Abstract
Neuraminidase (NA) is a glycoside hydrolase that has been proposed as a potential therapeutic target for influenza. Thus, the identification of compounds that modulate NA activity could be of great therapeutic importance. The aim of this study is to develop a drug discovery tool for the identification of novel modulators of NA from both compound libraries and natural plant extracts. NA was immobilized onto the surface of magnetic beads and the inherent catalytic activity of NA-functionalized magnetic beads was characterized. Based on the enzymatic activity (hydrolysis ratio), the inhibitory activities of 12 compounds from plant secondary metabolites were screened, and the desired anti-NA activities of flavonoids were certified. Ligand fishing with the immobilized enzyme was optimized using an artificial test mixture consisting of oseltamivir, lycorine and matrine prior to carrying out the proof-of-concept experiment with the crude extract of Flos Lonicerae. The combination of ligand fishing and HPLC-MS/MS identified luteolin-7-O-β-D-glucoside, luteolin, 3,5-di-O-caffeoylquinic acid and 3,4-di-O-caffeoylquinic acid as neuraminidase inhibitory ligands in Flos Lonicerae. This is the first report on the use of neuraminidase functionalized magnetic beads for the identification of active ligands from a botanical matrix, and it sets the basis for the de novo identification of NA modulators from complex biological mixtures.
Keywords: Enzyme immobilization, Neuraminidase, Magnetic beads, Screening inhibitors
1. Introduction
Influenza viruses cause annual epidemics and occasional pandemics that have claimed the lives of millions. A limiting issue is the extreme plasticity of viral strains which can lead to the emergence of new strains [1]. Although vaccines have been an effective prevention, their production via current methods cannot keep up with the constant introduction of new strains [2]. As a result, developing novel antiviral agents for influenza is highly important.
Influenza neuraminidase (NA), a glycoside hydrolase, can reduce the number of receptor binding sites for hemagglutinin (HA) on host cells as well as on progeny viruses. This allows the mature virus to detach from host cells during release and prevents self-aggregation mediated by HA. NA can also cleave the glycan structure of mucus and allow viruses to reach host cells. As one of the approved primary therapeutic targets for influenza, NA plays an increasing critical role for identifying novel anti-flu agents [2,3]. Neuraminidase inhibitors (NAIs) are presently the only effective antiviral drugs for treatment and chemoprophylaxis of influenza A and B infections, because of their high homology across all nine NA subtypes from influenza A and influenza B [4]. Two current anti-influenza agents being used to prophylaxis and treatment of influenza infections, i.e. oseltamivir and zanamivir, are neuraminidase inhibitors (NAIs). However, the low bioavailability of the zanamivir and the appearance resistance strains of oseltamivir revealed that novel NAIs with higher effectiveness and better tolerability are urgently needed [5–7].
Current methods for discovering novel NAIs include NA-inhibitory assays, cell culture-based assays, and structure-based design [8]. The cell culture-based assays can provide evidence for NAIs activity against infectious virus. However, the interpretation of results is complicated by several factors unique to NAIs [9], and successful applications were mostly dependent on the test virus and cell line used [10,11]. Computational docking represents a highly useful tool to find new lead compounds from available structure collections. However, subsequent biological assays are indispensable for evaluating the active molecules identified or designed by computational approaches, which increases the workload [12].
Of these methods, the NA-inhibitory assays have become the method of choice due to its reliability and standardization [8]. However conventional inhibitory assays require large quantities of enzyme, are labor intensive and time consuming. Most of all, they can be only applied for screening library of pure compounds. More recently, several groups developed a novel method by which enzyme was immobilized onto a solid support and then incubated with natural products, allowing direct isolation of active compounds from a complex matrix [13–21]. To date, the most widely used solid support for ligand fishing experiments has been para-magnetic beads (hereinafter referred to as magnetic beads (MBs)). In a pioneering work, human serum albumin immobilized magnetic beads were employed for isolating active compounds from a mixture of binders and non-binders [19]. It was quickly expanded to the screening of plant extracts, such as AChE inhibitors isolated from meldoinus fusiformis [20]; a number of novel SIRT6 inhibitors isolated from fenugreek seed extract [21]; α-Glucosidase inhibitors discovered from Eugenia catharinae [22] and three ligands isolated from oolong tea for pancreatic lipase [23]. These examples illustrate the potential application of magnetic beads-based NA-inhibitory assays for identifying novel NAIs from plant extracts.
Herein, NA was immobilized onto the surface of MBs, and the resulting NA-MBs were then validated with a test-mixture and subsequently applied for screening active NA modulators from pure compound libraries and natural products.
