4 isolates from travelers returning from the Island of Hispanola. Am J Trop Med Hyg 76, 1163–1165 (2007). [PubMed] [Google Scholar]
- 7.Nogrady N et al. Antimicrobial resistance and genetic characteristics of integron-carrier shigellae isolated in Hungary (1998–2008). J Med Microbiol 62, 1545–1551, doi:10.1099/jmm.0.058917-0 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Toth I, Svab D, Balint B, Brown-Jaque M & Maroti G Comparative analysis of the Shiga toxin converting bacteriophage first detected in Shigella sonnei. Infect Genet Evol 37, 150–157, doi:10.1016/j.meegid.2015.11.022 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Kozyreva VK et al. Recent Outbreaks of Shigellosis in California Caused by Two Distinct Populations of Shigella sonnei with either Increased Virulence or Fluoroquinolone Resistance. mSphere 1, doi:10.1128/mSphere.00344-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gray MD et al. Prevalence of Shiga toxin-producing Shigella species isolated from French travellers returning from the Caribbean: an emerging pathogen with international implications. Clin Microbiol Infect 21, 765 e769–765 e714, doi:10.1016/j.cmi.2015.05.006 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gray MD et al. Clinical isolates of Shiga toxin 1a-producing Shigella flexneri with an epidemiological link to recent travel to Hispaniola. Emerg Infect Dis 20, 1669–1677, doi:10.3201/eid2010.140292 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gray MD et al. Stx-Producing Shigella Species From Patients in Haiti: An Emerging Pathogen With the Potential for Global Spread. Open Forum Infect Dis 2, ofv134, doi:10.1093/ofid/ofv134 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pupo GM, Lan R & Reeves PR Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc. Natl. Acad. Sci. U.S.A 97, 10567–10572 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidt H Shiga-toxin-converting bacteriophages. Res Microbiol 152, 687–695 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Bekal S, Pilon PA, Cloutier N, Doualla-Bell F & Longtin J Identification of Shigella flexneri isolates carrying the Shiga toxin 1-producing gene in Quebec, Canada, linked to travel to Haiti. Can J Microbiol 61, 995–996, doi:10.1139/cjm2015-0538 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Nataro JBC; Fields P; Kaper J; Strockbine N Escherichia, Shigella, and Salmonella. Vol. Manual of Clinical Microbiology 603–626 (2011). [Google Scholar]
- 17.Arndt D et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res 44, W16–21, doi:10.1093/nar/gkw387 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Darling AE, Mau B & Perna NT progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5, e11147, doi:10.1371/journal.pone.0011147 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bruen TC, Philippe H & Bryant D A simple and robust statistical test for detecting the presence of recombination. Genetics 172, 2665–2681, doi:10.1534/genetics.105.048975 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huson DH & Bryant D Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23, 254–267, doi:10.1093/molbev/msj030 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Bryant D & Moulton V Neighbor-net: an agglomerative method for the construction of phylogenetic networks. Mol Biol Evol 21, 255–265, doi:10.1093/molbev/msh018 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH & Frost SD GARD: a genetic algorithm for recombination detection. Bioinformatics 22, 3096–3098, doi:10.1093/bioinformatics/btl474 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Croucher NJ et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res 43, e15, doi:10.1093/nar/gku1196 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar S, Stecher G & Tamura K MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol 33, 1870–1874, doi:10.1093/molbev/msw054 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xia X, Xie Z, Salemi M, Chen L & Wang Y An index of substitution saturation and its application. Mol Phylogenet Evol 26, 1–7 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Xia X & Xie Z DAMBE: software package for data analysis in molecular biology and evolution. J Hered 92, 371–373 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Xia X DAMBE6: New Tools for Microbial Genomics, Phylogenetics, and Molecular Evolution. J Hered 108, 431–437, doi:10.1093/jhered/esx033 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidt HA, Strimmer K, Vingron M & von Haeseler A TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18, 502–504 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Nguyen LT, Schmidt HA, von Haeseler A & Minh BQ IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32, 268–274, doi:10.1093/molbev/msu300 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vrieze SI Model selection and psychological theory: a discussion of the differences between the Akaike information criterion (AIC) and the Bayesian information criterion (BIC). Psychol Methods 17, 228–243, doi:10.1037/a0027127 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Minh BQ, Nguyen MA & von Haeseler A Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol 30, 1188–1195, doi:10.1093/molbev/mst024 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siek Jeremy G., L. L-Q, and Lumsdaine Andrew. The Boost Graph Library: User Guide and Reference Manual (http://www.boost.org/libs/graph/doc/index.html). Addison-Wesley, Pearson Education Inc., 2002. xxiv+321pp. ISBN 0–201-72914–8 [Google Scholar]
- 33.Ramachandran SP An Optimal Minimum Spanning Tree Algorithm. Journal of the Association for Computing Machinery 49 16–34 (2002). [Google Scholar]
- 34.Hudson RR, Boos DD & Kaplan NL A statistical test for detecting geographic subdivision. Mol Biol Evol 9, 138–151, doi:10.1093/oxfordjournals.molbev.a040703 (1992). [DOI] [PubMed] [Google Scholar]
- 35.Hudson RR, Slatkin M & Maddison WP Estimation of levels of gene flow from DNA sequence data. Genetics 132, 583–589 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hudson RR A new statistic for detecting genetic differentiation. Genetics 155, 2011–2014 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Slatkin M Isolation by Distance in Equilibrium and Non-Equilibrium Populations. Evolution 47, 264–279, doi:10.1111/j.1558-5646.1993.tb01215.x (1993). [DOI] [PubMed] [Google Scholar]
- 38.Pond SL, Frost SD & Muse SV HyPhy: hypothesis testing using phylogenies. Bioinformatics 21, 676–679, doi:10.1093/bioinformatics/bti079 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Daniel LHA, Clark G Principles of Population Genetics 4th Edition
- 40.Lindsey RL et al. Complete Genome Sequences of Two Shiga Toxin-Producing Escherichia coli Strains from Serotypes O119:H4 and O165:H25. Genome Announc 3, doi:10.1128/genomeA.01496-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huson DH Drawing rooted phylogenetic networks. IEEE/ACM Trans Comput Biol Bioinform 6, 103–109, doi:10.1109/TCBB.2008.58 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Spada E et al. Use of the minimum spanning tree model for molecular epidemiological investigation of a nosocomial outbreak of hepatitis C virus infection. J Clin Microbiol 42, 4230–4236, doi:10.1128/JCM.42.9.4230-4236.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Butler T Haemolytic uraemic syndrome during shigellosis. Trans R Soc Trop Med Hyg 106, 395–399, doi:10.1016/j.trstmh.2012.04.001 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Kaper JB & O’Brien AD Overview and Historical Perspectives. Microbiol Spectr 2, doi:10.1128/microbiolspec.EHEC-0028-2014 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kupczok A et al. Rates of Mutation and Recombination in Siphoviridae Phage Genome Evolution over Three Decades. Mol Biol Evol 35, 1147–1159, doi:10.1093/molbev/msy027 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.