Abstract
In eukaryotes, the nuclear envelope (NE) physically separates nuclear components and activities from rest of the cell. The NE also provides rigidity to the nucleus and contributes to chromosome organization. At the same time, the NE is highly dynamic; it must change shape and rearrange its components during development and throughout the cell cycle, and its morphology can be altered in response to mutation and disease. Here we focus on the NE of budding yeast, Saccharomyces cerevisiae, which has several unique features: it remains intact throughout the cell cycle, expands symmetrically during interphase, elongates during mitosis and, expands asymmetrically during mitotic delay. Moreover, its NE is safely breached during mating and when large structures, such as nuclear pore complexes and the spindle pole body, are embedded into its double membrane. The budding yeast NE lacks lamins and yet the nucleus is capable of maintaining a spherical shape throughout interphase. Despite these eccentricities, studies of the budding yeast NE have uncovered interesting, and likely conserved, processes that contribute to NE dynamics. In particular, we discuss the processes that drive and enable NE expansion and the dramatic changes in the NE that lead to extensions and fenestrations.
The nuclear envelope (NE) in all eukaryotes is a physical barrier separating the cytoplasm from chromosomes and other nuclear constituents. It comprised two lipid bilayers, the outer nuclear membrane (ONM) and the inner nuclear membrane (INM). The ONM is continuous with both the ER membrane and the INM; in fact, the NE can be viewed as a flattened ER sheet, with the lumen between the inner and ONMs being continuous with the ER lumen (Walters et al., 2012). Nuclear pore complexes (NPCs), which are large multiprotein structures of approximately 60–120 MDa (depending on the organism), span both outer and INMs and are responsible for selective transport of materials in and out of the nucleus (Capelson et al., 2011). Also traversing the NE are LINC complexes, which transduce mechanical signals between the nucleoskeleton (e.g., the nuclear lamina) and cytoskeleton, and contribute to the constant spacing between the inner and ONMs when the nucleus experiences mechanical stress (Cain and Starr, 2015; Chang et al., 2015). Underlying the INM is the nuclear lamina, a network of lamin intermediate filaments and other proteins, that provide structural rigidity to the nucleus and contributes to chromosome organization (Gruenbaum and Foisner, 2015). No lamina in the form of intermediate filaments has been identified in fungi or plants, although lamin-associated proteins are present and functionally important (Grund et al., 2008; Mekhail et al., 2008; Gonzalez et al., 2012).
In metazoans, the NE breaks down during mitosis (known as “open mitosis”). This process entails disassembling the macromolecular structures that make up the NE, including the NPC and the nuclear lamina, and retraction of the nuclear membranes into the endoplasmic reticulum (ER). NE breakdown is necessary to allow spindle microtubules, emanating from cytoplasmic centrosomes, to contact chromosomes, and segregate them into the future daughter cells. Dismantling the nuclear compartment also allows cytoplasmic factors to contribute to mitotic progression, but at the same time it necessitates the existence of mechanisms that prevent cellular components from interfering with spindle assembly and chromosome segregation (Smyth et al., 2012; Schlaitz et al., 2013; Schweizer et al., 2015). The NE reforms once chromosome segregation is complete at the end of mitosis, re-establishing the nuclear compartment.
In contrast to metazoans, the NE of certain fungi, such as the fission yeast Schizosaccharomyces pombe and the budding yeast Saccharomyces cerevisiae, does not breakdown during mitosis (known as “closed mitosis”) (Arnone et al., 2013). The centrosome equivalent in these yeasts, the spindle pole body, is embedded in the NE such that it nucleates intranuclear spindle microtubules, circumventing the need to disassemble and then reassemble the NE, and preventing cellular structures from getting in the way of the spindle and its associated chromosomes. However, this seemingly efficient arrangement poses challenges in other aspects of nuclear biology: during chromosome segregation when the nucleus and its NE have to elongate for chromosomes to move apart; during mating when the components of two nuclei must intermix by breaching the NE; and when new large and complex structures, such as the spindle pole bodies and NPCs, must be inserted into an intact NE without breaching its permeability barrier. In this review, we describe the dynamic nature of the budding yeast NE that allows it to remain intact yet malleable. We also discuss the many open questions regarding the dynamic nature of the NE in yeast and other systems.
NE Expansion During the Cell Cycle
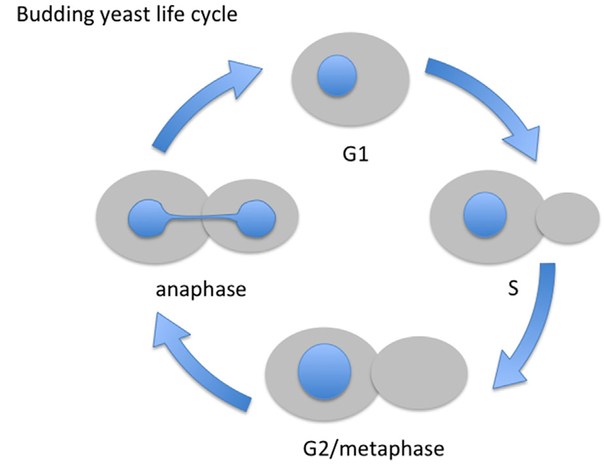
During interphase of vegetative growth, the budding yeast nucleus is typically round or slightly ovoid (Fig. 1). To enter a new cell cycle yeast must reach a critical size, after which budding, or the generation of a daughter cell, ensues (Futcher, 2006). During G1, S and G2 (which in budding yeast is short to nonexistent), cell size continues to increase, although most of the growth is confined to the daughter cell. Interestingly, in both budding and fission yeast, nuclear size scales with cell size throughout the cell cycle (Jorgensen et al., 2007; Neumann and Nurse, 2007), meaning that as the cell increases in volume, so does the nucleus. A constant ratio between nuclear and cell volume is seen in other organisms, and aberrant nucleocytoplasmic ratio is indicative of disease such as cancer (Edens et al., 2013). The mechanisms that regulate nuclear scaling in any organism are largely unknown.
Fig. 1.
The budding yeast life cycle. The cell cycle of budding yeast can be followed by bud size and nuclear shape (in blue). Cells are born in G1 without a bud. As the cell reaches a critical size it initiated DNA replication (S phase) and a bud forms. The bud continues to grow as cells traverse G2 and reach metaphase. At the metaphase to anaphase transition one copy of each of the sister chromatids begins to segregate to the bud (not shown) and the anaphase nucleus elongates to accommodate this segregation.
During interphase, the budding yeast nucleus remains within the mother cell and grows isometrically, maintaining its spherical shape. As yeast lack a nuclear lamina in the form of intermediate filaments such as lamins, there must be a different mechanism, currently unknown, that dictates nuclear shape. It has been proposed that the spherical nucleus, at least in fission yeast, is a consequence of minimizing the free energy of the NE (Lim et al., 2007). Moreover, a stiff nuclear lamina may be dispensable for maintaining nuclear shape in fungi and plants due to the presence of a cell wall that can protect the cell and its nucleus against deformation due to mechanical forces.
