Abstract
p53, the most commonly mutated tumor suppressor, is a transcription factor known to regulate proliferation, senescence and apoptosis. Compelling studies have found that p53 may prevent oncogenesis through effectors that are unrelated to these canonical processes and recent findings have uncovered ancient roles for p53 in the containment of mobile elements. Together, these developments raise the possibility that some p53-driven cancers could result from unrestrained transposons. Here, we explore evidence linking conserved features of p53 biology to the control of transposons. We also show how p53 deficient cells can be exploited to probe the behavior of transposons and illustrate how unrestrained transposons incited by p53 loss might contribute to human malignancies.
Keywords: p53 biology, transposons, retrotransposons, mobile elements, transrepression, cancer
p53 has a pivotal role in human tumorigenesis
p53 is a bona fide tumor suppressor and its role in human cancer has been well established. In 1990, the identification of inherited p53 mutations in patients with Li– Fraumeni syndrome (which predisposes to diverse tumor types) provided the association between p53 and human cancer [1–3]. This was further confirmed in 1992 by experiments showing that p53 knockout mice are prone to tumors [4, 5]. Almost thirty years later, the crucial role of p53 in cancer control is beyond any doubt, with it being the most studied gene in biology [6]. However, it is also clear that tumor suppressive mechanisms governed by p53 are not completely understood, since p53 retains cancer prevention activity even if decoupled from canonical effectors related to apoptosis, proliferative arrest and senescence [7–9]. Here we explore a novel hypothesis, suggesting that p53 acts through highly conserved mechanisms to restrict transposons and, in this way, might also restrict tumor formation.
p53 as a Suppressor of Retroelements
Transposons have invaded the genome of organisms across the tree of life. Unlike DNA transposons, which excise themselves from the genome and reinsert at a distal location, retrotransposons transpose through RNA intermediates, allowing them to increase their copy number within the host genome [10]. Thus, while DNA transposons comprise less than 3% of the human genome [11], retrotransposon derived sequences account for nearly 40% of the human genome [10, 11]. Retrotransposon expression has been shown to increase with age in both cell culture and mouse models, and retrotransposon derepression is strongly associated with human cancers [12–15]. While the presence of mobile elements creates complications for the host genome, they also provide a source of genetic innovation and, in this sense, they constitute a reservoir of genomic material that can be domesticated by the host to perform novel functions [16].
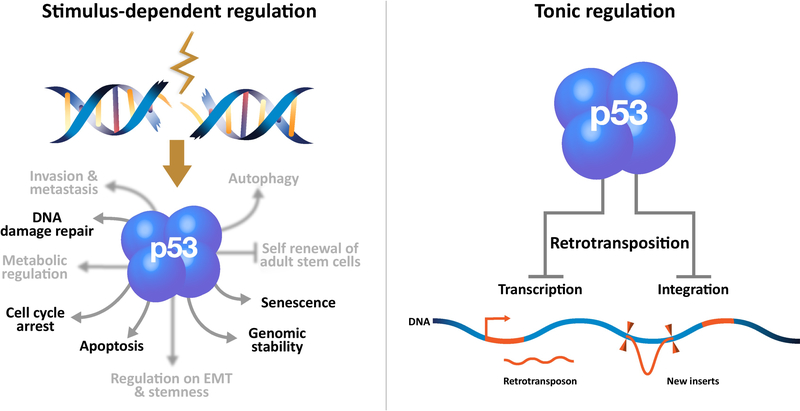
In vertebrates and invertebrates, p53 proteins respond to intrinsic and extrinsic stressors and can orchestrate a variety of cellular stress response programs including apoptosis [17]. It has been proposed that p53 may “sense” retrotransposon integration as DNA damage, since retrotransposition itself could provoke apoptosis in a p53 dependent manner [18–23]. This insight provided a simple and attractive mechanism for how p53 might restrict retroelements (See Glossary) exposing a previously unappreciated process by which p53 could potentially suppress cancer. However, emerging evidence indicates that stimulus-dependent p53 action restricts transposons in ways that are distinct from constitutive repression by p53, which is also imposed on these elements [24, 25]. In this view, p53 tonically acts to restrain retrotransposons but, as a fail-safe mechanism, stimulius-dependent p53 modalities (e.g. apoptosis, cell cycle arrest, senescence) are recruited as auxilliary countermeasures against potential eruptions of these elements (Figure 1).
Figure 1.
As a transcription factor p53 (four fused blue balls) governs numerous stress response programs. In the canonical view (left panel) signals provoked by genotoxic damage (brown jagged line) stimulate p53 binding and transactivation of targets that specify adaptive processes including apoptosis, cell cycle arrest, senescence, DNA repair and others not mentioned here (unfocused text) but comprehensively reviewed elsewhere [7, 56]. Recent studies indicate that cell cycle control, apoptosis and senescence do not adequately explain tumor suppression by p53 [8, 9]. The right panel illustrates that p53 tonically restrains mobile elements through activities that are not yet defined [24, 25]. Evidence outlined in this review suggests that, although historically understudied, transrepression by p53 is disease-relevant.
