Abstract
Introduction:
In head and neck squamous cell carcinoma (HNSCC) high numbers of tumor infiltrating CD8 T cells in the tumor microenvironment are associated with better outcome. However, no investigators have employed automated image analysis on whole slide images to permit CD8 scores for use in clinical practice. The aim of this study was to develop and validate an image analysis algorithm to automatically quantify CD8 T cells in patients with oropharyngeal HNSCC.
Materials and Methods:
Using brightfield image analysis results were cross-validated with fluorescence based quantification (AQUA™). A nuclear image algorithm designed to run on whole slide images was optimized to manual count. The algorithm was locked down and used on a cohort of whole tissue sections from HNSCC patients. Multivariate clinicopathologic parameters and outcomes were statistically correlated with image analysis results.
Results:
Linear correlation between manual counts and the customized CD8 algorithm was 0.943. A total of 74 oropharyngeal HNSCC cases were analyzed for CD8 immune cell infiltrate using this image analysis algorithm. A CD8 immune cell density above 136 cells/mm2 was associated with median survival of 18 years compared to 5 years. When multivariate modeling was performed, HPV infection was the only predictor of survival; however, when HPV was excluded only CD8 cell density predicts survival.
Conclusions:
We report the successful technical development and clinical validation of an image algorithm to automate CD8 immune cell density for oropharyngeal HNSCC. Employing brightfield image analysis on entire tumor sections instead of tumor subcompartments permits this strategy to be widely implemented.
Keywords: CD8, head and neck, HPV, image analysis, immunology, immunotherapy, quantification, score, squamous cell carcinoma, whole slide image
Introduction:
Tumor infiltrating lymphocytes (TILs) have become an important measurement in many organ sites as their presence correlates with prognosis and for some tumors TILs also have therapeutic implications[1]. Among these TILs, CD8 T cells have been shown to be tightly associated with cancer patient survival and the predominant effector for cancer immunotherapy [2, 3]. Cytotoxic CD8 T cells are able to directly kill neoplastic cells through direct lysis. Accordingly, many immunotherapy approaches have sought to target and enhance tumor-reactive CD8 T cells [3, 4].
For head and neck squamous cell carcinoma (HNSCC) high numbers of tumor infiltrating CD8 T cells in the tumor microenvironment (TME) are associated with oropharyngeal localization, limited tumor growth and a lower incidence of smoking [5, 6]. Patients with HNSCC that exhibit high infiltration of CD8 T cells also demonstrate a significantly better outcome [5, 7-16]. In HNSCC human papillomavirus (HPV) infection is associated with better prognosis, which appears in part to be due to enhanced immune activation in the TME, and especially enhanced infiltration of CD8 T cells [16-18]. Given the direct relationship between TILs and HNSCC, a direct method to assess an immune response within HNSCC would be desirable [19].
The assessment of the CD8 T cells in patients afflicted with HNSCC has been attempted using flow cytometry either on procured tissue biopsies or from peripheral blood [8, 20]. Attaining such a “CD8 score” may be of value to help determine prognosis and treatment before definitive surgical resection. An additional benefit of employing flow cytometry for this purpose is the multidimensional interrogation of immune cells that can be performed by this method. However, there are conflicting studies about whether systemic CD8 T cell levels correlate with tissue CD8 T cell infiltration.
Since there are no distinct morphologic features of CD8 T lymphocytes present in hematoxylin and eosin (H&E) stained tissues, an immunohistochemical (IHC) stain for CD8 T cells is necessary. The assessment of CD8 T cells within immunostained tissue sections has generally been reported in quantitative and semi-quantitative fashions. No specific method has yet been recommended to assess CD8 T cells and thus each published study assessing TILs in HNSCC has analyzed CD8 infiltrates using slightly different methods [19]. One method frequently exploited included counting the number of CD8 T cells in various high power fields and then averaging those values across these fields [7, 9-11, 13]. Many studies relied on manual counts from acquired still images or examining stained samples directly with a conventional light microscope. One group of authors utilized software (Count from Biomas) to perform cell counting [11, 21]. The output of prior quantification studies in TILs in HNSCC has typically been reported as an absolute count of T cells or percentage/ratio to other immune cells. Few studies have provided quantification in the form of density (i.e. number of CD8 T cells per area of measurement) [8, 22]. Using digital imaging, image analysis can be leveraged to perform this quantification [23].
Additionally, it is hard to extrapolate findings from different publications as these studies often quantified cells in different regions of tissue sections. The three main areas generally scored include the intratumoral/intraepithelial/tumor-host, peritumoral/tumor-stroma, and peripheral tumor compartments [5-7, 12]. Specific descriptions of these tissue areas are lacking in these studies.
Nevertheless, most investigations focused predominantly on the intratumoral component. Moreover, some of these studies were based on tissue microarrays while others used whole tissue sections. To the best of our knowledge, no investigators have employed automated image analysis of whole slide images to determine CD8 scores in oropharyngeal HNSCC.
The aim of this study was to develop and validate an image analysis algorithm to automatically quantify CD8 T cells in whole slide images acquired from immunohistochemically stained tissue sections from patients with HNSCC.
Methods:
This study was approved by the University of Pittsburgh’s Institutional review Board (IRB PRO 15070630) and Yale Human Investigation Committee protocol #9505008219 from Yale University.
The immunohistochemistry clone for CD8 was C8/144B (DAKO, Carpenteria, CA). Pre-treatment of tissue was performed and antibody was diluted to a concentration of 1:40. DAB (3,3’-diaminobenzidine) was used as the color reagent for visualization.