As budding yeast cells go through mitosis, a full complement of chromosomes must reach the bud. Therefore, the nucleus must elongate and the NE must increase in surface area to accommodate the movement of chromosomes into the bud (Fig. 1). Nuclear elongation is facilitated by the elongation of the intra-nuclear spindle into the bud, and because of the narrow opening at the bud neck the nucleus adopts an hourglass shape. However, the surface area of the nucleus increases even in the absence of a spindle (Walters et al., 2014), suggesting that NE expansion is regulated directly by the cell cycle machinery. The processes that drive and regulate the extent of NE expansion, for example, the source and amount of membrane added to the NE, are poorly understood. It may depend, at least in part, on the balance between phospholipids that give rise to membranes and neutral lipids that are stored in lipid droplets (Barbosaet al., 2015 and see below). Late in mitosis, when the chromosomes clear the bud neck and chromosome segregation is completed, the nucleus divides through an unknown mechanism to yield two spherical nuclei, one in the mother and one in the daughter (Fig. 1).
An interesting “spin” on the NE expansion problem was discovered in the yeast Schizosaccharomyces japonicus. The NE of this yeast does not disassemble during mitosis, nor does it expand. Instead, as the nucleus elongates during chromosome segregation the NE ruptures, creating visible holes. Importantly, these holes are not due to strain on the NE caused by spindle elongation because NE breaks appear even if nuclear elongation is blocked (Yam et al., 2013). The difference between fungal NEs that expand (e.g., budding and fission yeast) and those that do not appears to be in the way lipid synthesis is regulated by the cell cycle machinery (Makarova et al., 2016), lending further support to the idea that the increase in nuclear surface area in yeasts during mitosis is regulated by pathways controlling lipid synthesis. Indeed, activating a key regulator of lipid synthesis in budding yeast, Pah1 (formerly known as Smp2), which promotes the formation of neutral lipids at the expense of certain phospholipids, blocks NE expansion during mitosis (Santos-Rosa et al., 2005 and see below). Interestingly, lipid synthesis appears to be a major contributor to asymmetric NE expansion during a mitotic delay (Witkin et al., 2012), as will be discussed in the next section.
Mitosis is not the only budding yeast process during which the nucleus elongates; nuclear elongation is also observed when cells arrest in G1 in preparation for mating (Stone et al., 2000). Haploid budding yeast can have either of two mating types, a or α, and they can diploidize by mating with a cell of the opposite mating type. Mating is triggered when cells are exposed to mating pheromones from the opposite mating type (reviewed in Merlini et al., 2013). Under these conditions, cells arrest in G1 and develop a projection, or shmoo, toward the source of the mating pheromone. If two cells of opposite mating types are in close proximity, these cells will fuse, followed by the fusion of their nuclei, as will be discussed in a later section. The exposure to mating pheromone also induces nuclear elongation, but the mechanism and functional consequences of this elongation are not clear; while mitotic nuclear elongation is essential for chromosome segregation, it is not known whether and how nuclear elongation in response to pheromone contributes to the mating process.
Nuclear Flares: Asymmetric Expansion of the NE
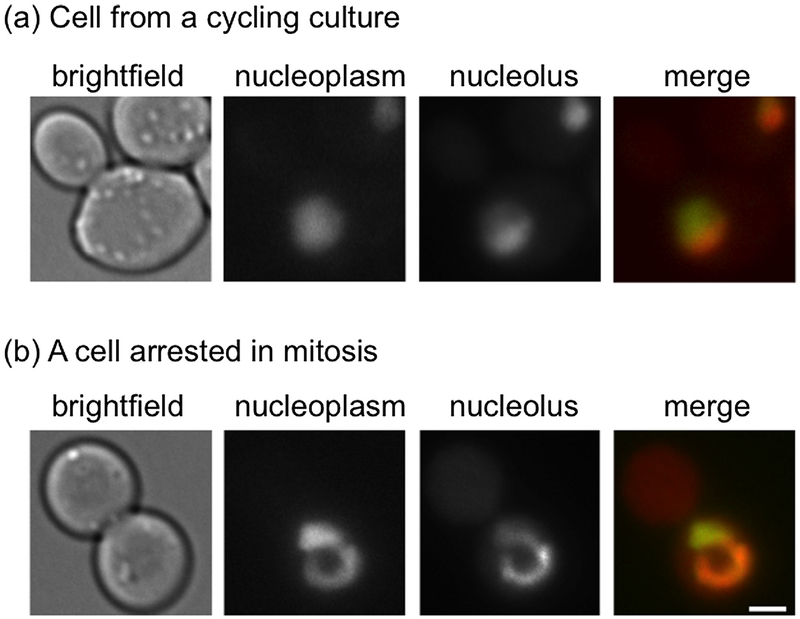
Cell cycle progression is subjected to regulation by various checkpoint pathways that delay cell cycle progression until proper structures have been assembled and certain processes have been faithfully completed. For example, failure to attach all paired sister chromatids to the mitotic spindle in a bipolar fashion activates the spindle assembly checkpoint (SAC) pathway, which blocks anaphase initiation until proper chromosome attachment is achieved (Musacchio, 2015). Interestingly, when budding yeast cells are arrested in mitosis by the SAC, the nucleus continues to expands, as it would in an unperturbed cell cycle, but the expansion is asymmetric, creating a single extension, or “flare” (Witkin et al., 2012) (Fig. 2). This flare is confined to the NE adjacent to the nucleolus, which in budding yeast forms a crescent shaped structure at the nuclear periphery (Fig. 2). What dictates this particular localization of the flare is not known, but the formation of the flare is independent of an increase in nucleolar volume (Walters et al., 2014). The confinement of the flare to the NE adjacent to the nucleolus may be due to the presence of a structure that prevents NE expansion away from the nucleolar region; alternatively, the NE adjacent to the nucleolus may have properties that make it more conducive to expansion (e.g., a unique lipid composition or the absence of a tethering factor), analogous to a weak patch on a balloon.
Fig. 2.
Flare formation during a mitotic delay. (a) In cycling cells, the nucleolus forms a crescent-shaped structure at the edge of the nucleus.(b) In cells delayed in mitosis the nucleolus forms a flare that extends away from the rest of the nucleus. The nucleoplasm was detected using Pus1-GFP. The nucleolus was detected using Nsr1-mCherry. Scale bar is 2 μm. Images were taken by Alison Walters (NIH, NIDDK).