2. Experimental
2.1. Chemicals and materials
Neuraminidase (25 UN in package, from Clostridium perfringens, Type V, lyophilized powder), and the substrate 2′-(4-methylumbelliferyl)-α-D-N-acetylneuraminic acid sodium salt hydrate (MUNANA) were purchased from Sigma–Aldrich (Steinheim, Germany). The product 4-methylumbelliferone (4-MU), coumarin, ammonium acetate, dimethyl sulfoxide, pyri-dine (99.8%), glutaraldehyde (50%), formic acid, galantamine, quercetin, kaempferol, kaempferide, isorhamnetin, myricitrin, hyperoside, matrine, lycorine, oxymatrine, ammothamnine, cyto-sine, thymine, guanosine, luteolin-7-O-β-D-glucoside, luteolin, 3,5-di-O-caffeoylquinic acid and 3,4-di-O-caffeoylquinic acid were purchased from Aladdin Chemistry Co. (Shanghai, China). Oseltamivir was obtained from a hospital with provided relevant certificates. HPLC grade methanol and acetonitrile were obtained from Merck (Darmstadt, Germany). Flos Lonicerae was purchased from a local drug store in China and authenticated by associate professor Tingting Zhang. One micrometer size Bc-Mag™, amine terminated magnetic beads (30 mg/mL) were purchased from Bio-clone Inc. (San Diego, CA). Deionized water was purified using a Milli-Q System (Millipore, Bedford, MA, USA).
2.2. Instrumentation
The analytes were separated using a Dionex UltiMate 3000 HPLC system, which consisted of an UltiMate 3000 RS pump, an UltiMate 3000 RS autosampler and an UltiMate 3000 RS column compartment (Dionex, Germering/Germany). The analytes were detected using an Applied Biosystems 3500 Qtrap linear ion trap quadrupole mass spectrometer with Analyst® software (Version1.6.3) equipped with a Turbo V ion source operated in ESI mode (AB SCIEX, Singapore). The confocal laser scanning microscope was operated by a Leica TCS SP8 system.
2.3. Preparation of NA -functionalized magnetic beads
NA was immobilized through the N-terminus onto the surface of modified MBs according to a previously published protocol with slight modification [20,21]. Briefly, 10 mg amine-terminated MBs were transferred into a 2 mL tube and magnetically separated from the supernatant using a magnetic separator. Then MBs were washed three times with 1 mL of coupling buffer (10 mM aqueous pyridine, pH 6.0). The MBs were then re-suspended with 1 mL of coupling buffer containing 5% glutaraldehyde and rotated in an orbital rotator for 3 h at 4°. After washing three times with 1 mL of coupling buffer, 10 mg of MBs were incubated with 125 μg of NA in 500 μL coupling buffer overnight at 4 °C with gentle rotation. The supernatant was then removed and the amount of free neuraminidase remaining in solution was determined using the Bradford assay. The neuraminidase functionalized magnetic beads (NA-MBs) were washed three times with 500 μL of Tris HCl (100 mM, pH 7.0) for end-capping. The resulting NA-MBs were finally washed with coupling buffer and stored in reaction buffer (ammonium acetate [15 mM, pH 5.0]) at 4°C before use.
The control magnetic beads (Control-MBs) were made in the same manner but without the addition of NA.
2.4. Evaluation and characterization of NA-MBs
2.4.1. LC–MS/MS conditions
An LC–MS/MS activity method based on the quantitative determination of the reaction product, 4-MU, was employed. The chromatographic separation of 4-MU, MUNANA and the internal standard was achieved by gradient elution on a Phenomenex Gemini C18 column (4.6 mm × 100 mm, 3 μm). The mobile phase consisted of solvent A (H2O with 0.1% formic acid) and solvent B (acetonitrile with 0.1% formic acid) with a gradient program as follows: 0 min, 30% B; 3 min, 30% B; 8 min, 90% B. Each run was followed by a 1 min postrun with 30% B. Flow rate was set at0.6 mL/min while the column oven temperature was controlled at 25°C. The mass spectrometer was operated under positive ionization mode using multiple reaction monitoring (MRM). The compound dependent parameters were summarized in Table 1. The optimized ion source dependent parameters were: ion spray voltage of 5500 V, temperature of 500°C, curtain gas and collision gas at 30 psig and 6 psig, ion source gas 1 at 45 psig and ion source gas 2 of 50 psig. Analyst® 1.6.3 software was used for all data analysis.
Table 1.
Lists of the retention time and the dependent parameters in HPLC-MS/MS analysis for each compound.a.
| Analytes | TR (min) |
Precursor Ion (m/z) |
Product Ion (m/z) |
CE (volts) |
DP (volts) |
EP (volts) |
|---|---|---|---|---|---|---|
| MUNANA | 3.99 | 468.1 | 292.1 | 12.58 | 92.97 | 10 |
| 4-MU | 5.91 | 177.1 | 103.0 | 35.95 | 110 | 10 |
| Coumarin (IS) | 6.96 | 147.1 | 103.0 | 21.45 | 91.75 | 10 |
a CE is collision energy, DP is declustering potential, and EP is entrance potential.
The optimized LC–MS/MS activity method was validated using parameters such as linearity, limit of detection (LOD), limit of quantification (LOQ). The calibration curve for 4-MU (10 concentrations in the range from 0.1 to 100 μM) was prepared in triplicate and 10 μM coumarin was used as the internal standard. All prepared samples (1 mL final volume) contain 3% DMSO (v/v). Standard sample solutions were prepared by serial dilution with the same solvent in the ratio of 1:2. Limit of detection (LOD) and limit of quantification (LOQ) were visually evaluated, defining concentrations with a peak height of 3- or 10-times the baseline noise, respectively.