The best approach to validate a new assay is to compare it to a previously proven assay, and to compare both assays to outcome. To verify that the algorithm results were accurate, the findings of brightfield analyses were compared to equivalent assessments obtained using a fluorescent-based AQUA™ system (NavigateBP, Carlsbad, CA) and to outcome in lung cancer [24]. For the standardization aspect of the study, sections from a Yale tissue microarray (YTMA) lung cancer cohort of 223 cases was stained with CD8 by immunohistochemistry at the University of Pittsburgh Medical Center (UPMC, Figure 1a). The YTMA was constructed as previously described [24]. The YTMA of non-small cell lung carcinoma cases had 177 intact cores for evaluation. TMA slides were digitized using an Aperio AT2 scanner at 40x magnification. Once uploaded in eSlideManager (eSM) the digital slide was de-arrayed. Some adjustment was needed for the de-array circles in order to ensure the sections from the cores were completely contained within the individual de-array images. Tissue cores in the TMA that had migrated out of the grid position and those that had incomplete edges were excluded from analysis. The number of CD8 stained cells in each core was manually counted, annotated within eSM, the annotations saved as XML files (Figure 1b). These annotations along with the whole slide image of the YTMA were used to develop our tailored algorithm. Due to technical issues, only 122 cores with both images and annotations could be evaluated. The parameters of this algorithm were locked down. Using this customized algorithm the density of CD8+ cells per mm2 was determined for all cores, based on the area of each de-arrayed image. CD8 scores were also determined on the de-arrayed images using unmodified Leica nuclear, cytoplasmic and membranous image analysis algorithms. Manual counts were compared to automated counts obtained from the standard algorithms as well the customized algorithm (Figure 1c).
Figure 1a:
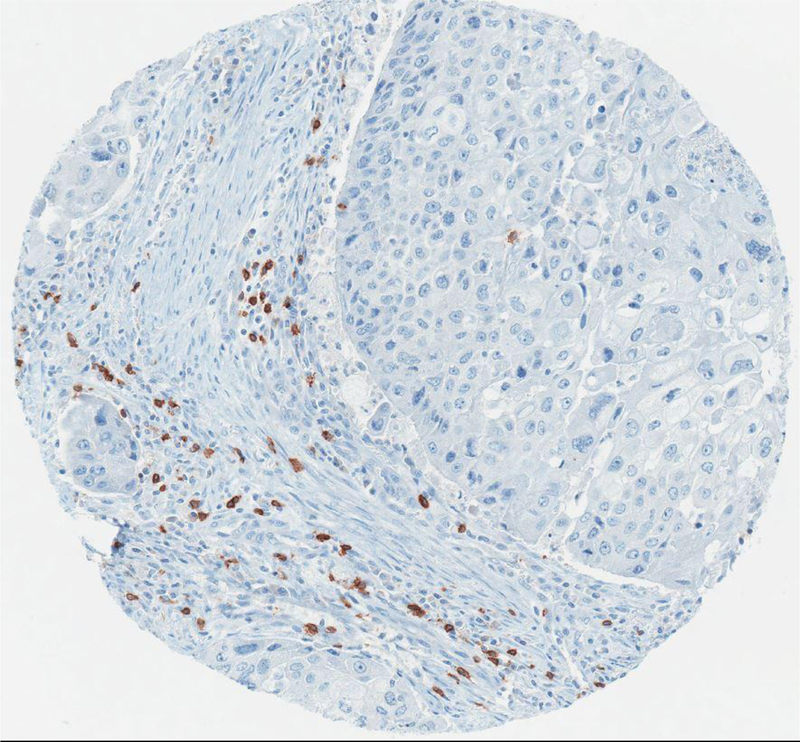
Example of a core from the YTMA stained by CD8 immunostain.
Figure 1b:
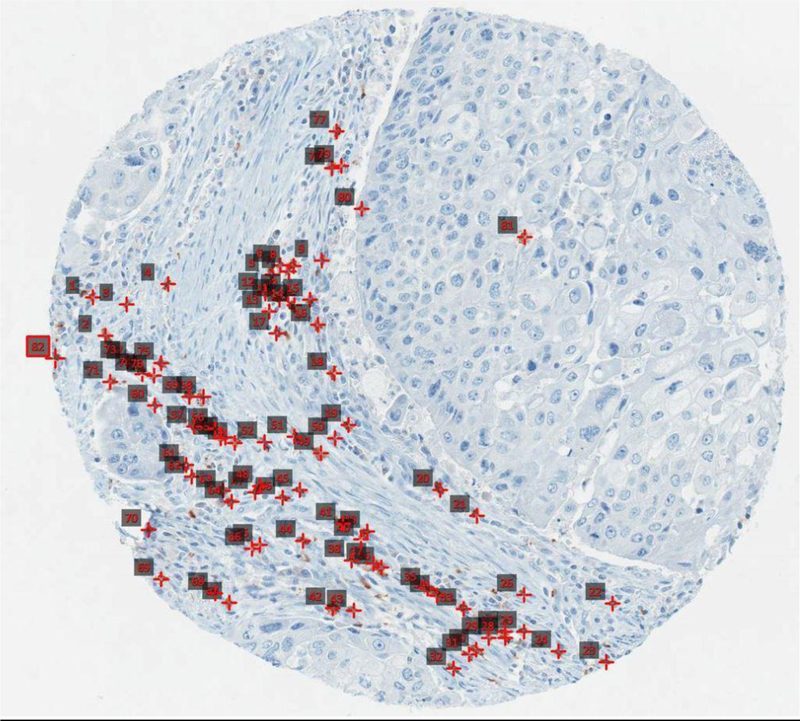
Example of a manual annotation of the same core as Fig 1a. Annotated red squares indicate individual CD8 T cells
Figure 1c:

Example of the same core as Fig 1a, annotated by the automated algorithm.