Why the flare forms is somewhat better understood: while SAC activation stops both cell cycle progression and cell growth, the NE continues to expand, likely because lipid synthesis is not affected by the SAC (Witkin et al., 2012). Membrane may flow directly from the peripheral ER to the NE, but the extent to which this happens and how this could be regulated remain to be determined. One key enzyme that may drive NE expansion in budding yeast is the conserved phosphatidic acid hydrolase, Pah1 (lipin in metazoans) (Santos-Rosa et al., 2005; Han et al., 2006; Siniossoglou, 2013). Pah1 converts phosphatidic acid to diacylglycerol, some of which is, in turn, converted to triacylglycerol and stored in lipid droplets. Phosphatidic acid is also a precursor for phospholipids. Thus, Pah1 activity contributes to the balance between membrane synthesis and lipid storage. The activity of Pah1 is regulated by multiple kinases, including the cell cycle kinase Cdk1, and by a phosphatase complex composed of Spo7 and Nem1 (Santos-Rosa et al., 2005). Moreover, Spo7/Nem1 are responsible for Pah1’s recruitment to the ER/NE, and during starvation Pah1 associates with the NE at sites of lipid droplets (Karanasios et al., 2013; Barbosa et al., 2015). Thus, ER and NE expansion could be controlled, at least in part, by Pah1’s activity as a molecular switch regulating the flow of lipids to either phospholipid synthesis (when Pah1 is inactive) or storage of neutral lipids (when Pah1 is active). Interestingly, when Pah1 is inactivated, cells exhibit an increase in the level of total phospholipids and the nucleus develops a single flare that localizes to the NE adjacent to the nucleolus, much like the mitotic flare (Siniossoglou et al., 1998; Santos-Rosa et al., 2005; Campbell et al., 2006).
Why, then, during a mitotic delay or in the presence of excess phospholipids, does the nucleus not simply expand isometrically, forming a larger, spherical nucleus? The reason for this may lie in the cell’s adherence to a constant nuclear/cell volume ratio: During a mitotic delay cell growth is halted. In order to expand the NE without increasing nuclear volume the NE must deform in some way, and the cell’s solution appears to be flare formation. In other words, the cell may use the NE adjacent to the nucleolus as a “membrane sink” (Witkin et al., 2012). Vesicle transport also appears to play a role in regulating NE morphology because combining certain vesicle transport mutants with mutations that inactivate Pah1 leads to the formation of multiple flares while maintaining its nuclear/cell volume ratio (Webster et al., 2010). Thus, the NE can deform under both normal conditions, such as a mitotic delay, and during an induced imbalance in lipid synthesis, perhaps to maintain a constant nuclear/cell volume ratio. The functional significance of these deformations, what directs the flare to form adjacent to the nucleolus, and the consequences to the cell if the ability to deform the NE was blocked, are not known.
Piecemeal Microautophagy of the Nucleus: Another Form of NE Extension
Because the NE remains intact throughout the yeast cell’s entire life, a mechanism must exist for removing misfolded and aggregated proteins from of the nucleus. One such mechanism likely involves proteosomal degradation, either within the nucleus or after being exported to the cytoplasm (Chen and Madura, 2014). Another mechanism is piecemeal microautophagy of the nucleus (PMN) that occurs under carbon- or nitrogen-limited conditions (Roberts et al., 2003, reviewed in Krick et al., 2014; Mijaljica et al., 2010). PMN is one of several bulk autophagic processes in the cell, in contrast to proteosomal degradation that requires specific targeting of individual proteins. In PMN, non-essential parts of the nucleus are selectively degraded and recycled by the vacuole, the lysosome equivalent in yeast. Recycling of the nuclear compartment in other organisms is sometimes referred to a nucleophagy. In mammalian cells, this processes involves autophagosomes that engulf nuclear components from outside the nucleus and target them to the lysosome (Mijaljica and Devenish, 2013). The involvement of autophagosomes qualifies this process as macroautophagy (Devenish and Klionsky, 2012). Unlike nucleophagy in mammalian cells, during PMN the transfer of material from the nucleus to the vacuole occurs through direct contact between the two organelles at the nuclear vacuole (NV) junction (Pan et al., 2000; Roberts et al., 2003; Kvam and Goldfarb, 2004; Kvam et al., 2005; Kvam and Goldfarb, 2006; Dawaliby and Mayer, 2010). Because material is engulfed directly by the vacuole, namely without the involvement of autophagosomes, this process is defined as microautophagy (Devenish and Klionsky, 2012). PMN is initiated by the formation of small, teardrop-shaped invaginations of the NE directly into the vacuole, forming a tri-membranous extension (the two NE membranes plus the vacuole membrane). These extensions are subsequently pinched off into the vacuole and degraded by vacuolar hydrolases (for review, see Mijaljica and Devenish, 2013). A core suite of autophagy genes also involved in macroautophagy have been shown to be required for final stages of PMN, though not for the initial formation of NV junctions (Krick et al., 2008). The NV junction is formed through the interactions of the NE protein Nvj1 and the vacuolar protein Vac8, and the size of the NV junction is dictated by the amount of Nvj1, which is regulated by nutrient availability (Pan et al., 2000; Roberts et al., 2003). Lipid biosynthesis proteins, and specifically the ER membrane-localized enoyl-CoA reductase Tsc13 and the oxysterol binding protein Osh1, are both targeted to the NV junction by Nvj1 and have been implicated in PMN, suggesting a role for very-long-chain fatty acid and sterols in the ability to form NE extensions into the vacuole (Kvam and Goldfarb, 2004; Kvam et al., 2005). Furthermore, inhibition of fatty acid synthesis by cerulenin also blocks the formation of NE extensions into the vacuole during PMN (Kvam et al., 2005), reminiscent of the requirement of phospholipid synthesis in mitotic flare formation described above. PMN also requires an electrochemical gradient generated by the vacuolar ATPase, both to create invaginations of the nucleus at NV junctions and for scission of the invaginations. However, this gradient is not necessary to maintain the nuclear invaginations once they are formed (Dawaliby and Mayer, 2010). There is still much to be learned about the precise mechanisms governing NE morphology during PMN. Moreover, unlike the mitotic flares described above, PMN is not confined to the NE adjacent to the nucleolus and is not specific to any cell cycle phase. Interestingly, NPCs are typically absent from the NV junctions and from the regions of the NE engulfed by the vacuole (Pan et al., 2000; Roberts et al., 2003). Given the high mobility of NPCs (Belgareh and Doye, 1997; Bucci and Wente, 1997), this observation points to the existence of a yet unknown mechanism preventing NPCs from entering the area of interaction between the nucleus and the vacuole, and raising the possibility that the NE may contain diffusion barriers.
