Abstract
Membrane curvature remodeling induced by amphipathic helices (AHs) is essential in many biological processes. Here we studied a model amphipathic peptide, M2AH, derived from influenza A M2. We are interested in how M2AH may promote membrane curvature by altering membrane physical properties. We used atomic force microscopy (AFM) to examine changes in membrane topographic and mechanical properties. We used electron paramagnetic resonance (EPR) spectroscopy to explore changes in lipid chain mobility and chain orientational order. We found that M2AH perturbed lipid bilayers by generating nanoscale pits. The structural data are consistent with lateral expansion of lipid chain packing, resulting in a mechanically weaker bilayer. Our EPR spectroscopy showed that M2AH reduced lipid chain mobility and had a minimal effect on lipid chain orientational order. The EPR data are consistent with the surface-bound state of M2AH that acts as a chain mobility inhibitor. By comparing results from different lipid bilayers, we found that cholesterol enhanced the activity of M2AH in inducing bilayer pits and altering lipid chain mobility. The results were explained by considering specific M2AH-cholesterol recognition and/or cholesterol-induced expansion of interlipid distance. Both AFM and EPR experiments revealed a modest effect of anionic lipids. This highlights that membrane interaction of M2AH is mainly driven by hydrophobic forces. Lastly, we found that phosphatidylethanolamine (PE) lipids inhibited the activity of M2AH. We explained our data by considering interlipid hydrogen-bonding that can stabilize bilayer organization. Our results of lipid-dependent membrane modulations are likely relevant to M2AH-induced membrane restructuring.
Graphical Abstract
1. Introduction
Many membrane-associated proteins and peptides contain a structural motif called amphipathic helix (AH) such that polar residues are segregated on one face and hydrophobic residues are clustered on the opposite face [1]. Examples include the Huntington's disease-related α-synuclein [2] and membrane lytic antimicrobial peptides [3]. One important consequence of the amphipathic characteristic is that AHs have a large preference for interacting with organellar and plasma membranes. In a membrane environment, the polar face of AHs prefers to be exposed to the aqueous medium, whereas the hydrophobic face of AHs prefers to interact with membrane's hydrocarbon core. The unique disposition of AHs has been attributed to physiological functions of many AH-containing proteins [4]. For example, the lipid biosynthetic enzyme, CTP:phosphocholine cytidylyltransferase (CCT), senses membrane deficiency of phosphatidylcholine; membrane binding of CCT is facilitated by folding the M domain into an AH [5].
Membrane surface binding AHs will cause an imbalanced stress exerted on lipid headgroup and acyl chains. (We use "surface binding" as a general term to describe that an AH is mainly located near the lipid headgroup region.) One physical manifestation is a modified membrane curvature. Enhancement of membrane curvature by AH-containing proteins plays an essential role in many biological functions. Secretory protein trafficking from the endoplasmic reticulum (ER) to the Golgi requires vesicle biogenesis; the N-terminal AH of Sar1p, one of five core COPII proteins involved in forming cargo-loaded vesicles, was found to sculpt ER membranes into highly curved structures [6]. Neural transmission requires synaptic vesicle retrieval from synaptic plasma membranes. Several proteins have been identified to use AH motifs to facilitate synaptic vesicle endocytosis by inducing membrane curvature. A few of them are endophilin [7-9], amphiphysin [10], and Epsin [11].
In addition to generating membrane curvature, AHs can also function as a membrane curvature sensor. N-terminal AHs of several nonstructural proteins of hepatitis C virus (HCV) were found to mediate virus binding to intracellular lipid droplets with high curvature – binding to lipid droplets is a critical step for the organization of the virus replication complex [12, 13]. Interestingly, the antiviral protein viperin used an AH motif to inhibit HCV by competitively binding to lipid droplets [14]. Lipid packing is affected by membrane curvature; larger curvature will result in more packing defects. The Golgi-associated protein ArfGAP1 was found to use an AH motif called amphipathic lipid packing sensor (ALPS) to distinguish between curved and flat membranes [15]
M2 of influenza A virus is a versatile protein composed of an extracellular domain, a transmembrane helix, and a cytoplasmic domain. M2 forms a low-pH activated proton channel [16]. The channel activity of M2 is essential for virus unpacking. At a later stage of the virus cycle, M2 was found to localize at the neck of budding virions, facilitating virion scission [17, 18]. M2 is also required for the formation of filamentous virion [19]. It seems that M2 is capable of modulating membrane curvature, so the budding neck can be narrowed below a threshold to complete virion release. Truncation of the M2 cytoplasmic domain caused a reduction in virion budding and release [20, 21]. The cytoplasmic domain of M2 contains a membrane-associated AH called M2AH that spans residues 47–62 [22]. Biophysical studies have shown that M2AH can cause budding of giant unilamellar vesicles (GUVs) [19]; the addition of M2AH to lipid assemblies in a cubic phase revealed that M2AH increased the negative Gaussian curvature [23]. M2AH can also generate positive Gaussian curvature as evidenced by the formation of vesicle buds and tubules [19, 24]. Conversely, mutation of M2AH was found to inhibit budding of transfected virus in vivo [25].
Membrane curvature modulation requires overcoming an energy penalty imparted by the curved geometry. In this paper, we studied how M2AH may compensate the curvature-associated energy penalty by altering membrane physical properties. In particular, we used high-resolution atomic force microscopy (AFM) and electron paramagnetic resonance (EPR) spectroscopy to explore changes in membrane material properties. Our AFM imaging experiment allows direct visualization of local membrane structures, and our AFM-based forced force spectroscopy experiment can reveal modulations of lipid membrane mechanical properties within a nanoscopic regime. Our EPR measurements probe lipid chain mobility and lipid chain orientational order near a spin probe. The obtained biophysical data highlight that M2AH may facilitate membrane curvature reorganization by altering membrane material properties. Viral pathogens exploit lipid complexity of host cell membranes [26]. Membranes with different lipid compositions can aid or negate AH recruitment by altering lipid packing defects [27]. To determine the role of different lipid species in mediating M2AH-membrane interactions, we selectively studied model lipid membranes containing cholesterol (Chol), anionic phosphatidylglycerol (PG), and phosphatidylethanolamine (PE) lipids. By determining the impacts of M2AH on physical properties of lipid bilayers containing different lipid species, our study provides useful insights into the role of various lipids in regulating M2AH-mediated membrane restructuring.
2. Materials and Methods
Lipids including 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylcholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylethanolamine (POPE), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1'-rac-glycerol) (POPG), 1,2-dihexanoyl-sn-glycero-3-phosphocholine (DHPC), 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC), and Chol were purchased from Avanti Polar Lipids (Alabaster, AL). 5-doxyl stearic acid (5-SASL) was purchased from Sigma-Aldrich (St. Louis, MO). The 16-residue peptide M2AH of sequence F47FKSIYRFFEHGLKRG62 was synthesized by Genscript. The vendor confirmed peptide purity >90% by HPLC and mass spectroscopy. Fresh peptide stock solution was prepared by dissolving lyophilized powder in 10 mM HEPES pH 7.4. Unless noted otherwise, all ratios and percentages are molar based in this paper.
2.1. Atomic force microscopy (AFM)
Appropriate ratios of lipid stock solutions were mixed to obtain different lipid mixtures. Organic solvents were removed by a gentle stream of nitrogen gas, followed by vacuum pumping for ~2 hours. Lipid films were hydrated in 10 mM HEPES pH 7.4. We used a Sonic Dismembrator and a 3-mm microprobe to produce small unilamellar vesicles (SUVs). A Multimode 8 AFM (Bruker, Santa Barbara, CA) was used for solution AFM imaging (room temperature). Experimental procedure has been reported elsewhere [28-30]. Mica-supported planar bilayers were prepared by injecting SUVs into a fluid cell. Planar bilayers were formed by the vesicle rupture and fusion mechanism [31, 32]. Excess SUVs that did not participate in bilayer formation were removed. The peak-force quantitative nanomechanics (QNM) mode was used for bilayer imaging. AFM height images were acquired using a DNP-S10 probe at a scan rate of ~1 Hz. The obtained height images were leveled by subtracting a polynomial background. Image analysis was performed using in-house developed Matlab scripts.