A serial section from the same YTMA lung cancer was stained for CD8 (C8/144B) and using a standard tyramide based assay as previously described [24]. The CD8 fluorescent intensity score was measured and previously shown to correlate with cell density. The manual counts, automated counts and AQUA score for CD8 cells were compared for each corresponding core.
Optimization of Aperio Nuclear v9 Algorithm
Aperio Nuclear v9 algorithm (Aperio Technologies, Inc., Vista USA) was chosen as the most appropriate algorithm to be used in this study in order to suitably detect the CD8 chromogenic staining and count the corresponding CD8-positive stained cells based on different classes of staining intensity (0, 1+, 2+ and 3+). The Aperio Nuclear v9 algorithm was firstly used to adjust (tune) the parameters in order to accurately match the manual count of CD8 stained cells, obtained as described above. The adjustment was performed by training the Aperio Nuclear v9 algorithm on 122 cores from the digitized TMA slide (for which the manual count of CD8-positive cells was available) and based on a three-step protocol, which encompassed (1) color separation, (2) nuclei segmentation, and (3) intensity scoring criteria definition. Color separation aimed to differentiate brown (DAB stain which was indicative of the protein of interest, i.e. CD8) from blue/purple (counterstain which was present in all cell structures, i.e. hematoxylin) colors on the tissue cores. This adjustment was achieved by using the smart training tool embedded in the Aperio Nuclear v9 algorithm which automatically identified each color by means of a deconvolution method, namely it automatically separates the image into the two channels corresponding to the actual colors of the stains used. Nuclei segmentation was, then, performed in order to identify both negative and positive cell nuclei. Nuclear objects were detected by intensity thresholding, smoothing, merging, and trimming. An additional step of nuclei exclusion was performed in order to filter nuclei based on their size and, hence, include only those nuclear objects matching the size criteria in the final analysis of the tissue cores. Scoring criteria were defined by adjusting intensity thresholds so that nuclei could be classified with regard to the presence of weak (1+), moderate (2+), and high (3+) staining of CD8. The set of tuned parameters was saved and used to run analysis on each tissue core. The analysis provided independent outputs for each tissue core, including count of nuclei stained positive for CD8 falling in the appropriate scoring intensity class (1+, 2+, or 3+) and measurement of the analyzed area (expressed as mm2).
CD8 Algorithm Validation
Once the customized CD8 algorithm had been cross-validated with the AQUA method, the algorithm parameters were no longer adjusted (“locked down”). A representative set of 80 oropharyngeal HNSCC whole slide tumor sections were then submitted for CD8 immunohistochemistry using the same clone as above. These whole sections were analyzed using the “locked down” CD8 algorithm run on both the entire digital slide, and separately annotated subcompartments (invasive carcinoma area, tumor margin/interface, and stroma area) selected by one pathologist to avoid inter-operator variability (Figures 1d-1f). The density of the CD8 immunostained T cells was determined by dividing the number of CD8 T cells (including cells stained with 3+, 2+, and 1+ intensity) by the area in mm2 examined. Measurements from all of the subgroups were also summed to evaluate whether subgroup analysis made any difference to the interpretation - this was labeled as total density to distinguish this analysis from the evaluation of the entire tissue section. Analyzed data for CD8 density was correlated with clinical, pathological and outcomes data. The following data elements were collected (Table 1): gender, race, smoking history, alcohol history, original diagnosis date, original histopathological diagnosis, histologic grade, perineural invasion, radiation therapy, chemotherapy, HPV status, P16 IHC status, most recent cancer status.
Figure 1d:

Tumor center subcompartment labeled with yellow boxes; green boxes are other annotated subcompartment regions
Figure 1e:

Tumor margin/interface subcompartment labeled with red boxes; green boxes are other annotated subcompartment regions
Figure 1f:

Tumor stroma subcompartment labeled with blue boxes; green boxes are other annotated subcompartment regions
Table 1:
Demographic data of oropharyngeal HNSCC patients enrolled in the study
| Parameter | HNSCC Cohort (n=74) |
| Gender | Male = 58; Female = 16 |
| Race | Caucasian = 69; African American = 5 |
| Smoking History | Yes = 46; No = 16 |
| Alcohol History | Yes = 44; No = 17 |
| Tumor Grade | Poor = 14; Moderate/Well = 26 |
| Perineural Invasion | No = 39; Yes = 13 |
| Radiation Therapy | No = 15; Yes = 57 |
| Chemotherapy | No = 33; Yes = 39 |
| HPV Status | No = 25; Yes = 48 |
| Most recent Cancer Status | No Evidence of Disease = 49; Recurrence = 22 |
| Overall Survival | Dead = 38; Alive = 36 |
| Pathologic T Stage | Unknown = 8 T1 = 27 T2 = 28 T3 = 9 T4 = 2 |
| Pathologic N Stage | Unknown = 13 N0 = 8 Any N = 53 |
Statistical analysis
The normality of the distributions of continuous variables were examined using the Shapiro-Wilk normality test. As the data were mostly not normally distributed (only age was normally distributed), non-parametric statistical tests were used. The Mann-Whitney test was used for comparisons with dichotomous variables, and the Kruskal-Wallis test was used for comparisons involving more than two variables. Kendall rank correlation was used to correlate the continuous variables.
