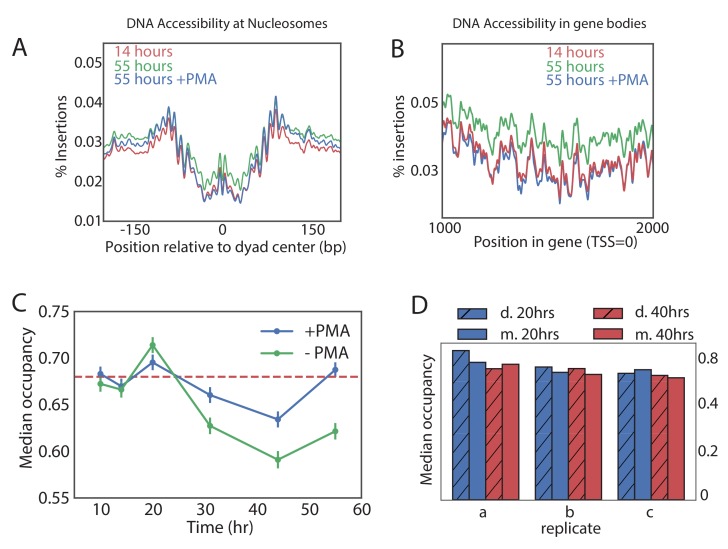
Figure 4. Global nucleosome occupancy remains nearly constant with aging.
(A,B) Tn5 insertion density around well-positioned nucleosomes (A) and 1000 bases downstream of TSS (B) at 14 hr, 55 hr and 55 hr after treatment with PMA. (C) Median nucleosomal occupancy estimated by NucleoATAC on well-positioned nucleosomes in open chromatin at different time points during time course. See Materials and methods for the method of estimation. Error bars represent five standard errors of occupancy. (D) Same as (C) but comparing mothers and daughters during aging time courses - three independent replicates are shown.
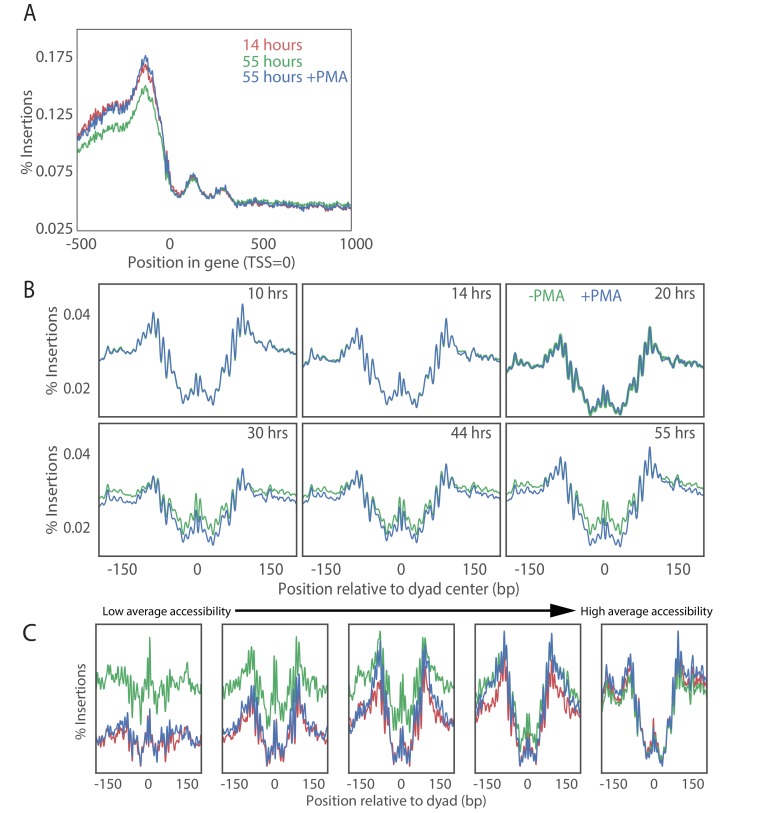
Figure 4—figure supplement 1. PMA treatment rescues young cell ATAC-seq profile at transcriptional start sites in old cell populations.
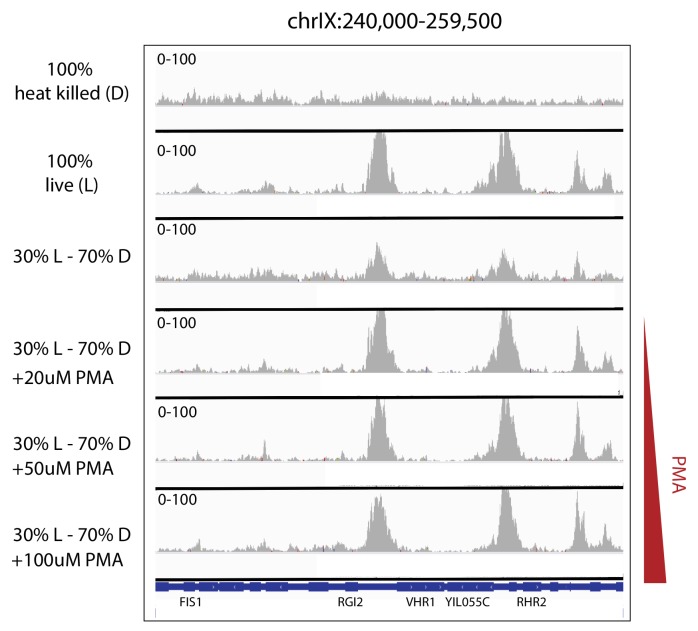
Figure 4—figure supplement 2. Efficacy of PMA in removing the ATAC-seq signature of heat-killed cells from mixed populations.
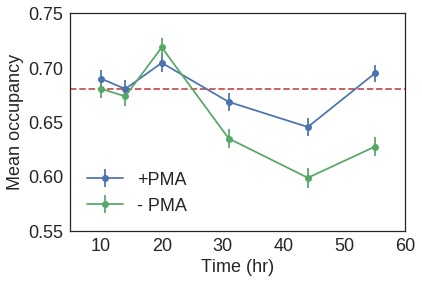
Figure 4—figure supplement 3. Mean nucleosomal occupancy estimated by NucleoATAC on well-positioned nucleosomes in open chromatin.