Abstract
β-Lactamases are hydrolytic enzymes capable of opening the β-lactam ring of antibiotics such as penicillin, thus endowing the bacteria that produce them with antibiotic resistance. Of particular medical concern are metallo-β-lactamases (MBLs), with an active site built around coordinated Zn cations. MBLs are pan-reactive enzymes that can break down almost all classes of β-lactams, including such last-resort antibiotics as carbapenems. They are not only broad-spectrum-reactive but are often plasmid-borne (e.g., the New Delhi enzyme, NDM), and can spread horizontally even among unrelated bacteria. Acquired MBLs are encoded by mobile genetic elements, which often include other resistance genes, making the microbiological situation particularly alarming. There is an urgent need to develop MBL inhibitors in order to rescue our antibiotic armory. A number of such efforts have been undertaken, most notably using the 3D structures of various MBLs as drug-design targets. Structure-guided drug discovery depends on the quality of the structures that are collected in the Protein Data Bank (PDB) and on the consistency of the information in dedicated β-lactamase databases. We conducted a careful review of the crystal structures of class B β-lactamases, concluding that the quality of these structures varies widely, especially in the regions where small molecules interact with the macromolecules. In a number of examples the interpretation of the bound ligands (e.g., inhibitors, substrate/product analogs) is doubtful or even incorrect, and it appears that in some cases the modeling of ligands was not supported by electron density. For ten MBL structures, alternative interpretations of the original diffraction data could be proposed and the new models have been deposited in the PDB. In four cases, these models, prepared jointly with the authors of the original depositions, superseded the previous deposits. This review emphasizes the importance of critical assessment of structural models describing key drug design targets at the level of the raw experimental data. Since the structures reviewed here are the basis for ongoing design of new MBL inhibitors, it is important to identify and correct the problems with ambiguous crystallographic interpretations, thus enhancing reproducibility in this highly medically relevant area.
Keywords: antibiotic resistance; metallo-β-lactamase; New Delhi Metallo-β-lactamase (NDM); drug design target; metal coordination; structural databases; data mining; Protein Data Bank (PDB); reproducibility, model validation; structure re-deposition
1. Introduction – reproducibility crisis in biomedical research
A number of recent reports have brought to the attention of the scientific community the uncomfortable fact that a noticeable fraction of biomedical research cannot be reproduced (Minor et al., 2016; Prinz et al., 2011). This is an alarming trend, and even more alarming outlook, calling for immediate action. Not only are large resources of time, manpower, and money (estimated at $28 billion a year in the US alone (Freedman et al., 2015)) invested in potentially useless endeavors, but also false hopes may be raised for patients and attention may be diverted in the erroneous directions. Of great concern is also the prospect of undermining the confidence in science and of corrupting the scientific system itself (Rupp et al., 2016). In particular, the “ripple effect” of false results can propagate very quickly in the scientific literature, as illustrated by Shabalin et al. (Shabalin et al., 2015).
An area of biomedical research that has been traditionally viewed as particularly solid and almost free of errors is protein crystallography. Its tremendous power lies in the fact that, on one hand, its models are based in each case on hundreds of thousands of very accurate physical measurements (the intensities of X-ray diffraction on protein crystals), and on the other hand each model can (and must) be confronted with a huge volume of prior structural information collected in the Protein Data Bank (PDB) (Berman et al., 2000). It must be realized, however, that the crystallographic models are only atomic interpretations of the electron density maps, which are the primary product of any X-ray diffraction experiment (Wlodawer et al., 2008, 2013). In most cases those interpretations are straightforward and unambiguous; however, with difficult structures, suboptimal data, and – especially – with an overdose of wishful thinking, the subjective element may lead on occasion to questionable results. Hence, every structure presented as a model for further work (e.g., drug design, biochemical analysis, etc.) needs to be carefully validated with all available tools, including inspection of electron density maps, ligand-omit maps, and so-called polder maps; monitoring of statistical parameters, such as Rfree with and without problematic ligands; and analysis of ligand interactions, such as H-bond distances, van der Waals contacts, etc. The very isolated cases of structure fabrication (discussed in (Wlodawer et al., 2013)) can be disregarded in this context, as they have been rapidly identified and eliminated, leading in fact to the development of much improved tools for structure validation and error detection (Read et al., 2011). More long-term harm is done by structural models deposited in the PDB that at face value do not raise red flags for the unsuspecting consumers (not necessarily familiar with structural biology), but on closer scrutiny demonstrate a number of problems. The most serious problems are related to modeling ligands without the support of electron density (Pozharski et al., 2013; Wlodawer et al., 2017). For example, in a recent paper (Shabalin et al., 2015) we showed that some protein complexes of cisplatin and carboplatin deposited in the PDB had troubling interpretational problems with modeling of the metal-coordination ligands. In another study, no experimental evidence could be found for the presence of ligands in a number of protein complexes, including antibody complexes (Wlodawer et al., 2017). In some cases, the structures could be significantly corrected (and re-deposited) and given an alternative interpretation, or at least the original interpretation had to be put in serious doubt. In the present review, we have critically assessed the PDB models of metallo-β-lactamases (MBLs), a group of enzymes that are of high clinical relevance.
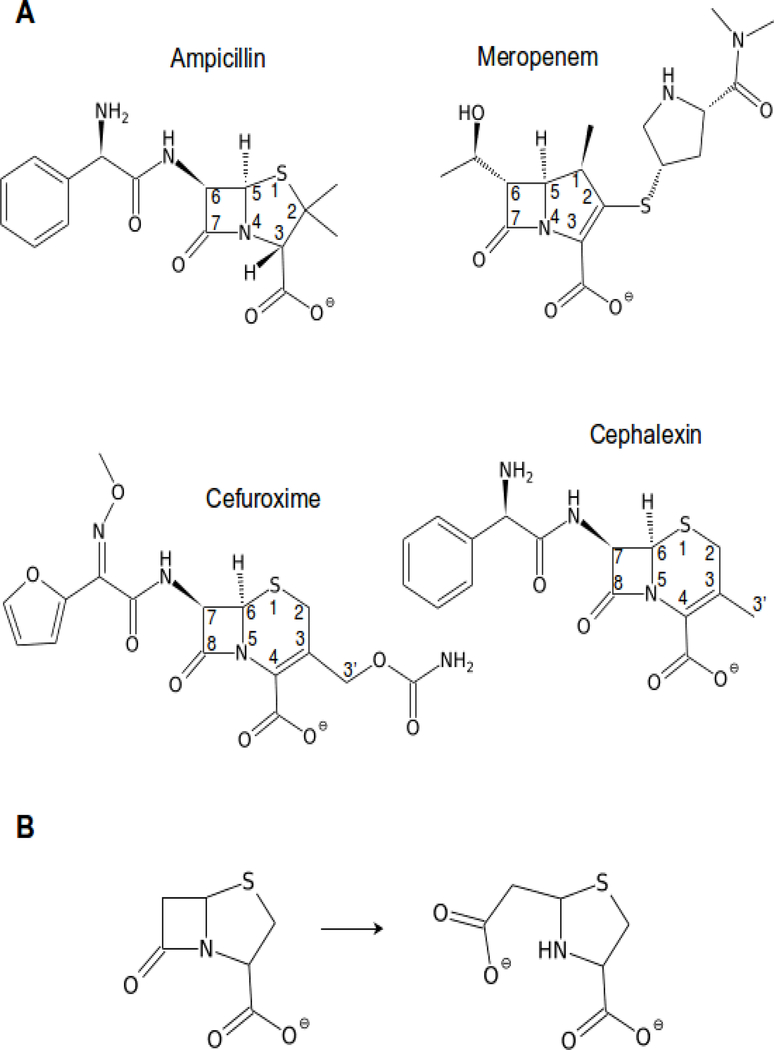
Metallo-β-lactamases are significantly contributing to the problem of antibiotic resistance, which has currently become one of the major health concerns (Davies and Davies, 2010; Frère et al., 2016; Mojica et al., 2016; Walsh et al., 2005; World Health Organization, 2017). MBLs are broad-spectrum hydrolases, capable of breaking down almost all β-lactam antibiotics, including carbapenems, which are considered the drugs of last resort (McKenna, 2013). Only monobactams are not hydrolyzed by MBLs. Fig. 1 shows a few examples of common β-lactams. The most straightforward approach to combating MBL-related antibiotic resistance would be to develop efficient inhibitors with as wide a spectrum as possible. Such inhibitors, used in combination with the existing β-lactam antibiotics, would rescue them for prolonged use. Toward this end, 235 MBL structures (as of December 4, 2017) have been determined in different laboratories and deposited in the PDB. In addition, references to those models, their classification, characterization, as well as literature sources, are being collected in a dedicated β-Lactamase Database (www.bldb.eu(Naas et al., 2017)).
Figure 1.
(A) Some of the common β-lactam antibiotics shown with systematic numbering of the β-lactam part and the fused ring. Ampicillin belongs to the penicillin class of β-lactams, cephalexin and cefuroxime are cephalosporins, while meropenem is an example of a carbapenem. (B) The hydrolysis reaction scheme, shown for a penicillin core example. In the case of unsaturated fused rings, tautomerization of the double bond can occur concurrently or following the hydrolysis.
In view of the above-mentioned problems with the quality of some structural models, we have analyzed over 150 crystallographic structures of class B β-lactamases (the B1, B2 and B3 subgroups). We applied stringent validation criteria to verify the original interpretation of the crystallographic data and to offer a revised interpretation for the most prominent cases. The encountered problems ranged from as severe as a seriously wrong structure (marked by impossible crystal packing, PDB ID 1ddk), or incorrectly traced protein backbone (3s0z), ligands placed without sufficient evidence (e.g., 1jt1, 2fu7, 4nq7, 5a5z), incorrectly modeled ligands (e.g., 1k07, 4exy, 4eyl, 4hky, 4rl0, 4rl2), to less severe cases of solvent molecules modeled in place of protein chain (3rkk), incorrect placement of side chains (e.g., 1sml), missed modifications of amino acid residues (e.g., cysteine oxidation, 3sfp), placement of a calcium cation instead of chloride anion (4awy), or the use of isotropic (instead of anisotropic) temperature factors (ADPs) in high-resolution refinement (1mqo). Proper review and assessment of crystallographic models is only possible in confrontation with the electron density maps, which in turn requires access to the original diffraction data. This is even more true if a given model needs correction. In our review of the structures of MBLs we ultimately chose ten representative models for re-refinement and PDB re-deposition. We only re-refined those models for which the experimental data were available, and we focused on structures with ligands bound at the Zn center. Our analysis should not only contribute to improved reproducibility in biomedical science but, by generating a thoroughly validated set of drug design targets, will provide more reliable structural information for the development of MBL inhibitors.