2.4.2. Kinetic and inhibition studies of NA-MBs
MUNANA (8 concentrations in the range from 1 to 500 μM) was incubated with 5 mg of NA-MBs (12.5 μg NA/mg) at 37 °C for 30 s before separation, collection. 10 μM of coumarin was added into 500 μL of supernatant and analysis by LC–MS/MS. Samples were prepared in triplicate and analyzed two times each. The data were evaluated using the double reciprocal plotting method and fitted to the Michaelis − Menten equation using Origin Pro 8.0 software (OriginLab, Northampton, MA, USA).
40 μM MUNANA was incubated together with a series of concentrations of oseltamivir (0.01–500 μM) and 5 mg NA-MBs, at 37 °C for 3 min. After magnetic separation, 10 μM of coumarin was added into 500 μL of supernatant, then injected into LC–MS/MS. Samples were prepared in triplicate and analyzed two times each. The data were calculated by GraphPad Prism 7.0 software (San Diego, CA).
2.5. Screening of a compound library
2.5.1. Establishment and validation of the screening assay
Galantamine (negative control) and oseltamivir (positive control) were used to establish and validate the screening assay. Three parallel assays, i.e. a control assay with control-MBs (presence of MUNANA and compound); an inhibition assay with NA-MBs (presence of MUNANA and compound); and a blank assay with NA-MBs (presence of MUNANA), were carried out for the evaluation of each compound. 1 mL reaction buffer consisting of 3% (v/v) DMSO, 40 μM MUNANA and 50 μM tested compound was incubated with 5 mg NA-MBs (12.5 μg of NA/mg) at 37 °C for 3 min. The blank sample was prepared in the same way but without adding the test compound in 1 mL reaction buffer. After incubation and magnetic separation, 500 μL of supernatants were collected and transferred to an autosampler vial containing 10 μM internal standard (IS) for LC–MS/MS analysis. The inhibitory percentage (%) for each analyte was calculated using equation as follows:
Where [S4−MU]C, [S4−MU]I and [S4−MU]B are concentrations of product obtained from control assay, inhibition assay and blank assay, respectively. All of them were converted from correct peak area of 4-MU via calibration curves.
2.5.2. Screening assay
12 compounds (kaempferol, kaempferide, isorhamnetin, quercetin, myricitrin, hyperoside, lycorine, oxymatrine, ammothamnine, cytosine, thymine and guanosine) were prepared as 10 mM primary stock solutions in DMSO. All compounds were diluted to a final concentration of 50 μM using reaction buffer (containing 3% (v/v) DMSO). The blank sample was prepared in the same way in 3% (v/v) DMSO without adding inhibitors. The mixture of 12 compounds was incubated with NA-MBs (5 mg) at 37 °C and 40 μM MUNANA for 3 min. After magnetic separation, 500 μL of supernatant was collected and 5 μL of 1 mM coumarin (IS) (10 μM) was added. The NA-MBs were regenerated by washing three times with 1 mL of reaction buffer containing 40 μM of substrate. The inhibitory percentage (%) for each compound was calculated via equation described in 2.5.1.
2.6. Ligand fishing assay from natural products
2.6.1. Establishment and validation of the ligand fishing assay
A model mixture comprising of oseltamivir, matrine and lycorine (0.1 mM each) was prepared in the reaction buffer (denoted as S0) and subjected to ligand fishing as follow. To 10 mg of NA-MBs (12.5 μg of NA/mg) were suspended in 1 mL of S0. After sharking and incubating at 37 °C for 1 h, the supernatant (named S1) was collected after magnetic separation. The NA-MBs were washed two times with 1 mL of reaction buffer (S2-S3) and one time with 1 mL solution of 1:1 reaction buffer/methanol (1/1, v/v) (S4). The bound material was then eluted from the NA-MBs with 1 mL of 100% methanol twice to release the active components. The eluent was carefully removed and saved as S5-S6. The solutions of mixture (S0) along with S1- S6 were analyzed by LC–MS/MS using an Agilent ZORBAX SB-C18 column (4.6 mm × 150 mm, 5 μm). The mobile phase consisted of solvent A (H2O with 0.1% formic acid) and solvent B (acetonitrile) with a gradient program as follows: 0 min, 10% B; 25 min, 90% B. Each run was followed by a 1 min postrun with 10% B. Flow rate was set at 0.5 mL/min while the column oven temperature was controlled at 25 °C.
2.6.2. Fishing potential ligands from a Flos Lonicerae extract
2.6.2.1. Preparation of Flos Lonicerae extract.
The dry flowers of Flos Lonicerae (5.0 g) were pulverized and refluxed three times with 70% ethanol for 1.5 h at 70 °C. The filtrate was filtered, collected and concentrated in a rotatory evaporation (EYELA, Janpan). The crude extracts (561.2 mg) were fractionated through solvent-solvent extraction to acquire petroleum ether extract fraction, ethyl acetate extract fraction and n-butanol extract fraction successively.2.0 mg of each extract fractions were dissolved in 1.0 mL of reaction buffer containing 3% DMSO (v/v) and then incubated with NA-MBs (5 mg) for 3 min at 37 °C. The inhibition ratio of each fraction was measured via the formula mentioned in 2.5.1.