Retrotransposons are a driving evolutionary force [10, 26]. In eukaryotes they can be divided into three classes: long terminal repeat (LTR) elements, long interspersed elements (LINEs) and short interspersed elements (SINEs). Although they can potentiate evolutionary change, new insertions can be disruptive, with damaging consequences to cells. Retrotransposons are associated with approximately 0.1% and 10% of de novo germline mutations in humans and mice respectively [27]. Insertions of the LTR endogenous retroviruses family (ERVs), mammalian apparent LTR retrotransposons (MaLRs) as well as SINE and LINE1 elements have been implicated in the pathogenesis of human diseases including cancer [28–30]. Hence, it is clearly adaptive for the host to silence these repetitive elements and numerous reports suggest that p53 can promote this process [18, 19, 24, 25, 31]. For example, p53-binding sites have been identified computationally in human element-LINE1s and, where tested, these conferred positive regulation in co-expression reporter assays [32]. Likewise, ERV-associated LTRs [33, 34] also contain p53 sites and, where studied, these showed evidence for both positive and negative control by p53 [35]. While the discrepancies could reflect true biological differences, these binding sites were excerpted from their native context and have not yet been interrogated through loss-of-function approaches (e.g. mutation in situ). Further evidence consistent with models favoring negative regulation is that DNA methylation seems to cooperate with p53 to silence retroelements [19]. For example, when cultured cells were treated with a demethylating agent, transcripts corresponding to SINEs, satellite repeats, and other long non-coding RNAs were substantially higher in cells lacking p53 [19].
In vivo studies provide further evidence for an antagonistic relationship between p53 and retrotransposons. For example, in the Drosophila and Zebrafish models, p53-deficient animals experienced widespread upregulation of different classes of transposon RNAs not seen in wild-type counterparts [24, 25, 36]. Furthermore, using a synthetic retrotransposon, it was found that de novo integrations of a synthetic retroelement were, likewise, far more frequent in p53- backgrounds [24]. Conversely, in these same in vivo studies, p53 was acutely sensitive to transposon activity. The highly conserved PIWI/piRNA pathway acts through small RNAs and Argonaute nucleases to control mobile elements and, using a p53 biosensor in mutants defective for the piRNA network, it was shown that hyperactive transposons could trigger persistent p53 activity without the need for exogenous stressors [37]. In these models, transposon restriction by p53 was clearly not associated with apoptosis, offering a powerful illustration for context-specific p53 responses that may contain mobile elements [37]. Together, these findings suggest that ancestral properties enable p53 to respond to – and restrain – mobile elements. Seen from this perspective, transposons embedded within the genome represent a powerful contrast to extrinsic insults, such as radiation or genotoxic compounds that are commonly used to activate p53. Consistent with this, genetic interactions were also observed between p53 and the PIWI/piRNA pathway [24]. Specifically, piRNA biogenesis was altered in p53 mutants and, furthermore, p53 interacted with core elements of the piRNA pathway encoded by aubergine (aub) and cutoff (cuff), as double mutants had a strong germ line phenotype [24].
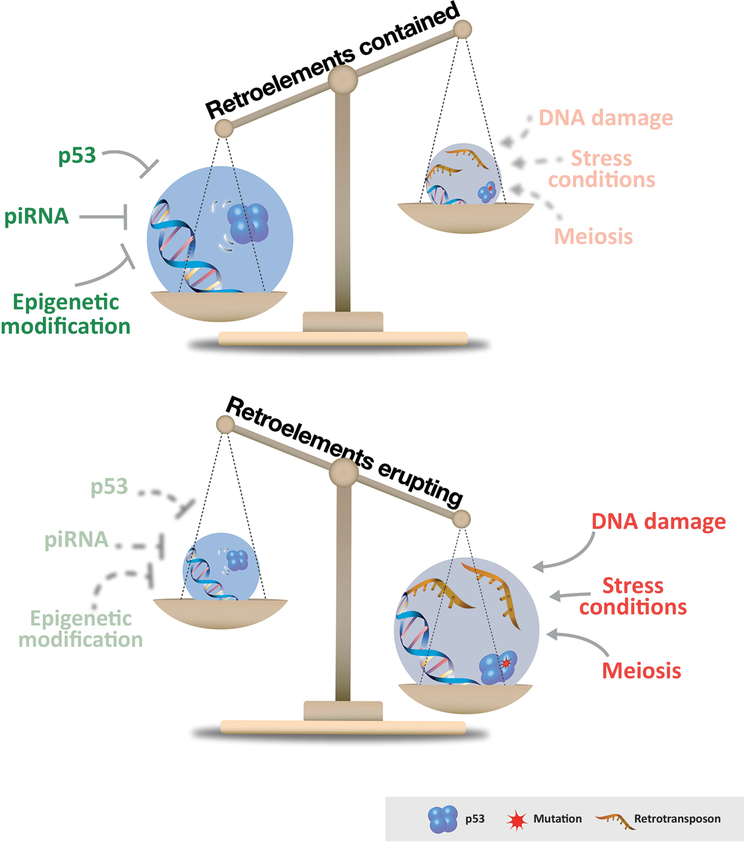
Collectively, these results expose how cross talk among the p53 and piRNA networks [24] cooperate to specify epigenetic features (e.g. methylation, histone marks) that restrain retrotransposon activity in animal germlines (Figure 2). A compelling illustration of this in vivo, is the example mentioned above when a p53 biosensor was used to show that p53 was persistently hyperactive in the germ line of piRNAs mutants [37]. Intriguingly, using this same biosensor, it was found that as every gamete is made, the first enzymatic step in the process of meiotic recombination actually provokes a transient burst of p53 activity [38]. Since this step involves the creation of a double stranded DNA break, this transient burst of p53 activity was originally attributed to double stranded DNA breaks during meiotic recombination [38]. However, now it seems equally possible that these p53 bursts could be attributed to relaxed transposon controls that accompany the meiotic process [39]. Indeed, meiotic double strand breaks are the proximal trigger for retrotransposon eruptions in animals lacking p53 [24].