Nuclear Fusion During Mating: The Two-Membrane Challenge
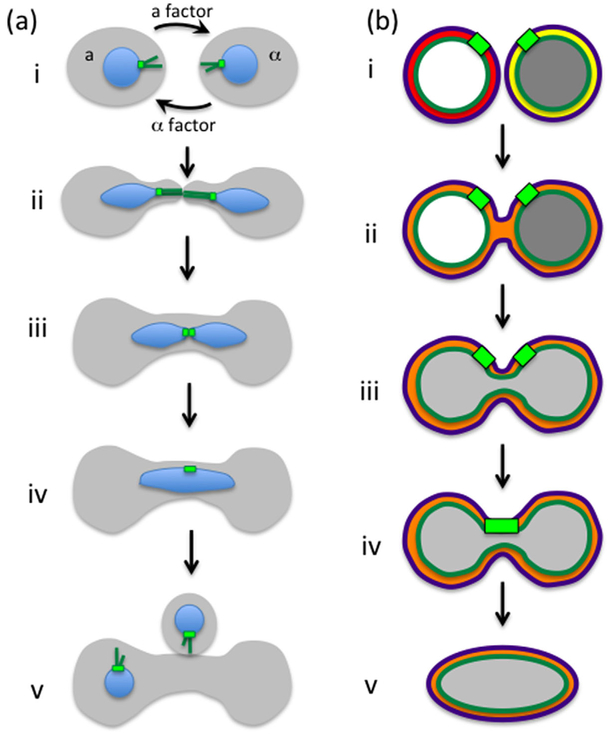
Fusion of membranes is a common occurrence in cellular biology, such as during vesicular trafficking, cell fusion during fertilization and muscle development, and cellular invasion by certain viruses. These fusion events are facilitated by protein complexes (e.g., SNAREs, Risselada and Grubmüller, 2012) and may also involve changes in membrane tension (Kozlov and Chernomordik, 2015). In all these cases, fusion is mediated between two bodies that are encapsulated by a single membrane. Nuclear fusion, however, requires the coordinated fusion of two membranes: the ONM and the INM. In yeast, nuclear fusion, also known as karyogamy, occurs as part of the mating process between cells of opposite mating type (for review, see Gibeaux and Knop, 2013). As described earlier, during mating, cells adhere to one another at the shmoo tip via agglutinins, at which point the cell walls break down, the plasma membranes come into contact and the cells fuse, forming a zygote (Fig. 3). Upon cell fusion, the two nuclei congress at the center of the zygote in a process mediated by microtubules that emanate from the outer-nuclear surface of the spindle pole body (SPB) of each nucleus. Once nuclear congression is completed, the NEs of both nuclei fuse to create a single diploid nucleus.
Fig. 3.
Mating and nuclear fusion (karyogamy). (a) Stages of mating: (i) cells of opposite mating types, a and a, secrete pheromones (a factor and a factor) that are received by the other cell type, activating the mating pathway. (ii) In response to pheromone, cells generate mating projections (shmoos) in the direction of the pheromone gradient. In addition, nuclei become elongated and orient toward the shmoo tip by the action of cytoplasmic microtubules (dark green) that emanate from the cytoplasmic face of the SPB (light green). (iii) Once the cells come in close contact they fuse, allowing microtubules from the two nuclei to interdigitate and pull the two nuclei toward each other. (iv) Once nuclei come in close contact they fuse to form a diploid nucleus. (v) The zygote re-enters the cell cycle, buds, and completes nuclear division in the subsequent mitosis. (b) Stages of nuclear fusion: (i) Nuclei before mating. The lumens of the nuclei (between the ONM in purple and the INM in dark green) are labeled either red or yellow. The nucleoplasms are labeled in either white or gray. (ii) Fusion of the ONMs results in mixing of the lumens (orange). (iii) Fusion of the INMs results in mixing of the nucleoplasms (light gray). (iv) The SPBs fuse. (v) The nucleus rounds up.
Genetic and electron microscopy studies, together with time-lapse fluorescence microscopy analyses that allowed the visualization of intermediate steps in the mating process, led to a step-wise model for NE fusion and identified some of the proteins involved in each step (Conde and Fink, 1976; Polaina and Conde, 1982; Kurihara et al., 1994; Beh et al., 1997; Melloy et al., 2007) (Fig. 3b). To date, however, the molecular function of most proteins involved in nuclear fusion is still unknown. The first step of NE fusion involves the fusion of the ONMs, resulting in the mixing the lumens of the perinuclear spaces of the two nuclei. This process depends on Prm3, and to a lesser extent on Kar5 (Heiman and Walter, 2000; Melloy et al., 2009; Shen et al., 2009). Following ONM fusion, the INMs fuse in a process that depends on Kar2, Kar5, and Kar8/Jem1 (Nishikawa and Endo, 1997; Melloy et al., 2009; Rogers and Rose, 2015). Interestingly, both Kar2 and Kar8/Jem1 are ER chaperones and several of the proteins involved in protein translocation to the ER (namely components of the Sec63 complex) are also needed for efficient nuclear fusion (Brizzio et al., 1999). Whether these protein translocation factors are needed directly to promote fusion or indirectly to facilitate the ER/NE localization of karyogamy proteins is not known. The fusion of the INMs allows the content of both nuclei to mix. Finally, the two spindle pole bodies, which were already in close proximity, fuse and form a single SPB. Notably, none of proteins listed above appear to have the characteristics of a protein that can mediate membrane fusion. This led Rogers et al. (2013) to purposefully screen through mutants in proteins known to be involved in membrane fusion for their possible involvement in nuclear fusion. Their screen yielded several SNARE proteins that appear to affect ONM fusion during karyogamy (Rogers et al., 2013). The exact steps that require SNARE activity remain to be elucidated.
While much remains to be discovered, yeast karyogamy is a tractable system to understand the process of nuclear fusion that occurs during fertilization in plants and in certain metazoans. For example, Kar5 is an evolutionarily conserved protein (Ning et al., 2013), and the zebrafish homolog of Kar5, Brambleberry, has been implicated in the fusion of karyomeres, which are nuclei that form around individual chromosomes during early embryogenesis (Abrams et al., 2012). Thus, knowledge gained from yeast karyogamy will likely have broader implications, not only in cell fusion during fertilization but perhaps also in other cell fusion events.
NE Fenestration: Inserting Large Complexes Into Intact Membranes
Because the NE of budding yeast remains intact throughout the cell cycle, insertion of large multiprotein structures, such as NPCs and the SPB, into the NE requires fenestration of the two nuclear membranes. In fact, given the evolutionary conservation in NPC structure and composition, the mechanism of NPC insertion in yeast likely resembles the mechanism of NPC insertion during interphase in metazoan cells, after NE reassembly at the end of mitosis has been completed. How can holes effectively be punched into the NE without the contents of the nucleus escaping, or unwanted cytoplasmic elements entering? And how are these complexes assembled in the NE? Clearly, fenestration and assembly must be carefully choreographed to avoid deleterious effects on the cell. As it turns out, the insertion of NPCs and the SPB into the NE takes place in a series of steps to minimize the risks associated with opening up the membrane.