Overall survival (OS) was obtained using the Kaplan-Meier method, and comparisons were made with the log-rank test. Cell density was classified as low or high density based on median split of the data for each density measure including tumor, margin, stroma, section, and their total (i.e. sum of tumor + margin + stroma). Patients who were alive at the most recent follow up date were classified as censored. For statistical purposes, patients who died with the disease were considered to have died of HNSCC.
Statistical significance was assumed at p<=0.05. Analyses were performed using IBM SPSS Statistics 22.
Results:
CD8 algorithm performance
Manual CD8 cell count for TMA cores ranged from 0 cells to 946 cells/core. The linear correlation between the manual count and the unadjusted cytoplasmic algorithm for the TMA was 0.861. The linear correlation between the manual count and the customized CD8 algorithm for the TMA was 0.943. The linear correlation between the customized CD8 algorithm performed on the digitized TMA slide at Leica in Dublin, Ireland and the same customized algorithm performed at UPMC on the TMA digital slide was 0.995. The YTMA lung cohort stained with immunofluorescence was compared to the total number of positive CD8 cell numbers/spot. Cross platform validation comparing TMA CD8 scores obtained from running the customized algorithm on an IHC-stained slide (brightfield) versus AQUA (fluorescence) showed reasonable correlation (Figure 2). The two tests were compared by determining their association with patient outcome. Figures 3a and 3b show such outcome analysis for each CD8 assessment method. Based on these results, we felt confident that the automated locked down CD8 algorithm generated accurate results. The automated algorithm used nuclear segmentation in order to evaluate for CD8 positive cells and so necrosis was excluded by the algorithm. No additional effort was taken to exclude tissue folds, however this did not affect the algorithm performance.
Figure 2:
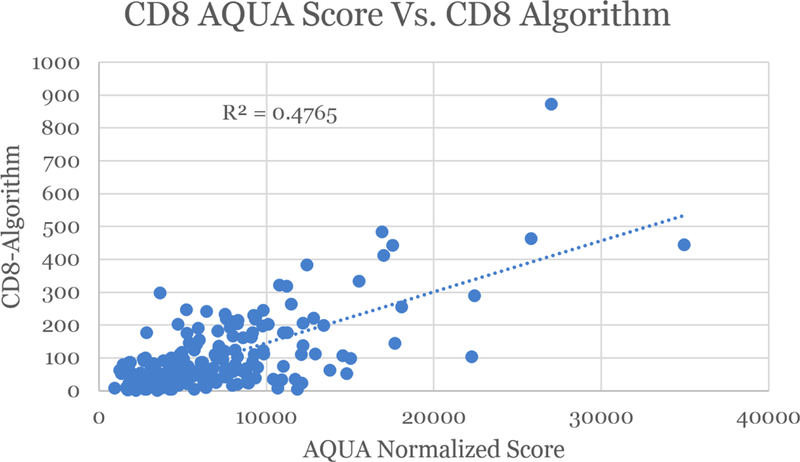
Regression between Immunofluorescence and Immunohistochemical CD8 algorithms; Comparison of IHC automated analysis to fluorescent analysis (AQUA) of same cores
Figure 3a:
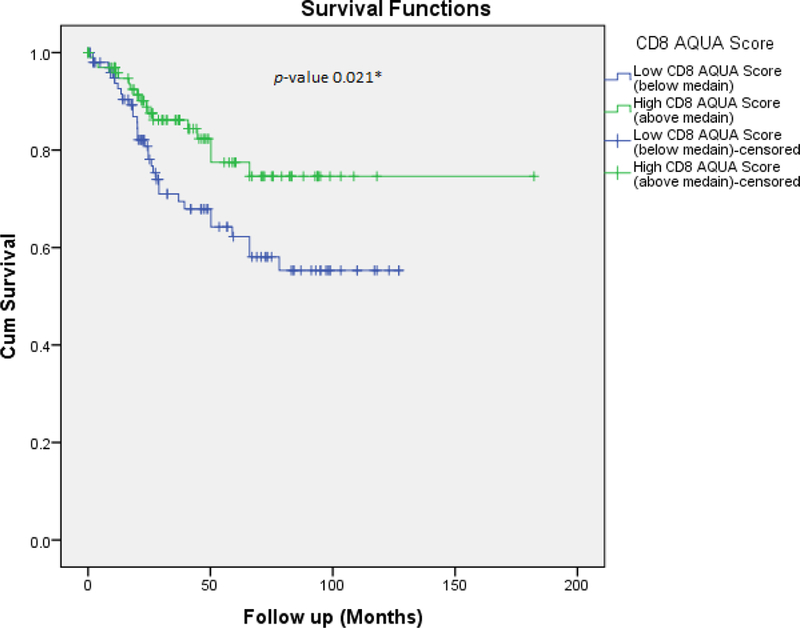
Overall survival based on AQUA analysis of CD8 on Yale TMA.