Figure 2.
Host pathways or signals that suppress (green) or provoke (red) retrotransposon activity are depicted. Depending on cellular conditions and extrinsic stimuli the illustrated balance weighs in favor of transposon silencing or stochastic eruptions.
Taken together, the evolutionary picture suggests that primordial effectors of p53 conserved in vertebrates and invertebrates act in the earliest stages of germ line development to contain mobile elements, thereby helping to insure the fidelity of genomic information across generations (Figure 2). Have these functions been retained by human p53 genes? Compelling experiments using a collection of flies expressing human p53 suggest that the answer is clearly yes [24, 25]. For example, using genetic complementation, human p53 (hp53) could effectively substitute for the fly counterpart and restrain retroelements [24] while “hotspot” missense alleles, which commonly emerge in cancer patients, starkly failed to repress retrotransposons in these assays [24, 25]. Hence, antagonizing retroelements is not only a conserved function in human p53, but variants associated with human cancers are also commonly disabled for this activity. These data raise the possibility that the ability of p53 to suppress tumors is coupled to its ability to suppress transposons. Since p53 still prevents cancer even when un-coupled from canonical effectors [7–9] this is an appealing idea. If true, the steps that might link unrestrained mobile elements to cellular transformation are an open question [25, 40] and numerous scenarios are possible (Box 1).
Box 1 Pathogenic scenarios associated with the LINE1 retrotransposition cycle.
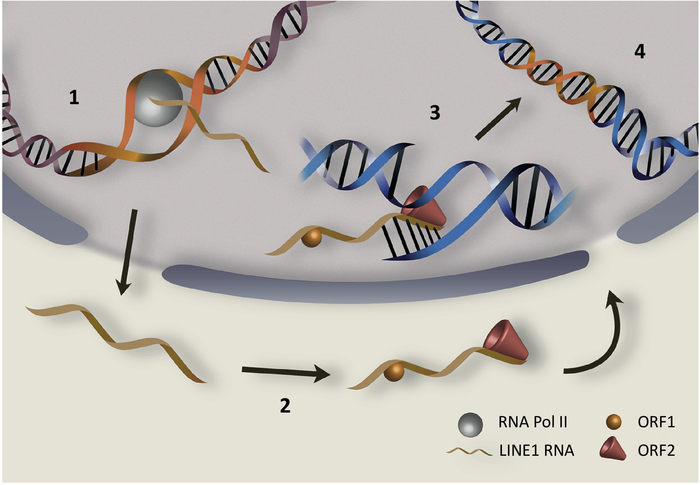
LINE1 (L1), the only autonomous transposable element in humans, comprises almost 17% of the whole human genome. It is upregulated in certain cancers, contributing to tumor genome dynamics [75, 76]. Figure I illustrates basic steps in the LINE1 retrotransposition cycle. After transcription (step1) LINE1 RNAs are translated (step 2) to produce two proteins, ORF1p and ORF2p, which are essential for mobility. ORF1p is an RNA binding protein with nucleic acid chaperone activity that binds to LINE1 transcript [77–79]. ORF2p possesses endonuclease and reverse transcriptase activities [80, 81]. Through reverse transcription of LINE1 transcripts (step 3), ORF1p and ORF2p can generate de novo insertions of LINE1 copies at new positions in the genome (step 4) [29] and, as a source of germ line mutations, these insertions can cause heritable disease [82, 83]. Likewise, as a source of new mutations in somatic tissues, retrotransposition may activate oncogenes or inactivate tumor suppressor genes and, in this way, drive cancers [83–85]. However, other steps in the cycle could also trigger damaging effects and, conceivably, derepressed retroelements may spur pathogenesis without generating new insertions. For example, open chromatin associated with highly expressed LINE1s (step 1) could stimulate homologous recombination between LINE1 copies, causing rearrangements and destabilized genomes, which are hallmarks of cancer [86]. Also, abnormally high levels of LINE1 RNAs (step 1) could promote the formation of repeat RNAs or unusual double stranded RNAs that are pro-inflammatory, leading to interferon responses and possible apoptosis [19]. Once translated (step 2) excessive LINE1 protein products might also incite potential harm. For example, ORF1p, an RNA binding protein, could potentially bind other cellular transcripts and interfere with their function. Likewise, excessive endonuclease ORF2p activity could cause sporadic DNA damage and, consistent with this, ORF2p was associated with mutagenesis and large genomic deletions [20, 53, 87]. Abnormal intermediates occurring in step 3 could also cause cellular injury. For example, excessive RNA:DNA hybrid molecules generated during reverse transcription could trigger aberrant inflammatory responses [88], which is yet another hallmark of cancer [89].