2.6.2.2. Procedure for fishing ligands of NA.
5.0 mg/mL S0 solution of the ethyl acetate extract fraction from Flos Lonicerae was prepared in reaction buffer containing 3% DMSO (v/v). 10 mg of NA-MBs(12.5 μg of NA/mg) was suspended in 1 mL of this solution (S0) and incubated at 37 °C for 1 h. The ligand fishing assay was carried out as optimized (2.6.1) and S0–6 were collected. All of the samples (S0-S6) were then analyzed with LC–MS/MS, following a previously reported method [24].
3. Results and discussion
3.1. Preparation of NA -MBs
NA, a therapeutic target for the treatment of influenza, was covalently immobilized onto the surface of magnetic beads via Schiff base formation. Glutaraldehyde was used as the cross-linker to link the amino groups on the beads and the N-terminal of the enzyme, similar to previously reported methods (Fig. S1) [19–21].
It has been reported that increasing amount of immobilized enzyme does not necessarily mean increased activity. Higher surface density may reduce the access of substrate to the immobilized enzyme. For example, C. Megías et al. demonstrated that increasing the amount of enzyme resulted in decreasing in activity [26]. Therefore, the optimal loading capacity of MBs was initially investigated by using fixed amount of NA (125 μg) and various amount of MBs (5, 10, 20 and 40 mg). The amount of immobilized NA was determined, by quantitating the amount of free NA remaining in supernatant using a BCA protein assay. As shown in Fig. S2, the highest yield was obtained with 10 mg MBs. In addition, the hydrolysis ratio of the substrate was 98.69% for the optimal NA-MBs, indicating that the protein was immobilized in its proper conformation and that an ideal loading capacity was obtained.
3.2. Validation and Characterization of NA-MBs
3.2.1. LC–MS/MS conditions
Currently, several fluorescent based assays are available to asses NA activity [25,28]. While fluorescent tags are very sensitive, they are limited due to being able to be only used for single compound analysis, as a result they are problematic for studying complex matrixes. Therefore, a LC–MS/MS based assay was developed for quantitatively determining the product 4-MU, as changes in the amount of 4-MU produced represents the inhibition of NA activity.
The chromatographic separation of 4-MU was optimized to resolve 4-MU from the remainder of the reaction components. Optimal conditions were found to be Gemini C18 column with small particle size (4.6 mm × 100 mm, 3 μm), using 0.1% formic acid-water/acetonitrile as the mobile phase, which favors the neutral state of the ionizable groups and improved their retention time. Reproducible separation was achieved, with MUNANA, 4-MU, and coumarin (IS) at 3.9, 5.9, and 6.9 min, respectively, in a 9 min run using a gradient mode (Fig. S3).
The MS signal was linear (y = 2.98907x + 0.00717; R2 = 0.998) over a range of concentrations (0.5–50 μM) sufficient to cover 4-MU formed in the wide variety of reactions performed (Fig. S4). LOQ and LOD for 4-MU are 100 nM (S/N = 10) and 20 nM (S/N = 3), respectively.
3.2.2. Validation of NA-MBs
Based on the LC–MS/MS activity method, the basic kinetic parameter Michaelis-Menten constant (Km) and inhibition parameter half maximal inhibitory concentration (IC50) of NA-MBs were tested to ensure that the immobilized enzyme retain its molecular integrity and inherent hydrolytic activity after immobilization.
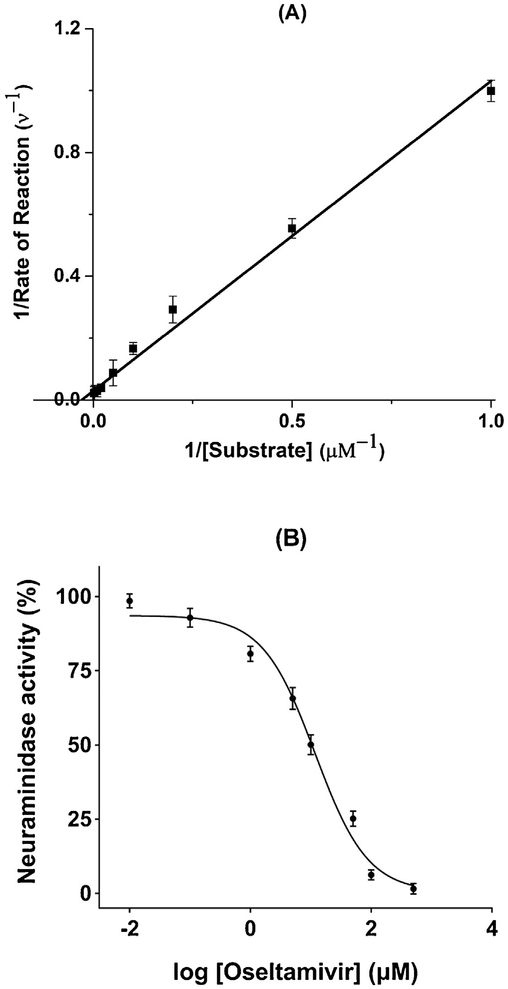
MUNANA, a commonly used substrate, was employed for determining Km. In order to limit the conversion rate to be less than 10% [28], the incubation time at this step was set at 30 s. The measured plots are shown in Fig. 1A. Based on the double reciprocal plot, Km on the NA-MBs was calculated as 46.49 ± 3.68 μM, which is lower than that measured on the NA-immobilized capillary microreactor (Km = 136.6 ± 10.8 μM) [28]. This observed increase in affinity of NA-MBs might be attributed to the size of MBs. Reducing the size of the material has been demonstrated to enhance the activity of immobilized enzyme due to the changes in mobility, curvature-induced associations and spatial confinement [29].