2. Model validation methods
No new experimental data were collected for this work and all analyses were based on data obtained by others and deposited in the PDB and/or https://proteindiffraction.org/. We reviewed over 150 deposited crystal structures of metallo-β-lactamases and we used the BLDB database (Naas et al., 2017) to identify the relevant structures of B1, B2, and B3 β-lactamases. Only the entries containing experimental diffraction data were subjected to our detailed review.
When available, electron density maps were downloaded from the Uppsala Electron Density Server EDS (Kleywegt et al., 2004); otherwise they were calculated from the structure factor data deposited together with the atomic coordinates in the PDB, using programs from the CCP4 package (Winn et al., 2011). Anomalous-difference electron density maps were calculated if the deposited structure factor files contained the required information. Structures selected for re-refinement were refined with REFMAC5 version 5.8 (Murshudov et al., 2011) launched from HKL-3000 (Minor et al., 2006) or CCP4 (Winn et al., 2011), using either the structure factors as deposited in the PDB or diffraction data reprocessed by us with HKL-3000 (Otwinowski and Minor, 1997) after downloading the original images from the http://www.proteindiffraction.org server (Grabowski et al., 2016). Whenever possible, reflections selected for Rfree testing (Brünger, 1992) were the same as in the originally deposited datasets. Modeling of molecular oscillations by TLS parameters was tested for all structures in the later stages of refinement using the TLSMD Server (Painter and Merritt, 2006). The TLS parameters were kept if confirmed by a significantly improved Rfree and the Hamilton R-factor ratio test (Merritt, 2012) as implemented in HKL-3000. Coot (Emsley et al., 2010) was used for the visualization of the electron density maps and correction of the atomic models. The re-refinement procedure usually resulted in the addition of more solvent molecules than originally present; however, it was always ensured that the number of solvent atoms was within reasonable limits (Wlodawer et al., 2008). When needed, we used the ACHESYM server (Kowiel et al., 2014) for standardized placement of the structural models within the crystal unit cell. Various tools offered by Coot, MolProbity (Chen et al., 2010), and the PDB (Berman et al., 2000) were used for structure validation and quality assessment. Metal-binding sites were validated using the CheckMyMetal server (Zheng et al., 2017, 2014). When possible, we used polder maps (Liebschner et al., 2017) from the PHENIX suite (Adams et al., 2010) to validate our findings about the presence or absence of ligands. Molstack (Porebski et al., 2017), a web-based publishing platform, was used to provide the reader with a tool for interactive inspection of electron density maps and assessment of the interpretations presented in this work in comparison with those of the original authors. A directory with all the Molstack figures can be found at http://molstack.bioreproducibility.org/c/AnAP/. For each re-refined entry, we contacted the original authors and proposed joint co-authorship of the new deposit. Some authors of the original structures agreed to that procedure, while others either did not agree or did not answer the invitation, in which case the structures were re-deposited in our names only but with a clear explanation about the utilization of the original experimental data.
3. Revised structures of selected metallo-β-lactamases
Ligands without sufficient evidence to support their presence
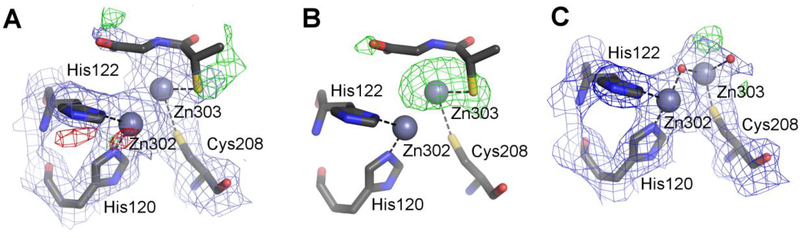
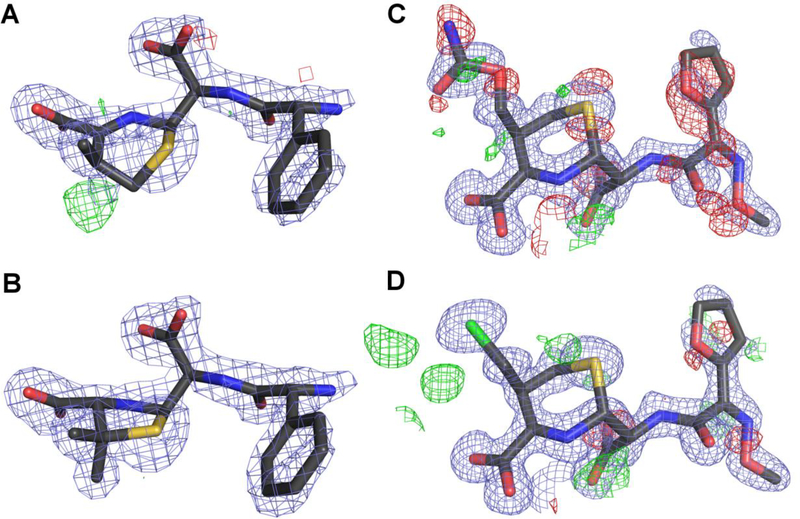
NDM-1 with tiopronin (PDB ID 5a5z)
A 2.6 Å structure of NDM-1, supposedly with the ligand tiopronin bound at the active site, was determined as part of a study entitled “Approved drugs containing thiols as inhibitors of metallo-β-lactamases: Strategy to combat multidrug-resistant bacteria” (Klingler et al., 2015). When this work was initially published, there was no accompanying PDB deposit. After our correspondence with the author and, subsequently, with the journal editor, the authors submitted the model coordinates and the experimental data to the PDB and published a correction to their paper, in which they list the PDB ID of their deposit.
As the title of the publication indicates, various drugs with thiol groups were tested for their MBL inhibitory properties. Tiopronin was found to be an inhibitor, and an X-ray structure of its complex with NDM-1 was presented as a proof of its direct binding to MBLs. Two ligand molecules, one per protein subunit, were modeled at 0.5 occupancy each. The PDB validation report shows RSR values of 0.48 and 0.32 and RSCC of 0.57 and 0.71 for chains A and C, respectively (the two independent protein chains were labeled A and C in the original structure, we have kept the labels for consistency). Moreover, the 2mFo-DFc map calculated in REFMAC5 without prior refinement does not show any discernible electron density for the ligand at the 1.0σ contour level next to any protein chain (Fig. 2A). The isotropic ADPs for the sulfur atoms of the originally modeled ligands are 154 and 125 Å2 with the all-ligand averages being 73.3 and 67.5 Å2, whereas the average values for all atoms within a 7 Å radius are 53.5 and 52.2 Å2 for chains A and C, respectively. Moreover, interactions of the ligand molecules, modeled in different conformations in the two protein chains, are suspicious from the chemical point of view. In both chains, the thiol group interacts with only one of the zinc ions while the typical MBL binding mode of thiol compounds is with the S atom located between the two active-site zinc ions. The carboxylate group of tiopronin in chain A does not form any hydrogen bonds or ionic contacts. In chain C the carboxylate interacts with Lys211, which is the major specificity-defining protein residue, other than the zinc centers.
Figure 2.
Various stages of modeling chain A of the structure of NDM-1 5a5z (PDB ID of the re-refined model: 5nbk). All maps are contoured at 1.0σ for 2mFo-DFc (blue) and +/−3.0σ for mFo-DFc (green/red). (A) The original coordinates with the electron density maps produced by REFMAC5 without prior refinement, with the putative tiopronin ligand visible at the top. (B) The original coordinates but with the omit map calculated from the final refinement phases by omitting only the water molecules modeled in the active site. (C) The re-refined entry 5nbk, with the final electron density maps. The ligands and the electron density maps, including omit maps, can be inspected using an interactive figure created with Molstack (http://molstack.bioreproducibility.org/project/view/2gniVO4HvvzPJnvb0Sd7/).
The authors state that the electron density map allowed for the placement of the ligand in several different conformations and, since it was not possible to distinguish them all, only the most prevalent was modeled. A figure in their paper (Fig. 3 in (Klingler et al., 2015)) shows an omit map to confirm the placement of the ligand. The presented map disagrees with the one calculated by us, which shows (at 3.0σ) only patches of positive difference density between the two Zn ions and at the sulfur atom of the ligand (Fig. 2B). Simply removing tiopronin from the model decreases Rfree by over 0.4% compared with the deposited coordinates refined using the same procedure.
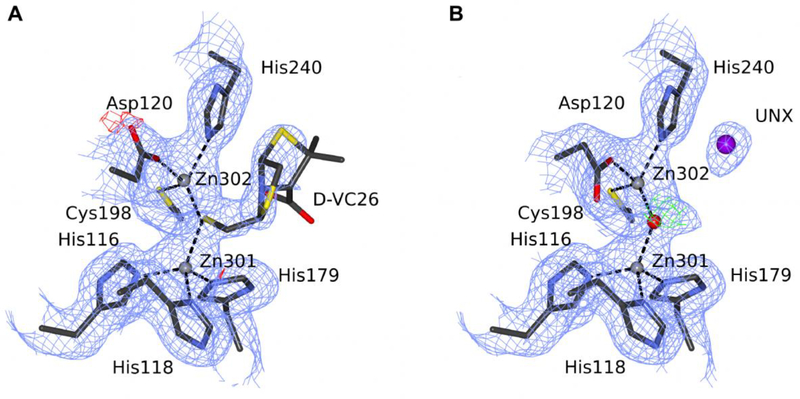
Figure 3.
The substrate-binding site of Bacillus cereus MBL. (A) The original 4nq7 deposit is modeled with compound D-VC26, and (B) the re-refined deposit 5w8w is modeled with an unknown atom. C atoms are shown in dark gray, N in blue, O in red (water as a sphere), S in yellow, Zn ions as gray spheres, and UNX atom as a magenta sphere. The electron density maps are contoured at 1.0σ for 2mFo-DFc (blue) and +/−3.0σ for mFo-DFc (green/red). Black dash lines represent coordination bonds. The ligands and the electron density maps, including omit maps, can be inspected using an interactive figure created with Molstack (http://molstack.bioreproducibility.org/project/view/ENLUIPQ0L5LVX6D4GI3B/).