We also used AFM to perform force spectroscopy measurements on the same bilayer used for AFM imaging. An automated force-ramping function was used for the force measurements as a function of the piezo z-position. A z-range of 100 nm and a tip speed of 200 nm/s were used. To describe the position of the AFM tip, we used a parameter s, which is the distance between the mica surface and the tip end position.
2.2. Electron paramagnetic resonance (EPR) spectroscopy
EPR measurements on liposomes.
Liposomes were prepared as described in Hope et al. [33] and Szoka et al. [34]. Lipids in chloroform were mixed in a glass tube and dried as thin films under a stream of nitrogen gas. 1–2 mol% of 5-SASL was added to the lipid mixture. Lipid films were further dried by using a vacuum pump for ~16 hours to remove the residual organic solvent. The lipids were resuspended in 10 mM HEPES pH 7.4 by vortexing for 1–2 min and then subjected to ≥6 freeze/thaw cycles. The lipid suspension was extruded ≥30 times through a mini-extruder with a 100-nm polycarbonate membrane (Avanti Polar Lipids).
EPR spectra were collected using a Bruker E680 spectrometer in X-band (9.5 GHz) with a high sensitivity cavity (ER 4119HS) and a variable temperature controller (ER 4111VT), all from Bruker BioSpin GmbH, Rheinstetten, Germany. The equipment is located at the National High Magnetic Field Laboratory (NHMFL). EPR spectra of liposomes were acquired with 100 kHz modulation frequency, 2.5 mW incident microwave power (18 dB), 0.16 mT modulation amplitude, 81.92 ms time constant, 81.92 ms conversion time, and 15 mT scan width. Samples were loaded in 0.6 mm or 0.9 mm inner diameter glass capillary tubes.
EPR measurements on bicelles.
The lipid composition of each preparation was 5.6 μmol of DMPC, 1.55 μmol of DHPC, and 50 nmol of 5-SASL (0.7% of the total lipids). The Q ratio (Q=[DMPC]/[DHPC]) for these bicelles was 3.6. The M2AH peptide was co-dried with the lipids to obtain the desired peptide-to-lipid (P/L) ratios relative to the DMPC concentration. The dried film was rehydrated in 22.5 μL of 50 mM HEPES pH 7.0, and the suspension was repeatedly vortexed and sonicated in ice-cold water for ~15 min. The solution was then subjected to 4 freeze/thaw cycles. The resulting bicelle solution appeared transparent and viscous.
We followed a reported procedure to align bicelles in the magnetic field [30]. Briefly, TmCl3·6H2O was mixed with a bicelle/peptide solution for a final Tm3+/lipids molar ratio of 0.2. A glass tube containing the bicelle solution was placed in the EPR cavity at room temperature (295 K). Under a magnetic field of 1350 mT, the sample temperature was slowly raised to 318 K (about 15 min) and then slowly lowered to the desired temperature of 308 K. The EPR spectra of bicelles were acquired with 100 kHz modulation frequency, 4.9 mW incident microwave power (16 dB), 0.10 mT modulation amplitude, 40.96 ms time constant, 81.92 ms conversion time, and 15 mT scan width. Changes in lipid chain orientational order were determined by the order parameter S2o[35, 36]. S20 describes the orientational order of lipid acyl chains relative to each other near the spin probe. A reduction in S20 upon peptide binding reflects a loss of chain order. To calculate S20, EPR spectra were simulated and fitted using the Multicomponent program (http://www.chemistry.ucla.edu/directory/hubbell-wayne-l) as described before [30]. Briefly, the experimental spectra of all P/L ratios were simulated using the same magnetic and motional parameters, while the orientational order parameter S20, calculated from the orienting potential coefficient c20, is varied to fit the spectra [30].
3. Results
3.1. AFM experiments on planar lipid bilayers
POPC.
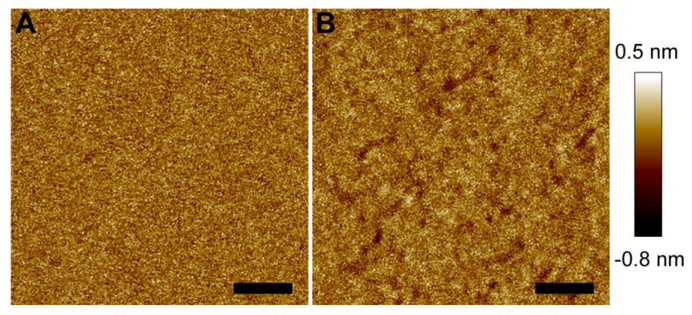
We conducted solution AFM imaging to visually detect structural changes of a POPC bilayer caused by M2AH. Figure 1A shows the topographic image of the control bilayer over an area of 500 nm × 500 nm. The bilayer exhibited a homogeneous structure. Height profile along an arbitrary path is shown in Fig. S1-A (Supplementary Material). The height of each pixel is within a range of ~−0.2–0.2 nm. This is further corroborated by the height probability distribution of the entire bilayer (Fig. S1-D). Gaussian curve fitting to the height distribution resulted in a full width at half maximum (FWHM) of 0.25 nm. Once the quality of the control bilayer was confirmed, we used a syringe pump to inject a 4 μM M2AH solution. Figure 1B shows the peptide-treated bilayer structure. The bilayer exhibited many small pits as indicated by the darker color (Fig. 1B). AFM probes lipid bilayer structure in a three-dimensional space. The sub-Å sensitivity along the bilayer normal direction enables us to assess the depth scale of the bilayer pits. Height profile along an arbitrary path revealed that the bilayer pits have a depth of ~0.2–0.5 nm. We also plotted the height probability distribution of the modified bilayer (Fig. S1-D, SI). The FWHM became 0.27 nm, a slightly larger value than that of the intact bilayer. An increase in the FWHM reflects that M2AH enhanced the roughness of the POPC bilayer, primarily contributed by the emerging pits.
Figure 1.
AFM height images of a POPC bilayer before (A) and after (B) being exposed to 4 μM M2AH. Height scale is indicated by the color bar at the right. Scale bars are 100 nm.
POPC/Chol.
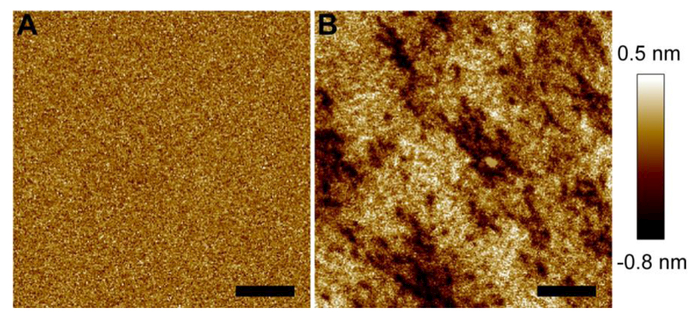
We used a POPC+30%Chol bilayer to study the role of Choi in mediating M2AH-bilayer interactions. AFM height image of the control bilayer is shown in Fig. 2A. Exposure to 4 μM M2AH resulted in significant changes of the bilayer topographic structure (Fig. 2B). Many pits were observed over the entire region. At selective locations where small pits overlap, larger pits with deeper penetration depth were generated. This is confirmed by the height profile along a path going over two larger pits (Fig. S2, SM). The larger pits have a depth of ~1 nm compared to <0.5 nm for the smaller ones. Calculation of the height distribution of the entire bilayer showed that the FWHM is 0.26 nm for the intact bilayer. The height distribution for the peptide-treated bilayer deviates from a Gaussian curve; nevertheless, it is clear that the width of the height distribution curve is increased significantly by M2AH. The results from POPC and POPC+30%Chol bilayers indicate that Chol augmented the capability of M2AH in inducing bilayer pits.
Figure 2.
AFM height images of a POPC+30%Chol bilayer before (A) and after (B) being exposed to 4 μM M2AH. Scale bars are 200 nm.
