Abstract
The objectives of the study are to develop a new way to assess stability and discrimination capacity of radiomic features without the need of test-retest or multiple delineations and to use information obtained to perform a preliminary feature selection. Apparent diffusion coefficient (ADC) maps were computed from diffusion-weighted magnetic resonance images (DW-MRI) of two groups of patients: 18 with soft tissue sarcomas (STS) and 18 with oropharyngeal cancers (OPC). Sixty-nine radiomic features were computed, using three different histogram discretizations (16, 32, and 64 bins). Geometrical transformations (translations) of increasing entity were applied to the regions of interest (ROIs), and the intra-class correlation coefficient (ICC) was used to compare the features computed on the original and modified ROIs. The distribution of ICC values for minimal and maximal entity translations (ICC10 and ICC100, respectively) was used to adjust thresholds of ICC (ICCmin and ICCmax) used to discriminate between good, unstable (ICC10 < ICCmin), and non-discriminative features (ICC100 > ICCmax). Fifty-four and 59 radiomic features passed the stability-based selection for all the three histogram discretizations for the OPC and STS datasets, respectively. The excluded features were similar across the different histogram discretizations (Jaccard’s index 0.77 ± 0.13 and 0.9 ± 0.1 for OPC and STS, respectively) but different between datasets (Jaccard’s index 0.19 ± 0.02). The results suggest that the observed radiomic features are mainly stable and discriminative, but the stability depends on the region of the body under observation. The method provides a way to assess stability without the need of test-retest or multiple delineations.
Electronic supplementary material
The online version of this article (10.1007/s10278-018-0092-9) contains supplementary material, which is available to authorized users.
Keywords: Apparent diffusion coefficient maps, Radiomics, Radiomic features stability, Magnetic resonance imaging, Intra-class correlation coefficient
Introduction
Radiomics is an emerging field in quantitative imaging that uses image features to objectively and quantitatively describe tumor phenotypes [1]. The underlying hypothesis of radiomics is that such features could capture information not currently available using simple radiological analysis [2]. Radiomic features are non-invasively obtained on images that are part of the process of tumor evaluation and treatment, such as computed tomography (CT), magnetic resonance imaging (MRI), and positron emission tomography (PET). Thus, radiomic analysis could be performed without the need of further specific exams. Moreover, traditional histological analysis based on tissue samples, obtained through biopsies, cannot capture the heterogeneity of the whole tumor. On the other hand, radiomics, analyzing the entire tumor, can provide a complete and quantitative description of tumor heterogeneity, which may have profound implications for drug therapy in cancer [3]. All of the previous advantages make radiomics a technique of interest for tumor characterization. As a matter of fact, radiomics has already found a wide range of possible applications [4–14] such as prediction of clinical outcomes and response to treatment, tumor staging, discrimination of different types of tumor tissues, and assessment of cancer genetics.
The number of features used in radiomic studies may range from just a few [15] to several hundred [6]. However, not all the hundreds of extracted features bring information: some may be irrelevant or unreliable for the clinical question of interest. A process of feature selection is therefore necessary.
Stability analysis, assessing the robustness of the features, is a preliminary step in the process of feature selection [6, 12, 16]. Radiomic features stability can be investigated in several ways: (1) test-retest [6, 12, 16–23]; (2) multiple delineations of the region of interest (ROI) representing the tumor [6, 18, 21]; (3) change in image reconstruction and automatic segmentation parameters in PET or CT studies [20–22, 24, 25]; (4) change in image acquisition techniques [20, 24]; (5) inter-machine reproducibility [20, 26]. The most common techniques that are used for preliminary feature selection are typically the first two [6, 12, 16]. However, there are several problems concerning the different types of stability analysis. Different acquisitions are required to perform a proper test-retest analysis and the same thing can be said for analyzing stability to acquisition parameters and inter-machine reproducibility. Such requirements make the implementation of those types of analyses in the clinical routine. The analysis of stability to multiple delineations does not need multiple image acquisition, but drawing multiple ROIs on the same set of images can be very time-consuming. To solve the latter problem, alternative approaches may be considered. For example, in [12], stability is assessed through small geometrical transformations of the ROIs, which are used to mimic multiple manual delineations. In [27], the stability analysis is performed by comparing radiomic features computed on the entire ROI, and on a “digital biopsy,” i.e., a small portion of the ROI that is large enough to capture the heterogeneity of the tumor. Last, comparison of radiomic features obtained with multiple initialization of a semi-automatic segmentation algorithm or with different segmentation algorithms (like in [28]) could potentially be used for stability assessment. Although these approaches strongly reduce the amount of manual work necessary for a stability analysis of the radiomic features, they cannot be used to evaluate the discriminative power.
In the current study, we perform an analysis similar to the one presented in [12], so that stability of radiomic features could be evaluated starting with just one acquisition and one ROI. In addition to ROI transformations that are small and thus can mimic errors due to manual delineation, we apply also large geometrical transformations to evaluate features discrimination capacity. Our hypothesis is that features that do not change their values for large transformations are irrelevant and should therefore be excluded.
In this study, diffusion-weighted MRI (DW-MRI) of two different tumor types (oropharyngeal cancers and soft tissue sarcomas) are analyzed. DW-MRI have been chosen because they can be used to compute maps of apparent diffusion coefficient (ADC), which have been shown to be very useful for tumor detection and characterization [11, 29, 30], evaluation of treatment response [5, 31], and tumor staging [8, 32]. Also, unlike other types of MRI, ADC maps have been shown to be useful to assess tumor cellularity, even across different scanners [33], provided that the same range of b values and the same field strength are used [34, 35].
The aim of the present study is to provide a method to perform a preliminary feature selection based on features stability. An innovative characteristic of the method is that it does not require either multiple acquisitions or multiple manual delineations.
Material and Methods
Study Population
In this study, two different datasets were retrospectively analyzed: the first one contains DW-MRI images of soft tissue sarcomas (STS); the second one contains DW-MRI images of oropharyngeal cancer (OPC). The two datasets are provided by the Fondazione IRCCS Istituto Nazionale dei Tumori (Milan, Italy).
Both datasets consisted of 18 patients who underwent an MRI acquisition before starting the treatment. Both studies were approved by the ethical committee of Fondazione IRCCS Istituto Nazionale dei Tumori (Milan, Italy) and conducted in accordance with the Helsinki Declaration; all patients gave their written informed consent. All patients’ data were anonymized prior to the analysis.
Image Acquisition
STS Dataset
DW-MRI images were acquired using Achieva 1.5 T system (Philips Medical system, Eindhoven, Netherlands)—5 patients—or a Magnetom Avanto 1.5 T system (Siemens Medical Solutions, Erlangen, Germany)—13 patients—both with a body-matrix coil and spine array coil for signal reception. The data were acquired axially by means of echo planar imaging. The sequences’ parameters (for both equipment) are reported in Table 1. Diffusion-weighted images (DWI) were acquired using four b values (50, 400, 800, and 1000 s/mm2).
Table 1.
MRI sequence parameters by MRI scanners
| Sequence parameter | STS database | OPC database | |
|---|---|---|---|
| Siemens Avanto (n = 13) | Philips Achieva (n = 5) | Siemens Avanto (n = 18) | |
| Sequence name | ep2d | dwi_ssh | ep2d |
| Matrix (pixels) | 192 × 192 | 255 × 255 | 132 × 132 |
| Resolution (voxel/mm) | 1.98 × 198 | 1.37 × 1.37 | 1.89 × 1.89 |
| Field of view (mm) | 380 × 380 | 350 × 350 | 250 × 250 |
| Repetition time (msec) | 5400 | 7410 | 3300 |
| Echo time (msec) | 78 | 63 | 64 |
| Slice thickness (mm) | 4 (no gap) | 5 (no gap) | 3 (gap 0.9) |
| Number of excitations | 4 | 3 | 3 |
OPC Dataset
DWI were acquired using Magnetom Avanto 1.5 T system (Siemens Medical Solutions, Erlangen, Germany). The sequence parameters are reported in Table 1. DWI images were acquired using ten b values 0, 10, 20, 50, 70, 100, 150, 200, 500, and 1000 (s/mm2).