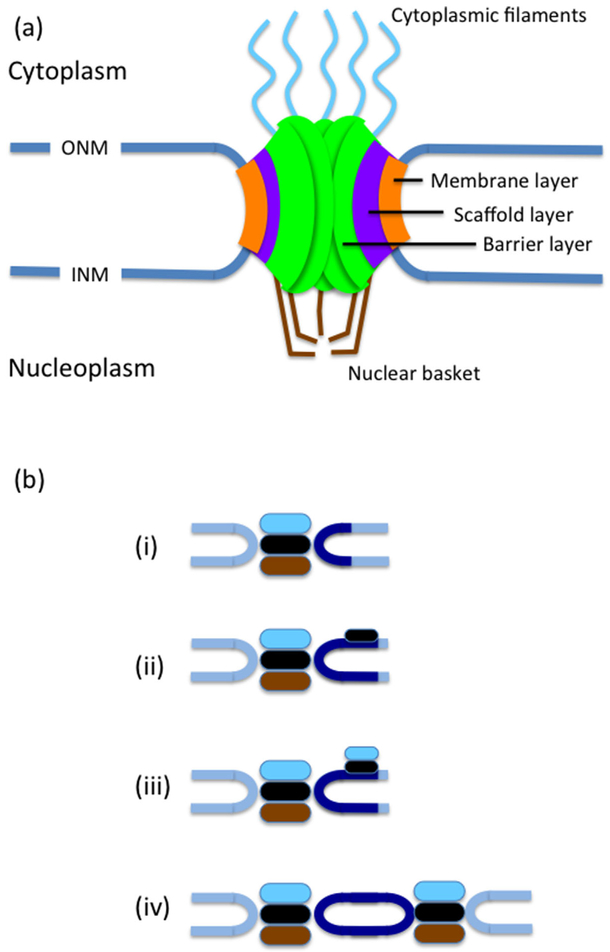
NPCs span the entirety of the NE and are surrounded by a highly curved membrane that forms via the fusion of the inner and outer nuclear membranes (Hetzer and Wente, 2009; Doucet and Hetzer, 2010) (Fig. 4a). All NPCs have at eightfold radial symmetry around a central pore through which trafficking occurs. The structure of the NPC can be broadly divided into three layers (starting from the outermost part): the membrane layer, which is embedded in the NE where the ONM and the INM meet, the scaffold layer, which stabilizes the pore, and the barrier layer, through which selective transport in and out of the nucleus occurs. Budding yeast NPCs are somewhat smaller than NPCs in vertebrates, being ~100 nm in diameter (compared to ~130 nm diameter in vertebrates), and they are built from multiples of 30 different proteins called nucleoporins (nups) (Aitchison and Rout, 2012). NPC biogenesis and insertion occurs throughout the cell cycle, leading to nearly double the number of nuclear pores in early mitosis as compared to G1 (Winey et al., 1997). It is also important to note that the composition of the NPC is not static and that NPC subunits can exchange with soluble subunits (see Tran and Wente, 2006). Moreover, unlike vertebrate NPCs which are largely immobile, perhaps due to anchorage by the nuclear lamina, yeast NPCs are highly mobile and move laterally within the NE (Bucci and Wente, 1997).
Fig. 4.
The NPC and SPB. (a) A simplified diagram of the NPC, showing the membrane layer embedded in the membrane connecting the outer and inner nuclear membranes, the scaffold layer and the barrier layer. In addition the NPC has a nuclear basket and cytoplasmic filaments. (b) Stages of SPB insertion: (i) The SPB is composed of 3 plaques, an outer plaque (light blue), central plaque (black) and inner plaque (brown). Next to the SPB is the half bridge (dark blue). (ii) The half bridge extends and the satellite assembles at its distal end.(iii) Outer plaque components are added to the satellite. (iv) The new SPB is inserted into the NE and components of the inner plaque are added.
How is the NE remodeled to allow NPC insertion? Studies both in yeast and animal models have shed some light on this process. Cell-free assays using nuclei assembled from Xenopus egg extracts have demonstrated that new NPCs are built in a stepwise fashion (D’Angelo et al., 2006). EM studies identified NE regions where the ONM and INM appear to move closer together and fuse (Goldberg et al., 1997), suggesting that these could be sites of NPC insertion. The mechanism of ONM and INM fusion is poorly understood, but it is likely facilitated by transmembrane NPC subunits called pore membrane proteins (Poms), which are present in the membrane layer, and by reticulons, which are ER-associated proteins that can directly bend membrane (Madrid et al., 2006; Voeltz et al., 2006; Dawson et al., 2009). Reticulons are not present in mature NPCs, suggesting that they are only needed for the initial membrane-bending step. The increasingly curved membranes are thought to be further stabilized by nucleoporins that form a core scaffold (the scaffold layer) and whose structures resemble those of clathrin, COPI and COPII membrane vesicle coating complexes (for review, see Onischenko and Weis, 2011). Membrane bending during NPC insertion may also involve a local modulation of membrane lipid composition (Schneiter et al., 1996; Scarcelli et al., 2007). Upon the stabilization of the membranes, the remainder of the nups can be successfully added to form the entire NPC.
Like the NPCs, the budding yeast SPB is embedded in the NE (Fig. 4b). The SPB is the fungal equivalent of the metazoan centrosome, and it is the site of nucleation for both cytoplasmic microtubules, which move the nucleus within the cell, and intranuclear microtubules, which form the mitotic spindle (Jaspersen and Winey, 2004). Following cell division, each daughter cell inherits only a single SPB. Thus, in order to form a functional bipolar spindle, cells must first duplicate the SPB once, and only once, and ensure that it is properly situated within the NE. The SPB duplicates conservatively, namely a new SPB is formed in the NE next to the old one. The SPBs are then moved away from each other by forces exerted by motors associated with interdigitating microtubules emanating from both SPBs (Jaspersen and Winey, 2004), ultimately forming a bipolar spindle.