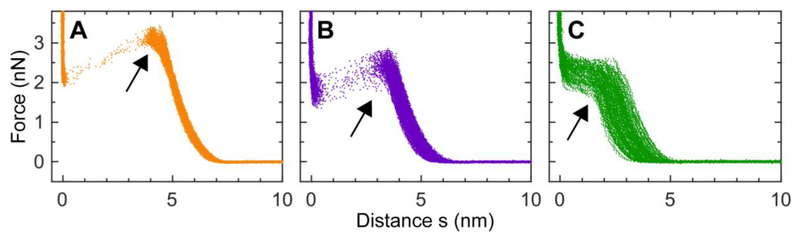
In parallel with the study of bilayer topographic structure, we used AFM-based force spectroscopy to explore mechanical properties of mica-supported planar bilayers. Figure 3 shows force curves for a POPC+30%Chol bilayer exposed to 0, 4, and 10 μM M2AH. The force exerted by an AFM tip increases as the tip indents into a mica-supported bilayer. A maximum force called the puncture force is reached when the bilayer is punctured by the tip (indicated by arrowheads in Fig. 3) [29]. For the intact bilayer, the puncture force is ~3 nN and the bilayer puncture takes place at a distance of ~4 nm away from the mica surface (located at zero distance). Exposure to M2AH caused a decrease in the puncture force and a shift of the puncture position. This is most evident at 10 μM M2AH. The puncture force became ~2 nN, and the puncture position was at ~2.5 nm. A smaller puncture force implies that the bilayer had weaker mechanical stability after treatment with M2AH. This conclusion agrees with the observation of many pits in the topographic image (Fig. 2).
Figure 3.
AFM force curves for a POPC+30%Chol bilayer exposed to 0 (A), 4 μM (B), and 10 μM (C) M2AH. Bilayer puncture is indicated by the black arrowheads.
POPC/POPG.
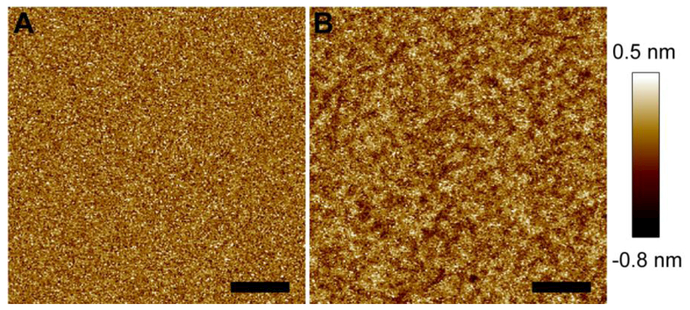
Host membranes contain many different species of anionic lipids, such as phosphatidylserine (PS), phosphatidylinositol (PI), and phosphatidic acid (PA). Because M2AH contains several basic residues, it is interesting to investigate how M2AH modulates anionic lipid bilayers. We used a POPC+20%POPG bilayer as a model system to evaluate the effect of anionic lipids in regulating M2AH-bilayer interactions. Figure 4 shows the AFM image of the bilayer before and after exposure to 4 μM M2AH. The intact bilayer has a smooth surface. Calculation of the height probability distribution revealed a FWHM of 0.31 nm. The addition of 4 μM M2AH caused many shallow pits, whose depth was ~0.2–0.5 nm (Fig. S3, SM). The shallow pits enhanced the roughness of the bilayer surface. This is evidenced by a larger FWHM value, 0.37 nm, obtained from the height probability distribution (Fig. S3, SM). Compared to the POPC bilayer, the presence of the anionic POPG lipid seems to slightly increase the degree of bilayer perturbation exerted by M2AH.
Figure 4.
AFM height images of a POPC+20%POPG bilayer before (A) and after (B) being exposed to 4 μM M2AH. Scale bars are 100 nm.
POPC/POPE.
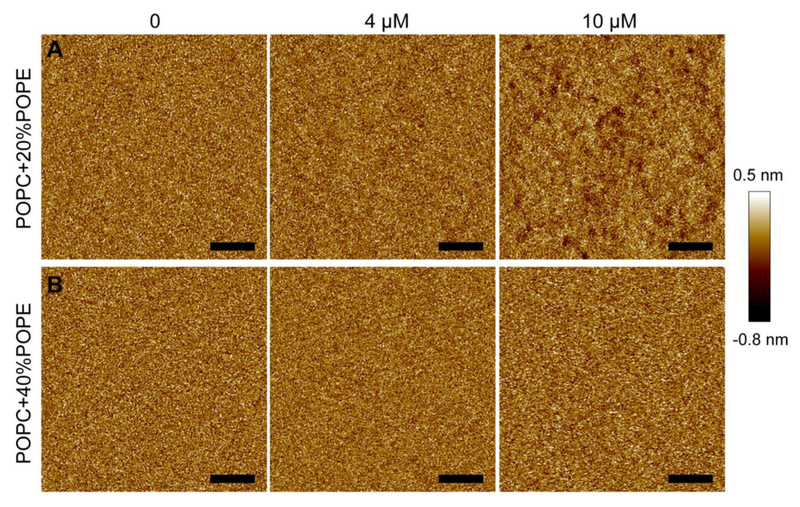
PE lipids have a conical shape owing to the smaller headgroup compared to the acyl chains. We used two POPC/POPE bilayers to study the role of PE lipids in modulating bilayer perturbations conferred by M2AH. Figure 5A shows AFM images of a POPC+20%POPE bilayer exposed to 0, 4, and 10 μM M2AH. The intact bilayer has a homogeneous topography. Height probability distribution resulted in a FWHM of 0.31 nm (Fig. S4, SM). Exposure to 4 μM M2AH did not cause noticeable changes in the bilayer structure. This is confirmed by height distribution analysis, which showed a FWHM of 0.31 nm. Increasing the peptide concentration to 10 μM yielded many small pits that are similar to those observed in the POPC bilayer treated with 4 μM M2AH. It seems that the presence of 20% POPE increased the concentration of the peptide that is required to generate bilayer pits. This statement is further supported by the study of a POPC+40%POPE bilayer (Fig. 5B). Increasing the peptide concentration to 10 μM did not yield discernable pits. The resistance of the POPC+40%POPE bilayer to M2AH treatment is also reflected by the height probability distributions (Fig. S5, SM). Collectively, the content of POPE in POPC/POPE bilayers correlates with the concentration of M2AH that is required to generate bilayer pits; higher contents of POPE suppressed the capability of M2AH in modulating bilayer structure.
Figure 5.
AFM height images of bilayers composed of POPC+20%POPE (A) and POPC+40%POPE (B) exposed to 0, 4, and 10 μM M2AH. Scale bars are 100 nm.
3.2. EPR spectroscopy experiments on lipid vesicles
Lipid chain mobility.
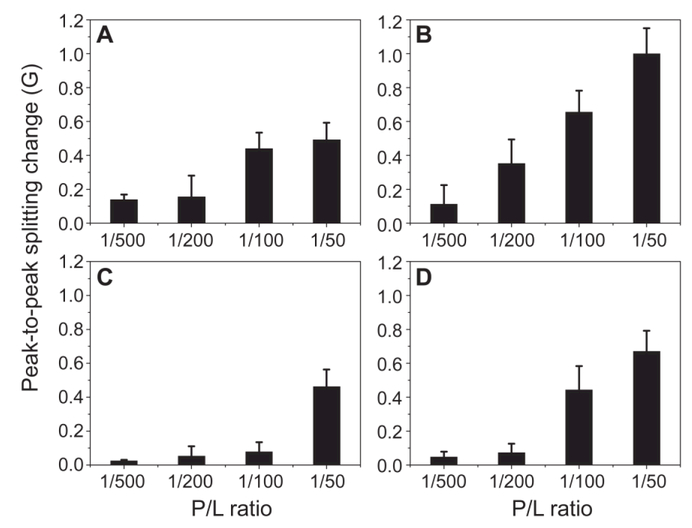
Next, we used EPR spectroscopy to investigate the effect of M2AH binding on lipid chain mobility. Chain mobility in a lipid bilayer can be influenced by the binding of peptides and proteins [37, 38]. The analysis of EPR spectral line-shape changes upon macromolecule binding represents a suitable tool to determine lipid chain mobility because EPR spectra are affected by molecular motion [39, 40]. In particular, an increase in the spectral peak-to-peak splitting and/or a broadening of the spectral line-shape indicates slower molecular motion. We used 5-SASL as a spin probe to assess the chain mobility changes induced by M2AH. 5-SASL is a suitable spin probe because position five of the acyl chain has been demonstrated to be the most sensitive to mobility changes in several molecular models [35, 41]. It is noteworthy that EPR spectroscopy relies on the spin probe; therefore, EPR spectral changes mainly reflect changes of the microenvironment near the spin probe. The lipid mobility changes are quantified using the motional parameter 2A//, i.e., the peak-to-peak splitting of the spectrum, upon peptide binding to lipid membranes with different lipid compositions and peptide-to-lipid (P/L) ratios. Liposomes with four lipid compositions were studied: POPC, POPC+30%Chol, POPC+40%POPE, and POPC+20%POPG. An example of typical EPR spectra is shown in Fig. S6 (SM). For all lipid compositions studied, the peak-to-peak splitting of the EPR spectrum was increased on the addition of M2AH (Fig. 6). The result can be interpreted as reduced mobility of lipid acyl chains. Moreover, it is evident that POPC+30%Chol liposomes showed greater mobility changes upon M2AH binding than that of POPC alone. Indeed, at P/L of 1/50, POPC liposomes displayed a 0.5±0.1 Gauss (G) splitting increase, whereas POPC+30%Chol liposomes displayed a 1.0±0.2 G increase. The 2A// value for POPC+20%POPG liposomes was also increased upon M2AH binding; the value was 0.7±0.1 G at P/L of 1/50, slightly larger than the increase for POPC liposomes. For POPC+40%POPE liposomes, the peak-to-peak splitting changes were much smaller than those of POPC liposomes; negligible increases were observed for P/L≤1/100, and a moderate increase was found at P/L=1/50. Collectively, the lipid chain mobility data are in good agreement with our AFM results.