Image Processing
For both the datasets, ADC maps were computed. The ADC was defined as the slope of the linear regression of the logarithm of the DWI exponential signal decay on the b values [36]. The calculation was performed pixel-wise using ITK 4.8 [3].
For the both datasets, the segmentation of the gross tumor volume (GTV) was performed by an expert radiologist on the DW-MRI computed with the lowest b value, where the tumor is the most visible. The preprocessing steps were performed using 3D Slicer [37].
Radiomic Feature Extraction
In this study, 69 radiomic features were computed, pertaining to two main categories: (1) intensity-based and (2) texture-based. The complete list is reported in Table 2.
Table 2.
Radiomic features analyzed in this study, divided by category
| First-order statistics (FOS) | ||
| -Signal energy -Signal kurtosis -Signal mean absolute deviation (MAD) -Signal maximum -Signal mean -Signal median -Signal minimum -Signal quantile 0.01 -Signal quantile 0.1 -Signal quantile 0.2 -Signal quantile 0.3 -Signal quantile 0.4 -Signal quantile 0.5 -Signal quantile 0.6 -Signal quantile 0.7 |
-Signal quantile 0.8 -Signal quantile 0.9 -Signal quantile 0.99 -Signal range -Signal root mean square (RMS) -Signal skewness -Signal standard deviation (SD) -Signal variance -Histogram entropy -Histogram kurtosis -Histogram mean absolute deviation (MAD) -Histogram maximum -Histogram mean |
-Histogram median -Histogram minimum -Histogram range -Histogram root mean square (RMS) -Histogram skewness -Histogram standard deviation (SD) -Histogram variance -Histogram uniformity -Histogram total frequency |
| Gray-level co-occurrence matrix (GLCM) | ||
| -Autocorrelation -Cluster prominence -Cluster shade -Cluster tendency -Contrast -Correlation -Difference entropy -Dissimilarity |
-Energy -Entropy -Homogeneity -Homogeneity 2 -Information measure of correlation 1 (IMOC1) -Information measure of correlation 2 (IMOC2) |
-Inverse difference moment -Inverse difference moment 2 -Inertia -Inverse variance -Max probability -Sum average -Sum entropy |
| Gray-level run length matrix (GLRLM) | ||
| -Gray-level non-uniformity -High gray-level emphasis -Long run emphasis -Long run high gray-level emphasis |
-Long run low gray-level emphasis -Low gray-level emphasis -Run length non-uniformity -Run percentage |
-Short run emphasis -Short run high gray-level emphasis -Short run low gray-level emphasis |
Features belonging to the intensity-based group (first-order statistics or FOS) included statistical information about the signal intensity and histogram distribution of the pixels in the ROI. The histogram was evaluated between 0 and 4000 *10−6 mm2/s using N bins. In this study, three values of N were tested (16, 32, and 64 bins) to evaluate whether the bin number affects the stability of the features.
Texture-based features were computed on the gray-level co-occurrence matrix (GLCM) [38] and the gray-level run length matrix (GLRLM) [39]. For a given direction α, the GLCM is a NxN matrix, whose (i, j) element is the counting of pixels of gray intensity level i which are adjacent (within a distance ρ) to pixels of the gray intensity level j. The GLRLM is an NxN matrix whose (i, j) element counts the number of runs of pixels of gray level i (run step 1) and run length j in a given direction. The same bin numbers (16, 32, and 64) used for FOS analysis were used for textural features computation. Range of ADC values for histogram creation was also the same (0–4000 *10−6 mm2/s). A distance ρ = 1 was used to create the GLCMs and GLRLMs.
For each patient, GLCMs and GLRLMs were created on 13 different directions. Textural features of Table 2 were computed on each matrix and the results averaged across all angles, thus obtaining two sets of features, one for the GLCM and one for the GLRLM. This average of the 13 different value is already been used in literature (see supplementary material of [6]) and it allows to deal with a lower dimensional features space (only one feature is considered instead of 13). All the algorithms were implemented in ITK 4.8 [3, 40].
Globally, 37 FOS, 21 GLCM-based, and 11 GLRLM-based features (69 in total) were considered for this analysis. Fifty-seven features out of 69 were bin-dependent and thus were computed three times, one for each histogram discretization.
Stability and Discrimination Capacity Analysis
We developed a framework to assess features stability and discrimination capacity that is based on geometrical transformations (translations in particular) of the ROIs representing the GTV. The entire workflow was implemented in MATLAB 2016b (Mathworks, Natick, MA, USA).
First, small entity translations were applied to the ROIs, along both the x (medial-lateral) and y (antero-posterior) directions. By small entity, we mean translations of ± 10% of the length of the bounding box surrounding the ROI in the direction of interest (Fig. 1a). We will also refer to this type of translation as minimal entity translation. We assume the variability due to such transformations to be comparable to the ones that could appear in a multiple delineations test. In total, for each ROI, four minimal entity translations were applied (one positive and one negative for both the x and y directions) and thus four transformed ROIs were obtained. The radiomic features were computed on the four transformed ROIs and compared to the ones obtained with the original one (the one segmented by the radiologist). Radiomic features were then compared using two similarity indexes: (1) percentage variation and (2) intra-class correlation coefficient (ICC).
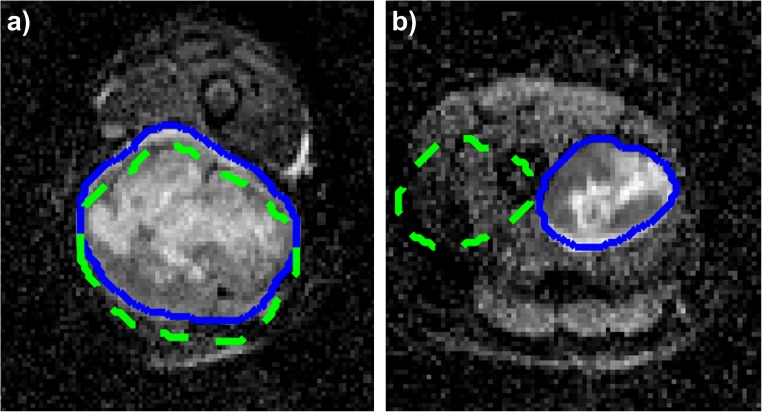
Fig. 1.
Example of translations applied to the regions of interest (ROIs). a Example of small entity translation in the y direction. b Example of maximal entity translation in the x direction. Continuous lines represent the contours of the original ROIs, while the dashed lines represent the contours of the modified ones
For each comparison, the absolute percentage variation with respect to the reference was computed as follows:
| 1 |
being FTransf and FOriginal the features computed on the transformed and original ROIs, respectively.
The ICC was computed as in [41, 42]: it measures the bivariate relation of variables representing different measurement classes and can be used to assess the agreement between data. The maximum value of ICC is 1, which indicates perfect agreement. The lower the ICC, the lower the similarity among the elements of the groups. In this study, a two-way mixed effect model was used (since the effect of the transformations is fixed and the variability for the different ROIs is random) [42].
For each feature, it is possible to compute 72 percentage variations (18 ROIs with 4 translations each) and 4 ICCs (one for each translation) and to compute the mean and standard deviation for both the distributions. Let us call the mean values obtained with such procedure ICCmean and Diff%mean.
We repeat the above-described steps for increasing translation entities ranging from 10% (minimal entity translations) to 100% (maximal entity translations) with a step of 10%, and we computed the ICCmean and Diff%mean of the features for each translation, to evaluate how the similarity varies with the entity of the translations. In Fig. 1b, an example of maximal entity (± 100%) translation is represented. As it can be seen, this situation is far from the error range obtainable with multiple delineations. This type of transformation was used to evaluate discrimination capacity because, as previously stated, the underlying hypothesis is that if a feature remains constant independently on the entity of the translation, that feature is not going to be a good clinical descriptor.