Similar to the process of inserting NPCs, insertion of the budding yeast SPB happens in stages (Jaspersen and Ghosh, 2012). SPB duplication begins during late G1, when a new SPB begins to assemble near the site of the preexisting SPB (Byers and Goetsch, 1975). Initially, a satellite, composed of cytoplasm-localized SPB components, is formed on the distal tip of a NE region called the half bridge, adjacent to the old SPB. Next, the satellite and half-bridge expand to form a duplication plaque, still on the cytoplasmic face of the NE. The duplication plaque is inserted into the NE from the cytoplasmic side in a process that depends on INM protein Nbp1(Kupke et al., 2011), resulting full insertion into the NE. Nbp1 contains amphipathic α-helices (Kupke et al., 2011), which can sense high membrane curvature and are also present in several NPC subunits of the scaffold layer (Doucet and Hetzer, 2010). The insertion of the duplication plaque allows the remaining nuclear components of the SPB to be added. Interestingly one of the insertion factors, Ndc1, is shared with the NPC insertion machinery suggesting that insertion of NPCs and the SPB may follow similar principles. SPB insertion requires at least two additional proteins, Mps2 and its interacting protein Bbp1, but their exactly contribution to the process is not known. The mechanism of SPB insertion must require fusion of the inner and outer nuclear membranes in a process that includes proteins that stabilize high curvature membranes. The exact nature of these proteins remains to be determined; the structure of the SPB was reported to be abnormal in mutants lacking reticulons and Yop1, which are membrane-bending proteins also involved in NPC insertion (Casey et al., 2012).
However, cells lacking reticulons and Yop1 are viable, indicating that these proteins are not necessary for SPB insertion. It has also been suggested that the lipid composition of the membrane at the site of insertion may be modified to facilitate high membrane curvature in both budding yeast and fission yeast (Witkin et al., 2010; Friederichs et al., 2011; Tamm et al., 2011).
Conclusions and Open Questions
The NE of budding yeast is a dynamic membrane system that is capable of expanding throughout the cell cycle, opening up for new NPC and SPB insertion, and merging with the NE of a mating partner during karyogamy. Although we know much about the components that facilitate these processes, the molecular mechanisms governing these structural changes are mostly obscure. For example, what allows the NE, and in particular the nuclear membranes to expand? Regulation of lipid synthesis clearly plays an important role in allowing NE expansion, but the spatial and temporal regulation of this process, as well as the possible involvement of other pathways for membrane expansion, still remains to be resolved. The asymmetric expansion of the NE, for example, during a mitotic arrest, hints to the possible existence of diffusion barriers within the NE. Indeed, NPCs and other proteins are excluded from the NV junction, giving further credence of the idea that the NE is a heterogeneous environment. The pathways driving this heterogeneity remain to be discovered; in metazoans the underlying lamina can be and is used to restrict movement of NE protein. In the absence of a lamina, diffusion barriers in the budding yeast NE may be associated with organelles and structures that come in contact with the NE, such as the nucleolus, lipid droplets and the vacuole. Another important question that remains to be resolved is what facilitates nuclear membrane bending. This occurs on a small scale, yet with very high curvature, between the inner and outer nuclear membranes when NPCs or the SPB are inserted into the NE, and on a much larger scale when the NE protrudes into the vacuole during PMN and during the formation of a mitotic flare. Finally, the membranes of the NE undergo a variety of fusion events: the ONM with the INM during fenestration and the homotypic ONM-to-ONM and INM-to-INM fusions during mating. In addition, some sort of fusion must also occur during nuclear division in a process that is completely unknown. Given that the NE is part of the ER, some principles in membrane dynamics and remodeling may be shared, but some are likely unique to the NE. Regardless, understanding NE dynamics will reveal fundamental principles in membrane homeostasis, and it is likely that features of NE malleability will apply to our overall understanding of membrane-bound organelle dynamics.
Acknowledgments
We thank Alison Walters for critical reading and comments on the manuscript. The authors were supported by an intramural grant from the National Institute of Diabetes and Digestive and Kidney Diseases.
Literature Cited
- Abrams EW, Zhang H, Marlow FL, Kapp L, Lu S, Mullins MC. 2012. Dynamic assembly of brambleberry mediates nuclear envelope fusion during early development. Cell 150:521–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitchison JD, Rout MP. 2012. The yeast nuclear pore complex and transport through it.Genetics 190:855–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnone JT, Walters AD, Cohen-Fix O. 2013. The dynamic nature of the nuclear envelope. Nucleus 4:261–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa AD, Sembongi H, Su WM, Abreu S, Reggiori F, Carman GM, Siniossoglou S. 2015. Lipid partitioning at the nuclear envelope controls membrane biogenesis. Mol Bio Cell 26:3641–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beh CT, Brizzio V, Rose MD. 1997. KAR5 encodes a novel pheromone-inducible protein required for homotypic nuclear fusion. J Cell Biol 139:1063–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgareh N, Doye V. 1997. Dynamics of nuclear pore distribution in nucleoporin mutant yeast cells. J Cell Biol 136:747–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brizzio V, Khalfan W, Huddler D, Beh CT, Andersen SS, Latterich M, Rose MD. 1999. Genetic interactions between KAR7/SEC71, KAR8/JEM1, KAR5, and KAR2 during nuclear fusion in Saccharomyces cerevisiae. Mol Biol Cell 10:609–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucci M, Wente SR. 1997. In vivo dynamics of nuclear pore complexes in yeast. J Cell Biol 136:1185–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B, Goetsch L. 1975. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J Bacteriol 124:511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain NE, Starr DA. 2015. SUN proteins and nuclear envelope spacing. Nucleus 6:2–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JL, Lorenz A, Witkin KL, Hays T, Loidl J, Cohen-Fix O. 2006. Yeast nuclear envelope subdomains with distinct abilities to resist membrane expansion. Mol Biol Cell 17:1768–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelson M, Doucet C, Hetzer MW. 2011. Nuclear pore complexes: Guardians of the nuclear genome. Cold Spring Harbor Symp Quant Biol 75:585–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey AK, Dawson TR, Chen J, Friederichs JM, Jaspersen SL, Wente SR. 2012. Integrity and function of the saccharomyces cerevisiae spindle pole body depends on connections between the membrane proteins Ndc1, Rtn1, and Yop1. Genetics 192:441–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W, Worman HJ, Gundersen GG. 2015. Accessorizing and anchoring the LINC complex for multifunctionality. J Cell Biol 208:11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Madura K. 2014. Degradation of specific nuclear proteins occurs in the cytoplasm in Saccharomyces cerevisiae. Genetics 197:193–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde J, Fink GR. 1976. A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc Natl Acad Sci 73:3651–3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo MA, Anderson DJ, Richard E, Hetzer MW. 2006. Nuclear pores form de novo from both sides of the nuclear envelope. Science 312:440–443. [DOI] [PubMed] [Google Scholar]