The authors of the original deposit used the polder omit map from the PHENIX suite to guide ligand placement. We calculated such a map from our final coordinates after replacing active-site water molecules with the tiopronin ligand copied from the original coordinates (in both chains) and selecting it as the omit region. The resultant map, shown as an interactive Molstack figure (http://molstack.bioreproducibility.org/project/view/2gniVO4HvvzPJnvb0Sd7/), covers a large region of the protein pocket; however, it is inconsistent with the shape of the ligand. The most likely interpretation is that the map shows either bulk solvent density or noise, and this is also suggested by the PHENIX log file.
In addition to removing the ligand, we found that the Zn ions bound at site 2 (also called the CDH site) have isotropic ADPs of 101.6 and 82.8 Å2 in chains A and C, compared to 46.3 and 36.4 Å2 for Zn1 (the HHH site). Examination of the anomalous difference map showed a peak of 3.5/4.0σ at Zn2, compared to 6.5/7.0σ (chain A/C) for Zn1. In our model, the Zn2 ions were refined with 0.5 occupancy. We also included TLS parameters and added local NCS restraints (Table S1). These changes resulted in R/Rfree factors of 18.2/22.9%, which are significantly better than the values of 20.8/25.2% reported in the original PDB entry 5a5z (Table 1).
Table 1.
Selected parameters of the originally deposited (left column) and re-refined (right column) MBL structures described in the section ‘Ligands without sufficient evidence to support their presence’. Data quality statistics are according to REFMAC5 (Murshudov et al., 2011) output in the PDB file (Bijovet pairs merged). Ramachandran analysis and clashscores were calculated with the MolProbity server (Chen et al., 2010). Values in parentheses are for the last resolution shell.
| Protein | NDM-1 | Bacillus cereus 569/H BcII | Legionella gormanii FEZ-1 | |||
|---|---|---|---|---|---|---|
| PDB accession code | 5a5z | 5nbk | 4nq7 | 5w8w | 1jt1 | 5w90 |
| Resolution [Å] | 76.40–2.60 (2.67–2.60) | 76.40–2.60 (2.67–2.60) | 35.40–2.25 (2.31–2.25) | 35.40–2.25 (2.31–2.25) | 38.08–1.78 (1.78 – 1.88) | 38.08–1.78 (1.88 – 1.78) |
| No. of reflections refinement/Rfree | 16540/871 | 16540/871 | 8356/915 | 8356/915 | 20581/2334 | 20581/2334 |
| Completeness [%] | 99.9(100.0) | 99.9(100.0) | 87.4 (80.1) | 87.4 (80.1) | 91.5 (46.8) | 91.5 (46.8) |
| <Ι/σ> in the highest resolution shell | 1.89 | 1.89 | 2.0 | 2.0 | 8.4 | 8.4 |
| R / Rfree‡ [%] | 20.8/25.1 | 18.1/22.9 | 18.9/24.7 | 15.2/21.9 | 16.1/19.0 | 11.2/15.5 |
| Protein molecules in ASU | 2 | 2 | 1 | 1 | 1 | 1 |
| NCS restraints | Local | - | - | |||
| ADP parametrization | Isotropic | Isotropic | Isotropic | Isotropic+TLS | Isotropic | [sotropic+TLS |
| No. atoms | ||||||
| Protein | 3520 | 3441 | 1654 | 1668 | 2056 | 2066 |
| Ligands | 24 | 61 | 18 | 5 | 43 | 26 |
| Water | 70 | 109 | 19 | 108 | 255 | 425 |
| Ramachandran statistics [%] | ||||||
| Favored/outliers | 96.15/0.64 | 99.12/0.00 | 92.52/1.87 | 97.20/0.00 | 96.54/0.77 | 96.55/0.88 |
| R.m.s. deviations from target values | ||||||
| Bond lengths [Å] | 0.013 | 0.014 | 0.019 | 0.013 | 0.020 | 0.013 |
| Bond angles [°] | 0.95 | 1.53 | 2.0 | 1.60 | 1.90 | 1.5 |
| Clashscore/percentile | 4.88/99* | 2.31/100th | 7.54/97th | 0.89/100th | 3.15/98th | 1.69/97th |
where Fo and Fc are the observed and calculated structure factors, respectively. Rfree is calculated analogously for a subset of randomly selected reflections excluded from the refinement, as defined in (Brünger, 1992).
We modeled two water molecules in the active site and they refined well. There is still some positive electron density in the mFo-DFc map calculated for the re-refined coordinates with an empty active site (Fig. 2C). However, our attempts to model any ligand there, either tiopronin or such buffer components as tartrate or glycyl-glycine, have all failed, resulting in high ADPs for some atoms and a complete lack of 2mFo-DFc electron density even at a very low contour level. We conclude that the observed electron density does not support modeling any ligand there and is best interpreted as two water molecules, as represented in the new PDB deposit 5nbk.
Bacillus cereus Zn-dependent metallo-β-lactamase at pH 7 with compound D-VC26 (PDB ID 4nq7)
The structure 4nq7 was deposited in the PDB in 2013, without an accompanying publication. Our re-refinement of this structure resulted in a decrease of Rfree by 2.8%, partly owing to the use of TLS parametrization and the addition of 89 water molecules (Tables 1, S1). We corrected the conformation of the side chains of several residues, as well as mainchain (MC) torsion angles for others: 6, 7, 12–13 (MC), 15, 28, 31 (MC), 48, 54, 90 (active site residue), 91, 137, 153, 173, and 218 (numbering as in the original deposit). As in some other entries, we adjusted the sequence numbering to match the numbering used in Uniprot (P04190) and in more recent structures of the same protein (e.g., 5fqa). In addition, we replaced a potassium cation (A303) with a sulfate anion (Table S1).
The original deposit has one protein molecule in the ASU with the heterocyclic compound D-VC26 bound to the active-center Zn ions via a sulfur atom (Fig. 3). However, the ligand is placed in very weak electron density (Fig. 3A), and its ADP values are twice as high as in the environment, despite 0.6 occupancy. These observations prompted us to remove the ligand from the model. The resulting 2mFo-DFc map showed that there is insufficient evidence for the presence of a ligand in this position, as there was only a small patch of electron density present. A polder map, calculated from our final coordinates with the original ligand placed, revealed some similarity to the ligand, and it was reported by the PHENIX program as likely to show the ligand. However, the similarity of the polder map to the ligand is quite inconclusive (http://molstack.bioreproducibility.org/project/view/ENLUIPQ0L5LVX6D4GI3B/). During our correspondence with the authors of the original model, several additional arguments were laid why the ligand might be bound with low occupancy. Ultimately, however, it has been mutually agreed that the small electron density peak at the active site should be modeled as an unknown atom (UNX) (Fig. 3B), as the atom makes no contacts to prototropic groups, meaning that it is unlikely to be a water molecule. Most likely, it reflects a small fraction of the protein molecules that do contain D-VC26 or some other ligand in this location. The unknown atom UNX provides a good explanation of the electron density, at the same time alerting potential users of the model that there is something uncertain about the interpretation of the active site.
With the agreement and co-authorship of the original depositors, we superseded the original PDB entry 4nq7 with the improved one, 5w8w. The original deposit lacked crucial data processing statistics with multiple NULL entries. Unfortunately, neither the original diffraction images nor the data processing log files could now be located. Therefore, the structure was re-deposited with the same limited scaling statistics as the original one. The case of the PDB deposit 4nq7 represents an example of a very productive and mutually beneficial collaboration with the original depositors, resulting in a better model re-deposited in the PDB. The problem with locating original data shows the importance of resources that describe, organize, and maintain the original diffraction experiments (Grabowski et al., 2016).
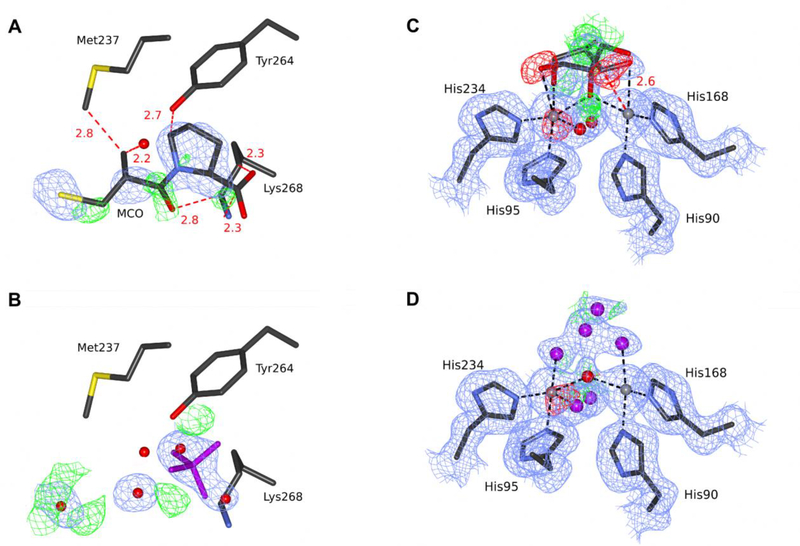
FEZ-1 from Legionella gormanii with D-captopril (PDB ID 1jt1)
The structure 1jt1 was deposited in 2001 with an accompanying paper published in 2003 (García-Sáez et al., 2003). The D-captopril ligand occupies a different position than in other captopril-bound MBL structures and does not interact with the zinc ions (the closest distance is 4.3 Å). As noted in the original publication, it is unusual when compared with other MBL complexes with thiol-containing ligands, which bind to the zinc ions via the thiol group. In addition, the ligand fits the electron density poorly (Fig. 4A, B), as also noted and diligently addressed by the authors: “due to the observed disorder, an occupancy of 1.0 was assigned to the ring of the D-proline, main-chain atoms and thiol group, and an occupancy of 0.0 for all the other atoms”. However, in the PDB model, there are six steric clashes between hydrophobic carbon and hydrophilic atoms of the protein and the ligand (Fig. 4A, B), suggesting that this mode of binding is highly unlikely.
Figure 4.
The substrate-binding site in two structures of FEZ-1 from Legionella gormanii. (A) The original 1jt1 deposit is modeled with D-captopril, and (B) the re-refined deposit 5w90 is modeled with an unknown ligand. (C) The original 1k07 deposit is modeled with a glycerol molecule in two conformations, and (D) the re-refined deposit 5wck is modeled with a set of unknown atoms. C atoms are shown in dark gray, N in blue, O in red (water as a sphere), S in yellow, Zn ions as gray spheres, UNX in magenta. The electron density maps are contoured at 1.0σ for 2mFo-DFc (blue) and +/−3.0σ for mFo-DFc (green/red). Black dash lines represent coordination bonds, red dash lines with distance labeled in Å represent steric clashes between the D-captopril molecule and surrounding residues. The ligands and the electron density maps, including omit maps, can be inspected using interactive figures created with Molstack (http://molstack.bioreproducibility.org/project/view/xpzboZ6XwokZ2DvYO9sH/http://molstack.bioreproducibility.org/project/view/xpzboZ6XwokZ2DvYO9sH/for 1jt1/5w90 and http://molstack.bioreproducibility.org/project/view/lSoPdEAwzOw4y8jthr2Z/for1k07/5wck).