Figure 3b:

Overall survival based on Leica algorithm analysis of CD8 immunostain on Yale TMA
CD8 algorithm validation findings
A total of 74 (92.5%) out of 80 oropharyngeal HNSCC cases enrolled had sufficient neoplastic tissue on deeper sections and adequate CD8 immunostaining. The density of CD8 cells identified after analyzing the entire digital slide for this series of patients ranged from18 cells/mm2 to 669 CD8 cells/mm2. No correlation was found between CD8 density and gender, smoking history, alcohol history, radiation therapy or chemotherapy (Table 2).
Table 2:
Results of subcompartment, whole slide and combined subcompartment univariate analysis. Please note that entire section density refers to the entire tissue section while total density is the summation of tumor density, margin density and stroma density.
| Mann-Whitney test | n | Tumor Density | Margin Density | Stroma Density | Entire Section Density | Total Density (tumor + margin + stroma) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mdn | U | P value |
Mdn | U | P value |
Mdn | U | P value |
Mdn | U | P value |
Mdn | U | P value |
|||
| Gender | Male | 58 | 256.5 | 426.0 | 0.618 | 665.1 | 458.0 | 0.937 | 101.4 | 445.0 | 0.803 | 136.0 | 442.0 | 0.773 | 1197.7 | 441.0 | 0.763 |
| Female | 16 | 132.6 | 678.0 | 157.2 | 119.2 | 1036.7 | |||||||||||
| Race | Caucasian | 69 | 258.0 | 82.0 | 0.051 | 713.1 | 53.0 | 0.010 | 109.7 | 136.0 | 0.432 | 140.8 | 100.0 | 0.118 | 1198.6 | 61.0 | 0.016 |
| African American | 5 | 48.8 | 152.8 | 74.4 | 66.2 | 252.8 | |||||||||||
| Smoking History | Yes | 46 | 254.7 | 339.0 | 0.641 | 718.3 | 342.0 | 0.676 | 153.2 | 349.0 | 0.760 | 153.5 | 367.0 | 0.987 | 1154.4 | 345.0 | 0.711 |
| No | 16 | 387.6 | 765.0 | 136.5 | 171.2 | 1392.8 | |||||||||||
| Alcohol History | Yes | 44 | 254.7 | 357.0 | 0.785 | 765.7 | 348.0 | 0.676 | 125.3 | 331.0 | 0.489 | 179.6 | 312.0 | 0.319 | 1336.8 | 374.0 | 1.000 |
| No | 17 | 352.6 | 632.5 | 227.0 | 133.0 | 1287.1 | |||||||||||
| Grade | Poor | 14 | 372.1 | 113.0 | 0.050 | 690.1 | 126.0 | 0.112 | 64.6 | 172.0 | 0.777 | 105.7 | 161.0 | 0.552 | 1370.6 | 128.0 | 0.126 |
| Moderate/ Well | 26 | 81.9 | 369.8 | 89.0 | 83.2 | 665.1 | |||||||||||
| Perineural Invasion | No | 39 | 397.1 | 151.0 | 0.030 | 817.1 | 190.0 | 0.180 | 177.8 | 182.0 | 0.131 | 147.7 | 222.0 | 0.506 | 1830.4 | 163.0 | 0.056 |
| Yes | 13 | 84.5 | 723.5 | 95.0 | 159.3 | 939.5 | |||||||||||
| Radiation Therapy | No | 15 | 280.5 | 418.0 | 0.895 | 614.1 | 413.0 | 0.841 | 123.9 | 331.0 | 0.181 | 191.3 | 390.0 | 0.603 | 1287.1 | 408.0 | 0.787 |
| Yes | 57 | 254.4 | 713.1 | 95.0 | 139.1 | 1196.7 | |||||||||||
| Chemo- therapy | No | 33 | 379.5 | 520.0 | 0.163 | 570.3 | 570.0 | 0.406 | 89.3 | 548.0 | 0.280 | 131.3 | 596.0 | 0.591 | 1110.3 | 625.0 | 0.834 |
| Yes | 39 | 193.2 | 755.7 | 176.1 | 147.5 | 1198.6 | |||||||||||
| HPV | Negative | 25 | 76.9 | 295.0 | <0.001 | 287.2 | 243.0 | <0.001 | 79.8 | 447.0 | 0.075 | 76.9 | 303.0 | 0.001 | 523.4 | 237.0 | <0.001 |
| Positive | 48 | 337.7 | 800.1 | 153.2 | 179.6 | 1548.5 | |||||||||||
| pl6 IHC | Negative | 22 | 78.1 | 295.0 | 0.003 | 295.8 | 232.0 | <0.001 | 90.8 | 407.0 | 0.126 | 77.7 | 301.0 | 0.004 | 589.8 | 228.0 | <0.001 |
| Positive | 48 | 321.5 | 800.1 | 144.3 | 174.1 | 1548.5 | |||||||||||
| Most Recent Cancer Status | No evidence of cancer | 49 | 322.9 | 312.0 | 0.005 | 783.2 | 282.0 | 0.001 | 160.6 | 276.0 | 0.001 | 163.4 | 251.0 | 0.019 | 1603.0 | 233.0 | <0.001 |
| Evidence of cancer | 22 | 81.9 | 317.7 | 42.6 | 98.2 | 561.6 | |||||||||||
| Event | Deceased | 38 | 110.8 | 376.0 | 0.001 | 394.2 | 287.0 | <0.001 | 50.9 | 326.0 | <0.001 | 92.7 | 348.0 | <0.001 | 696.8 | 247.0 | <0.001 |
| Alive | 36 | 388.3 | 1051.8 | 200.6 | 192.1 | 1892.8 | |||||||||||
Mdn = Median; U = Mann-Whitney U test
Two histologic features demonstrated correlation with CD8 T cell density within the tumor center subcompartment. Tumors with poor differentiation had higher tumor density (median=372.1 cells/mm2) than moderate or well-differentiated cancers (median=81.9 cells/mm2), U=113.0, p=0.05. Also, tumors with no perineural invasion had higher tumor density (median=397.1 cells/mm2) than those with perineural invasion (median=84.5 cells/mm2), U=151.0, p=0.03. Tumors that were positive for HPV had higher tumor CD8 cell density and margin/interface CD8 cell density than those patients whose tumors were negative for HPV (Table 2), when analyzing both the entire tumor section and combining tumor region subcompartments. Similar findings were also identified with tumors that had a positive p16 immunohistochemical result demonstrating higher CD8 density within the tumor and at the tumor margin (Table 2), irrespective of whether the entire tumor section or combined tumor subcompartments were analyzed. Of note, the stroma-associated CD T cell density did not correlate with either HPV status or p16 status. No evidence of cancer at the most recent follow-up was associated with higher CD8 cellular density for each category (tumor, margin, stroma, and for both entire s ections and total of tumor subcompartments) than patients who had evidence of cancer (Table 2).