- Dawaliby R, Mayer A. 2010. Microautophagy of the nucleus coincides with a vacuolar diffusion barrier at nuclear-vacuolar junctions. Mol Biol Cell 21:4173–4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson TR, Lazarus MD, Hetzer MW, Wente SR. 2009. ER membrane-bending proteins are necessary for de novo nuclear pore formation. J Cell Biol 184:659–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devenish RJ, Klionsky DJ. 2012. Autophagy: Mechanism and physiological relevance”brewed” from yeast studies. Front Biosci (Schol Ed) 4:1354–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet CM, Hetzer MW. 2010. Nuclear pore biogenesis into an intact nuclear envelope. Chromosoma 119:469–477. [DOI] [PubMed] [Google Scholar]
- Edens LJ, White KH, Jevtic P, Li X, Levy DL. 2013. Nuclear size regulation: From single cells to development and disease. Trends Cell Biol 23:151–159. [DOI] [PubMed] [Google Scholar]
- Friederichs JM, Ghosh S, Smoyer CJ, McCroskey S, Miller BD, Weaver KJ, Delventhal KM, Unruh J, Slaughter BD, Jaspersen SL. 2011. The SUN protein Mps3 is required for spindle pole body insertion into the nuclear membrane and nuclear envelope homeostasis. PLoS Genet 7:e1002365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futcher B 2006. Metabolic cycle, cell cycle, and the finishing kick to Start. Genome Biology 7:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibeaux R, Knop M. 2013. When yeast cells meet, karyogamy! Nucleus 4:182–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg MW, Wiese C, Allen TD, Wilson KL. 1997. Dimples, pores, star-rings, and thin rings on growing nuclear envelopes: Evidence for structural intermediates in nuclear pore complex assembly. J Cell Sci 110:409–420. [DOI] [PubMed] [Google Scholar]
- Gonzalez Y, Saito A, Sazer S. 2012. Fission yeast Lem2 and Man1 perform fundamental functions of the animal cell nuclear lamina. Nucleus 3:60–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum Y, Foisner R. 2015. Lamins: Nuclear intermediate filament proteins with fundamental functions in nuclear mechanics and genome regulation. Annu Rev Biochem 84:131–164. [DOI] [PubMed] [Google Scholar]
- Grund SE, Fischer T, Cabal GG, Ant unez O, P erez-Ortín JE, Hurt E. 2008. The inner nuclear membrane protein Src1 associates with subtelomeric genes and alters their regulated gene expression. J Cell Biol 182:897–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G-S, Wu W-I, Carman GM. 2006. The Saccharomyces cerevisiae Lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J Biol Chem 281:9210–9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman MG, Walter P. 2000. Prm1p, a pheromone-regulated multispanning membrane protein, facilitates plasma membrane fusion during yeast mating. J Cell Biol 151:719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer MW, Wente SR. 2009. Border control at the nucleus: Biogenesis and organization of the nuclear membrane and pore complexes. Dev Cell 17:606–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen SL, Ghosh S. 2012. Nuclear envelope insertion of spindle pole bodies and nuclear pore complexes. Nucleus 3:226–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen SL, Winey M. 2004. The budding yeast spindle pole body: Structure, duplication, and function. Annu Rev Cell Dev Biol 20:1–28. [DOI] [PubMed] [Google Scholar]
- Jorgensen P, Edgington NP, Schneider BL, Rupes I, Tyers M, Futcher B. 2007. The size of the nucleus increases as yeast cells grow. Mol Biol Cell 18:3523–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanasios E, Barbosa AD, Sembongi H, Mari M, Han GS, Reggiori F, Carman GM,Siniossoglou S. 2013. Regulation of lipid droplet and membrane biogenesis by the acidic tail of the phosphatidate phosphatase Pah1p. Mol Biol Cell 24:2124–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov MM, Chernomordik LV. 2015. Membrane tension and membrane fusion. Curr OpStruct Biol 33:61–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krick R, Muehe Y, Prick T, Bremer S, Schlotterhose P, Eskelinen E-L, Millen J, Goldfarb DS,Thumm M. 2008. Piecemeal microautophagy of the nucleus requires the core macroautophagy genes. Mol Biol Cell 19:4492–4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krick R, Mühe Y, Prick T, Bredschneider M, Bremer S, Wenzel D, Eskelinen E-L, Thumm M.2014. Piecemeal microautophagy of the nucleus: Genetic and morphological traits. Autophagy 5:270–272. [DOI] [PubMed] [Google Scholar]
- Kupke T, Di Cecco L, Muller H-M, Neuner A, Adolf F, Wieland F, Nickel W, Schiebel E. 2011. Targeting of Nbp1 to the inner nuclear membrane is essential for spindle pole body duplication. EMBO J 30:3337–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara LJ, Beh CT, Latterich M, Schekman R, Rose MD. 1994. Nuclear congression and membrane fusion: Two distinct events in the yeast karyogamy pathway. J Cell Biol 126:911–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvam E, Goldfarb DS. 2004. Nvj1p is the outer-nuclear-membrane receptor for oxysterol-binding protein homolog Osh1p in Saccharomyces cerevisiae. J Cell Sci 117:4959–4968. [DOI] [PubMed] [Google Scholar]
- Kvam E, Goldfarb DS. 2006. Structure and function of nucleus-vacuole junctions: Outer-nuclear-membrane targeting of Nvj1p and a role in tryptophan uptake. J Cell Sci 119:3622–3633. [DOI] [PubMed] [Google Scholar]
- Kvam E, Gable K, Dunn TM, Goldfarb DS. 2005. Targeting of Tsc13p to nucleus-vacuole junctions: A role for very-long-chain fatty acids in the biogenesis of microautophagic vesicles. Mol Biol Cell 16:3987–3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim HWG, Huber G, Torii Y, Hirata A, Miller J, Sazer S. 2007. Vesicle-like biomechanics governs important aspects of nuclear geometry in fission yeast. PLoS ONE 2:e948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid AS, Mancuso J, Cande WZ, Weis K. 2006. The role of the integral membrane nucleoporins Ndc1p and Pom152p in nuclear pore complex assembly and function. J Cell Biol 173:361–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova M, Gu Y, Chen J-S, Beckley JR, Gould KL, Oliferenko S. 2016. Temporal regulation of lipin activity diverged to account for differences in mitotic programs. Curr Biol 26:237–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhail K, Seebacher J, Gygi SP, Moazed D. 2008. Role for perinuclear chromosome tethering in maintenance of genome stability. Nature 456:667–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melloy P, Shen S, White E, Rose MD. 2009. Distinct roles for key karyogamy proteins during yeast nuclear fusion. Mol Biol Cell 20:3773–3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melloy P, Shen S, White E, McIntosh JR, Rose MD. 2007. Nuclear fusion during yeast mating occurs by a three-step pathway. J Cell Biol 179:659–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlini L, Dudin O, Martin SG. 2013. Mate and fuse: How yeast cells do it. Open Biol 3:130008–130008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijaljica D, Devenish RJ. 2013. Nucleophagy at a glance. J Cell Sci 126:4325–4330. [DOI] [PubMed] [Google Scholar]