Figure 6.
EPR spectroscopy revealed M2AH-induced changes of lipid chain mobility for liposomes composed of (A) POPC, (B) POPC+30%Chol, (C) POPC+40%POPE, and (D) POPC+20%POPG. For each lipid composition, four P/L ratios were examined. Positive changes of the peak-to-peak splitting (2A//) reflect lower lipid chain mobility.
Lipid chain orientational order.
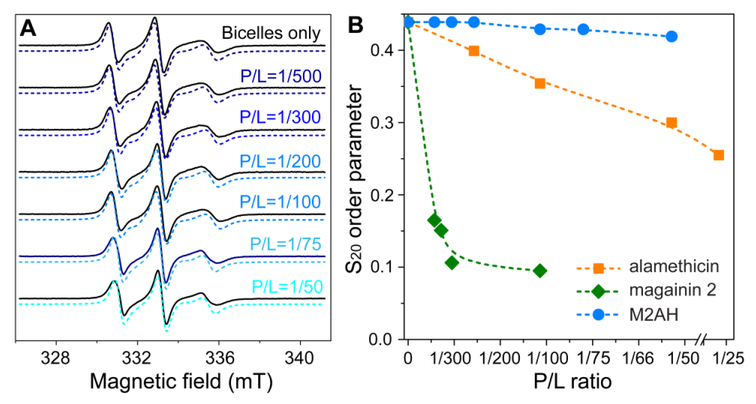
We used EPR and magnetically aligned DMPC/DHPC bicelles to study how M2AH perturbs lipid chain orientational order [30]. Importantly, the configuration of M2AH in bicelles can be deduced by comparing the order parameter S20 from M2AH and reference peptides. Again, we note that the obtained EPR data mainly reflect the effect of M2AH on the microenvironment near the spin probe. The result is summarized in Fig. 7. For aligned bicelles without the peptide, EPR spectrum showed three broad peaks that are typical for a nitroxide spin probe partially oriented in the magnetic field with an orientational order parameter S20 of 0.439. The addition of M2AH at P/L ratios from 1/500 to 1/50 resulted in minuscule changes in spectral line-shape and the order parameter S20. The lowest S20 value was found to be 0.419 at P/L of 1/50. The trend of S20 induced by M2AH is in stark contrast to those of the two reference peptides: alamethicin and magainin 2 [35, 42]. Alamethicin is known to form voltage-dependent barrel-stave pores [43]. This type of ordered pores caused a slow and modest decrease of S20. Magainin 2 induces more substantial membrane perturbations by forming toroidal pores and acting ultimately as a detergent [44-46]; as such, magainin 2 caused a rapid decrease of S20. Compared to the two reference peptides, M2AH had a minimal effect on the orientational order parameter, suggesting that M2AH interacts with lipid membranes without forming transmembrane pores.
Figure 7.
Lipid chain orientational order analysis upon M2AH binding studied using EPR and bicelles. (A) Experimental (solid lines) and simulated (dashed lines) EPR spectra of DMPC/DHPC bicelles containing the spin label 5-SASL. The bilayer normal of the bicelles is aligned parallel to the magnetic field at 308 K. (B) Plot of the orientational order parameter S20 as a function of the P/L ratio. For comparison, the reported values for alamethicin and magainin 2 are also displayed.
4. Discussion
4.1. Mechanism of membrane perturbation by M2AH.
M2 of influenza A virus is a multifaceted protein that is required for ion channel activity and virion release. M2AH is located at the intercellular domain of M2. The relevance of M2AH in virus cycle is highlighted by the finding that M2AH is necessary and sufficient to cause membrane scission of progeny virions [25]. Here we used high-resolution AFM imaging to explore structural remodeling of lipid bilayers induced by M2AH. We studied several lipid bilayers composed of different lipid species. A common theme is the observation of shallow pits on bilayer surfaces with a depth scale of ~0.2–1 nm. M2AH is unstructured in aqueous solution; it converts into an α-helix once bound to lipid membranes [24]. Helix analysis using a web server [47] predicted that M2AH has a hydrophobicity of 0.442, a hydrophobic moment of 0.630, and a net charge is +3. The amphipathic characteristic represents a driving force for membrane association of M2AH. Unlike the amphipathic peptide melittin, which produced transmembrane defects (or pores) [28], we did not observe transmembrane defects induced by M2AH. This leads to the proposition that M2AH is oriented parallel to the bilayer surface. Binding of M2AH to the lipid headgroup region is also supported by the lack of changes in the orientational order parameter S20 derived from our EPR measurements of magnetically aligned bicelles. Insertion of M2AH is likely facilitated by interactions with lipid headgroup through the polar face of M2AH, whereas partial intercalation into bilayer hydrocarbon core is driven by hydrophobic interactions through the hydrophobic face of M2AH. The wedge-shaped insertion of M2AH expands lipid packing laterally, resulting in a smaller bilayer thickness locally surrounding the peptide. It is noteworthy that bilayer thinning by surface-bound AHs has been widely reported [48-50]. When there are several M2AH molecules located nearby, the cooperative effect of bilayer thinning due to multiple M2AH can cause the development of shallow pits observed in our AFM images. Note that M2 forms a tetramer in lipid membranes. The proximity of four copies of M2AH in a tetramer assembly makes the cooperative bilayer thinning effect even more appealing. Expansion of lipid packing laterally may increase the probability of lipid bulging from a flat bilayer, thus changing bilayer curvature.
In addition to AFM imaging, we used AFM-based force spectroscopy to explore changes in membrane mechanical properties induced by M2AH. The obtained force spectroscopy data revealed that the bilayer became weaker as manifested by a smaller bilayer puncture force. The magnitude of the bilayer puncture force serves as a fingerprint for bilayer mechanical stability [29]. The reduced bilayer puncture force by M2AH highlights a mechanically weaker bilayer. The impaired bilayer mechanical stability is also reflected by a shift of the position where bilayer puncture takes place. A similar position shift was reported for bilayers treated with membrane destabilizer ethanol [51]. The bending energy of a lipid bilayer is dependent on the bending rigidity [52]. Because M2AH caused a mechanically weaker (softer) bilayer, the corresponding bending rigidity is likely decreased [29]. Therefore, M2AH may assist membrane bending by affecting membrane mechanical properties. Our mechanical perspective agrees with other reports. For example, a reduction in bilayer surface tension was used to explain endophilin A1-induced membrane shape transition [53].