Fig. 1.
Lineweaver − Burk reciprocal plot (A) and Dose-response curve plot of inhibition percentage for oseltamivir (B).
Oseltamivir, a well characterized NA inhibitor, was selected to validate the inhibitory activity of the NA-MBs (Fig. 1B). The IC50 value of oseltamivir was measured as 11.82 ± 2.28 μM, which is comparable to literature reported values of 2.29 μM and 1.06 [H9262]M [30,31].
These results demonstrate that the molecular integrity and inherent enzymatic activity of NA is still maintained after immobilization.
3.2.3. Characterization of NA-MBs
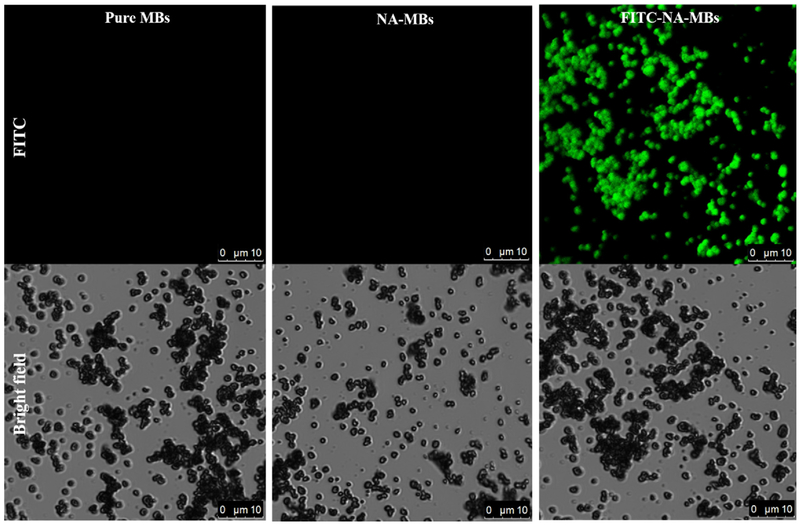
FITC (fluorescein isothiocyanate) labeled NA was employed to characterize the synthesis and morphology of NA-MBs. A FITC labelled NA was prepared following a previously described protocol with slight modifications [27]. A comparison between control MBs, NA-MBs and FITC-labeled NA-MBs was carried out. As shown in Fig. 2, fluorescence can only be observed for the FITC-NA-MBs, confirming the immobilization of NA. Further, the data also demonstrates the uniformity of the NA-functionalized MBs.
Fig. 2.
Images captured by confocal laser scanning microscope for monitoring the pure MBs, NA modified MBs and FITC-NA-MBs in the FITC fluorescence channel and bright field.
3.3. Screening NAIs from a compound library
3.3.1. Assay development and validation
Under the optimized LC–MS/MS conditions, the NA-MBs were used to investigate the inhibition potential of a series of compounds. The incubation time and reaction buffer system were first optimized. NA-MBs were incubated with MUNANA (40 μM) for various time (1–15 min). Fig. S5 shows that the amount of 4-MU significantly increases with increasing the incubation time from 1 to 3 min. Further increasing the incubation time to 15 min, the trend is slowed down gradually. Considering the detection sensitivity and analysis time, the curves exhibited linear in the first 3 min indicating an initial reaction stage, set as the following incubation time. For the buffer system, ammonium acetate [15 mM, pH 5.0] was selected based on the former research, while in order to improve the solubility of the tested compounds, less than 3% DMSO was added, which has no interference on neuraminidase activity (Fig. S6). Depending on these optimal conditions, three parallel assays, i.e. a control assay with control-MBs; an inhibition assay with NA-MBs; and a blank assay with NA-MBs, were carried out for the systematic evaluation inhibitory percentage (I%) of each compound. Further, oseltamivir and galantamine were used as the positive control and negative control to validate the screening assay, the results was shown in Table 2.
Table 2.