ICCmean was used to select the features with properties of stability and discrimination capacity. For this purpose, two ICC thresholds were used: a lower threshold for the ICC for the minimal entity translations (ICCmin) and an upper ICC threshold for the maximal entity translations (ICCmax). A feature is considered stable if the ICCmean for the minimal entity translations (ICC10) is larger than ICCmin (ICC10 ≥ ICCmin), and it is considered discriminative if the mean ICCmean for the maximal entity translations (ICC100) is lower than ICCmax (ICC100 ≤ ICCmax).
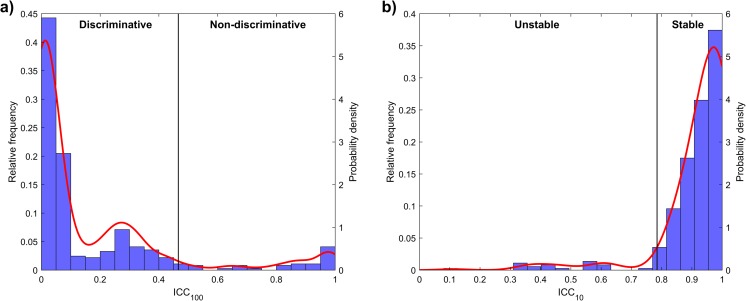
The two thresholds were set using information about the distributions of ICC10 and ICC100. The values of ICC100 for both the datasets and for all the bin discretizations are put together in the same histogram and, from this histogram, a continuous probability distribution is obtained (see Fig. 2). In particular, the probability distribution is a non-parametric kernel distribution fitted using MATLAB function fitdist (normal kernel, bandwidth 0.05). The value 0.05 was chosen as a good tradeoff to guarantee both smoothness of the curve and quality of the fitting (p > 0.05 for a χ2 test). ICCmax was defined as the quantile 0.9 of the continuous distribution previously defined. A similar procedure was used to define the ICCmin threshold starting from the histogram of all the ICC10, with the difference that the quantile used as a reference was 0.1.
Fig. 2.
Continuous distribution fitted on the values of ICC100 (a) and ICC10 (b). In both cases, the reference quantile is marked with a line that divides the plot in two sections (discriminative/non-discriminative and stable/unstable respectively in a and b)
The stability and discrimination capacity analysis is repeated 3 times, using 3 different bin numbers (16, 32, and 64 bins), to assess the effect of histogram discretization on the features. Jaccard’s index [43] was used to evaluate the similarity between the sets of excluded features for the different histogram discretizations, but also to compare excluded features in the two datasets.
Results
The identified thresholds for ICCmin and ICCmax that were identified with the method explained in the previous section were 0.78 and 0.46, respectively.
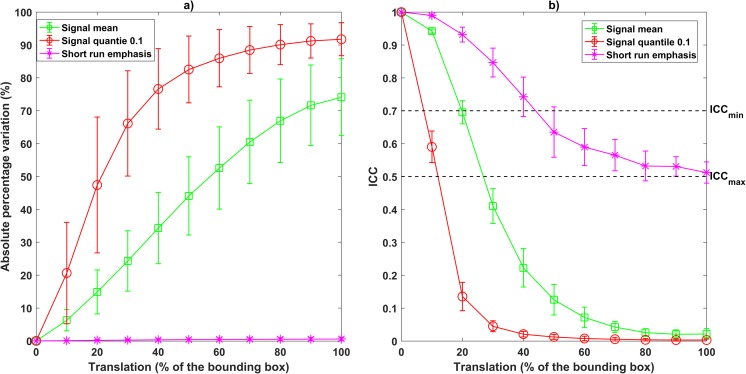
The heat maps in Figs. 3, 4, 5, 6, 7, and 8 show how the level of ICCmean varies with the entity of the translations in the two datasets. Figures 3, 4, and 5 show the ICCmean maps related to the OPC dataset using the three different histogram subdivisions, while Figs. 6, 7, and 8 show the ICCmean maps for the STS dataset. In Fig. 9a, examples of Diff%mean plot (with 95% confidence interval) for an unstable feature (signal quantile 0.1), a non-discriminative feature (short run emphasis), and a feature that is selected by the algorithm (signal mean) in the STS dataset can be seen. In Fig. 9b, the plot of ICCmean (with 95% confidence interval) for the same features can be seen. Since it is not possible to represent all the values of percentage variations and ICC, we refer to Tables 1–20 in the online resources, containing all the values of ICC10 and ICC100, together with the corresponding percentage variations.
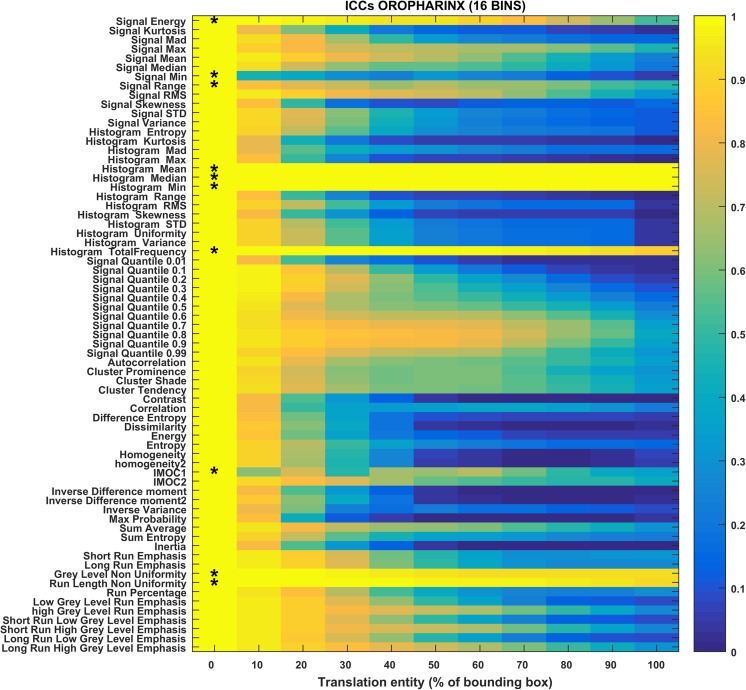
Fig. 3.
Heat map of the mean ICCmean displayed according to features (rows) and entity of the translations (columns). The heat map refers to the oropharyngeal cancers (OPC) dataset and to the radiomic features computed with the 16-bin discretization. The features removed by the ICC-based feature selection technique are marked with an asterisk in the first column
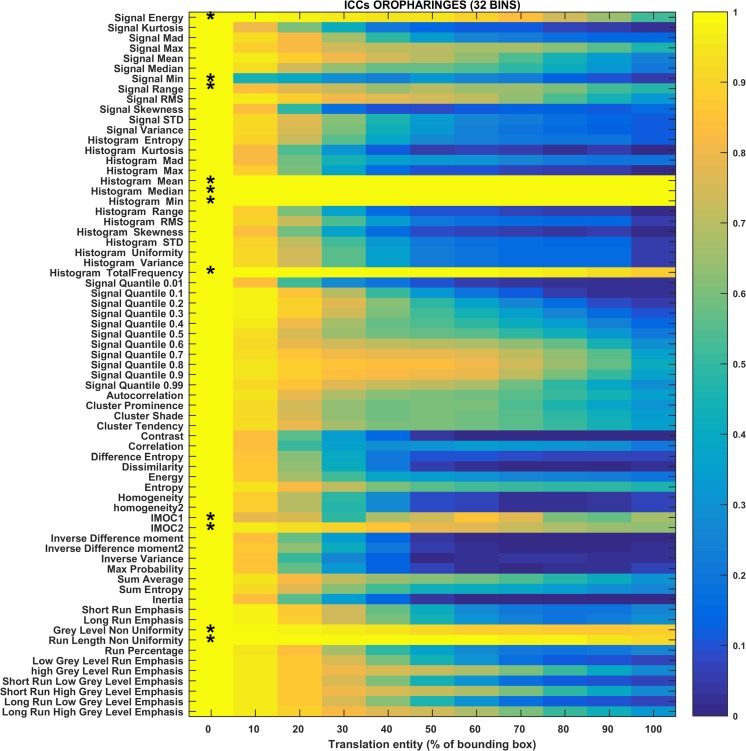
Fig. 4.