We used EPR spectroscopy to examine lipid chain mobility modulated by M2AH. An increase in the spectral peak-to-peak splitting was observed for almost all lipid compositions – the lipid-dependent effect is discussed below. This suggests that M2AH decreases lipid chain mobility after inserting into lipid bilayers. The reduced chain mobility can be understood by considering that membrane surface-bound M2AH acts as a mobility barrier; lipid chains in the vicinity of M2AH has reduced mobility owing to steric interactions. As a result, the overall chain mobility is decreased. It is conceivable that the impact on lipid chain mobility will be different when AHs are orientated parallel to the bilayer normal because the number of neighboring lipids is different. The membrane surface binding state of M2AH is corroborated by our EPR measurements on magnetically aligned bicelles. We showed that compared to the two reference peptides, M2AH had a minimal effect on the orientational order parameter S20. Because both barrel-stave and toroidal pores caused substantial changes of S20, we suggest that a surface-bound state of M2AH is likely to exert little impact on lipid chain orientational order.
4.2. The effect of Choi.
Chol plays an important role in influenza virus cycle; the level of Chol was found to affect virus infectivity [54]. Chol is an effective modulator of membrane material properties. The modified membrane thickness and mechanical aspects have been attributed to Chol-induced protein sorting [55]. Chol also plays an important role in forming ordered membrane domains called rafts [56]. Influenza virion was suggested to bud and pinch off at a budozone that is formed by coalescing rafts of the plasma membrane [57]. This view is supported by findings that newly formed viral membranes are enriched in raft lipids. Interestingly, HIV particles also have higher contents of raft-associated Chol and sphingolipids [58]. To facilitate influenza virion budding and scission, M2 is sequestered at the edge of the budozone. Interaction with Chol was suggested to be responsible for governing M2 localization. Indeed, M2 prepared from infected cells contained 0.5–0.9 Chol per monomer [59].
Biophysical studies have shown that Chol immobilizes M2 in lipid bilayers; Chol-stabilized M2 has a larger helical content [60]. Sequence analysis revealed that M2AH contains a consensus Chol-binding CRAC motif [59]. However, removal of the CRAC motif did not affect virus replication, suggesting that other factors dominate M2-Chol association [61, 62]. Similarly, alteration of the M2 CRAC motif did not change virus replication but does decrease the virulence in a mouse model [63]. Although the CRAC motif may not be responsible for M2-Chol binding, several studies indicated that M2AH is the primary region where Chol associates with M2. For example, M2AH-induced GUV budding is dependent on Chol content [25]. Examination of M2AH in lipid bilayers without and with Chol revealed that Chol suppressed the dynamics of M2AH [64]. More recently, nuclear magnetic resonance (NMR) spectroscopy determined strong interactions between Chol and hydrophobic residues of M2AH [65]. Collectively, M2AH is responsible for M2 binding to Chol, although the transmembrane domain of M2 may also participate in Chol binding by hydrophobic interactions [66].
We used AFM imaging and ERP spectroscopy to investigate the effect of M2AH on Chol-containing lipid bilayers. Both measurements showed that Chol enhanced the activity of M2AH in modulating lipid bilayer structure and organization. Specifically, bilayer pits became more pronounced in terms of the size and depth when 30% Chol was added to POPC bilayers. In parallel, the decrease in lipid chain mobility by M2AH was markedly enhanced by the addition of 30% Chol to POPC liposomes. An earlier study showed that GUV budding induced by M2AH was dependent on Chol content [25]. We examined the effect of the Chol content by using a POPC+17%Chol bilayer. AFM topographic images of the planar bilayer exposed to 4 and 10 μM M2AH are shown in Fig. S7 (SM). Shallow pits were observed at 4 μM MAH; the depth of the pits became larger at 10 μM M2AH. By comparing AFM images of POPC, POPC+17%Chol, and POPC+30%Chol exposed to 4 μM MAH, we conclude that the resulting bilayer pits by 4 μM M2AH are not linearly related to the Chol content; bilayer pits are similar at 0 and 17% Chol, while bilayer pits are markedly elevated at 30% Chol.
The enhanced activity of M2AH in the presence of 30% Chol can be caused by Chol binding to hydrophobic residues of M2AH [65]. Chol-M2AH recognition increases the binding affinity of M2AH to the bilayer surface, accompanied by a reduced peptide dynamics [64]. Alternatively, Chol expands lipid headgroup-headgroup spacing, resulting in larger packing voids in the headgroup region; the induced packing voids can augment the insertion of M2AH [67]. Note that the induced packing voids do not have a linear relationship with the Chol content. The umbrella model predicted that lipid headgroups shield Chol (and lipid acyl chains) from the aqueous medium [68]. At low Chol content, the bilayer can shield Chol by straightening lipid acyl chains. Therefore, lipid headgroup-headgroup spacing remains similar. The chain straightening effect is saturated at a certain Chol level [69], after which the bilayer is no longer able to shield Chol completely. The additional Chol expands lipid headgroup-headgroup spacing, causing more packing voids in the headgroup region to allow M2AH binding and insertion.
The effect of Chol on M2AH-induced vesicle budding has been reported earlier [25]. It was found that M2AH caused vesicle budding at a Chol content ≤17%, whereas higher Chol contents inhibited vesicle budding. The authors also found that inward and outward budding was induced at low and high peptide concentrations, respectively. Moreover, M2AH induced vesicle leakage at low Chol contents. In general, the reported study showed that the activity of M2AH in inducing vesicle budding is enhanced by low Chol contents and inhibited by high Chol contents. The switch is at 17% Chol. This result is different from our observation that 30% Chol increased the activity of M2AH in generating bilayer pits (AFM data) and changing lipid chain mobility (ERP data). We do not have a clear answer to the difference. A possible explanation is that vesicle budding does not correlate with bilayer pits or lipid chain mobility. For example, bilayers can have modified pits and chain mobility without undergoing shape transition.
4.3. The effect of anionic lipids.
M2AH has a net charge of +3, contributed by two lysine, two arginine, and one glutamate residues. The presence of anionic lipids in lipid membranes might enhance M2AH binding by electrostatic interactions. The significance of anionic lipids has been well documented for many membrane-associated proteins. One example is α-synuclein, which contains an AH motif; the binding affinity of α-synuclein to lipid membranes was significantly enhanced in the presence of anionic lipids [70]. We investigated the role of anionic lipids in mediating M2AH-membrane interactions by using a POPC+20%POPG bilayer. Our AFM measurements showed that POPG caused a slightly larger degree of bilayer perturbation induced by 4 μM M2AH. This is evidenced by the distribution of bilayer pits, as well as the bilayer surface roughness. A similar result was obtained from our EPR measurements, which showed that at the same P/L ratio, lipid chain mobility change is comparable for POPC and POPC/POPG liposomes. Together, the anionic lipid does not seem to have the ability of significantly impacting M2AH-induced membrane modulation. The lack of dramatic changes between POPC and POPC/POPG indicate that M2AH binding to lipid bilayers is mainly driven by hydrophobic interactions [24]. The dominant role of the hydrophobic driving force is supported by alanine substitution study of M2AH; the mutated peptide with a smaller hydrophobicity was more dynamic and penetrated less into lipid bilayers [71]. Another possible explanation for the mild impact of anionic lipids in our study is that regardless of initial peptide binding, which could be larger for anionic lipid-containing bilayers, the determinant for changes in bilayer pits and lipid chain mobility is the availability of bilayer packing voids to accommodate M2AH insertion. It is possible that both POPC and POPC/POPG bilayers have similar packing voids; therefore, the resulting bilayer changes caused by M2AH are similar.
4.4. The effect of PE lipids.
PE lipids have a conical shape owing to their smaller amino headgroup. Concomitantly, lipid assemblies (e.g., monolayer and bilayer) composed of PE lipids have a negative spontaneous curvature [72]. When PE lipids are confined in a flat bilayer, there is an energy penalty called "curvature strain energy"; protein activity can be regulated by altering the stored curvature strain energy [73]. The smaller PE headgroup also yields more packing voids in the headgroup region compared to PC bilayers. The difference in packing voids (or defects) has been attributed to different binding behaviors of several AHs, such as the H0 helix of endophilin [74] and the ALPS motif [75]. Another important feature of PE lipids is that the amine group is a good hydrogen bond donor. Interlipid hydrogen-bonding initiated by the amine moiety of PE lipids contributes to enhanced bilayer stability [76, 77]. This is manifested by a smaller area per PE lipid than that of the PC counterpart [78]. Similarly, the gel-to-fluid phase transition temperature is higher for PE than for PC bilayers (https://avantilipids.com/). Hydrogen-bonding renders PE-containing membranes more stable against membrane modulators. For instance, PE bilayers are less susceptible to ethanol-facilitated lipid desorption [79]. Our recent study of an amphipathic peptide derived from the Prion protein also showed that the peptide exhibited distinctive perturbations on PC and PE lipid vibrational dynamics [30].