NA inhibitory percentage (I%) for 12 plant secondary metabolites, also the galantamine (negative control) and oseltamivir (positive control).
| Compound | Inhibitory ratio (%) | |
|---|---|---|
| Flavonoids | Isorhamnetin | 46.8 ± 5.8 |
| Myricitrin | 29.7 ± 4.1 | |
| Hyperoside | 32.3 ± 2.7 | |
| Kaempferide | 43.6 ± 4.9 | |
| Kaempferol | 51.2 ± 6.1 | |
| Quercetin | 48.1 ± 3.4 | |
| Nucleosides | Guanosine | < 1% |
| Cytosine | < 1% | |
| Thymine | < 1% | |
| Ammothamnine | < 1% | |
| Alkaloids | Lycorine | < 1% |
| Oxymatrine | < 1% | |
| Negative control | Galantamine | < 1% |
| Positive control | Oseltamivir | 74.9 ± 3.5 |
For the compound library screening, the NA-MBs could be reused. The reusability of the NA-MBs for screening NAIs was assessed using oseltamivir as a test analyte. The RSD for the inhibition efficiency of oseltamivir was 2.8% for 10 consecutive runs (Fig. S7), which demonstrate good reusability of the NA-MBs.
3.3.2. Screening from a compound library
In order to evaluate the applicability of the NA-MBs for screening enzyme inhibitors from a compound library, 12 plant secondary metabolites, including six flavonoids, two alkaloids and four nucleosides, were selected for the test. The inhibitory percentage (I%) of total 12 compounds were measured and listed in Table 2. The results show that all flavonoids have moderate inhibitory activity against NA, with kaempferol being the most potent. The observed potent inhibitory activity of flavonoids against NA is not unexpected as it has been previously reported that certain flavonoids apigenin, baicalein, gossypetin and herbacetin are NA inhibitors [32,33]. It is worth noting that the flavones exhibit much higher activity in comparison with their corresponding glycosides, such as quercetin (I% = 48.1 ± 3.4) and hyperoside (I% = 32.3 ± 2.7). This observation is consistent with previous reports, where the 4′ OH, 7−OH, C4 = O and C2 = C3 groups (the structure shown in Fig. S8) were considered the essential for NA inhibition activity [34]. The glycosyl group on the 4′ −OH, 7 OH will reduce the inhibition significantly [34]. No inhibitory activities against NA were observed for two alkaloids and four nucleosides, which is not surprising as only palmatine and berberine were reported to show inhibition of bacterial NA [35]. These results demonstrate that the established NA-MBs based assay could distinguish and identify NA inhibitors from pure compound library.
3.4. Ligand fishing assay for natural products
3.4.1. Assay development and validation
According to the bioaffinity chromatography theory, the protein-functionalized magnetic beads are immersed directly into the extract and any compounds with an affinity for the immobilized protein are retained while non-binders remain in the supernatant [13,19–22]. The combination of this technique with chromatographic and MS/MS techniques allows for the detection and identification of new potential lead compounds is known as ligand fishing.
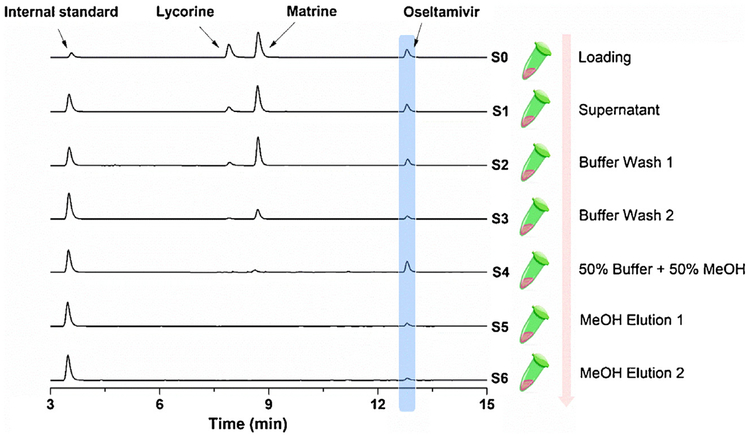
In order to evaluate the applicability of the NA-MBs for fishing NA inhibitors from natural products, the incubation time, washing steps and elution steps were first optimized using known active/inactive compounds for NA, i.e. oseltamivir (known binder), matrine and lycorine (reported non-binders) [28]. The mixture of three compounds was incubated with the NA-MBs for various time (data not shown). The washing and eluting solutions at each steps were collected and analyzed by LC–MS/MS in order to achieve direct isolation of binders from a complex matrix. Finally, a condition, i.e. incubated for 1 h, then washing two times with reaction buffer (ammonium acetate [15 mM, pH 5.0]); one time with reaction buffer containing 50% methanol (MeOH); final eluted two times with MeOH was selected. As can be seen in Fig. 3, under the optimized conditions, the NA binder (oseltamivir) retained until the MeOH elution step, while the non-binders (lycorine, matrine) were eluted in the washing steps.
Fig. 3.
LC–MS/MS chromatograms of the different solutions (S0-S6) from a model mixture via developed NA-MBs based ligand fishing approach. Experimental conditions: column: Agilent ZORBAX SB-C18 column (4.6 mm × 150 mm, 5 μm); mobile phase: solvent A (H2O with 0.1% formic acid) and solvent B (acetonitrile), gradient, 0 min, 10% B; 25 min, 90% B, each run was followed by a 1 min postrun with 10% B; column temperature: 25 °C; injection volume: 5 μL; flowrate: 0.5 mL/min; detection: TIC in ESI+ mode; compounds: mixture of the 3 standards.