Our re-refinement resulted in a decrease of Rfree by 3.5%, partly owing to almost doubling the number of water molecules and the use of TLS parametrization (Tables 1, S1). As for some other entries, we adjusted the sequence numbering to match the numbering used in Uniprot (Q9K578). Other than that, only small corrections were applied by us because the original protein model was in good agreement with the electron density. Notably, the signal-to-noise ratio in the outer resolution shell of the diffraction data is very high (8.4), but the original diffraction images were not available for re-processing and possible extension of resolution.
A polder map (http://molstack.bioreproducibility.org/project/view/xpzboZ6XwokZ2DvYO9sH/), calculated for our final coordinates with the original ligand placed, revealed some similarity to the ligand, but it was reported by PHENIX as more likely to show bulk solvent or noise. The omit electron density maps calculated after removal of D-captopril and structure re-refinement indicated the presence of two water molecules and an uninterpretable blob of density. The blob has some features of a sulfate ion, consistent with the presence of 0.2 M ammonium sulfate in the crystallization buffer. However, as the identification was only tentative, we modeled an unknown ligand (UNL) with 50% occupancy and two alternate water molecules. After inspection of the updated coordinates, the primary author of the original deposit noted that the modeled UNL does not fully reproduce the electron density, and that there are probably multiple fractional conformations (and multiple entities) present. Nevertheless, since we found it impossible to interpret the density in a better way, it was mutually agreed that UNL was a safe interpretation.
A sulfate ion bound to the catalytic zinc ions was present in the active site of the original structure, but it had an intermolecular contact at the correct distance (~2.0 Å) with only one zinc ion, rendering this interpretation implausible. We renamed this sulfate to UNL as well. In order to account for residual difference map peaks, we changed its occupancy to 0.5 and added four partial water molecules as an alternative entity. All changes to the model were agreed with the original author, who also suggested adding the N-terminal residue Ala20 (Table S1). The re-refined structure has been re-deposited jointly as 5w90, superseding the original deposit. This example serves as another illustration of a productive collaboration with the original depositors, resulting in a better model re-deposited in the PDB.
Potentially incorrect ligands
NDM-1 with hydrolyzed cephalexin (PDB ID 4rl2)
This structure was described in a study entitled “Structural and mechanistic insights into NDM-1 catalyzed hydrolysis of cephalosporins” (Feng et al., 2014). According to the authors, the ligand is a hydrolysis intermediate where tautomerization of the dihydrothiazine ring has occurred and the double bond shifted from C3=C4 to C4=N5, with the C3 carbon becoming protonated (see Fig. 1 for dihydrothiazine ring numbering). The evidence presented for this intermediate is that “electron density of the ligand clearly reveals complete sp3 hybridization of C3, suggestive of full protonation of cephalexin in this structure”.
Altogether, this 2 Å structure is of good quality with the R/Rfree values of 14.6/19.3%. However, the PDB validation report highlights several bond length and angle outliers for the ligand dihydrothiazine ring even though the electron density fit criteria of the ligand are in the acceptable range. A closer inspection of the coordinates and electron density maps shows that the ligand is well ordered and fits the electron density properly except for the dihydrothiazine ring (Fig. 5A). The CAT atom seems to be sticking out of the electron density and the sixmembered ring looks squeezed into a too-small space. There is also a positive difference density peak in a position below the ring plane opposite to the methyl group. We calculated an omit map and found that hydrolyzed ampicillin fits perfectly into the electron density (Fig. 5B). Ampicillin has an identical side chain as cephalexin but differs by the identity of the ring fused to the β-lactam ring: instead of the six-membered unsaturated dihydrothiazine ring, it has a five-membered thiazolidine ring, which is fully saturated. The authors themselves noticed this similarity in structural superpositions and stated that “cephalexin perfectly overlays with ampicillin”. In chain A, the electron density for the second methyl group is less pronounced but still clearly visible. Based on this evidence, the most likely identity of the bound ligand is a hydrolyzed ampicillin molecule and this is what we included in our rerefined entry 5o2f. Detailed refinement statistics are presented in Table 2, and a detailed list of the applied changes is in Table S1. An interactive Molstack figure can be found at: http://molstack.bioreproducibility.org/project/view/m2wmZN7nuxjBGXCzWIS9/.
Figure 5.
Putative cephalosporin hydrolysis intermediates from selected NDM-1 complexes. (A) The original version of the ligand in structure 4rl2, modeled as hydrolyzed cephalexin, and (B) hydrolyzed ampicillin in the final electron density maps (5o2f). (C) Hydrolyzed cefuroxime ligand with attached carbamate group as modeled in the original PDB file 4rl0, and (D) the final model of the ligand (5o2e) with a Cl- ion modeled in place of the carbamate group and labeled UNL. The 2mFo-DFc electron density maps (blue) are contoured at 1.3σ (A and B) or at 1.2σ (C and D), and the mFo-DFc maps (green/red) are contoured at +/−3.0σ. The ligands and the electron density maps, including omit maps, can be inspected using interactive figures created with Molstack (http://molstack.bioreproducibility.org/project/view/m2wmZN7nuxjBGXCzWIS9/http://molstack.bioreproducibility.org/project/view/m2wmZN7nuxjBGXCzWIS9/ for 4rl2/5o2f, and http://molstack.bioreproducibility.org/project/view/6qfvCzkxsxZ8QGpNWkG6/http://molstack.bioreproducibility.org/project/view/6qfvCzkxsxZ8QGpNWkG6/for 4rl0/5o2e).
Table 2.
Selected parameters of the originally deposited (left column) and re-refined (right column) MBL structures described in the section ‘Potentially incorrect ligands’ Data prepared as for Table 1.
| Protein | NDM-1 | NDM-1 | Legionella gormanii FEZ-1 | |||
|---|---|---|---|---|---|---|
| PDB accession code | 4rl2 | 5o2f | 4exy | 5n0i | 1k07 | 5wck |
| Resolution [Å] | 66.92–2.01 (2.06–2.01) | 66.92–2.01 (2.06–2.01) | 75.82–1.47 (1.51–1.47) | 75.82–1.47 (1.51–1.47) | 50.00–1.65 (1.69–1.65) | 50.00–1.65 (1.69–1.65) |
| No. of reflections refinement/Rfree | 25437/1345 | 25437/1345 | 87106/4592 | 87106/4592 | 55140/6172 | 55140/6172 |
| Completeness [%] | 97.71(92.68) | 97.71(92.68) | 99.62(96.74) | 99.62(96.74) | 97.29 (89.93) | 97.29 (89.93) |
| <Ι/σ> in the highest resolution shell | 2.9 | 2.9 | 3.0 | 3.0 | 19 | 19 |
| R / Rfree [%] | 15.1/18.9 | 13.2/18.2 | 14.0/18.2 | 10.9/14.7 | 17.4/19.8 | 12.3/15.8 |
| Protein molecules in ASU | 2 | 2 | 2 | 2 | 2 | 2 |
| NCS restraints | - | - | - | Local | ||
| ADP parametrization | Isotropic | Isotropic | Isotropic | Anisotropic | Isotropic | Isotropic |
| No. atoms | ||||||
| Protein | 3594 | 3644 | 3506 | 3594 | 4148 | 4153 |
| Ligands | 54 | 87 | 12 | 36 | 72~^ | 78 |
| Water | 438 | 433 | 369 | 568 | 501 | 866 |
| Ramachandran statistics [%] | ||||||
| Favored/outliers | 97.91/0.21 | 98.55/0.00 | 97.21/0.21 | 98.50/0.00 | 95.96/0.66 | 95.98/0.22 |
| R.m.s. deviations from target values | ||||||
| Bond lengths [Å] | 0.011 | 0.011 s | 0.017 | 0.014 | 0.014 | 0.009 |
| Bond angles [°] | 1.23 | 1.45 | 2.10 J | 1.65 | 1.62 | 1.40 |
| Clashscore/percentile | 6.13/95th | 1.77/100th | 4.75/93th | 2.5/99th | 4.69/96th | 1.19/97th |
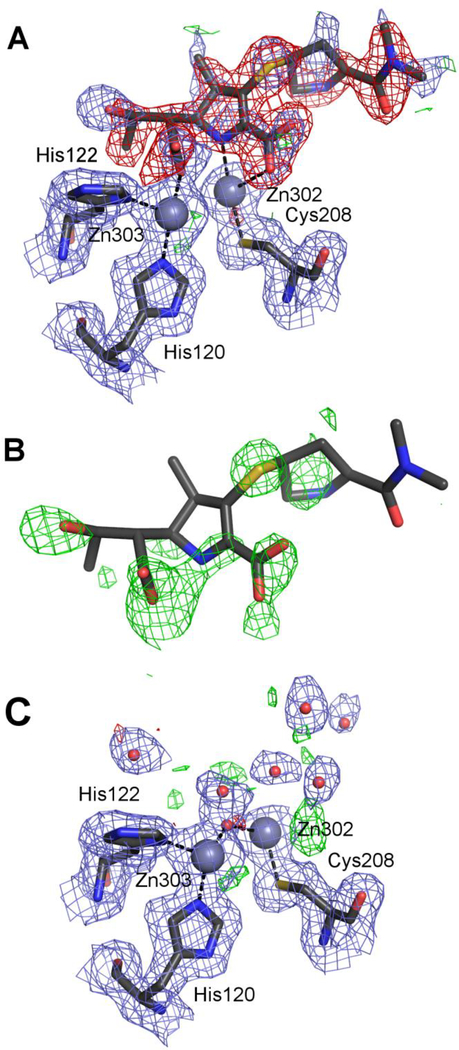
NDM-1 with ethylene glycol (PDB ID 4exy)
The 4exy structure of NDM-1 was solved at 1.47 Å resolution and described as a complex with ethylene glycol (EDO). It is part of the same study (King et al., 2012) as the structure 4eyl but it was not described in any detail in the original paper. Apart from a number of Ramachandran outliers and a slightly elevated clashscore, the PDB validation report does not indicate any major problems. The EDO molecules were modeled at full occupancy in spite of the partial occupancy of some of the Zn ions. The electron density maps downloaded from the EDS server reveal a 5.5σ positive difference peak at the oxygen atom coordinating both zinc ions in chain A, and a corresponding 4.2σ peak in chain B. The isotropic ADPs for these atoms were 6.4 and 4.7 Å2, with the average ADPs for the whole molecules of 18.6 and 16.8 Å2 for chains A and B, respectively.