Here we used POPC/POPE bilayers to examine the effect of PE lipids on M2AH-induced bilayer perturbation. Our AFM measurements showed that the degree of bilayer perturbation is inversely related to the PE content in the POPC/POPE binary mixtures. This is evidenced by the diminished bilayer pits at higher POPE contents. Similarly, our EPR measurements revealed a much weaker impact of M2AH on lipid chain mobility in the presence of 40% POPE (P/L≤1/100). Because the available packing voids in POPC/POPE bilayers are proportional to the PE content, our experimental data highlight that packing voids are not the determinant in regulating M2AH insertion into POPC/POPE bilayers. Interlipid hydrogen bonding facilitated by PE lipids creates a network that likely negates the binding and insertion of M2AH. This view is in line with the observation that PE lipids conferred a reduced capability of ethanol in extracting lipid molecules [79]. Studies of three influenza strains revealed that the PE content of virion envelopes is significantly elevated compared to the host cell [80]. The rise in PE content in newly released virions suggests that PE lipids are sequestered at the budozone. Because M2AH interacts unfavorably with PE lipids, it is reasonable to argue that M2AH does not cluster in the budozone during virion release. This view is supported by the finding that in progeny virions, the number of M2 is markedly lower than the budozone-enriched glycoproteins (e.g., hemagglutinin and neuraminidase) [17]. PE-induced exclusion of M2 from the budozone may be important for M2 to cluster at the budozone edge during virion release.
5. Conclusions
In this paper, we used high-resolution AFM and EPR spectroscopy measurements to investigate membrane modulations caused by an amphipathic peptide, M2AH, which is located at the intracellular domain of influenza A M2 protein. We used lipid bilayers with several lipid compositions to explore different lipid species in regulating M2AH-membrane interactions. We found that M2AH perturbed lipid bilayer structures by forming nanoscale pits, yielding a rough texture of the bilayer surfaces. A summarization of lipid bilayer height probability distributions before and after exposure to 4 μM M2AH is shown in Fig. S8 (SM). The obtained results are explained by considering that the wedge-type insertion of M2AH expands lipid packing locally. We also used AFM-based force spectroscopy to examine modulations of membrane mechanical properties. M2AH caused a mechanically weaker bilayer as reflected by a smaller bilayer puncture force and a shift of the bilayer puncture position. Our EPR measurements showed that M2AH decreased lipid chain mobility and had a minimal effect on the lipid chain orientational order. The EPR data are consistent with the surface-bound state of M2AH that acts as a mobility inhibitor. By comparing results from different lipid bilayers, we found that membrane modulation by M2AH is significantly enhanced by the presence of Chol. This is evidenced by the formation of more extensive bilayer pits and a larger decrease in lipid chain mobility. The effect of Chol can be explained by specific Chol-M2AH recognition and/or Chol-induced expansion of lipid-lipid distance. Both our AFM and EPR experiments revealed a modest effect of the anionic POPG lipid in mediating M2AH-induced bilayer perturbation. This result implies that membrane binding of M2AH is mainly driven by hydrophobic interactions [24]. Lastly, we found an inhibitory effect of the cone-shaped POPE lipid on M2AH-induced membrane remodeling. We explained our data by considering interlipid hydrogen-bonding that stabilizes bilayer organization. The obtained changes in bilayer structural and mechanical properties may play a role in M2AH-assisted membrane curvature remodeling that is required during virion budding and scission.
Supplementary Material
Highlights.
Amphipathic helices are important membrane curvature modulators
M2AH perturbs lipid bilayer by forming nanoscale pits
M2AH alters lipid chain mobility and chain orientational order
Lipid species determine membrane modulations caused by M2AH
Acknowledgements
This work is partly supported by the National Institutes of Health Award Number 1R15GM117531-01 to J. P.; the NHMFL UCGP Grant Number 5080 and the National Institutes of Health Award Number 5R03AI122860 to L. S. The EPR data were collected at the NHMFL supported by the NSF DMR-1157490 and the State of Florida.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Segrest JP, Deloof H, Dohlman JG, Brouillette CG, Anantharamaiah GM, Amphipathic Helix Motif - Classes and Properties, Proteins-Structure Function and Genetics, 8 (1990) 103–117. [DOI] [PubMed] [Google Scholar]
- [2].Jao CC, Hegde BG, Chen J, Haworth IS, Langen R, Structure of membrane-bound alpha-synuclein from site-directed spin labeling and computational refinement, P Natl Acad Sci USA, 105 (2008) 19666–19671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Dathe M, Wieprecht T, Structural features of helical antimicrobial peptides: their potential to modulate activity on model membranes and biological cells, Bba-Biomembranes, 1462 (1999) 71–87. [DOI] [PubMed] [Google Scholar]
- [4].Drin G, Antonny B, Amphipathic helices and membrane curvature, Febs Lett, 584 (2010) 1840–1847. [DOI] [PubMed] [Google Scholar]
- [5].Cornell RB, Membrane lipid compositional sensing by the inducible amphipathic helix of CCT, Bba-Mol Cell Biol L, 1861 (2016) 847–861. [DOI] [PubMed] [Google Scholar]
- [6].Lee MCS, Orci L, Hamamoto S, Futai E, Ravazzola M, Schekman R, Sar1p N-terminal helix initiates membrane curvature and completes the fission of a COPII vesicle, Cell, 122 (2005) 605–617. [DOI] [PubMed] [Google Scholar]
- [7].Farsad K, Ringstad N, Takei K, Floyd SR, Rose K, De Camilli P, Generation of high curvature membranes mediated by direct endophilin bilayer interactions, J Cell Biol, 155 (2001) 193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Gallop JL, Jao CC, Kent HM, Butler PJG, Evans PR, Langen R, McMahon HT, Mechanism of endophilin N-BAR domain-mediated membrane curvature, Embo J, 25 (2006) 2898–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Jao CC, Hegde BG, Gallop JL, Hegde PB, McMahon HT, Haworth IS, Langen R, Roles of Amphipathic Helices and the Bin/Amphiphysin/Rvs (BAR) Domain of Endophilin in Membrane Curvature Generation, J Biol Chem, 285 (2010) 20164–20170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJG, Evans PR, McMahon HT, BAR domains as sensors of membrane curvature: The amphiphysin BAR structure, Science, 303 (2004) 495–499. [DOI] [PubMed] [Google Scholar]
- [11].Ford MGJ, Mills IG, Peter BJ, Vallis Y, Praefcke GJK, Evans PR, McMahon HT, Curvature of clathrin-coated pits driven by epsin, Nature, 419 (2002) 361–366. [DOI] [PubMed] [Google Scholar]
- [12].Brass V, Bieck E, Montserret R, Wolk B, Hellings JA, Blum HE, Penin F, Moradpour D, An amino-terminal amphipathic alpha-helix mediates membrane association of the hepatitis C virus nonstructural protein 5A, J Biol Chem, 277 (2002) 8130–8139. [DOI] [PubMed] [Google Scholar]
- [13].Elazar M, Liu P, Rice CM, Glenn JS, An n-terminal amphipathic helix in hepatitis C virus (HCV) NS4B mediates membrane association, correct localization of replication complex proteins, and HCV RNA replication, J Virol, 78 (2004) 11393–11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hinson ER, Cresswell P, The antiviral protein, viperin, localizes to lipid droplets via its N-terminal amphipathic alpha-helix, P Natl Acad Sci USA, 106 (2009) 20452–20457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Drin G, Casella JF, Gautier R, Boehmer T, Schwartz TU, Antonny B, A general amphipathic alpha-helical motif for sensing membrane curvature, Nat Struct Mol Biol, 14 (2007) 138–146. [DOI] [PubMed] [Google Scholar]