Heat map of the mean ICCmean displayed according to features (rows) and entity of the translations (columns). The heat map refers to the oropharyngeal cancers (OPC) dataset and to the radiomic features computed with the 32-bin discretization. The features removed by the ICC-based feature selection technique are marked with an asterisk in the first column
Fig. 5.
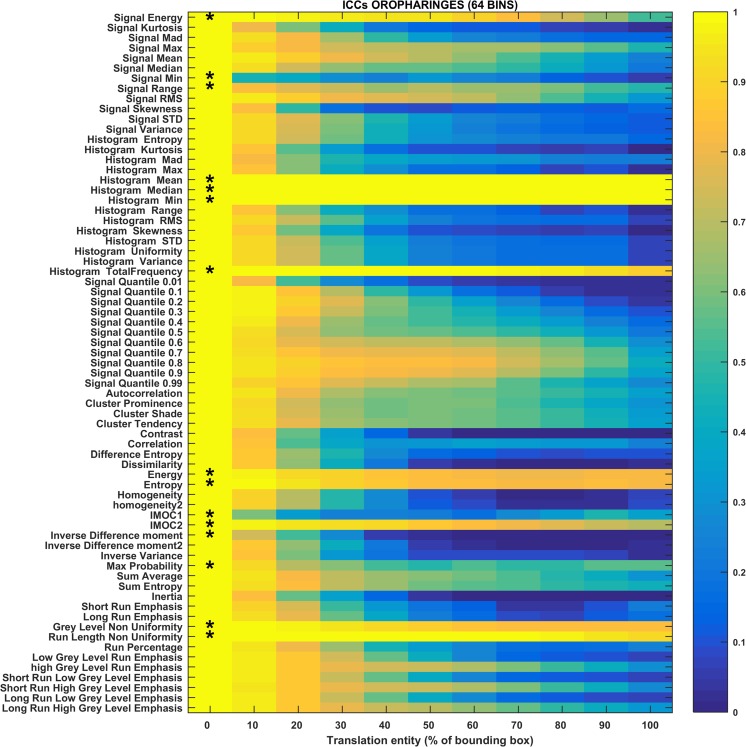
Heat map of the mean ICCmean displayed according to features (rows) and entity of the translations (columns). The heat map refers to the oropharyngeal cancers (OPC) dataset and to the radiomic features computed with the 64-bin discretization. The features removed by the ICC-based feature selection technique are marked with an asterisk in the first column
Fig. 6.
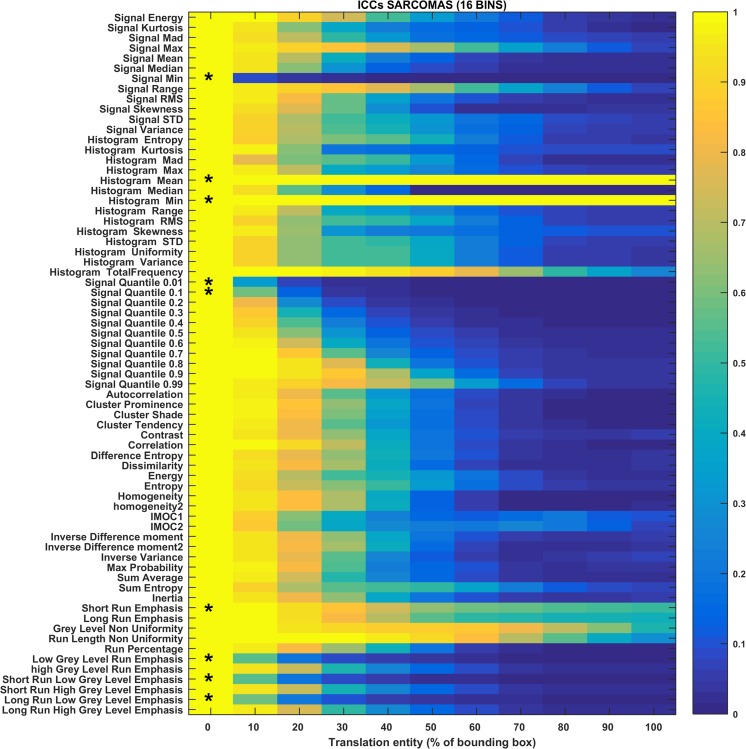
Heat map of the mean ICCmean displayed according to features (rows) and entity of the translations (columns). The heat map refers to the soft tissue sarcoma (STS) dataset and to the radiomic features computed with the 16-bin discretization. The features removed by the ICC-based feature selection technique are marked with an asterisk in the first column
Fig. 7.
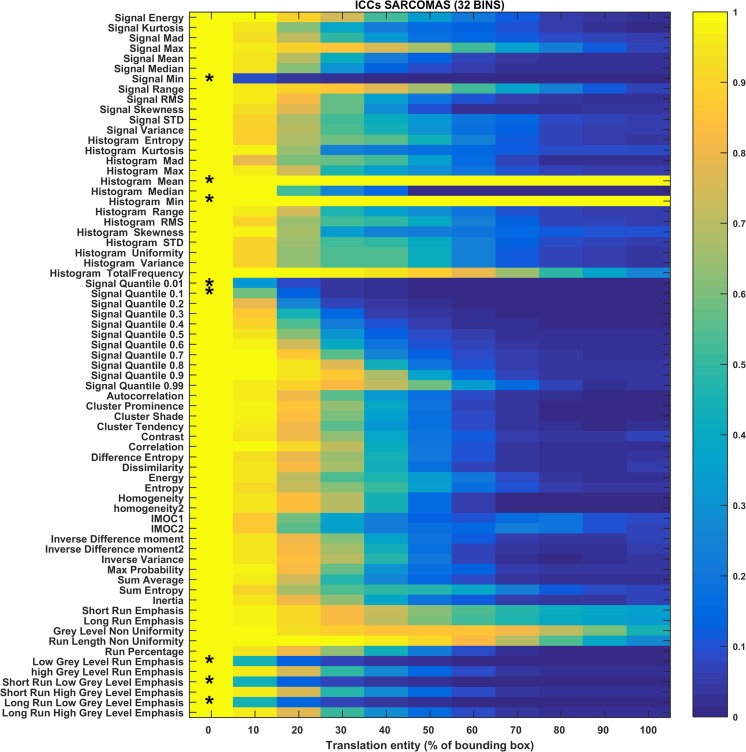
Heat map of the mean ICCmean displayed according to features (rows) and entity of the translations (columns). The heat map refers to the soft tissue sarcoma (STS) dataset and to the radiomic features computed with the 32-bin discretization. The features removed by the ICC-based feature selection technique are marked with an asterisk in the first column
Fig. 8.