3.4.2. Ligand fishing from Flos Lonicerae
The optimized conditions for the ligand fishing assay were then applied to Flos Lonicerae, a commonly used herb medicine in traditional Chinese medicine (TCM) for the treatment of exopathogenic wind-heat or the early stages of epidemic febrile diseases [34,36].
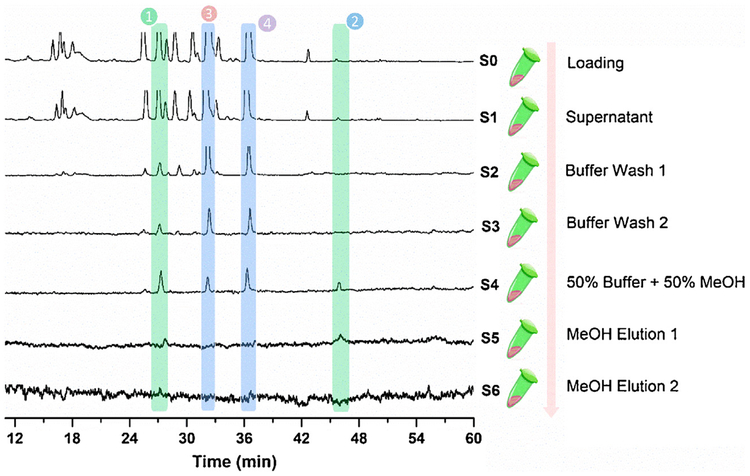
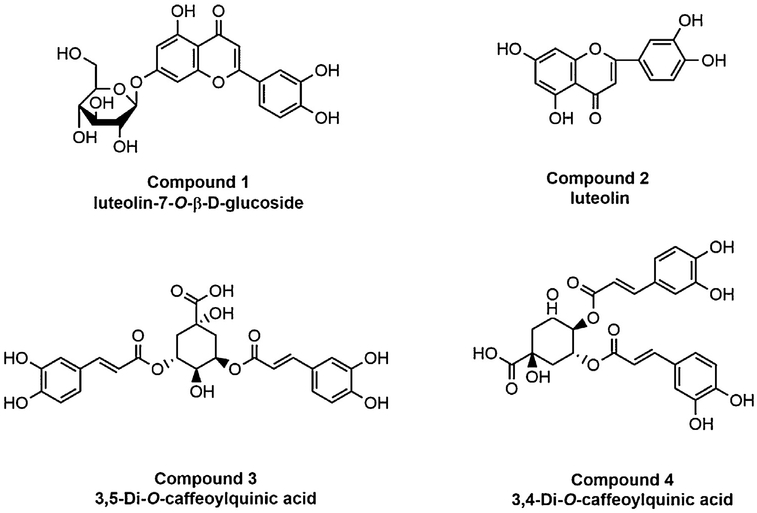
Before the ligand fishing experiment, the NA inhibition activity of different polar fractions from Flos Lonicerae were tested. The ethyl acetate fraction showed the highest inhibitory activity (data not shown). As a result, the ethyl acetate extract of Flos Lonicerae was chosen for further fishing experiments to identify potentially active NAIs using the NA-MBs. The ligand fishing results are presented in Fig. 4, where four compounds were retained until the final elution steps. Based on the HPLC-MS/MS data and literature [24,37], these four compounds were tentatively identified as luteolin-7-O-β-D-glucoside (compound 1), luteolin (compound 2), 3,5-di-O-caffeoylquinic acid (compound 3), and 3,4-di-O-caffeoylquinic acid (compound 4), shown in Fig. 5. Their structure elucidation and inhibition activity were further verified using commercially available reference standards. The luteolin (compound 2), appears to be a low abundant compound in the matrix, which demonstrates one of the greatest advantages of using magnetic particles is the possibility of identification of low-affinity ligands, which have been usually overlooked, when using other screening techniques, as well as low abundant secondary metabolites which have high affinity for the targeted protein [13]. As binding to the bound protein does not necessarily translate into activity, each compound was individually tested for activity as a NAI.
Fig. 4.
LC–MS/MS chromatograms of the different solutions (S0-S6) from the ethyl acetate extract of Flos Lonicerae via the NA-MBs based ligand fishing approach. Experimental conditions: column: Agilent ZORBAX SB-C18 column (4.6 mm × 150 mm, 5 μm); mobile phase: solvent A (H2O with 0.1% formic acid) and solvent B (acetonitrile), gradient, 0 min, 10% B, 20 min, 17% B; 32 min, 21% B; 40 min, 26% B; 49 min, 36% B; 54 min, 36% B; 59 min, 37% B; 65 min, 58% B; 70 min, 61% B; 75 min, 65% B, each run was followed by a 1 min postrun with 10% B; column temperature: 25 °C; injection volume: 5 μL; flowrate: 1 mL/min; detection: TIC in ESI+ mode.
Fig. 5.
Structures of Compounds 1–4 identified in Flos Lonicerae.