- [16].Pinto LH, Holsinger LJ, Lamb RA, Influenza-Virus M2 Protein Has Ion Channel Activity, Cell, 69 (1992) 517–528. [DOI] [PubMed] [Google Scholar]
- [17].Leser GP, Lamb RA, Influenza virus assembly and budding in raft-derived microdomains: A quantitative analysis of the surface distribution of HA, NA and M2 proteins, Virology, 342 (2005) 215–227. [DOI] [PubMed] [Google Scholar]
- [18].Rossman JS, Lamb RA, Influenza virus assembly and budding, Virology, 411 (2011) 229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Rossman JS, Jing XH, Leser GP, Balannik V, Pinto LH, Lamb RA, Influenza Virus M2 Ion Channel Protein Is Necessary for Filamentous Virion Formation, J Virol, 84 (2010) 5078–5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].McCown MF, Pekosz A, Distinct domains of the influenza A virus M2 protein cytoplasmic tail mediate binding to the M1 protein and facilitate infectious virus production, J Virol, 80 (2006) 8178–8189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Iwatsuki-Horimoto K, Horimoto T, Noda T, Kiso M, Maeda J, Watanabe S, Muramoto Y, Fujii K, Kawaoka Y, The cytoplasmic tail of the influenza A virus M2 protein plays a role in viral assembly, J Virol, 80 (2006) 5233–5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sharma M, Yi MG, Dong H, Qin HJ, Peterson E, Busath DD, Zhou HX, Cross TA, Insight into the Mechanism of the Influenza A Proton Channel from a Structure in a Lipid Bilayer, Science, 330 (2010) 509–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Schmidt NW, Mishra A, Wang J, DeGrado WF, Wong GCL, Influenza Virus A M2 Protein Generates Negative Gaussian Membrane Curvature Necessary for Budding and Scission, J Am Chem Soc, 135 (2013) 13710–13719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Martyna A, Bahsoun B, Badham MD, Srinivasan S, Howard MJ, Rossman JS, Membrane remodeling by the M2 amphipathic helix drives influenza virus membrane scission, Sci Rep-Uk, 7 (2017) 44695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Rossman JS, Jing XH, Leser GP, Lamb RA, Influenza Virus M2 Protein Mediates ESCRT-Independent Membrane Scission, Cell, 142 (2010) 902–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].van der Meer-Janssen YPM, van Galen J, Batenburg JJ, Helms JB, Lipids in host-pathogen interactions: Pathogens exploit the complexity of the host cell lipidome, Prog Lipid Res, 49 (2010) 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Vanni S, Hirose H, Barelli H, Antonny B, Gautier R, A sub-nanometre view of how membrane curvature and composition modulate lipid packing and protein recruitment, Nat Commun, 5 (2014) 4916. [DOI] [PubMed] [Google Scholar]
- [28].Pan JJ, Khadka NK, Kinetic Defects Induced by Melittin in Model Lipid Membranes: A Solution Atomic Force Microscopy Study, J Phys Chem B, 120 (2016) 4625–4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Khadka NK, Teng P, Cai JF, Pan JJ, Modulation of lipid membrane structural and mechanical properties by a peptidomimetic derived from reduced amide scaffold, Bba-Biomembranes, 1859 (2017) 734–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Pan JJ, Sahoo PK, Dalzini A, Hayati Z, Aryal CM, Teng P, Cai JF, Gutierrez HR, Song LK, Membrane Disruption Mechanism of a Prion Peptide (106-126) Investigated by Atomic Force Microscopy, Raman and Electron Paramagnetic Resonance Spectroscopy, J Phys Chem B, 121 (2017) 5058–5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Reviakine I, Brisson A, Formation of supported phospholipid bilayers from unilamellar vesicles investigated by atomic force microscopy, Langmuir, 16 (2000) 1806–1815. [Google Scholar]
- [32].Leonenko ZV, Carnini A, Cramb DT, Supported planar bilayer formation by vesicle fusion: the interaction of phospholipid vesicles with surfaces and the effect of gramicidin on bilayer properties using atomic force microscopy, Bba-Biomembranes, 1509 (2000) 131–147. [DOI] [PubMed] [Google Scholar]
- [33].Hope MJ, Bally MB, Webb G, Cullis PR, Production of Large Unilamellar Vesicles by a Rapid Extrusion Procedure - Characterization of Size Distribution, Trapped Volume and Ability to Maintain a Membrane-Potential, Biochim Biophys Acta, 812 (1985) 55–65. [DOI] [PubMed] [Google Scholar]
- [34].Szoka F, Olson F, Heath T, Vail W, Mayhew E, Papahadjopoulos D, Preparation of Unilamellar Liposomes of Intermediate Size (0.1-0.2-Mumm) by a Combination of Reverse Phase Evaporation and Extrusion through Polycarbonate Membranes, Biochim Biophys Acta, 601 (1980) 559–571. [DOI] [PubMed] [Google Scholar]
- [35].Bortolus M, Dalzini A, Toniolo C, Hahm KS, Maniero AL, Interaction of hydrophobic and amphipathic antimicrobial peptides with lipid bicelles, J Pept Sci, 20 (2014) 517–525. [DOI] [PubMed] [Google Scholar]
- [36].Budil DE, Lee S, Saxena S, Freed JH, Nonlinear-least-squares analysis of slow-motion EPR spectra in one and two dimensions using a modified Levenberg-Marquardt algorithm, J Magn Reson Ser A, 120 (1996) 155–189. [Google Scholar]
- [37].Ge MT, Freed JH, An Electron-Spin-Resonance Study of Interactions between Gramicidin-a' and Phosphatidylcholine Bilayers, Biophys J, 65 (1993) 2106–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kiricsi M, Horvath LI, Dux L, Pali T, Spin label EPR studies of the effect of gramicidin S on lipid chain dynamics, Journal of Molecular Structure, 563 (2001) 469–475. [Google Scholar]
- [39].Hubbell WL, Mcconnel.Hm, Molecular Motion in Spin-Labeled Phospholipids and Membranes, J Am Chem Soc, 93 (1971) 314–326. [DOI] [PubMed] [Google Scholar]
- [40].Hubbell WL, Hm Mcconnel, Orientation and Motion of Amphiphilic Spin Labels in Membranes, P Natl Acad Sci USA, 64 (1969) 20–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Bortolus M, Parisio G, Maniero AL, Ferrarini A, Monomeric Fullerenes in Lipid Membranes: Effects of Molecular Shape and Polarity, Langmuir, 27 (2011) 12560–12568. [DOI] [PubMed] [Google Scholar]
- [42].Bortolus M, De Zotti M, Formaggio F, Maniero AL, Alamethicin in bicelles: Orientation, aggregation, and bilayer modification as a function of peptide concentration, Bba-Biomembranes, 1828 (2013) 2620–2627. [DOI] [PubMed] [Google Scholar]
- [43].Fox RO, Richards FM, A Voltage-Gated Ion Channel Model Inferred from the Crystal-Structure of Alamethicin at 1.5-a Resolution, Nature, 300 (1982) 325–330. [DOI] [PubMed] [Google Scholar]
- [44].Matsuzaki K, Magainins as paradigm for the mode of action of pore forming polypeptides, Bba-Rev Biomembranes, 1376 (1998) 391–400. [DOI] [PubMed] [Google Scholar]
- [45].Papo N, Shai Y, Exploring peptide membrane interaction using surface plasmon resonance: Differentiation between pore formation versus membrane disruption by lytic peptides, Biochemistry-Us, 42 (2003) 458–466. [DOI] [PubMed] [Google Scholar]
- [46].Li C, Salditt T, Structure of magainin and alamethicin in model membranes studied by x-ray reflectivity, Biophys J, 91 (2006) 3285–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Gautier R, Douguet D, Antonny B, Drin G, HELIQUEST: a web server to screen sequences with specific alpha-helical properties, Bioinformatics, 24 (2008) 2101–2102. [DOI] [PubMed] [Google Scholar]