p53 offers a sensitized model to track retroelement behavior
Most mutations that relax transposon control cause pronounced tissue atrophy, sterility or lethality [41], precluding direct visualization of dysregulated elements. However, the gonads of p53- flies, fish and mice can produce functional gametes despite derepressed retroelements [24, 25] and, consequently, p53- germ lines offer uniquely sensitized systems to interrogate fundamental properties of mobile elements that are otherwise undetectable. For example, the migratory behavior of retroelement RNAs was recently visualized during oogenesis in p53 deficient ovaries [36]. Surprisingly, in both flies and fish, it was observed that certain classes of retroelement RNAs migrated to a specialized region within the oocyte called the germ plasm, which is composed of RNA factors that specify embryonic germ cells in the next generation. In the fly model, retroelements localized to this specialized region by mimicking oskar RNA, a pioneer factor that actually initiates germ plasm assembly. In this way, retroelement transcripts can be considered “stow aways” inside RNA transport granules and can gain passage to the germ plasm, even when germ plasms are ectopically positioned [36]. By invading the germ line before it is actually formed in subsequent generations, these studies exposed novel adaptive strategies that are presumably used by retroelements to elude protective host defenses [42] and insure transgenerational inheritance. Furthermore, since germ plasm components evolved independently in Drosophila [43] and fish [44], the ability of retrotransposons to mimic these structures in the context of oogenesis also reflects curious examples of convergent evolution. In mammals, oocytes evidently lack a germ plasm and, hence, retrotransposons may have adopted analogous - but mechanistically distinct - tactics that achieve the same purpose. From this perspective, it is intriguing that oocyte attrition in mammals is tightly linked with increased retroelement expression [45] and, likewise, p53 function impacts gamete formation [38], general fertility [46] and even implantation [47].
Retroelements are restrained by p53 in cancers
The ancestral role of p53 as a suppressor of mobile elements very likely extends to somatic tissues in long-lived animals. This is perhaps most apparent in the transformed state where p53 alterations were highly correlated with transposon derepression. For example, in mouse models of myc-driven carcinomas, LINE1 and intracisternal A-type particle (IAP) retrotransposons were both dramatically derepressed in p53- mice, compared to their wild type counterparts [24]. Likewise, in human cancers, LINE1 eruptions stratified with p53 loss in anaplastic Wilms tumor and equally compelling trends were observed in colon, lung and ovarian carcinomas as well as secondary glioblastomas [24, 25, 48, 49]. More recently, it was shown that LINE1 eruptions integrate to produce de novo insertions and that new insertions were highly correlated with mutant p53 status in stomach cancers [50]. While these correlations are clearly intriguing, two additional points are worth noting here. First, though more severe in p53-driven tumors, transposon derepression is a general feature shared among many cancers. Second, DNA hypomethylation, a ubiquitous feature of carcinogenesis [51], is also associated with an increased expression of repetitive elements and other transcripts [19, 52–55]. Hence, relationships between p53 and retroelements manifested in oncogenic samples could represent global effects on methylation landscapes, rather than activities directed specifically to retrotransposons (see below). On the other hand, missense alterations in p53 appear far more commonly than truncations or frameshifted alleles in human cancers, and at least some of these are associated with gain-of-function phenotypes [56, 57]. Therefore, it is possible that these alleles could incite transposon eruptions more severely relative to variants that simply eliminate p53 function.
Potential mechanisms
So how does p53 actually restrain retrotransposons? Possible mechanisms fall under two general models. The first involves quality control processes that purge cells suffering from transposon eruptions. As discussed earlier, this hypothetical scenario invokes canonical apoptogenic responses mediated by p53 and provoked by DNA breaks that occur as new insertions are generated [18–20, 23, 58, 59]. Consistent with this, it was shown that cells overexpressing a synthetic retrotransposon [60] were able to evade apoptosis when p53 was absent [22]. Therefore, hyperactive transposons in tissues wild type for p53 could potentially lead to the death of cancer cells rather than enabling their growth [19]. However, most cancers lack p53 and, as mentioned earlier, several compelling lines of evidence show that p53 dependent repression of retrotransposons does not depend upon the apoptotic pathway. First, in developing flies and fish, p53 retroelement repression occurs in the absence of apoptosis, long before the onset of programmed cell death [24]. Second, chk2 mutants, like p53 mutants, are defective for stress-induced apoptosis but, despite this phenotype, they clearly retain normal restrictions on transposons [24]. Third, in zebrafish embryos, p53 dependent repressive epigenetic marks were deposited onto synthetic retroelements well before the first signs of programmed cell death [24]. Hence, while apoptotic purging may be engaged as an auxiliary process, the primary means by which p53 restrains retroelements is probably unrelated to cell death.