Survival Analysis
The median split with respect to patient survival for analyses attained after examining entire tumor sections was identified at a CD8 density of 136 cells/mm2. A log-rank test was run to determine if there were differences in the survival distribution between low-density and high-density CD8 cells in the entire section. The survival distributions for the two entire section density groups were statistically significantly different (Chi-square = 11.413, p=0.001), demonstrating that the overall survival of patients with a high-density of CD8 cells in entire tumor section cellularity was significantly longer than patients with low-density cellularity. The median survival was 18 years for the high-density section group (95% CI 9.1-26.9 years), compared to 5 years for the low-density section group (95% CI 0.6-9.4 years) (Figure 4). The Kaplan-Meier survival curves shown in Figure 4 demonstrate that patients with low-density CD8 T cell cellularity were at greater risk of earlier death than patients with high-density CD8 cellularity (log rank p value = 0.001). Patients who were alive as of the most recent follow-up also had higher CD8 T cell density for each category (tumor, margin, stroma, and for both entire sections and total of tumor subcompartments) than patients who had died.
Figure 4:
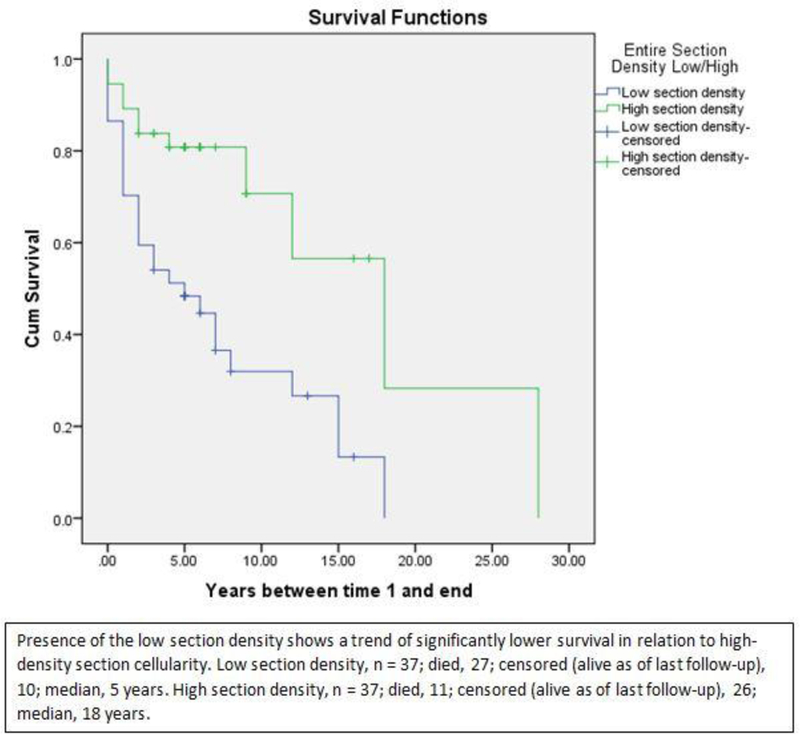
Kaplan-Meier survival curve for median cutoff of 136 cells/mm2. The survival distributions for the two entire section density groups were statistically significantly different (Chi-square = 11.413, p=0.001), demonstrating that the overall survival of patients with high-density entire section cellularity was significantly longer than patients with low-density section cellularity. The median survival (the smallest time at which the survival probability drops to at or below 50%) was 18 years for the high- density section group (95% CI 9.1-26.9 years), compared to 5 years for the low-density section group (95% CI 0.6-9.4 years). The Kaplan-Meier survival curves shown below demonstrates that patients with low-density section cellularity were at a greater risk of earlier death than patients with high-density section cellularity.
Given that CD8 infiltration is significantly associated with HPV status and that HPV status is a well-known positive prognostic marker, a decision tree analysis (specifically a Chi-square Automatic Interaction Detector) was created to build a predictive model with the variables (CD8 section density, HPV status, gender, race, alcohol history, radiation, chemotherapy, and survival as of most recent follow-up). The approach was to build a model that included HPV, and then to build another without HPV. When HPV status was included in the model, it proved to be the only variable that has an impact on survival. When excluded from the model, CD8 T cell density of the entire tumor section is the only variable that has an impact on survival.