According to the Methods section in the original paper, β-mercaptoethanol (BME) was present at 2 mM concentration in the crystallization buffer (Protein Expression and Purification section in Supporting Information). BME differs from EDO by just one atom, S in place of one of the O atoms. Refinement of BME in the active site (at 0.8 occupancy, the same as for one of the Zn ions) with the S atom bound between the Zn ions and overlapping with the positive difference density peak, resulted in clear electron density around the ligand and in isotropic ADPs for the S atoms of 14.1 and 11.9 Å2, with the whole-molecule averages of 19.2 and 16.5 Å2 for chains A and B, respectively. This overwhelming crystallographic evidence clearly suggests that the active site is occupied by β-mercaptoethanol in both protein chains, rather than by ethylene glycol. Thiol groups have a very high affinity for Zn ions and thiol compounds are well-known inhibitors of metallo-β-lactamases (Brem et al., 2016, 2014; Goto et al., 1997; Greenlee et al., 1999; Siemann et al., 2003), their primary interaction with the enzyme being thiol-Zn coordination. Goto et al. (1997) determined the inhibition constant of BME for the metallo-β-lactamase produced by Serratia marcesens to be 12 μM. It seems safe to assume that BME would be bound with a much higher affinity than EDO, in spite of the high concentration of the latter compound in the crystallization solution. Therefore, we conclude that the bound ligand is β-mercaptoethanol and not ethylene glycol.
Our corrections also included changing the protein backbone conformation at Asn220 in chain B and modeling anisotropic displacement parameters for all atoms (Table S1). All these changes resulted in an improvement of R/Rfree from 14.0/18.2% for the original coordinates to 11.0/14.7% for the re-refined model (Table 2), which has been deposited in the PDB as 5n0i together with the authors of the original entry. An interactive Molstack figure can be viewed at http://molstack.bioreproducibility.org/project/view/eS9jfnc50FXzwl4GwoVe/.
Native FEZ-1 from Legionella gormanii (PDB ID 1k07)
The native structure of FEZ-1 was refined at 1.65 Å resolution and deposited as 1k07; it was described in the same paper as 1jt1 (García-Sáez et al., 2003). Our re-refinement of this structure resulted in a decrease of Rfree by 4.0%, mostly due to the addition of 365 water molecules and improvements of the refinement software (Tables 2, S1). We adjusted the sequence numbering to match the numbering used in Uniprot (Q9K578) and corrected the side-chain conformations of several residues. The occupancy of multiple side chains had been originally set to extremely low values, e.g., 0.05. We changed the occupancy of all protein atoms to 1.0, unless double conformation was modeled, in which case the sum of the alternative occupancies was set to 1.0. Interestingly, the signal-to-noise ratio in the outermost resolution shell is very high (19), but the original diffraction images were not available for re-processing and possible extension of resolution.
Glycerol molecules modeled in two alternate conformations in both subunits were included in the active site of the original model. However, these molecules have poor electron density fit and a short contact between one conformer and a zinc ion (Fig. 4C). A polder map (http://molstack.bioreproducibility.org/project/view/lSoPdEAwzOw4y8jthr2Z/) calculated for our final coordinates with the original glycerol molecules placed, revealed only remote similarity to the ligands, and it was reported by PHENIX as more likely to show bulk solvent or noise. We tried to model various conformations of a glycerol molecule, a sulfate ion, as well as fragments of common antibiotics (to account for the slight possibility of sample contamination) but none of these interpretations resulted in an acceptable electron density fit. The primary author of the original deposit noted that glycerol is a plausible interpretation (in part, according to ligand identification by PHENIX), but not a confident one. Therefore, we mutually concluded that a set of unknown atoms is the safest interpretation (Fig. 4D). All changes to the model were agreed with the original author, who also suggested adding the N-terminal residue Ala20, as well as adjusting some side chains (Table S1). The re-refined structure has been re-deposited jointly as 5wck, superseding the original deposit.
Incorrect treatment of ligands
NDM-1 with hydrolyzed meropenem (PDB ID 4eyl)
The 1.9 Å structure 4eyl is part of a comprehensive study “New Delhi Metallo-β-lactamase: Structural insights into β-lactam recognition and inhibition” (King et al., 2012) that describes six structures of NDM-1 complexed with various ligands. The key parts of this particular structure are two hydrolyzed meropenem molecules bound at the active site in both protein subunits (A and B) present in the asymmetric unit. A quick look at the entry’s validation report reveals potential problems as both ligand molecules are marked as outliers by electron density fit criteria. After downloading the coordinates and map coefficients from the EDS server, we noticed strong negative difference density peaks around the modeled hydrolyzed meropenem moieties in both protein molecules. Upon closer inspection, we found that all atoms of the ligand molecules have isotropic ADPs exactly equal to 20 Å2, confirming that they were not refined, which explains the strong negative difference density peaks (Fig. 6A).
Figure 6.
The active site of chain B in the structure 4eyl (new PDB ID 5n0h). (A) The original coordinates and electron density maps calculated by REFMAC5 without prior refinement. Negative difference density peaks covering the meropenem ligand are clearly visible. (B) The meropenem molecule shown in an omit map calculated with the final refinement phases, excluding the active site water molecules. (C) The final re-refined coordinates 5n0h with water molecules modeled in place of the original meropenem ligand. The electron density maps are contoured at 1.0σ for 2mFo-DFc (blue) and +/−3.0σ for mFo-DFc (green/red). The ligands and the electron density maps, including omit maps, can be inspected using an interactive figure created with Molstack (http://molstack.bioreproducibility.org/project/view/YcQYlF9oKqfpEzeMlPxc/).
After our corrections (see Table S1 for details), which resulted in an improvement of R/Rfree from 18.4/22.4% to 15.5/20.0% (Table 2), a hydrolyzed meropenem moiety could be refined only in chain A (at 0.6 occupancy and with an average isotropic ADP of 28 Å2). We also modeled partial water molecules overlapping with the carboxylate groups of this ligand, one between the two Zn ions and one interacting with the main-chain N atom of Asn220 and the side chain of Lys211. In chain B, we observed in the omit map contoured at 3.0σ some residual density for the carboxylate groups, the ring N atom, and the S atom. We judged this electron density (Fig. 6B) to be insufficient for modeling any kind of ligand at this site. Instead, the difference density peaks were filled with a few water molecules (Fig. 6C). Our observations are in contrast to the statement from the paper describing the original data that “all of the hydrolyzed β-lactam products display clear 2mFo-DFc electron density in both protein chains in the asymmetric unit”. While traces of the ligand are probably still bound in the crystal, in the majority of the unit cells it is absent from the binding site of protein subunit B. We also calculated polder maps for the ligand separately for each protein subunit and show the maps in our Molstack figure at http://molstack.bioreproducibility.org/project/view/YcQYlF9oKqfpEzeMlPxc/. The conclusion from the polder map is that the ligand is bound in subunit A, but not in subunit B.
In the original structure, hydrolyzed meropenem has the ligand ID 0rv and its description in the PDB contains two double bonds, C1=C2 and C3=N4. By comparison, the substrate contains only one double bond, C2=C3 (Fig. 1). It has been reported that the dihydropyrrole ring of carbapenem antibiotics (such as meropenem) may undergo tautomerization following hydrolysis and it was observed spectroscopically for imipenem and BcII (Tioni et al., 2008). However, the possible tautomers have only one double bond within the dihydropyrrole moiety, either C2=C3 (as in the substrate, Δ2 tautomer), or N4=C3 (Δ1 tautomer). Nowhere in the literature were we able to find any description of a hydrolyzed carbapenem compound with two double bonds (C1=C2 and C3=N4) within the dihydropyrrole ring as in the PDB library. In the 4eyl entry the electron density for the ligand is rather weak but seems to be in better agreement with an sp2 C2 carbon atom. As this is also the most commonly described form of this compound, we created a new ligand ID 8yl with just one double bond, C2=C3. With this approach, in our new deposit 5n0h we do not make any additional assumptions about the ligand and provide a reasonable explanation of the electron density. The new deposit was submitted jointly with the authors of the original entry.
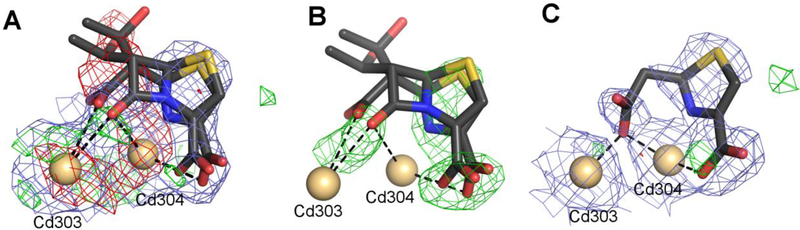
NDM-1 with Cd2+ and faropenem (PDB ID 4hky)
The 2.0 Å structure 4hky was determined after incubation of the NDM-1 protein with CdCl2 and faropenem (Kim et al., 2013). Cadmium was found in place of the Zn ions and two forms of the ligand, hydrolyzed and unhydrolyzed, were originally modeled. The authors claim that in the presence of Cd2+ ions the enzymatic reaction is slow enough for the unhydrolyzed antibiotic to be observed in the crystals.
The structure has a high clashscore (9) and Rfree of 25.1%. The Cd ions that are coordinated in the characteristic Zn sites fit the electron density very well, with RSR values between 0.07 and 0.11 and RSCC between 0.98 and 0.99. The same cannot be said about the β-lactam molecules, which all have RSR above 0.30 and RSCC in the range of 0.55–0.86. In the electron density maps downloaded from the EDS server, a patch of negative difference density overlaps the unhydrolyzed β-lactam part of the ligand. The same situation is observed in the electron density maps calculated by REFMAC5 without prior refinement (Fig. 7A). An omit map shows only unconnected patches of density (Fig. 7B).
Figure 7.