A second more direct model proposes that p53 exerts transrepressive functions, which normally prevent expression from transposon sequences (Figure 1). Consistent with this, there is substantial evidence indicating that p53 can directly bind retroelements. For example, predicted p53 consensus binding motifs exist in the promoter region of the human LINE1 retroelement and in non-autonomous Alu sequences [32, 61, 62]. Alu elements are one of the most common retrotransposable elements in human genome. In the presence of DNA damage, p53 suppresses Alu expression, highlighting the protective ability of p53 [19]. Similarly, p53-like binding sites were reported in mouse B1, a member of SINE family of retroelements in mouse genome [61, 62]. These p53-like binding sites were able to compete with the consensus p53-binding site in vitro suggesting that they may indeed be recognized by p53 in vivo [61, 62]. Likewise, flies humanized for p53 alleles that are common in cancer patients failed to repress retrotransposons and, since these alleles impact DNA binding, this activity was needed to restore retrotransposon repression by gene complementation [24]. Finally, chromatin immunoprecipitation (ChIP) studies undertaken in human cell lines show extensive p53 enrichment within repetitive regions of the genome [33] and, as noted earlier, one such binding region is within the promoter region of the retrotransposon element LINE1 [32, 63]. Notably, none of these sites have been functionally interrogated by targeted mutagenesis in vivo and, consequently, the precise function of p53 binding sites at these elements is unclear. Therefore, it seems both reasonable and plausible to connect this activity with epigenetic features (e.g. DNA methylation and histone H3K9me2/3) that are known to be potent retrotransposon repressors. Consistent with this, mutations in DNA methyltransferases or histone H3K9 methyltransferases caused selective derepression of mobile elements in mammalian systems [64–69] and, likewise, drug-induced DNA hypomethylation caused hyperactivation of certain retrotransposons, but only if p53 was absent [19]. Together these data suggest that p53 likely cooperates with histone and DNA methylation to silence specific retroelements. Indeed, in the zebrafish model, it was shown that p53-dependent H3K9me3 methylation existed in the promoter region of a synthetic human LINE1 element [24] and, intriguingly, this H3K9me3 peak mapped to a known p53-binding site [32]. In flies, which lack extensive regulatory DNA methylation [70], H3K9 methylation is likely more pivotal in this context. Combined, these studies suggest that p53 may directly bind and recruit a variety of repressive features to retroelements – but how? Some evidence in human cell lines suggests that p53 can physically interact with both H3K9 tri-methyltransferases and DNA methyltransferases [71, 72]. Furthermore, it was shown that, in basal stress-free conditions, unacetylated p53 is pre-bound to many target genes together with a repressor protein, SET, which mediates repression of p53 target genes [73]. Additionally, p53 as a master regulator of transcription might regulate gene expression of key epigenetic or piRNA factors. This latter hypothesis is supported by data showing an upregulation of piRNA precursors in p53 null fly ovaries, a phenotype which mimics loss of piRNA processing proteins [24]. Future studies may validate one or more of these scenarios and perhaps expose additional mechanisms in parallel. Nevertheless, in our view, it seems likely that p53 exerts multiple activities to repress mobile and ensure that they are contained.
Concluding Remarks
An expanding body of literature continues to highlight how relaxed transposon control is a common feature in age-related diseases, including cancer. The p53 regulatory network constitutes one of several host defenses that, when compromised, promotes conditions that are permissive for retrotransposon eruptions (Figure 2). As the centerpiece of an ancestral regulatory network, primordial forms of p53 probably evolved to safeguard the germ line, where mobile elements exert powerful selective pressures. In long-lived animals, this germ line activity was likely retained [38] and expanded to ensure that transposons are restrained in somatic tissues as well. To the extent that unrestrained transposons may contribute as drivers of cancer, this scenario offers attractive explanations for how, in mammals, p53 was evolutionarily co-opted for tumor suppression and perhaps as a more general safeguard against “transposopathies”. Historically appreciated as a stimulus-dependent transactivator [7, 56] there is now growing appreciation for p53 as a tonic transcriptional repressor and that, whatever these suppressed targets are, they may provide useful clues related to tumor suppression [74]. Though various models have been proposed, no mechanistic consensus has yet emerged for how p53 operates as a transrepressor or how its targets are specified (see Outstanding Questions). Here we’ve argued that retrotransposons qualify as prominent – and possibly pathogenic - targets of transrepression by p53. As non-unique sequences, retrotransposons are often filtered in next-generation sequencing pipelines and, consequently, they are routinely overlooked. However, in the final analysis, these potential genome destabilizers and inflammatory threats deserve far more attention.
Figure I.
Highlights of the human LINE1 retrotransposition cycle begins with transcription (step1), followed by its translation into ORF1p and ORF2p proteins (step 2), which bind to LINE1 RNA. The final step is in the nucleus where ORF2p cuts the DNA and reverse transcribes LINE1 (step 3) leading to its integration (step 4). The numbers highlight steps in the retrotransposition cycle that, if unrestrained, could potentially incite pathogenic effects. Since many of these occur prior to integration, transposopathies may occur in the absence of new insertions as detailed in Box 1.
Highlight.
p53-mediated tumor suppression was co-opted from ancestral functions that preceded the evolutionary need to prevent cancers.
p53 triggered apoptosis, cell cycle arrest and senescence cannot adequately explain p53-mediated tumor suppression [1, 2], highlighting additional cancer prevention modalities that are currently obscure.
Normal p53 restrains transposons but cancer-associated variants do not, raising the possibility that derepressed retroelements contribute to p53-driven diseases.
p53 mutant backgrounds provide sensitized systems that can expose otherwise undetectable features of transposon biology.
Mechanisms of p53 retroelement repression may uncover new translational opportunities for biomarker development and therapies to treat “transposopathies”.
Outstanding questions.
How exactly does p53 restrain the activity of retroelements?