Discussion:
Numerous studies have revealed that the infiltration of CD8 T cells within tumor sections in many organ system tumors, including HNSCC, has both prognostic and potentially therapeutic implications. One impediment to the routine evaluation of infiltrating of CD8 T cells within tumors is the availability of a reproducible and reliable method to quantify the amount of CD8 T cells within tissue. Different techniques of scoring CD8 T cell density (e.g. flow cytometry, western blots, morphological assessment, immunohistochemistry) may further impact the quantification of TILs and hence the outcome of results [19]. To the best of our knowledge, we report for the first time a quantification strategy utilizing image analysis applied to the whole slides images to determine CD8 scores in oropharyngeal HNSCC tumor tissue sections.
The current study represents a cross method validation of automated IHC-stained CD8 T cell counting using a laboratory developed customized brightfield image algorithm with the AQUA™ method of fluorescence-based assessment. A comparison of the two methods showed good correlation, and more importantly, both methods showed similar associations with outcome in a lung cancer coh ort. Using the CD8 brightfield algorithm, our data showed that increased CD8 T cell density in HNSCC tumors correlated with Caucasian race, poor tumor differentiation, absent perineural invasion, p16 positivity, HPV infection, and longer patient survival. Several studies have reported similar prognostic findings, including high numbers of CD8 T cells present at the invasive margin correlating significantly with prolonged survival [25]. To date, no standard cut-off point for TIL quantification has been reported in published studies [19]. Whilst this study uncovered a wide range of CD8 T cell densities in the tumors studied, a CD8 density score of 136 cells/mm2 was shown to significantly split patients into two populations where prolonged survival was associated with a high CD8 score.
It is plausible that CD8 T cell density may represent another surrogate marker of HPV positivity in HNSCC. However, we found that even in the HPV negative patients an increased CD8 T cell density may be of value confirming, because when HPV infection was excluded from the predictive model in our analysis CD8 T cell density was found to be the only variable with an impact on survival. Other investigators have shown that in HPV-negative HNSCC patients, especially those with more advanced stage disease, there are higher proportions of CD8 T cells present than is seen in HPV-positive patients [26].
TILs can reside in association with tumor cells, their stroma or both compartments. Reports about the impact of TILs in these various tumor compartments are somewhat disparate. While current dogma assumes that those TILs that interact directly with tumor cells are most important, prior publications advocate scoring stromal TILs probably as this has been found to be a more reproducible parameter [19]. We evaluated several possible methods to assess the CD8 T cell density within tumor sections. Although the subcompartment analysis (particularly for tumor center) demonstrated statistical significance with respect to patient survival, so too did quantitative image analysis of entire tumor sections. Given the variability that can be introduced by selection bias related to how tumor subcompartments are determined and annotated by varied end-users, the whole tissue section analysis is recommended as the best method by which to routinely assess CD8 T cell density in HNSCC.
There are several limitations of this study. In this series of oropharyngeal HNSCC, the cases were skewed toward an HPV-positive group of patients. In a larger study, perhaps a stronger relationship between CD8 T cell density and prognosis and/or therapy could be demonstrated. Additionally, the significance of CD8 T cell density with regard to immune therapy is unknown at this time. Future studies are needed to address these factors and better understand how automated quantitative image analysis of TILs will influence therapeutic decisions.
Conclusion:
In summary, we report the development and validation of an image analysis algorithm that can be utilized to automate CD8 T cell density evaluation for oropharyngeal HNSCC. Employing brightfield analysis and immunohistochemistry of entire tumor sections instead of specific tumor subcompartments permits this strategy to be easily adopted in routine clinical practice by many pathology laboratories.
Research Highlights:
1.- Development and validation of a brightfield quantitative image analysis algorithm to perform automated CD8 quantification based on whole slide images
2.- Demonstrated that a cutoff value greater than 136 CD8 cells/mm2 is associated with a better prognosis in oropharyngeal head and neck squamous cell carcinoma
3.- Automated tumor infiltrating lymphocyte assessment by whole slide image analysis has prognostic value and may be useful for clinical decision support
Acknowledgements:
Colleen Vrbin (President of Analytical Insights, LLC) for performing the statistical analysis for the HNSCC cohort.
Head and Neck SPORE (P50 CA097190), and UPMC Hillman Cancer Center Support Grant (P30 CA047904).
Lindsey Seigh for her technical help in the UPMC Image Analysis Laboratory.
This work was made possible by grant funding including the Yale SPORE in Lung Cancer, P50-CA196530, the Yale Cancer Center Support Grant, P30-CA016359, and a sponsored research agreement from Ultivue to Dr. Rimm.