- Mijaljica D, Prescott M, Devenish RJ. 2010. The intricacy of nuclear membrane dynamics during nucleophagy. Nucleus 1:213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A 2015. The molecular biology of spindle assembly checkpoint signaling dynamics. Curr Biol 25:R1002–R1018. [DOI] [PubMed] [Google Scholar]
- Neumann FR, Nurse P. 2007. Nuclear size control in fission yeast. J Cell Biol 179:593–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning J, Otto TD, Pfander C, Schwach F, Brochet M, Bushell E, Goulding D, Sanders M, Lefebvre PA, Pei J, Grishin NV, Vanderlaan G, Billker O, Snell WJ. 2013. Comparative genomics in Chlamydomonas and Plasmodium identifies an ancient nuclear envelope protein family essential for sexual reproduction in protists, fungi, plants, and vertebrates. Genes Dev 27:1198–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa SI, Endo T. 1997. The yeast JEM1p is a DnaJ-like protein of the endoplasmic reticulum membrane required for nuclear fusion. J. Biol. Chem 272:12889–12892. [DOI] [PubMed] [Google Scholar]
- Onischenko E, Weis K. 2011. Nuclear pore complex—A coat specifically tailored for the nuclear envelope. Curr Op Cell Biol 23:293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Roberts P, Chen Y, Kvam E, Shulga N, Huang K, Lemmon S, Goldfarb DS. 2000. Nucleus-vacuole junctions in Saccharomyces cerevisiae are formed through the direct interaction of Vac8p with Nvj1p. Mol Biol Cell 11:2445–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polaina J, Conde J. 1982. Genes involved in the control of nuclear fusion during the sexual cycle of Saccharomyces cerevisiae. Mol Gen Genet 186:253–258. [DOI] [PubMed] [Google Scholar]
- Risselada HJ, Grubmüller H. 2012. How SNARE molecules mediate membrane fusion: Recent insights from molecular simulations. Curr Op Struct Biol 22:187–196. [DOI] [PubMed] [Google Scholar]
- Roberts P, Moshitch-Moshkovitz S, Kvam E, O’Toole E, Winey M, Goldfarb DS. 2003. Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol Biol Cell 14:129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers JV, Rose MD. 2015. Kar5p is required for multiple functions in both inner and outer nuclear envelope fusion in Saccharomyces cerevisiae. G3 5:111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers JV, Arlow T, Inkellis ER, Koo TS, Rose MD. 2013. ER-associated SNAREs and Sey1p mediate nuclear fusion at two distinct steps during yeast mating. Mol Biol Cell 24:3896–3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H, Leung J, Grimsey N, Peak-Chew S, Siniossoglou S. 2005. The yeast lipinSmp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J 24:1931–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarcelli JJ, Hodge CA, Cole CN. 2007. The yeast integral membrane protein Apq12 potentially links membrane dynamics to assembly of nuclear pore complexes. J Cell Biol 178:799–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaitz A-L, Thompson J, Wong CCL, Yates JR,III, Heald R. 2013. REEP3/4 ensure endoplasmic reticulum clearance from metaphase chromatin and proper nuclear envelope architecture. Dev Cell 26:315–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiter R, Hitomi M, Ivessa AS, Fasch EV, Kohlwein SD, Tartakoff AM. 1996. A yeast acetyl coenzyme A carboxylase mutant links very-long-chain fatty acid synthesis to the structure and function of the nuclear membrane-pore complex. Mol Cell Biol 16:7161–7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer N, Pawar N, Weiss M, Maiato H. 2015. An organelle-exclusion envelope assists mitosis and underlies distinct molecular crowding in the spindle region. J Cell Biol 210:695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Tobery CE, Rose MD. 2009. Prm3p is a pheromone-induced peripheral nuclear envelope protein required for yeast nuclear fusion. Mol Biol Cell 20:2438–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S 2013. Phospholipid metabolism and nuclear function: Roles of the lipin family of phosphatidic acid phosphatases. Bioch Biophys Acta 1831:575–581. [DOI] [PubMed] [Google Scholar]
- Siniossoglou S, Santos-Rosa H, Rappsilber J, Mann M, Hurt E. 1998. A novel complexof membrane proteins required forformation of a spherical nucleus. EMBO J 17:6449–6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth JT, Beg AM, Wu S, Putney JW Jr., Rusan NM. 2012. Phosphoregulation of STIM1 leadsto exclusion of the endoplasmic reticulum from the mitotic spindle. Curr Biol 22:1487–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone EM, Heun P, Laroche T, Pillus L, Gasser SM. 2000. MAP kinase signaling induces nuclear reorganization in budding yeast. Curr Biol 10:373–382. [DOI] [PubMed] [Google Scholar]
- Tamm T, Grallert A, Grossman EPS, Alvarez-Tabares I, Stevens FE, Hagan IM. 2011. Brr6 drives the Schizosaccharomyces pombe spindle pole body nuclear envelope insertion/extrusion cycle. J Cell Biol 195:467–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran EJ, Wente SR. 2006. Dynamic nuclear pore complexes: Life on the edge. Cell 125:1041–1053. [DOI] [PubMed] [Google Scholar]
- Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. 2006. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 124:573–586. [DOI] [PubMed] [Google Scholar]
- Walters AD, Bommakanti A, Cohen-Fix O. 2012. Shaping the nucleus: Factors and forces. J Cell Biochem 113:2813–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters AD, May CK, Dauster ES, Cinquin BP, Smith EA, Robellet X, D’Amours D, Larabell CA, Cohen-Fix O. 2014. The yeast polo kinase Cdc5 regulates the shape of the mitotic nucleus. Curr Biol 24:2861–2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster MT, McCaffery JM, Cohen-Fix O. 2010. Vesicle trafficking maintains nuclear shape in Saccharomyces cerevisiae during membrane proliferation. J Cell Biol 191:1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Yarar D, Giddings TH, Mastronarde DN. 1997. Nuclear pore complex number and distribution throughout the Saccharomyces cerevisiae cell cycle by three-dimensional reconstruction from electron micrographs of nuclear envelopes. Mol Biol Cell 8:2119–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkin KL, Chong Y, Shao S, Webster MT, Lahiri S, Walters AD, Lee B, Koh JLY, Prinz WA, Andrews BJ, Cohen-Fix O. 2012. The budding yeast nuclear envelope adjacent to the nucleolus serves as a membrane sink during mitotic delay. Curr Biol 22:1128–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkin KL, Friederichs JM, Cohen-Fix O, Jaspersen SL. 2010. Changes in the nuclear envelope environment affect spindle pole body duplication in Saccharomyces cerevisiae. Genetics 186:867–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yam C, Gu Y, Oliferenko S. 2013. Partitioning and remodeling of the Schizosaccharomyces japonicus mitotic nucleus require chromosome tethers. Curr Biol 23:2303–2310. [DOI] [PubMed] [Google Scholar]