Does p53 employ a single strategy to repress divergent classes of retroelements?
How does p53 “sense” transposon activity?
How frequently do transposon eruptions occur in vivo?
Are retroelement eruptions uniquely initiated by distinct copies in different tissues?
Are p53 mutations permissive for cancer because they are, in part, permissive for retroelement eruptions?
Can retrotransposon eruptions incite pathogenesis without new de novo integrations?
Acknowledgements
This work was supported by the CPRIT training grant to UT Southwestern (to B.T), an ACS fellowship (to A.J.), NIH grants (R01GM072124, R01GM115682) a CPRIT grant (RP170086) and a Welch Foundation award (I-1865) to J.M.A. We are also grateful to Filipa Ferreira and Fernando Augusto for help with the text and artwork.
Glossary
- Epigenetic
Effects upon gene expression without changes in DNA sequence, which often involve DNA methylation or histone modifications and are, therefore, not conventionally heritable
- Germ plasm
A specialized compartment in the oocyte, which contains determinants that specify germ cells in the resulting embryo
- Molecular mimicry
An adaptive phenomenon where pathogens or foreign-derived factors are benefited by sharing similarities at the structural and functional levels with host factors
- Retrotransposition
The mobilization process by which retrotransposon copies integrate at new genomic locations via RNA intermediates that are reverse transcribed
- Retroelement
The class of mobile genetic elements associated with retrotransposition
- Tonic Regulation
Stimulus-independent regulation that occurs in a basal condition
- Transactivation
The process whereby one or more factors directly mediate elevated expression of a target gene
- Transrepression
The process whereby one or more factors directly prevent or reduce expression of a target gene
- Transposopathy
A disease associated with – and possibly incited by-eruptions of transposon activity
- Tumor suppressor
A gene or protein that protects cells from oncogenic transformation that, if mutated, may help cause cancer
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Baker SJ et al. (1989) Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science 244 (4901), 217–21. [DOI] [PubMed] [Google Scholar]
- 2.Malkin D et al. (1990) Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science 250 (4985), 1233–8. [DOI] [PubMed] [Google Scholar]
- 3.Srivastava S et al. (1990) Germ-line transmission of a mutated p53 gene in a cancer-prone family with Li-Fraumeni syndrome. Nature 348 (6303), 747–9. [DOI] [PubMed] [Google Scholar]
- 4.Donehower LA et al. (1992) Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356 (6366), 215–21. [DOI] [PubMed] [Google Scholar]
- 5.Lowe SW et al. (1993) p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature 362 (6423), 847–9. [DOI] [PubMed] [Google Scholar]
- 6.Dolgin E (2017) The most popular genes in the human genome. Nature 551 (7681),427–431. [DOI] [PubMed] [Google Scholar]
- 7.Mello SS and Attardi LD (2018) Deciphering p53 signaling in tumor suppression. Curr Opin Cell Biol 51, 65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valente LJ et al. (2013) p53 efficiently suppresses tumor development in the complete absence of its cell-cycle inhibitory and proapoptotic effectors p21, Puma, and Noxa. Cell Rep 3 (5), 1339–45. [DOI] [PubMed] [Google Scholar]
- 9.Li T et al. (2012) Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell 149 (6), 1269–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cordaux R and Batzer MA (2009) The impact of retrotransposons on human genome evolution. Nat Rev Genet 10 (10), 691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lander ES et al. (2001) Initial sequencing and analysis of the human genome. Nature 409 (6822), 860–921. [DOI] [PubMed] [Google Scholar]
- 12.De Cecco M et al. (2013) Transposable elements become active and mobile in the genomes of aging mammalian somatic tissues. Aging (Albany NY) 5 (12), 867–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Cecco M et al. (2013) Genomes of replicatively senescent cells undergo global epigenetic changes leading to gene silencing and activation of transposable elements. Aging Cell 12 (2), 247–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Belancio VP et al. (2010) Somatic expression of LINE-1 elements in human tissues. Nucleic Acids Res 38 (12), 3909–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pal S and Tyler JK (2016) Epigenetics and aging. Sci Adv 2 (7), e1600584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jurka J et al. (2007) Repetitive sequences in complex genomes: structure and evolution. Annu Rev Genomics Hum Genet 8, 241–59. [DOI] [PubMed] [Google Scholar]
- 17.Lu WJ et al. (2009) p53 ancestry: gazing through an evolutionary lens. Nat Rev Cancer 9 (10), 758–62. [DOI] [PubMed] [Google Scholar]
- 18.Levine AJ et al. (2016) P53 and the defenses against genome instability caused by transposons and repetitive elements. Bioessays 38 (6), 508–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leonova KI et al. (2013) p53 cooperates with DNA methylation and a suicidal interferon response to maintain epigenetic silencing of repeats and noncoding RNAs. Proceedings of the National Academy of Sciences of the United States of America 110 (1), E89–E98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gasior SL et al. (2006) The human LINE-1 retrotransposon creates DNA double-strand breaks. J Mol Biol 357 (5), 1383–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farkash EA and Prak ETL (2006) DNA Damage and L1 Retrotransposition. Journal of Biomedicine and Biotechnology 2006, 37285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haoudi A et al. (2004) Retrotransposition-Competent Human LINE-1 Induces Apoptosis in Cancer Cells With Intact p53. J Biomed Biotechnol 2004 (4), 185–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morrish TA et al. (2002) DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat Genet 31 (2), 159–65. [DOI] [PubMed] [Google Scholar]