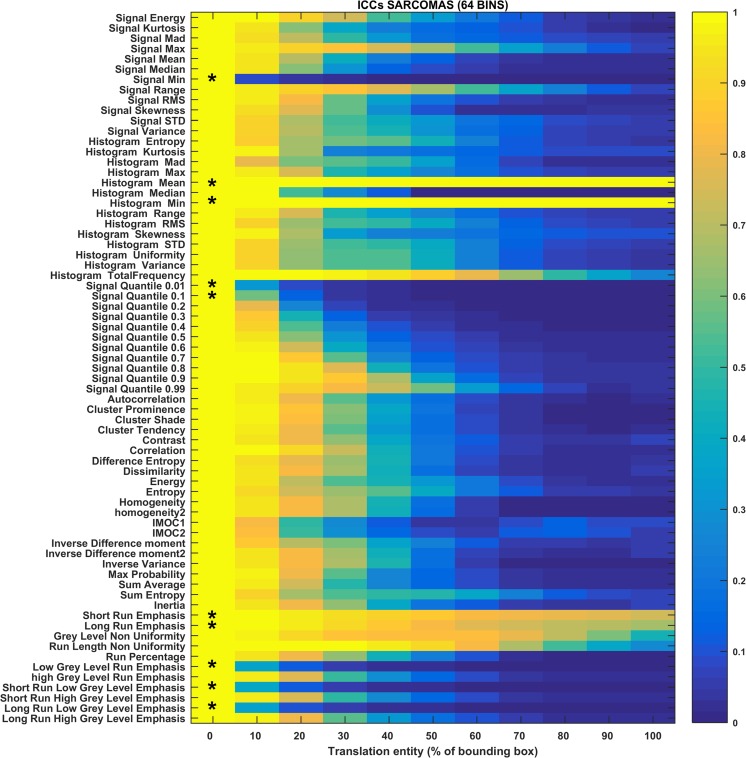
Heat map of the mean ICCmean displayed according to features (rows) and entity of the translations (columns). The heat map refers to the soft tissue sarcoma (STS) dataset and to the radiomic features computed with the 64-bin discretization. The features removed by the ICC-based feature selection technique are marked with an asterisk in the first column
Fig. 9.
Plot representing the variation with respect to entity of translation for 3 different radiomic features measured on the soft tissue sarcoma (STS) dataset, with 16-bin discretization. a Absolute percentage variation plot. b ICC variation. One representative of each group of feature is represented: signal mean (squared markers) is both stable and discriminative; signal quantile 0.1 (circular markers) is unstable; short run emphasis (asterisks) is non-discriminative. Both mean values and 95% confidence interval are shown
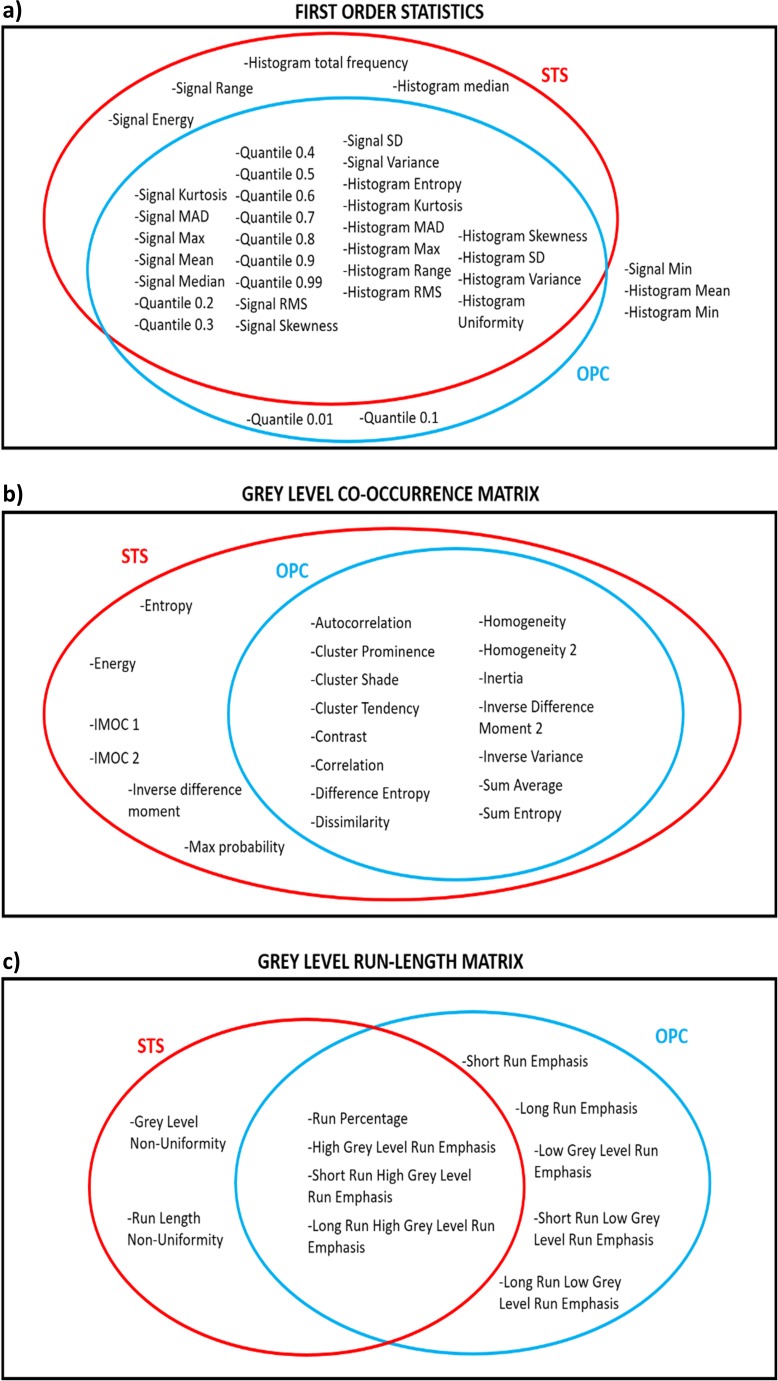
Table 3 lists the features removed with our ICC-based feature selection method. The six boxes show the results in the two datasets with each of the three histogram discretizations. The ICC-based feature selection method removes 8–15 features. If we consider the features that are stable for all the three histogram discretizations, the method selects 54 features out of 69 for the OPC dataset and 59 features out of 69 for the STS dataset. Such features, divided by groups, are shown in the Euler-Venn diagrams in Fig. 10. If we take into account the three subsets of the excluded features for the three histogram subdivisions and we compute the Jaccard’s similarity index for the three possible combinations, we obtain a value of 0.77 ± 0.13 for the OPC dataset and 0.9 ± 0.1 for the STS dataset. If we compare the set of excluded features for the OPC and STS dataset for each of the three histogram discretizations, we get a Jaccard’s index of 0.17 ± 0.03.
Table 3.
Features removed by the ICC-based feature selection algorithm
| 16 bins | 32 bins | 64 bins |
|---|---|---|
| OPC dataset | ||
| -Signal energy -Signal minimum -Signal range -Histogram mean -Histogram median -Histogram minimum -Histogram total frequency -Information measure of correlation 1 (IMOC1) -Gray-level non-uniformity -Run length non-uniformity |
-Signal energy -Signal minimum -Signal range -Histogram mean -Histogram median -Histogram minimum -Histogram total frequency -Information measure of correlation 1 (IMOC1) -Information measure of correlation 2 (IMOC2) -Gray-level non-uniformity -Run length non-uniformity |
-Signal energy -Signal minimum -Signal range -Histogram mean -Histogram median -Histogram minimum -Histogram total frequency -Energy -Entropy -Information measure of correlation 1 (IMOC1) -Information measure of correlation 2 (IMOC2) -Inverse difference moment -Max probability -Gray-level non-uniformity -Run length non-uniformity |
| STS dataset | ||
| -Signal minimum -Signal quantile 0.01 -Signal quantile 0.1 -Histogram mean -Histogram minimum -Short run emphasis -Low gray-level run emphasis -Short run low gray-level emphasis -Long run low gray-level emphasis |
-Signal minimum -Signal quantile 0.01 -Signal quantile 0.1 -Histogram mean -Histogram minimum -Low gray-level run emphasis -Short run low gray-level emphasis -Long run low gray-level emphasis |
-Signal minimum -Signal quantile 0.01 -Signal quantile 0.1 -Histogram mean -Histogram minimum -Short run emphasis -Long run emphasis -Low gray-level run emphasis -Short run low gray-level emphasis -Long run low gray-level emphasis |
Fig. 10.