- [48].Mecke A, Lee DK, Ramamoorthy A, Orr BG, Holl MMB, Membrane thinning due to antimicrobial peptide binding: An atomic force microscopy study of MSI-78 in lipid bilayers, Biophys J, 89 (2005) 4043–4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Pfefferkorn CM, Heinrich F, Sodt AJ, Maltsev AS, Pastor RW, Lee JC, Depth of alpha-Synuclein in a Bilayer Determined by Fluorescence, Neutron Reflectometry, and Computation, Biophys J, 102 (2012) 613–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Faust JE, Desai T, Verma A, Ulengin I, Sun TL, Moss TJ, Betancourt-Solis MA, Huang HW, Lee T, McNew JA, The Atlastin C-terminal Tail Is an Amphipathic Helix That Perturbs the Bilayer Structure during Endoplasmic Reticulum Homotypic Fusion, J Biol Chem, 290 (2015) 4772–4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Stetter FWS, Hugel T, The Nanomechanical Properties of Lipid Membranes are Significantly Influenced by the Presence of Ethanol, Biophys J, 104 (2013) 1049–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Helfrich W, Elastic Properties of Lipid Bilayers - Theory and Possible Experiments, Z Naturforsch C, C 28 (1973) 693–703. [DOI] [PubMed] [Google Scholar]
- [53].Shi Z, Baumgart T, Membrane tension and peripheral protein density mediate membrane shape transitions, Nat Commun, 6 (2015) 5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Sun XJ, Whittaker GR, Role for influenza virus envelope cholesterol in virus entry and infection, J Virol, 77 (2003) 12543–12551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Lundbaek JA, Andersen OS, Werge T, Nielsen C, Cholesterol-induced protein sorting: An analysis of energetic feasibility, Biophys J, 84 (2003) 2080–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Pike LJ, Rafts defined: a report on the Keystone Symposium on Lipid Rafts and Cell Function, J Lipid Res, 47 (2006) 1597–1598. [DOI] [PubMed] [Google Scholar]
- [57].Scheiffele P, Rietveld A, Wilk T, Simons K, Influenza viruses select ordered lipid domains during budding from the plasma membrane, J Biol Chem, 274 (1999) 2038–2044. [DOI] [PubMed] [Google Scholar]
- [58].Brugger B, Glass B, Haberkant P, Leibrecht I, Wieland FT, Krausslich HG, The HIV lipidome: A raft with an unusual composition, P Natl Acad Sci USA, 103 (2006) 2641–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Schroeder C, Heider H, Moncke-Buchner E, Lin TI, The influenza virus ion channel and maturation cofactor M2 is a cholesterol-binding protein, Eur Biophys J Biophy, 34 (2005) 52–66. [DOI] [PubMed] [Google Scholar]
- [60].Liao SY, Fritzsching KJ, Hong M, Conformational analysis of the full-length M2 protein of the influenza A virus using solid-state NMR, Protein Sci, 22 (2013) 1623–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Thaa B, Tielesch C, Moller L, Schmitt AO, Wolff T, Bannert N, Herrmann A, Veit M, Growth of influenza A virus is not impeded by simultaneous removal of the cholesterol-binding and acylation sites in the M2 protein, J Gen Virol, 93 (2012) 282–292. [DOI] [PubMed] [Google Scholar]
- [62].Thaa B, Siche S, Herrmann A, Veit M, Acylation and cholesterol binding are not required for targeting of influenza A virus M2 protein to the hemagglutinin-defined budozone, Febs Lett, 588 (2014) 1031–1036. [DOI] [PubMed] [Google Scholar]
- [63].Stewart SM, Wu WH, Lalime EN, Pekosz A, The cholesterol recognition/interaction amino acid consensus motif of the influenza A virus M2 protein is not required for virus replication but contributes to virulence, Virology, 405 (2010) 530–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Kim SS, Upshur MA, Saotome K, Sahu ID, McCarrick RM, Feix JB, Lorigan GA, Howard KP, Cholesterol-Dependent Conformational Exchange of the C-Terminal Domain of the Influenza A M2 Protein, Biochemistry-Us, 54 (2015) 7157–7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Ekanayake EV, Fu RQ, Cross TA, Structural Influences: Cholesterol, Drug, and Proton Binding to Full-Length Influenza A M2 Protein, Biophys J, 110 (2016) 1391–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Elkins MR, Williams JK, Gelenter MD, Dai P, Kwon B, Sergeyev IV, Pentelute BL, Hong M, Cholesterol-binding site of the influenza M2 protein in lipid bilayers from solid-state NMR, P Natl Acad Sci USA, 114 (2017) 12946–12951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Egashira M, Gorbenko G, Tanaka M, Saito H, Molotkovsky J, Nakano M, Handa T, Cholesterol modulates interaction between an amphipathic class A peptide, Ac-18A-NH2, and phosphatidylcholine bilayers, Biochemistry-Us, 41 (2002) 4165–4172. [DOI] [PubMed] [Google Scholar]
- [68].Huang JY, Feigenson GW, A microscopic interaction model of maximum solubility of cholesterol in lipid bilayers, Biophys J, 76 (1999) 2142–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Pan JJ, Tristram-Nagle S, Nagle JF, Effect of cholesterol on structural and mechanical properties of membranes depends on lipid chain saturation, Phys Rev E, 80 (2009) 021931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Davidson WS, Jonas A, Clayton DF, George JM, Stabilization of alpha-synuclein secondary structure upon binding to synthetic membranes, J Biol Chem, 273 (1998) 9443–9449. [DOI] [PubMed] [Google Scholar]
- [71].Herneisen AL, Sahu ID, McCarrick RM, Feix JB, Lorigan GA, Howard KP, A Budding-Defective M2 Mutant Exhibits Reduced Membrane Interaction, Insensitivity to Cholesterol, and Perturbed Interdomain Coupling, Biochemistry-Us, 56 (2017) 5955–5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Kollmitzer B, Heftberger P, Rappolt M, Pabst G, Monolayer spontaneous curvature of raft-forming membrane lipids, Soft Matter, 9 (2013) 10877–10884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Davies SMA, Epand RM, Kraayenhof R, Cornell RB, Regulation of CTP: Phosphocholine cytidylyltransferase activity by the physical properties of lipid membranes: An important role for stored curvature strain energy, Biochemistry-Us, 40 (2001) 10522–10531. [DOI] [PubMed] [Google Scholar]
- [74].Cui HS, Lyman E, Voth GA, Mechanism of Membrane Curvature Sensing by Amphipathic Helix Containing Proteins, Biophys J, 100 (2011) 1271–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Vanni S, Vamparys L, Gautier R, Drin G, Etchebest C, Fuchs PFJ, Antonny B, Amphipathic Lipid Packing Sensor Motifs: Probing Bilayer Defects with Hydrophobic Residues, Biophys J, 104 (2013) 575–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Boggs JM, Lipid Intermolecular Hydrogen-Bonding - Influence on Structural Organization and Membrane-Function, Biochim Biophys Acta, 906 (1987) 353–404. [DOI] [PubMed] [Google Scholar]
- [77].Pink DA, McNeil S, Quinn B, Zuckermann MJ, A model of hydrogen bond formation in phosphatidylethanolamine bilayers, Bba-Biomembranes, 1368 (1998) 289–305. [DOI] [PubMed] [Google Scholar]
- [78].Kucerka N, van Oosten B, Pan JJ, Heberle FA, Harroun TA, Katsaras J, Molecular Structures of Fluid Phosphatidylethanolamine Bilayers Obtained from Simulation-to-Experiment Comparisons and Experimental Scattering Density Profiles, J Phys Chem B, 119 (2015) 1947–1956. [DOI] [PubMed] [Google Scholar]
- [79].Slater SJ, Ho C, Taddeo FJ, Kelly MB, Stubbs CD, Contribution of Hydrogen-Bonding to Lipid Lipid Interactions in Membranes and the Role of Lipid Order - Effects of Cholesterol, Increased Phospholipid Unsaturation, and Ethanol, Biochemistry-Us, 32 (1993) 3714–3721. [DOI] [PubMed] [Google Scholar]
- [80].Ivanova PT, Myers DS, Milne SB, McClaren JL, Thomas PG, Brown HA, Lipid Composition of the Viral Envelope of Three Strains of Influenza Virus-Not All Viruses Are Created Equal, Acs Infect Dis, 1 (2015) 435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.