Ligand molecules in the structure 4hky, with Cd2+ ions shown as beige spheres. (A) The original coordinates containing two alternative states of the faropenem ligand. Negative difference density peaks are visible, especially at the unhydrolyzed β-lactam moiety. (B) The original coordinates shown in an omit map after final re-refinement. (C) The final model (6ex7) with a truncated, hydrolyzed version of faropenem. The electron density maps are contoured at 1.0σ for 2mFo-DFc (blue) and +/−3.0σ for mFo-DFc (green/red). The ligands and the electron density maps, including omit maps, can be inspected using an interactive figure created with Molstack (http://molstack.bioreproducibility.org/project/view/FU0iFwAkG0vsoOtUDZd5/).
We reprocessed the original X-ray diffraction data (https://proteindiffraction.org/project/apc105101_4HKY/) in the anomalous mode using corrections for radiation decay and anisotropic diffraction, built a missing fragment of the protein backbone (residues 31–40 of chain A), added two polyethylene glycol molecules, and refined anisotropic temperature factors for the Cd2+ ions. We also modeled a truncated version of the faropenem ligand in the hydrolyzed state at full occupancy and called it UNL to indicate that the electron density is weak, and the identity of the ligand is uncertain (Fig. 7 C).
An interactive Molstack figure is presented at http://molstack.bioreproducibility.org/project/view/FU0iFwAkG0vsoOtUDZd5/. Table S1 presents a detailed list of the applied changes. These corrections resulted in an improved PDB model (Tables 3, S2), characterized by lower R/Rfree (19.5/22.6% vs 20.4/25.1%) and better MolProbity score (0.98 vs 2.08). We submitted the new model to the PDB under the accession code 6ex7 together with the authors of the original entry, who agreed with the implemented changes.
Table 3.
Selected parameters of the originally deposited (left column) and re-refined (right column) MBL structures described in the sections ‘Incorrect treatment of ligands’ and ‘Improvement of structural models after re-processing of the diffraction images’. Data prepared as for Table 1.
| Protein | NDM-1 | NDM-1 | NDM-1 | Bradyrhizobium diazoefficiens BJP-1 | ||||
|---|---|---|---|---|---|---|---|---|
| PDB accession code | 4eyl | 5n0h | 4hky | 6ex7 | 4rl0 | 5o2e | 3m8t | 5wcm |
| Resolution [Å] | 74.95–1.90 (1.95–1.90) | 74.95–1.90 (1.95–1.90) | 74.23–2.00 (2.06–2.00) | 74.20–1.951 (2.00–1.95) | 67.86–1.30 (1.33–1.30) | 67.86–1.30 (1.33–1.30) | 22.21–1.33 (1.36–1.33) | 50.00–1.20* (1.22 – 1.20) |
| No. of reflections refinement/Rfree | 38308/2030 | 38308/2030 | 27161/1446 | 27008/1353 | 88894/4618 | 88894/4618 | 97262/5123 | 117432/6083 |
| Completeness [%] | 95.79(90.94) | 95.79(90.94) | 99.50(97.73) | 91.72(66.00) | 91.38(84.16) | 91.38(84.16) | 91.85 (89.24) | 81.32 (26.78) |
| <Ι/σ> in the highest resolution shell | 2.1 | 2.1 | 2.3 | 2.2 | 2.6 | 2.6 | 6.9 | 4.4 |
| R / Rfree [%] | 18.7/22.1 | 15.5/19.8 | 24.1/26.9 | 19.4/22.6 | 15.7/16.5 | 10.1/13.3 | 14.3/17.7 | 13.2/16.0 |
| Protein molecules in the ASU | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| NCS restraints | Local | Local | - | - | ||||
| ADP parametrization | Isotropic | Isotropic | Isotropic+TLS | Isotropic+TLS | Isotropic | Anisotropic | Anisotropic+TLS# | Anisotropic |
| No. atoms | ||||||||
| Protein | 3500 | 3520 | 3487 | 3498 | 3499 | 3705 | 4043 | 4031 |
| Ligands | 58 | 66 | 77 | 55 | 64 | 63 | 41 | 37 |
| Water | 161 | 398 | 136 | 178 | 645 | 676 | 690 | 829 |
| Ramachandran statistics [%] | ||||||||
| Favored/outliers | 97.43/0.43 | 98.29/0.00 | 97.41/0.65 | 99.16/0.00 | 99.13/0.00 | 98.73/0.00 | 96.36/0.38 | 95.98/0.38 |
| R.m.s. deviations from target values | ||||||||
| Bond lengths [Å] | 0.017 | 0.018 | 0.016 | 0.014 | 0.016 | 0.013 | 0.015 | 0.009 |
| Bond angles [°] | 1.35 | 1.80 | 1.80 | 1.58 | 2.58 | 1.62 | 1.61 | 1.40 |
| Clashscore/percentile | 7.42/90th | 2.54/99th | 9.15/86th | 1.73/100th | 7.15/70th | 2.28/99th | 3.08/97th | 0.62/99th |
The dataset was reprocessed with HKL-3000, and the resolution was extended. Auto-correction mode was used in data scaling which resulted in the elimination of non-informative reflections.
This was a redundant combination of ADP parameters.
NDM-1 with hydrolyzed cefuroxime (PDB ID 4rl0)
The structure of NDM-1 in complex with a product of cefuroxime hydrolysis was determined at the resolution of 1.33 Å (Feng et al., 2014). The overall quality of the structure appears to be good, with R/Rfree of 13.1/15.7%. The PDB validation report reveals an elevated clashscore and seven bond length and seven bond angle outliers for each hydrolyzed β-lactam molecule. Cefuroxime contains a carbamic substituent at C3’ (Fig. 1A). For cephalosporins with a good leaving group at C3’, tautomerization of the dihydrothiazine ring is thought to cooccur with the formation of a double bond at C3-C3’ and dissociation of the leaving group, concurrently with or following the hydrolysis reaction (Faraci and Pratt, 1984). In this example, the authors state that “density for the carbamoyl group was surprisingly present and well connected with the rest of the ligand”. The ligand is described as a reaction intermediate where the double bond is shifted from C3=C4 to C4=N5. Each ligand molecule contains four bond angle outliers within the small carbamate fragment. We examined the electron density maps in this region, first by downloading the coefficients from the EDS and then by recalculating them from the deposited coordinates. In the latter maps, in both protein chains the electron density looks unconvincing; for example, there are negative difference density peaks at the oxygen and nitrogen atoms within the carbamate group (Fig. 5C, see also Fig. 1, cefuroxime, for a scheme of the carbamate group). The group also has improbable isotropic ADP statistics, with values for the heteroatoms of ~30 Å2 and ~10 Å2 for the central C atom. The C3 carbon atom within the dihydrothiazine ring shows tetrahedral geometry which is consistent with the suspected shift of the double bond to C4=N5.
We re-refined this structure with anisotropic temperature factors, and also built a few missing backbone fragments and modeled additional solvent and buffer components. The electron density suggested that one of the Zn ions is only present at 0.9 occupancy in both protein subunits. We modeled the ligand occupancy accordingly. Our corrections resulted in R/Rfree drop from 13.1/15.7% to 10.4/13.5% (Table 3). Refinement of the ligand with no group attached at the C3’ atom results in the appearance of a 20σ positive difference density peak at ~1.8 Å from C3’ in both chains. When a water molecule is refined at this position it has lower isotropic ADP than the C3 or C3’ atoms and there are positive difference peaks in its vicinity. Chloride ions refined at the same (0.9) occupancy as the β-lactam ligands resulted in negative difference density peaks and much higher ADP values. In the re-deposited structure 5o2e, we modeled Cl- ions at 0.8 occupancy and annotated them as UNL. This change provides a good explanation of the electron density (Fig. 5D), while at the same time not implicating any specific interpretation. A list of the applied changes is provided in Table S1. An interactive Molstack figure is presented at http://molstack.bioreproducibility.org/project/view/6qfvCzkxsxZ8QGpNWkG6/.
Improvement of structural models after re-processing of diffraction images
Class B3 β-lactamase BJP-1 complexed with 4-nitrobenzene-sulfonamide (PDB ID 3m8t)
This 1.33 Å structure contains 4-nitrobenzene-sulfonamide, a potential inhibitor, bound in the active site and interacting with the Zn ions (Docquier et al., 2010). The reported <I/σ> value in the highest resolution shell was 6.9, suggesting a possibility of extending the resolution limit. Upon our request, we received the original diffraction images for this entry from the original depositor (https://proteindiffraction.org/project/BJP1_3m8t/). We reprocessed the images, extending the resolution limit to 1.20 Å by including the areas in the corners of the detector. Because of the nature of this extension, data completeness in the highest resolution shell is relatively low, but the total number of unique reflections has increased by 21% (Tables 3, S2). In addition, the data file accompanying the original deposit did not include Bijvoet pairs. After re-processing with HKL-3000 in the anomalous mode using corrections for radiation decay and anisotropic diffraction, the anomalous signal was used to locate the Zn ions in the model, resulting in the addition of three non-catalytic Zn ions. Furthermore, we remodeled several side chains (e.g., Asp144A/B, Glu185A, Lys192A/B, Lys256A/B, Asp257B, Lys265A/B, Lys282A, Glu292A/B), added OXT to chain B, deleted a DMSO molecule (not supported by electron density and with packing clashes), deleted a formate molecule (not present in the crystallization mixture), modeled 140 additional water molecules, and standardized the placement of the model within the unit cell. The clashscore improved from 3.08 to 0.62. The original depositors agreed with the changes (Table S1), and the structure has been re-deposited jointly as 5wcm, superseding the original deposit. This example can serve as yet another illustration of a fruitful and gratifying collaboration with the original depositors, resulting in a better model and data re-deposited in the PDB.
4. The ripple effect of sub-optimal and incorrect structural models
Crystallographic structural data, once deposited in the PDB and reported in the literature, tend to be treated by the non-structural community with full trust and their correctness is rarely questioned. The “ripple effect” of structural models (good and bad!) is very significant, given that many other fields of science use these models as the foundation for their research. Protein crystallography gives scientists an unprecedented power to investigate life at sub-microscopic levels, by revealing the 3D structure of biological molecules and explaining how to identify and treat molecular causes of infection and disease. In this context, chemical crystallography is critical for drug discovery and the correctness of structural models as potential drug targets is of high importance.
Recent advances in crystallographic software have allowed for almost automatic data processing, structure determination, and refinement, with an option for obtaining a molecular model with minimal investment of time and effort. An unfortunate side effect of this development is that neither formal crystallographic training nor experience in dealing with crystallographic data are now required for obtaining and publishing crystallographic results. This shift of professional focus is already visible in cases of suboptimal X-ray data processing and/or model refinement and completion, with further consequences in suboptimal, inaccurate, or even wrong structural models, especially in the regions of the sensitive interface where small molecules interact with the macromolecules. Some of the structures discussed in this review were determined and deposited many years ago, when modern tools were not yet available, making the interpretation, refinement, and validation much more difficult and a really pioneering kind of work. For these structures, significant improvements can be easily achieved with the application of modern methodology and software.