Euler-Venn diagram representing the accepted features divided by group. a First-order statistics. b Gray-level co-occurrence matrix. c Gray-level run length matrix. Selected features are grouped by dataset: the soft tissue sarcoma (STS) dataset and oropharyngeal cancer (OPC) dataset
Discussion
The assessment of features stability is an important preliminary step in any radiomic analysis. In this study, we developed a new method to assess the stability and the discrimination capacity of radiomic features computed from medical images (in this case DW-MRI images). In particular, we proposed a fast way to assess features stability and discrimination capacity without the need of multiple acquisitions or multiple delineations, thus performing a preliminary step of feature selection.
Both in STS and OPC datasets, features can be divided in three groups: (I) features whose ICC decreases gradually but constantly; (II) features whose ICC sharply decreases; (III) features that remain similar for all translations. These three groups can be approximately considered as (I) the stable and discriminative features, (II) unstable features, and (III) stable and non-discriminative features, respectively.
In the STS dataset, the ICC-based feature selection removes the features in group II (unstable features) and many of the ones of group III (non-discriminative features). However, there are some features for which ICC100 is slightly under the threshold that are therefore not considered as non-discriminant (histogram total frequency and some GLRLM-based features matrix). Some of these features are removed for some of the histogram discretizations (e.g., short and long run emphasis).
Something similar can be said for the features in the OPC dataset in Figs. 3, 4, and 5. There are features, like signal energy, gray-level non-uniformity, and run length non-uniformity, that are removed because they remain very similar inside and outside the tumor. There are also features, like signal minimum, that are too unstable and drastically change even for small translations. Some features, like the information measures of correlation, present an ICC that is very close to the threshold and therefore they are excluded just for some histogram discretizations. Two features (entropy and energy) strongly change their behavior according to histogram discretization. It can be seen that for 16-bin discretization, the ICC level for those features decreases quite gradually, and the features are accepted according to our method. Using the 64-bin discretization, their values of ICC remain almost constant and the features are considered non-discriminative. The increase in entropy with the number of bins is predictable: more bins means more gray levels and more disorder. However, the fact that the change in the measured ICC is so high, it is worth noting. The fact that both energy and entropy have high dependency on the histogram discretization is also reported in [44]. Max probability also changes its stability behavior for the 64-bin discretization, similarly to what happens for entropy. Last, ICC10 for inverse difference moment is close to the threshold of stability and the feature is labeled as unstable when the 64-bin discretization is used.
Although the behavior of some features, like energy and entropy, is highly dependent on the number of bins used, in general, the results of the ICC-based feature selection do not depend on histogram discretization. The type of tumor, instead, strongly affects the excluded features. There are only three common features between the datasets. Signal minimum is unstable as it can be expected since it is an extreme value of a distribution. Histogram mean is always constant throughout all the translation because it only depends on the number of bins. Histogram minimum is 0 when there is at least one empty bin in the histogram, which is very common; therefore, the feature is non-discriminative. This is true at least for the histogram subdivisions that were used in this study.
To our knowledge, this is the first time that both small and large translations of the ROI are used to evaluate fatures stability and discriminative power respectively. It is also the first time in which the thresholds of ICC used to distinguish the type of feature (stable, unstable, or non-discriminative) are not empirically set.
The values of ICC for small transformations computed for the radiomic features analyzed in this study are around 0.9 (median 0.94, quartiles 0.89 and 0.97). In [12], similar values of ICC are found for the stable features (median 0.97, quartiles 0.92 and 0.99). The Mann-Whitney test reveals no significant difference between the ICC values of the stable features identified in the current study and in [12] (p = 0.92). However, a smaller number of features is actually stable (18 out of 79). This could depend from the fact that in the present study and [12], the features set used is not the same.
Compared to a study in which features stability is assessed through multiple manual delineation, like [18], the values of ICC found for small translations are higher than the ones found for multiple delineations (median 0.94 vs median 0.89, Mann-Whitney test p < 0.01). The initial assumption that the low entity translations are equivalent to multiple delineations in terms of evaluating stability seems to be rejected, even though the differences in the ICC values could also depend on the different imaging technique (MRI vs PET) and in the different region of the body analyzed (lung vs limbs and head and neck). According to such findings, our method is potentially less restrictive for the assessment of stability, but for this reason, we can be sure that the features that we identify as unstable are indeed unstable. Moreover, if a more restrictive method is required, the translation considered for stability analysis could be increased to 15–20% of the bounding box.
In this paper, as opposed to [12], we presented only translations of the ROIs and we did not show the effect of rotation, dilatation, and shrinking. Those types of transformations were also applied in our investigation but their use did not influence the results of the ICC-based feature selection method, and therefore they were not reported (for further details, refer to the Tables 21–60 of the online resources).
The method presented in this study has some advantages over other methods of literature. Compared to [27], it does not need a digital biopsy, which requires a further segmentation step, although a digital biopsy takes less time to be segmented than a normal ROI. Compared to a method based on [28], it requires no segmentation algorithm, which can be difficult to design for oropharyngeal tumors. Last, the presented method allows to evaluate not only stability, but also the discriminative power of the features, which is something that, to the knowledge of the authors, was never considered before.
This study highlights the difference in stability of the radiomic features for tumors in different regions of the body, which is not typically done. As a matter of fact, the majority of the studies on stability of radiomic features focuses on tumors in a specific region of the body: esophagus [17], liver [19], brain [12], lung [22], or kidney [23]. A study analyzing multiple body regions exists [24], but even though the data come from multiple sources, they are analyzed all together and differentiation in the stability behavior for the different body regions is not explored. In this paper, we observed that radiomic features from tumors in the head and neck region (OPC dataset) present in general lower stability to small translations than tumors in the limbs (STS dataset). In fact, the values of ICCs for small translations are significantly higher in the STS dataset (Wilcoxon signed rank test p < 0.01; see also online resources, Tables 1–20). This result could come from the fact that sarcomas have larger volume and small translations have less effect on features that are computed on the entire ROI. The opposite happens when we consider the ICCs for large transformations (Wilcoxon signed rank test p < 0.01; see also online resources, Tables 1–20). This could depend from the fact that the contrast between tumoral and healthy tissue in ADC images is different for the two types of cancer. As a matter of fact, sarcomas have higher contrast and are much easier to distinguish, rather than head and neck tumors.
We think that the presented study could provide a better understanding of radiomic features stability for DW-MRI. It is worth underlining that this methodology should be used just as a preliminary feature selection. In fact, of the 69 radiomic features that were analyzed, only 8–15 are excluded by our algorithm, which is about 10–20% of the total number features. In order to further reduce the number of selected features, a possible approach could be to add a correlation-based (as shown in [16]) or a wrapper feature selection method after the ICC-based analysis. A limitation of this approach is that it cannot be used for geometrical features like shape and size or location (which are also used in [16]) since the shape and size of each ROI are kept constant throughout all the experiment, while the ROI location is changed. A possible solution to this could be to apply random combination of geometrical transformations to mimic the effects of random multiple delineations or ROI registration, and we plan to investigate this in further studies.
Conclusion
In this study, a method to assess the stability and the discrimination capacity of the radiomic features has been developed, using small and large translations of the ROI. The method was applied to two independent datasets containing DW-MRI images of different tumors (oropharyngeal tumors and sarcomas). The proposed method excluded 10–20% of the original features set.
We think that the presented study could provide a better understanding of radiomic features stability and discrimination capacity for DW-MRI, providing a way to assess features stability without the need of multiple acquisitions or delineations.
Electronic Supplementary Material
(DOCX 166 kb)
Compliance with Ethical Standards
Both studies were approved by the ethical committee of Fondazione IRCCS Istituto Nazionale dei Tumori and conducted in accordance with the Helsinki Declaration; all patients gave their written informed consent. All patients’ data were anonymized prior to the analysis.