- 24.Wylie A et al. (2016) p53 genes function to restrain mobile elements. Genes Dev 30 (1), 64–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wylie A et al. (2016) p53 in the game of transposons. Bioessays 38 (11), 1111–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kazazian HH Jr. (2004) Mobile elements: drivers of genome evolution. Science 303 (5664), 1626–32. [DOI] [PubMed] [Google Scholar]
- 27.Maksakova IA et al. (2006) Retroviral elements and their hosts: insertional mutagenesis in the mouse germ line. PLoS Genet 2 (1), e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun W et al. (2018) Pathogenic tau-induced piRNA depletion promotes neuronal death through transposable element dysregulation in neurodegenerative tauopathies. Nat Neurosci 21 (8), 1038–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scott EC and Devine SE (2017) The Role of Somatic L1 Retrotransposition in Human Cancers. Viruses 9 (6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katoh I and Kurata S (2013) Association of endogenous retroviruses and long terminal repeats with human disorders. Front Oncol 3, 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tasnim S and Kelleher ES (2018) p53 is required for female germline stem cell maintenance in P-element hybrid dysgenesis. Dev Biol 434 (2), 215–220. [DOI] [PubMed] [Google Scholar]
- 32.Harris CR et al. (2009) p53 responsive elements in human retrotransposons. Oncogene 28 (44), 3857–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang T et al. (2007) Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc Natl Acad Sci U S A 104 (47), 18613–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoh J et al. (2002) The p53MH algorithm and its application in detecting p53-responsive genes. Proc Natl Acad Sci U S A 99 (13), 8467–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang NT et al. (2007) The transcriptional activity of HERV-I LTR is negatively regulated by its cis-elements and wild type p53 tumor suppressor protein. J Biomed Sci 14 (2), 211–22. [DOI] [PubMed] [Google Scholar]
- 36.Tiwari B et al. (2017) Retrotransposons Mimic Germ Plasm Determinants to Promote Transgenerational Inheritance. Curr Biol 27 (19), 3010–3016 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wylie A et al. (2014) p53 activity is selectively licensed in the Drosophila stem cell compartment. Elife 3, e01530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu WJ et al. (2010) Meiotic recombination provokes functional activation of the p53 regulatory network. Science 328 (5983), 1278–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zamudio N et al. (2015) DNA methylation restrains transposons from adopting a chromatin signature permissive for meiotic recombination. Genes Dev 29 (12), 1256–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sciamanna I et al. (2016) The Reverse Transcriptase Encoded by LINE-1 Retrotransposons in the Genesis, Progression, and Therapy of Cancer. Front Chem 4, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toth KF et al. (2016) The piRNA Pathway Guards the Germline Genome Against Transposable Elements. Adv Exp Med Biol 886, 51–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kelleher ES (2017) Retrotransposons: Stowaways in the Primordial Germline. Curr Biol 27 (19), R1066–R1068. [DOI] [PubMed] [Google Scholar]
- 43.Lynch JA et al. (2011) The phylogenetic origin of oskar coincided with the origin of maternally provisioned germ plasm and pole cells at the base of the Holometabola. PLoS Genet 7 (4), e1002029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bontems F et al. (2009) Bucky ball organizes germ plasm assembly in zebrafish. Curr Biol 19 (5), 414–22. [DOI] [PubMed] [Google Scholar]
- 45.Malki S et al. (2014) A role for retrotransposon LINE-1 in fetal oocyte attrition in mice. Dev Cell 29 (5), 521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. https://www.jax.org/strain/002101.
- 47.Hu W et al. (2007) p53 regulates maternal reproduction through LIF. Nature 450 (7170), 721–4. [DOI] [PubMed] [Google Scholar]
- 48.Morikawa T et al. (2012) Tumor TP53 expression status, body mass index and prognosis in colorectal cancer. Int J Cancer 131 (5), 1169–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rodic N et al. (2014) Long interspersed element-1 protein expression is a hallmark of many human cancers. Am J Pathol 184 (5), 1280–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jung H et al. (2018) Immune signatures correlate with L1 retrotransposition in gastrointestinal cancers. Genome Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ehrlich M (2009) DNA hypomethylation in cancer cells. Epigenomics 1 (2), 239–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alves G et al. (1996) Differential methylation of human LINE-1 retrotransposons in malignant cells. Gene 176 (1–2), 39–44. [DOI] [PubMed] [Google Scholar]