The most serious problems encountered in our review are overinterpretation or misinterpretation of the results of the diffraction experiment. These include placing a ligand where there is no significant electron density to support it, or claiming the observation of a reaction intermediate when in fact the electron density is unclear, indicates a different compound, or the ligand is disordered. A similar category of errors applies to cases where a ligand was modeled in all independent protein subunits while there is no experimental evidence for its presence in some of them. Another group of problems arise from the lack of proper stereochemical restraints in structure modeling and refinement, or even in the PDB ligand definition (Jaskolski, 2013). Such errors decrease the quality of the final model and increase the difficulty of correct interpretation of the key structural elements, and in extreme cases may lead to incorrect interpretations. This is especially true of structures determined at low-to-medium resolution.
The proteins analyzed in this review play a major role in antimicrobial resistance, and many of them are potential drug targets. Studying them, with a strong focus on enzyme-product or enzyme-inhibitor interactions and on how the enzyme structure adjusts to accommodate the small-molecule ligands, provides the insight needed for designing effective therapeutic agents. In particular, such information may help in achieving the desired selectivity. However, inaccurate or even incorrect structural results have the opposite effect by confusing the subject, elevating the noise level, and subsequently affecting the drug discovery process.
Four of our re-refined structures contain products of the hydrolysis reaction and their specific interactions with the protein are of direct biological relevance. The re-refined structure 5n0i (original PDB ID 4exy) presents a complex of NDM-1 with β-mercaptoethanol, which is a known inhibitor of metallo-β-lactamases. This structure confirms the usual binding mode of thiol compounds to MBLs with the thiol group inserted between the Zn ions, and shows that just this interaction is sufficient to keep the ligand bound in place. This evidence also strengthens our argument about the problems with ligands in the PDB entries 5a5z and 1j31, which had thiol compounds modeled in unusual biding modes. Crystallization and buffer agents are known to often interfere with enzymatic activities (Majorek et al., 2014) and if such a molecule is found in the active site, it requires the attention of the experimenter to decipher the potential implications of the observed interactions for other experiments using the same compounds.
The consequences of uncontrolled propagation of substandard or incorrect structural models are especially severe for structures of medical relevance. For example, the structure 4rl2, which supposedly contained a new tautomer of hydrolyzed cephalexin (see above), served as the starting point for QM/MM simulations of cephalexin hydrolysis by NDM-1 (Das and Nair, 2017). That study also analyzed meropenem hydrolysis using structure 4eyl as the initial model, and was concluded with an explanation of two different hydrolysis mechanisms for these compounds that is highly dependent on the structural data used as the starting point for the simulations. This case highlights the problem of how easily inaccurate structural data can propagate in the literature, with new results accumulating on top of it. Another example is provided by an excellent recent paper that reviews the status of β-lactamase inhibitor discovery and development (Docquier and Mangani, 2018). In that review, a table summarizing the availability of structural data for MBLs and their complexes lists all relevant structures, including six structures identified in the present study as having problems with modeling of the primary ligands (5a5z, 4nq7, 1jt1, 4eyl, 4rl0, 4hky). Even though the authors of the review did not discuss most of those structures explicitly (although they did mention unusual ligand binding in one of the structures, 1jt1), the table may be perceived by unaware readers as a summary of completely reliable data.
We should again emphasize that the improvements in refinement statistics and model quality for the structures re-refined by us are partly due to the development of new methodologies, refinement software, and validation tools that have revolutionized structure determination/validation process in recent years. This is especially true for older structural models. Availability of raw diffraction images (unfortunately only in some cases) was crucial for improvement of the corresponding structures, showing the importance of resources that store the original diffraction experiments (Grabowski et al., 2016).
In the course of our project we have encountered a variety of reactions from the original authors, ranging from a complete lack of answer, to lack of interest, rejection of our conclusions, discussions questioning the validity of the new results, and finally, and most commonly, to mutual and harmonious work on the improvement of the final results. Our goal has always been the benefit of science.
Problems with misinterpretations or irreproducible results are present in all areas of science. It is important to always look at scientific results, including our own results, with a critical eye, in order to spot any inconsistencies, while at the same time not falling in the opposite trap of unreasonable suspicions. In crystallography, it is relatively easy to objectively verify the presented results, as long as the experimental data are available. Therefore, we encourage all crystallographers to thoroughly check our results, as well as those of others, because such an independent validation preserves the integrity of scientific research and is one of the most important aspects of scientific progress.
Supplementary Material
Synopsis:
Metallo-β-lactamases (MBLs) are important drug design targets for combating antibiotic resistance. Because inhibition of β-lactamases is so important medically, their crystal structures deposited in the Protein Data Bank (PDB) have been reviewed with focus on various ligands found in the active site and on the catalytic metal cations. In some cases, the models have been revised and re-deposited in the PDB as to provide a bias-free basis for the design of MBL inhibitors as rescue drugs for the existing arsenal of β-lactam antibiotics.
Acknowledgments
We wish to thank Dr. Bernhard Rupp for valuable comments on one of the cases (5a5z). We also acknowledge, with thanks, the cooperation and joint authorship of the revised PDB deposits on the part of some of the original authors, especially Dr. Andrzej Joachimiak, Dr. Youngchang Kim, Dr. Natalie Strynadka, Dr. Dustin King, Dr. Alejandro Vila, Dr. Javier Gonzalez, Dr. Stefano Mangani, Dr. Isabel Garcia-Saez, Dr. Jean-Marie Frere, and Dr. Otto Dideberg. The work of MJ and JR was supported by the DesInMBL grant from the National Center for Research and Development within the JPIAMR initiative. The work of IS and WM was funded by federal funds from the National Institute of General Medical Sciences under grant GM117325, National Institutes of Health BD2K program under grant HG008424, and the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services under contracts HHSN272201200026C and HHSN272201700060C. AW was supported by the Intramural Research Program of the National Cancer Institute, Center for Cancer Research.
Abbreviations:
- ADP
atomic displacement parameters (B factors)
- ASU
asymmetric unit
- BLDB
β-Lactamase Data Base
- BME
β-mercaptoethanol
- EDO
ethylene glycol
- EDS
Electron Density Server
- MBL
metallo-β-lactamase
- MC
main chain
- NCS
non-crystallographic symmetry
- NDM
New Delhi Metallo-β-lactamase
- QM/MM
quantum mechanics/molecular mechanics
- PDB
Protein Data Bank
- RSCC
real space correlation coefficient
- RSR
real space R-factor
- TLS
translation/libration/screw
Footnotes
Dedication: As a birthday tribute, this work is dedicated to Dr. Zbigniew Dauter, an untiring advocate of the highest quality in structural research
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH, 2010. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr 66, 213–221. https://doi.org/10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE, 2000. The Protein Data Bank. Nucleic Acids Res 28, 235–242. https://doi.org/10.1093/nar/28.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem J, van Berkel SS, Aik W, Rydzik AM, Avison MB, Pettinati I, Umland K-D, Kawamura A, Spencer J, Claridge TDW, McDonough MA, Schofield CJ, 2014. Rhodanine hydrolysis leads to potent thioenolate mediated metallo-β-lactamase inhibition. Nat. Chem 6, 1084–1090. https://doi.org/10.1038/nchem.2110 [DOI] [PubMed] [Google Scholar]
- Brem J, Van Berkel SS, Zollman D, Lee SY, Gileadi O, McHugh PJ, Walsh TR, McDonough MA, Schofield CJ, 2016. Structural basis of metallo-β-lactamase inhibition by captopril stereoisomers. Antimicrob. Agents Chemother 60, 142–150. https://doi.org/10.1128/AAC.01335-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger AT, 1992. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355, 472. [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC, 2010. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr 66, 12–21. https://doi.org/10.1107/S0907444909042073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das CK, Nair NN, 2017. Hydrolysis of cephalexin and meropenem by New Delhi metallo-β-lactamase: the substrate protonation mechanism is drug dependent. Phys. Chem. Chem. Phys 19, 13111–13121. https://doi.org/10.1039/C6CP08769H [DOI] [PubMed] [Google Scholar]
- Davies J, Davies D, 2010. Origins and Evolution of Antibiotic Resistance. Microbiol. Mol. Biol. Rev 74, 417–433. https://doi.org/10.1128/MMBR.00016-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docquier J-D, Mangani S, 2018. An update on β-lactamase inhibitor discovery and development. Drug Resist. Updat 36, 13–29. https://doi.org/10.1016/j.drup.2017.11.002 [DOI] [PubMed] [Google Scholar]
- Docquier JD, Benvenuti M, Calderone V, Stoczko M, Menciassi N, Rossolini GM, Mangani S, 2010. High-resolution crystal structure of the subclass B3 metallo-β-lactamase BJP-1: Rational basis for substrate specificity and interaction with sulfonamides. Antimicrob. Agents Chemother 54, 4343–4351. https://doi.org/10.1128/AAC.00409-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K, 2010. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr 66, 486–501. https://doi.org/10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraci WS, Pratt RF, 1984. Elimination of a good leaving group from the 3’-position of a cephalosporin need not be concerted with β-lactam ring opening: TEM-2 β-lactamase-catalyzed hydrolysis of pyridine-2-azo-4’-(“,N’-dimethylaniline) cephalosporin (PADAC) and of cephaloridine. J. Am. Chem. Soc 106, 1489–1490. [Google Scholar]