References
- 1.Yip SSF, Aerts HJWL. Applications and limitations of radiomics. Phys. Med. Biol. 2016;61:R150–R166. doi: 10.1088/0031-9155/61/13/R150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, Van Stiphout RGPM, Granton P, Zegers CML, Gillies R, Boellard R, Dekker A, Aerts HJW. Radiomics: extracting more information from medical images using advanced feature analysis. Eur. J. Cancer. 2012;48:441–446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fisher R, Pusztai L, Swanton C. Cancer heterogeneity: implications for targeted therapeutics. Br. J. Cancer. 2013;108:479–485. doi: 10.1038/bjc.2012.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang H, Tan S, Chen W, Kligerman S, Kim G, D’Souza WD, Suntharalingam M, Lu W. Modeling pathologic response of esophageal cancer to chemoradiotherapy using spatial-temporal 18F-FDG PET features, clinical parameters, and demographics. Int. J. Radiat. Oncol. Biol. Phys. 2014;88:195–203. doi: 10.1016/j.ijrobp.2013.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lambrecht M, Van Calster B, Vandecaveye V, De Keyzer F, Roebben I, Hermans R, Nuyts S. Integrating pretreatment diffusion weighted MRI into a multivariable prognostic model for head and neck squamous cell carcinoma. Radiother. Oncol. 2014;110:429–434. doi: 10.1016/j.radonc.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 6.Aerts HJWL, Velazquez ER, Leijenaar RTH, Parmar C, Grossmann P, Cavalho S, Bussink J, Monshouwer R, Haibe-Kains B, Rietveld D, Hoebers F, Rietbergen MM, Leemans CR, Dekker A, Quackenbush J, Gillies RJ, Lambin P: Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 5, 2014 [DOI] [PMC free article] [PubMed]
- 7.Ganeshan B, Skogen K, Pressney I, Coutroubis D, Miles K. Tumour heterogeneity in oesophageal cancer assessed by CT texture analysis: preliminary evidence of an association with tumour metabolism, stage, and survival. Clin. Radiol. 2012;67:157–164. doi: 10.1016/j.crad.2011.08.012. [DOI] [PubMed] [Google Scholar]
- 8.Kierans AS, Rusinek H, Lee A, Shaikh MB, Triolo M, Huang WC, Chandarana H. Textural differences in apparent diffusion coefficient between low- and high-stage clear cell renal cell carcinoma. Am. J. Roentgenol. 2014;203:W637–W644. doi: 10.2214/AJR.14.12570. [DOI] [PubMed] [Google Scholar]
- 9.Mu Wei, Chen Zhe, Liang Ying, Shen Wei, Yang Feng, Dai Ruwei, Wu Ning, Tian Jie. Staging of cervical cancer based on tumor heterogeneity characterized by texture features on18F-FDG PET images. Physics in Medicine and Biology. 2015;60(13):5123–5139. doi: 10.1088/0031-9155/60/13/5123. [DOI] [PubMed] [Google Scholar]
- 10.Xu Rui, Kido Shoji, Suga Kazuyoshi, Hirano Yasushi, Tachibana Rie, Muramatsu Keiichiro, Chagawa Kazuki, Tanaka Satoshi. Texture analysis on 18F-FDG PET/CT images to differentiate malignant and benign bone and soft-tissue lesions. Annals of Nuclear Medicine. 2014;28(9):926–935. doi: 10.1007/s12149-014-0895-9. [DOI] [PubMed] [Google Scholar]
- 11.Wibmer A, Hricak H, Gondo T, Matsumoto K, Veeraraghavan H, Fehr D, Zheng J, Goldman D, Moskowitz C, Fine S, Reuter VE, Eastham J, Sala E, Vargas HA. Haralick texture analysis of prostate MRI: utility for differentiating non-cancerous prostate from prostate cancer and differentiating prostate cancers with different Gleason scores. Eur. Radiol. 2016;25:2840–2850. doi: 10.1007/s00330-015-3701-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gevaert Olivier, Mitchell Lex A., Achrol Achal S., Xu Jiajing, Echegaray Sebastian, Steinberg Gary K., Cheshier Samuel H., Napel Sandy, Zaharchuk Greg, Plevritis Sylvia K. Glioblastoma Multiforme: Exploratory Radiogenomic Analysis by Using Quantitative Image Features. Radiology. 2014;273(1):168–174. doi: 10.1148/radiol.14131731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutman DA, Dunn WD, Grossmann P, Cooper LAD, Holder CA, Ligon KL, Alexander BM, Aerts HJWL. Somatic mutations associated with MRI-derived volumetric features in glioblastoma. Neuroradiology. 2015;57:1227–1237. doi: 10.1007/s00234-015-1576-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corino VDA, Montin E, Messina A, Casali PG, Gronchi A, Marchianò A, Mainardi LT. Radiomic analysis of soft tissues sarcomas can distinguish intermediate from high-grade lesions. J. Magn. Reson. Imaging. 2017;47:829–840. doi: 10.1002/jmri.25791. [DOI] [PubMed] [Google Scholar]
- 15.King Ann D., Chow Kwok-Keung, Yu Kwok-Hung, Mo Frankie Kwok Fai, Yeung David K. W., Yuan Jing, Bhatia Kunwar S., Vlantis Alexander C., Ahuja Anil T. Head and Neck Squamous Cell Carcinoma: Diagnostic Performance of Diffusion-weighted MR Imaging for the Prediction of Treatment Response. Radiology. 2013;266(2):531–538. doi: 10.1148/radiol.12120167. [DOI] [PubMed] [Google Scholar]
- 16.Balagurunathan Yoganand, Gu Yuhua, Wang Hua, Kumar Virendra, Grove Olya, Hawkins Sam, Kim Jongphil, Goldgof Dmitry B., Hall Lawrence O., Gatenby Robert A., Gillies Robert J. Reproducibility and Prognosis of Quantitative Features Extracted from CT Images. Translational Oncology. 2014;7(1):72–87. doi: 10.1593/tlo.13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tixier F, Hatt M, Le Rest CC, Le Pogam A, Corcos L, Visvikis D. Reproducibility of tumor uptake heterogeneity characterization through textural feature analysis in 18F-FDG PET. J. Nucl. Med. 2012;53:693–700. doi: 10.2967/jnumed.111.099127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leijenaar RTH, Carvalho S, Velazquez ER, Van Elmpt WJC, Parmar C, Hoekstra OS, Hoekstra CJ, Boellaard R, Dekker ALAJ, Gillies RJ, Aerts HJWL, Lambin P. Stability of FDG-PET radiomics features: an integrated analysis of test-retest and inter-observer variability. Acta Oncol. (Madr). 2013;52:1391–1397. doi: 10.3109/0284186X.2013.812798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Velden FHP, Nissen IA, Jongsma F, Velasquez LM, Hayes W, Lammertsma AA, Hoekstra OS, Boellaard R. Test-retest variability of various quantitative measures to characterize tracer uptake and/or tracer uptake heterogeneity in metastasized liver for patients with colorectal carcinoma. Mol. Imaging Biol. 2014;16:13–18. doi: 10.1007/s11307-013-0660-9. [DOI] [PubMed] [Google Scholar]
- 20.Hunter Luke A., Krafft Shane, Stingo Francesco, Choi Haesun, Martel Mary K., Kry Stephen F., Court Laurence E. High quality machine-robust image features: Identification in nonsmall cell lung cancer computed tomography images. Medical Physics. 2013;40(12):121916. doi: 10.1118/1.4829514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Velden FHP, Kramer GM, Frings V, Nissen IA, Mulder ER, de Langen AJ, Hoekstra OS, Smit EF, Boellaard R. Repeatability of radiomic features in non-small-cell lung cancer [18F]FDG-PET/CT studies: impact of reconstruction and delineation. Mol. Imaging Biol. 2016;18:788–795. doi: 10.1007/s11307-016-0940-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao, B., Tan, Y., Tsai, W.Y., Qi, J., Xie, C., Lu, L., Schwartz, L.H.: Reproducibility of radiomics for deciphering tumor phenotype with imaging. Sci. Rep. 6, 1–7 (2016). [DOI] [PMC free article] [PubMed]