Footnotes
Conflicts of Interest:
Dr. Liron Pantanowitz is on the medical Advisory board for Leica.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- 1.Taube JM, et al. , Implications of the tumor immune microenvironment for staging and therapeutics. Mod Pathol, 2018. 31(2): p. 214–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ovarian Tumor Tissue Analysis, C., et al. , Dose-Response Association of CD8+ Tumor-Infiltrating Lymphocytes and Survival Time in High-Grade Serous Ovarian Cancer. JAMA Oncol, 2017. 3(12): p. e173290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jackson SR, Yuan J, and Teague RM, Targeting CD8+ T-cell tolerance for cancer immunotherapy. Immunotherapy, 2014. 6(7): p. 833–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klebanoff CA, Gattinoni L, and Restifo NP, CD8+ T-cell memory in tumor immunology and immunotherapy. Immunol Rev, 2006. 211: p. 214–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balermpas P, et al. , CD8+ tumour-infiltrating lymphocytes in relation to HPVstatus and clinical outcome in patients with head and neck cancer after postoperative chemoradiotherapy: A multicentre study of the German cancer consortium radiation oncology group (DKTK-ROG). Int J Cancer, 2016. 138(1): p. 171–81. [DOI] [PubMed] [Google Scholar]
- 6.Cho YA, et al. , Relationship between the expressions of PD-L1 and tumor-infiltrating lymphocytes in oral squamous cell carcinoma. Oral Oncol, 2011. 47(12): p. 1148–53. [DOI] [PubMed] [Google Scholar]
- 7.Balermpas P, et al. , Tumour-infiltrating lymphocytes predict response to definitive chemoradiotherapy in head and neck cancer. Br J Cancer, 2014. 110(2): p. 501–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wansom D, et al. , Infiltrating lymphocytes and human papillomavirus-16--associated oropharyngeal cancer. Laryngoscope, 2012. 122(1): p. 121–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nordfors C, et al. , CD8+ and CD4+ tumour infiltrating lymphocytes in relation to human papillomavirus status and clinical outcome in tonsillar and base of tongue squamous cell carcinoma. Eur J Cancer, 2013. 49(11): p. 2522–30. [DOI] [PubMed] [Google Scholar]
- 10.Nasman A, et al. , Tumor infiltrating CD8+ and Foxp3+ lymphocytes correlate to clinical outcome and human papillomavirus (HPV) status in tonsillar cancer. PLoS One, 2012. 7(6): p. e38711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pretscher D, et al. , Distribution of immune cells in head and neck cancer: CD8+ T-cells and CD20+ B-cells in metastatic lymph nodes are associated with favourable outcome in patients with oro- and hypopharyngeal carcinoma. BMC Cancer, 2009. 9: p. 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watanabe Y, et al. , Tumor-infiltrating lymphocytes, particularly the balance between CD8(+) T cells and CCR4(+) regulatory T cells, affect the survival of patients with oral squamous cell carcinoma. Oral Surg Oral Med Oral Pathol Oral Radiol Endod, 2010. 109(5): p. 744–52. [DOI] [PubMed] [Google Scholar]
- 13.Zancope E, et al. , Differential infiltration of CD8+ and NK cells in lip and oral cavity squamous cell carcinoma. J Oral Pathol Med, 2010. 39(2): p. 162–7. [DOI] [PubMed] [Google Scholar]
- 14.de Ruiter EJ, et al. , The prognostic role of tumor infiltrating T-lymphocytes in squamous cell carcinoma of the head and neck: A systematic review and meta-analysis. Oncoimmunology, 2017. 6(11): p. e1356148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Meulenaere A, et al. , Prognostic markers in oropharyngeal squamous cell carcinoma: focus on CD70 and tumour infiltrating lymphocytes. Pathology, 2017. 49(4): p. 397–404. [DOI] [PubMed] [Google Scholar]
- 16.Saber CN, et al. , Immune cells and prognosis in HPV-associated oropharyngeal squamous cell carcinomas: Review of the literature. Oral Oncol, 2016. 58: p. 8–13. [DOI] [PubMed] [Google Scholar]
- 17.Chen X, et al. , Immunological network analysis in HPV associated head and neck squamous cancer and implications for disease prognosis. Mol Immunol, 2018. 96: p. 28–36. [DOI] [PubMed] [Google Scholar]
- 18.Bersani C, et al. , A model using concomitant markers for predicting outcome in human papillomavirus positive oropharyngeal cancer. Oral Oncol, 2017. 68: p. 53–59. [DOI] [PubMed] [Google Scholar]
- 19.De Meulenaere A, et al. , TILs in Head and Neck Cancer: Ready for Clinical Implementation and Why (Not)? Head Neck Pathol, 2017. 11(3): p. 354–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Partlova S, et al. , Distinct patterns of intratumoral immune cell infiltrates in patients with HPV- associated compared to non-virally induced head and neck squamous cell carcinoma. Oncoimmunology, 2015. 4(1): p. e965570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tabachnyk M, et al. , Radiochemotherapy induces a favourable tumour infiltrating inflammatory cell profile in head and neck cancer. Oral Oncol, 2012. 48(7): p. 594–601. [DOI] [PubMed] [Google Scholar]
- 22.Wolf GT, et al. , Tumor infiltrating lymphocytes (TIL) and prognosis in oral cavity squamous carcinoma: a preliminary study. Oral Oncol, 2015. 51(1): p. 90–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lloyd MC, et al. , Image Analysis of the Tumor Microenvironment. Adv Exp Med Biol, 2016. 936: p. 1–10. [DOI] [PubMed] [Google Scholar]
- 24.Schalper KA, et al. , Objective measurement and clinical significance of TILs in non-small cell lung cancer. J Natl Cancer Inst, 2015. 107(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng Z, et al. , Multiparametric immune profiling in HPV- oral squamous cell cancer. JCI Insight, 2017. 2(14). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poropatich K, et al. , Comprehensive T-cell immunophenotyping and next-generation sequencing of human papillomavirus (HPV)-positive and HPV-negative head and neck squamous cell carcinomas. J Pathol, 2017. 243(3): p. 354–365. [DOI] [PubMed] [Google Scholar]