- Feng H, Ding J, Zhu D, Liu X, Xu X, Zhang Y, Zang S, Wang DC, Liu W, 2014. Structural and mechanistic insights into NDM-1 catalyzed hydrolysis of cephalosporins. J. Am. Chem. Soc 136, 14694–14697. https://doi.org/10.1021/ja508388e [DOI] [PubMed] [Google Scholar]
- Freedman LP, Cockburn IM, Simcoe TS, 2015. The economics of reproducibility in preclinical research. PLoS Biol. 13, 1–9. https://doi.org/10.1371/journal.pbio.1002165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frère J-M, Sauvage E, Kerff F, 2016. From « An enzyme able to destroy penicillin » to carbapenemases: 70 years of β-lactamase misbehavior. Curr. Drug Targets 17, 974–982. [DOI] [PubMed] [Google Scholar]
- García-Sáez I, Mercuri PS, Papamicael C, Kahn R, Frère JM, Galleni M, Rossolini GM, Dideberg O, 2003. Three-dimensional structure of FEZ-1, a monomeric subclass B3 metallo-β-lactamase from Fluoribacter gormanii, in native form and in complex with D-captopril. J. Mol. Biol 325, 651–660. https://doi.org/10.1016/S0022-2836(02)01271-8 [DOI] [PubMed] [Google Scholar]
- Goto M, Takahashi T, Yamashita F, Koreeda A, Mori H, Ohta M, Arakawa Y, 1997. Inhibition of the metallo-beta-lactamase produced from Serratia marcescens by thiol compounds. Biol. Pharm. Bull 20, 1136–1140. [DOI] [PubMed] [Google Scholar]
- Grabowski M, Langner KM, Cymborowski M, Porebski PJ, Sroka P, Zheng H, Cooper DR, Zimmerman MD, Elsliger MA, Burley SK, Minor W, 2016. A public database of macromolecular diffraction experiments. Acta Crystallogr. Sect. D Struct. Biol 72, 1181–1193. https://doi.org/10.1107/S2059798316014716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenlee ML, Laub JB, Balkovec JM, Hammond ML, Hammond GG, Pompliano DL, Epstein-Toney JH, 1999. Synthesis and SAR of thioester and thiol inhibitors of IMP-1 metallo-β-lactamase. Bioorganic Med. Chem. Lett 9, 2549–2554. https://doi.org/10.1016/S0960-894X(99)00425-4 [DOI] [PubMed] [Google Scholar]
- Jaskolski M, 2013. On the propagation of errors. Acta Crystallogr. Sect. D Biol. Crystallogr 69, 1865–1866. https://doi.org/10.1107/S090744491301528X [DOI] [PubMed] [Google Scholar]
- Kim Y, Cunningham MA, Mire J, Tesar C, Sacchettini J, Joachimiak A, 2013. NDM-1, the ultimate promiscuous enzyme: Substrate recognition and catalytic mechanism. FASEB J. 27, 1917–1927. https://doi.org/10.1096/fj.12-224014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King DT, Worrall LJ, Gruninger R, Strynadka NCJ, 2012. New Delhi Metallo-β-lactamase: Structural insights into β-lactam recognition and inhibition. J. Am. Chem. Soc 134, 11362–11365. https://doi.org/10.1021/ja303579d [DOI] [PubMed] [Google Scholar]
- Kleywegt GJ, Harris MR, Zou JY, Taylor TC, Wählby A, Jones TA, 2004. The Uppsala Electron-Density Server. Acta Crystallogr. Sect. D Biol. Crystallogr 60, 2240–2249. https://doi.org/10.1107/S0907444904013253 [DOI] [PubMed] [Google Scholar]
- Klingler FM, Wichelhaus TA, Frank D, Cuesta-Bernal J, El-Delik J, Müller HF, Sjuts H, Göttig S, Koenigs A, Pos KM, Pogoryelov D, Proschak E, 2015. Approved drugs containing thiols as inhibitors of metallo-β-lactamases: Strategy to combat multidrug-resistant bacteria. J. Med. Chem 58, 3626–3630. https://doi.org/10.1021/jm501844d [DOI] [PubMed] [Google Scholar]
- Kowiel M, Jaskolski M, Dauter Z, 2014. ACHESYM: An algorithm and server for standardized placement of macromolecular models in the unit cell. Acta Crystallogr. Sect. D Biol. Crystallogr 70, 3290–3298. https://doi.org/10.1107/S1399004714024572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebschner D, Afonine PV, Moriarty NW, Poon BK, Sobolev OV, Terwilliger TC, Adams PD, 2017. Polder maps: Improving OMIT maps by excluding bulk solvent. Acta Crystallogr. Sect. D Struct. Biol 73, 148–157. https://doi.org/10.1107/S2059798316018210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majorek KA, Kuhn ML, Chruszcz M, Anderson WF, Minor W, 2014. Double trouble - Buffer selection and his-tag presence may be responsible for nonreproducibility of biomedical experiments. Protein Sci 23, 1359–1368. https://doi.org/10.1002/pro.2520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna M, 2013. Antibiotic resistance: The last resort. Nature 499, 394–396. https://doi.org/10.1038/499394a [DOI] [PubMed] [Google Scholar]
- Merritt EA, 2012. To B or not to B: A question of resolution? Acta Crystallogr. Sect. D Biol. Crystallogr 68, 468–477. https://doi.org/10.1107/S0907444911028320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor W, Cymborowski M, Otwinowski Z, Chruszcz M, 2006. HKL-3000: The integration of data reduction and structure solution - From diffraction images to an initial model in minutes. Acta Crystallogr. Sect. D Biol. Crystallogr 62, 859–866. https://doi.org/10.1107/S0907444906019949 [DOI] [PubMed] [Google Scholar]
- Minor W, Dauter Z, Helliwell JR, Jaskolski M, Wlodawer A, 2016. Safeguarding structural data repositories against bad apples. Structure 24, 216–220. https://doi.org/10.1016/j.str.2015.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica MF, Bonomo RA, Fast W, 2016. B1-Metallo-β-Lactamases: Where Do We Stand? Curr. Drug Targets 17, 1029–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Skubák P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA, 2011. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. Sect. D Biol. Crystallogr 67, 355–367. https://doi.org/10.1107/S0907444911001314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naas T, Oueslati S, Bonnin RA, Dabos ML, Zavala A, Dortet L, Retailleau P, Iorga BI, 2017. Beta-lactamase database (BLDB)–structure and function. J. Enzyme Inhib. Med. Chem 32, 917–919. https://doi.org/10.1080/14756366.2017.1344235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W, 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326. https://doi.org/10.1016/S0076-6879(97)76066-X [DOI] [PubMed] [Google Scholar]
- Painter J, Merritt EA, 2006. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. Sect. D Biol. Crystallogr 62, 439–450. https://doi.org/10.1107/S0907444906005270 [DOI] [PubMed] [Google Scholar]
- Porebski PJ, Sroka P, Zheng H, Cooper DR, Minor W, 2017. Molstack—Interactive visualization tool for presentation, interpretation, and validation of macromolecules and electron density maps. Protein Sci. n/a--n/a. https://doi.org/10.1002/pro.3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozharski E, Weichenberger CX, Rupp B, 2013. Techniques, tools and best practices for ligand electron-density analysis and results from their application to deposited crystal structures. Acta Crystallogr. Sect. D Biol. Crystallogr 69, 150–167. https://doi.org/10.1107/S0907444912044423 [DOI] [PubMed] [Google Scholar]
- Prinz F, Schlange T, Asadullah K, 2011. Believe it or not: How much can we rely on published data on potential drug targets? Nat. Rev. Drug Discov 10, 712–713. https://doi.org/10.1038/nrd3439-c1 [DOI] [PubMed] [Google Scholar]
- Read RJ, Adams PD, Arendall WB, Brunger AT, Emsley P, Joosten RP, Kleywegt GJ, Krissinel EB, Lütteke T, Otwinowski Z, Perrakis A, Richardson JS, Sheffler WH, Smith JL, Tickle IJ, Vriend G, Zwart PH, 2011. A new generation of crystallographic validation tools for the Protein Data Bank. Structure 19, 1395–1412. https://doi.org/10.1016/j.str.2011.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp B, Wlodawer A, Minor W, Helliwell JR, Jaskolski M, 2016. Correcting the record of structural publications requires joint effort of the community and journal editors. FEBS J. 283, 4452–4457. https://doi.org/10.1111/febs.13765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalin I, Dauter Z, Jaskolski M, Minor W, Wlodawer A, 2015. Crystallography and chemistry should always go together: A cautionary tale of protein complexes with cisplatin and carboplatin. Acta Crystallogr. Sect. D Biol. Crystallogr 71, 1965–1979. https://doi.org/10.1107/S139900471500629X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siemann S, Clarke AJ, Viswanatha T, Dmitrienko GI, 2003. Thiols as classical and slow-binding inhibitors of IMP-1 and other binuclear metallo-β-lactamases. Biochemistry 42, 1673–1683. https://doi.org/10.1021/bi027072i [DOI] [PubMed] [Google Scholar]
- Tioni MF, Llarrull LI, Poeylaut-Palena AA, Marti MA, Saggu M, Periyannan GR, Mata EG, Bennett B, Murgida DH, Vila AJ, 2008. Trapping and Characterization of a Reaction Intermediate in Carbapenem Hydrolysis by B. cereus Metallo-β-lactamase. J. Am. Chem. Soc 130, 15852–15863. https://doi.org/10.1007/s11103-011-9767-z.Plastid [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh TR, Toleman MA, Poirel L, Nordmann P, 2005. Metallo-β-lactamases: The quiet before the storm? Clin. Microbiol. Rev 18, 306–325. https://doi.org/10.1128/CMR.18.2.306-325.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS, 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr. Sect. D Biol. Crystallogr 67, 235–242. https://doi.org/10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodawer A, Dauter Z, Porebski PJ, Minor W, Stanfield R, Jaskolski M, Pozharski E, Weichenberger CX, Rupp B, 2017. Detect, Correct, Retract: How to manage incorrect structural models . FEBS J. in press. https://doi.org/10.1111/FEBS.14320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodawer A, Minor W, Dauter Z, Jaskolski M, 2013. Protein crystallography for aspiring crystallographers or how to avoid pitfalls and traps in macromolecular structure determination. FEBS J. 280, 5705–5736. https://doi.org/10.1111/febs.12495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodawer A, Minor W, Dauter Z, Jaskolski M, 2008. Protein crystallography for non-crystallographers, or how to get the best (but not more) from published macromolecular structures. FEBS J. 275, 1–21. https://doi.org/10.1111/j.1742-4658.2007.06178.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization, 2017. Prioritization of pathogens to guide discovery, research and development of new antibiotics for drug-resistant bacterial infections, including tuberculosis.
- Zheng H, Chordia MD, Cooper DR, Chruszcz M, Müller P, Sheldrick GM, Minor W, 2014. Validation of metal-binding sites in macromolecular structures with the CheckMyMetal web server. Nat. Protoc 9, 156–170. https://doi.org/10.1038/nprot.2013.172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Cooper DR, Porebski PJ, Shabalin IG, Handing KB, Minor W, 2017. CheckMyMetal: A macromolecular metal-binding validation tool. Acta Crystallogr. Sect. D Struct. Biol 73, 223–233. https://doi.org/10.1107/S2059798317001061 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.