- 23.Antunes J, Viswanath S, Rusu M, Valls L, Hoimes C, Avril N, Madabhushi A. Radiomics analysis on FLT-PET/MRI for characterization of early treatment response in renal cell carcinoma: a proof-of-concept study. Transl. Oncol. 2016;9:155–162. doi: 10.1016/j.tranon.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galavis PE, Hollensen C, Jallow N, Paliwal B, Jeraj R. Variability of textural features in FDG PET images due to different acquisition modes and reconstruction parameters. Acta Oncol. (Madr). 2010;49:1012–1016. doi: 10.3109/0284186X.2010.498437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He L, Huang Y, Ma Z, Liang C, Liang C, Liu Z. Effects of contrast-enhancement, reconstruction slice thickness and convolution kernel on the diagnostic performance of radiomics signature in solitary pulmonary nodule. Sci. Rep. 2016;6:34921. doi: 10.1038/srep34921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mackin D, Fave X, Zhang L, Fried D, Yang J, Taylor B, Rodriguez-Rivera E, Dodge C, Jones AK, Court L. Measuring computed tomography scanner variability of radiomics features. Invest. Radiol. 2015;50:757–765. doi: 10.1097/RLI.0000000000000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Echegaray S, Nair V, Kadoch M, Leung A, Rubin D, Gevaert O, Napel S. A rapid segmentation-insensitive “digital biopsy” method for radiomic feature extraction: method and pilot study using CT images of non–small cell lung cancer. Tomography. 2016;2:283–294. doi: 10.18383/j.tom.2016.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalpathy-Cramer J, Zhao B, Goldgof D, Gu Y, Wang X, Yang H, Tan Y, Gillies R, Napel S. A comparison of lung nodule segmentation algorithms: methods and results from a multi-institutional study. J. Digit. Imaging. 2016;29:476–487. doi: 10.1007/s10278-016-9859-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holzapfel K, Duetsch S, Fauser C, Eiber M, Rummeny EJ, Gaa J. Value of diffusion-weighted MR imaging in the differentiation between benign and malignant cervical lymph nodes. Eur. J. Radiol. 2009;72:381–387. doi: 10.1016/j.ejrad.2008.09.034. [DOI] [PubMed] [Google Scholar]
- 30.Fruehwald-Pallamar J, Czerny C, Holzer-Fruehwald L, Nemec SF, Mueller-Mang C, Weber M, Mayerhoefer ME. Texture-based and diffusion-weighted discrimination of parotid gland lesions on MR images at 3.0 Tesla. NMR Biomed. 2013;26:1372–1379. doi: 10.1002/nbm.2962. [DOI] [PubMed] [Google Scholar]
- 31.Sun Ying-Shi, Zhang Xiao-Peng, Tang Lei, Ji Jia-Fu, Gu Jin, Cai Yong, Zhang Xiao-Yan. Locally Advanced Rectal Carcinoma Treated with Preoperative Chemotherapy and Radiation Therapy: Preliminary Analysis of Diffusion-weighted MR Imaging for Early Detection of Tumor Histopathologic Downstaging. Radiology. 2010;254(1):170–178. doi: 10.1148/radiol.2541082230. [DOI] [PubMed] [Google Scholar]
- 32.Vandecaveye V, De Keyzer F, Vander Poorten V, Dirix P, Verbeken E, Nuyts S, Hermans R. Head and neck squamous cell carcinoma: value of diffusion-weighted MR imaging for nodal staging. Radiology. 2009;251:134–146. doi: 10.1148/radiol.2511080128. [DOI] [PubMed] [Google Scholar]
- 33.Jafar MM, Parsai A, Miquel ME. Diffusion-weighted magnetic resonance imaging in cancer: reported apparent diffusion coefficients, in-vitro and in-vivo reproducibility. World J. Radiol. 2016;8:21–49. doi: 10.4329/wjr.v8.i1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Belli G, Busoni S, Ciccarone A, Coniglio A, Esposito M, Giannelli M, Mazzoni LN, Nocetti L, Sghedoni R, Tarducci R, Zatelli G, Anoja RA, Belmonte G, Bertolino N, Betti M, Biagini C, Ciarmatori A, Cretti F, Fabbri E, Fedeli L, Filice S, Fulcheri CPL, Gasperi C, Mangili PA, Mazzocchi S, Meliadò G, Morzenti S, Noferini L, Oberhofer N, Orsingher L, Paruccini N, Princigalli G, Quattrocchi M, Rinaldi A, Scelfo D, Freixas GV, Tenori L, Zucca I, Luchinat C, Gori C, Gobbi G. Quality assurance multicenter comparison of different MR scanners for quantitative diffusion-weighted imaging. J. Magn. Reson. Imaging. 2016;43:213–219. doi: 10.1002/jmri.24956. [DOI] [PubMed] [Google Scholar]
- 35.Ye XH, Gao JY, Yang ZH, Liu Y. Apparent diffusion coefficient reproducibility of the pancreas measured at different MR scanners using diffusion-weighted imaging. J. Magn. Reson. Imaging. 2014;40:1375–1381. doi: 10.1002/jmri.24492. [DOI] [PubMed] [Google Scholar]
- 36.Padhani AR, Liu G, Mu-Koh D, Chenevert TL, Thoeny HC, Takahara T, Dzik-Jurasz A, Ross BD, Van Cauteren M, Collins D, Hammoud DA, Rustin GJS, Taouli B, Choyke PL. Diffusion-weighted magnetic resonance imaging as a cancer biomarker: consensus and recommendations. Neoplasia. 2009;11:102–125. doi: 10.1593/neo.81328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin JC, Pujol S, Bauer C, Jennings D, Fennessy F, Sonka M, Buatti J, Aylward S, Miller JV, Pieper S, Kikinis R. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn. Reson. Imaging. 2012;30:1323–1341. doi: 10.1016/j.mri.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haralick RM. Statistical and structural approaches to texture. Proc. IEEE. 1979;67:786–804. doi: 10.1109/PROC.1979.11328. [DOI] [Google Scholar]
- 39.Tang X. Texture information in run-length matrices. IEEE Trans. Image Process. 1998;7:1602–1609. doi: 10.1109/83.725367. [DOI] [PubMed] [Google Scholar]
- 40.Yoo TS. Insight into images: principles and practice for segmentation, registration, and image analysis. Natick, MA: AK Peters; 2004. [Google Scholar]
- 41.Shrout PE, Fleiss JL. Intraclass correlations: uses in assessing rater reliability. Psychol. Bull. 1979;86:420–428. doi: 10.1037/0033-2909.86.2.420. [DOI] [PubMed] [Google Scholar]
- 42.Mcgraw KO. Forming inferences about some intraclass correlation coefficients. Psychol. Methods. 1996;1:30–46. doi: 10.1037/1082-989X.1.1.30. [DOI] [Google Scholar]
- 43.Jaccard Paul. THE DISTRIBUTION OF THE FLORA IN THE ALPINE ZONE.1. New Phytologist. 1912;11(2):37–50. doi: 10.1111/j.1469-8137.1912.tb05611.x. [DOI] [Google Scholar]
- 44.Leijenaar RTH, Nalbantov G, Carvalho S, Van Elmpt WJC, Troost EGC, Boellaard R, Aerts HJWL, Gillies RJ, Lambin P. The effect of SUV discretization in quantitative FDG-PET radiomics: the need for standardized methodology in tumor texture analysis. Sci. Rep. 2015;5:1–10. doi: 10.1038/srep11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 166 kb)