- 53.Belancio VP et al. (2010) All y’all need to know ‘bout retroelements in cancer. Semin Cancer Biol 20 (4), 200–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kitkumthorn N and Mutirangura A (2011) Long interspersed nuclear element-1 hypomethylation in cancer: biology and clinical applications. Clin Epigenetics 2 (2), 315–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kreimer U et al. (2013) HERV-K and LINE-1 DNA Methylation and Reexpression in Urothelial Carcinoma. Front Oncol 3, 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kastenhuber ER and Lowe SW (2017) Putting p53 in Context. Cell 170 (6), 1062–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oren M and Rotter V (2010) Mutant p53 gain-of-function in cancer. Cold Spring Harb Perspect Biol 2 (2), a001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Belgnaoui SM et al. (2006) Human LINE-1 retrotransposon induces DNA damage and apoptosis in cancer cells. Cancer Cell Int 6, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haoudi A et al. (2004) Retrotransposition-Competent Human LINE-1 Induces Apoptosis in Cancer Cells With Intact p53. Journal of Biomedicine and Biotechnology 2004 (4), 185–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moran JV et al. (1996) High frequency retrotransposition in cultured mammalian cells. Cell 87 (5), 917–27. [DOI] [PubMed] [Google Scholar]
- 61.Cui F et al. (2011) Impact of Alu repeats on the evolution of human p53 binding sites. Biol Direct 6, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zemojtel T et al. (2009) Methylation and deamination of CpGs generate p53-binding sites on a genomic scale. Trends Genet 25 (2), 63–6. [DOI] [PubMed] [Google Scholar]
- 63.Botcheva K and McCorkle SR (2014) Cell Context Dependent p53 Genome Wide Binding Patterns and Enrichment at Repeats. PLoS ONE 9 (11), e113492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu S et al. (2014) Setdb1 is required for germline development and silencing of H3K9me3-marked endogenous retroviruses in primordial germ cells. Genes Dev 28 (18), 2041–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li Z et al. (2015) Distinct roles of DNMT1-dependent and DNMT1-independent methylation patterns in the genome of mouse embryonic stem cells. Genome Biol 16, 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Barau J et al. (2016) The DNA methyltransferase DNMT3C protects male germ cells from transposon activity. Science 354 (6314), 909–912. [DOI] [PubMed] [Google Scholar]
- 67.Rangan P et al. (2011) piRNA production requires heterochromatin formation in Drosophila. Curr Biol 21 (16), 1373–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Olivieri D et al. (2010) An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. Embo j 29 (19), 3301–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang XA et al. (2013) A major epigenetic programming mechanism guided by piRNAs. Dev Cell 24 (5), 502–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bewick AJ et al. (2017) Evolution of DNA Methylation across Insects. Mol Biol Evol 34 (3), 654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang J et al. (2010) G9a and Glp methylate lysine 373 in the tumor suppressor p53. J Biol Chem 285 (13), 9636–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang YA et al. (2005) DNA methyltransferase-3a interacts with p53 and represses p53-mediated gene expression. Cancer Biol Ther 4 (10), 1138–43. [DOI] [PubMed] [Google Scholar]
- 73.Wang D et al. (2016) Acetylation-regulated interaction between p53 and SET reveals a widespread regulatory mode. Nature 538 (7623), 118–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Akdemir F et al. (2007) p53 directs focused genomic responses in Drosophila. Oncogene 26 (36), 5184–93. [DOI] [PubMed] [Google Scholar]
- 75.Ardeljan D et al. (2017) The Human Long Interspersed Element-1 Retrotransposon: An Emerging Biomarker of Neoplasia. Clin Chem 63 (4), 816–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xiao-Jie L et al. (2016) LINE-1 in cancer: multifaceted functions and potential clinical implications. Genet Med 18 (5), 431–9. [DOI] [PubMed] [Google Scholar]
- 77.Martin SL et al. (2003) Trimeric structure for an essential protein in L1 retrotransposition. Proc Natl Acad Sci U S A 100 (24), 13815–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Martin SL et al. (2005) LINE-1 retrotransposition requires the nucleic acid chaperone activity of the ORF1 protein. J Mol Biol 348 (3), 549–61. [DOI] [PubMed] [Google Scholar]
- 79.Martin SL et al. (2000) Deletion analysis defines distinct functional domains for protein-protein and nucleic acid interactions in the ORF1 protein of mouse LINE-1. J Mol Biol 304 (1), 11–20. [DOI] [PubMed] [Google Scholar]
- 80.Dombroski BA et al. (1994) An in vivo assay for the reverse transcriptase of human retrotransposon L1 in Saccharomyces cerevisiae. Mol Cell Biol 14 (7), 4485–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Feng Q et al. (1996) Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 87 (5), 905–16. [DOI] [PubMed] [Google Scholar]
- 82.Hancks DC and Kazazian HH Jr. (2012) Active human retrotransposons: variation and disease. Curr Opin Genet Dev 22 (3), 191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hancks DC and Kazazian HH Jr. (2016) Roles for retrotransposon insertions in human disease. Mob DNA 7, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shukla R et al. (2013) Endogenous retrotransposition activates oncogenic pathways in hepatocellular carcinoma. Cell 153 (1), 101–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Morse B et al. (1988) Insertional mutagenesis of the myc locus by a LINE-1 sequence in a human breast carcinoma. Nature 333 (6168), 87–90. [DOI] [PubMed] [Google Scholar]
- 86.Hanahan D and Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144 (5), 646–74. [DOI] [PubMed] [Google Scholar]
- 87.Wallace NA et al. (2008) L1 mobile element expression causes multiple types of toxicity. Gene 419 (1–2), 75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Choi J et al. (2018) Interplay between RNASEH2 and MOV10 controls LINE-1 retrotransposition. Nucleic Acids Res 46 (4), 1912–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hanahan D and Weinberg RA (2000) The hallmarks of cancer. Cell 100 (1), 57–70. [DOI] [PubMed] [Google Scholar]