The IC50 values of four compounds were determined as 76.5 ± 3.9 μM (luteolin-7-O-β-D-glucoside), 53.2 ± 5.2μm (luteolin), 61.2 ± 1.5 μM (3,5-di-O-caffeoylquinic acid) and 68.3 ± 3.1 μM (3,4-di-O-caffeoylquinic acid), respectively. Luteolin-7-O-β-D-glucoside and luteolin represent an important chemical scaffold phenyl-benzo-pyranes, which can be used for designing neuraminidase inhibitors [38]. 3,5-Di-O-caffeoylquinic acid and 3,4-di-O-caffeoylquinic acid are reported main bioavailability components in Flos Lonicerae. This class of chlorogenic acid derivatives has been previously reported to be active against the influenza enzyme [26,39]. These results demonstrated that the NA-MBs based ligand fishing assay can be profitably exploited to identify novel NA ligands from complex natural extracts.
4. Conclusions
Herein, we report for the first time the synthesis and characterization of NA functionalized magnetic beads and their application as a drug discovery tool for screening NAIs from both pure compound libraries and plant extracts. The NA-MBs based screening assay was developed and applied for compound library screening. These results not only demonstrated high enzymatic activity and satisfactory stability of this tool, but also verified the desired anti-NA activity of flavonoids. Compared to conventional assays, the method was effective and maintained enzymatic activity demonstrating the reusability of NA-MBs. A ligand fishing assay was optimized and it was clearly demonstrated that the NA-MBs were able to distinguish between known NA ligands and non-ligands. The method was then applied to ‘fishing’ NAIs from Flos Lonicerae. Luteolin-7-O-β-D-glucoside, luteolin, 3,5-di-O-caffeoylquinic acid, and 3,4-di-O-caffeoylquinic acid were isolated and demonstrated to be active (IC50 = 76.5 ± 3.9 μM, 53.2 ± 5.2 μM, 61.2 ± 1.5 μM and 68.3 ± 3.1 μM). To the best of our knowledge, this is the first time that NA inhibitors were isolated and identified from natural complex extracts using NA-MBs. This clearly demonstrates that advantages of the ligand fishing method over conventional methods, as it offers in a significant promise and more efficient approach to drug discovery for profiling complex matrices.
Supplementary Material
Acknowledgements
We gratefully appreciate the financial support by the National Natural Science Foundation of China (81673391), the 111 Project (No. B13038), the Science and Technology Planning Project of Guangdong Province, China (2016A040403056) and the International Science and Technology Cooperation Program of Guangzhou, China (201807010022). This work was also supported in part by the Intramural Research Program of the National Institute on Aging, NIH.
Footnotes
Appendix A. Supplementary data
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.chroma.2018.07.031.
References
- [1].Neumann G, Noda T, Kawaoka Y, Emergence and pandemic potential of swine-origin H1N1 influenza virus, Nature 459 (2009) 931–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Moscona A, Neuraminidase inhibitors for influenza, N. Engl. J. Med 353 (2005) 1363–1373. [DOI] [PubMed] [Google Scholar]
- [3].Russell RJ, Haire LF, Stevens DJ, Collins PJ, Lin YP, Blackburn GM, Hay AJ, Gamblin SJ, Skehel JJ, The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design, Nature 443 (2006) 45–49. [DOI] [PubMed] [Google Scholar]
- [4].von Itzstein M, Wu WY, Kok GB, Pegg MS, Dyason JC, Jin B, Phan TV, Smythe ML, White HF, Oliver SW, Colman PM, Varghese JN, Ryan DM, Woods JM, Bethell RC, Hotham VJ, Cameron JM, Penn CR, Rational design of potent sialidase-based inhibitors of influenza virus replication, Nature 363 (1993) 418–423. [DOI] [PubMed] [Google Scholar]
- [5].Gubareva LV, Kaiser L, Hayden FG, Influenza virus neuraminidase inhibitors, Lancet 355 (2000) 827–835. [DOI] [PubMed] [Google Scholar]
- [6].Baz M, Abed Y, Papenburg J, Bouhy X, Hamelin MÈ, Boivin G, Emergence of oseltamivir-resistant pandemic H1N1 virus during prophylaxis, N. Engl. J. Med 361 (2009) 2296–2297. [DOI] [PubMed] [Google Scholar]
- [7].de Jong MD, Thanh TT, Khanh TH, Hien VM, Smith GJD, Chau NV, Cam BV, Qui PT, Ha DQ, Guan Y, Peiris JSM, Hien TT, Farrar J, Oseltamivir resistance during treatment of influenza A (H5N1) infection, N. Engl. J. Med 353 (2005) 2667–2672. [DOI] [PubMed] [Google Scholar]
- [8].Grienke U, Schmidtke M, von Grafenstein S, Kirchmair J, Liedl KR, Rollinger JM, Influenza neuraminidase: a druggable target for natural products, Nat. Prod. Rep 29 (2012) 11–36. [DOI] [PubMed] [Google Scholar]
- [9].Tisdale M, Monitoring of viral susceptibility: new challenges with the development of influenza NA inhibitors, Rev. Med. Virol 10 (2000) 45–55. [DOI] [PubMed] [Google Scholar]
- [10].Gubareva LV, Molecular mechanisms of influenza virus resistance to neuraminidase inhibitors, Virus Res. 103 (2004) 199–203. [DOI] [PubMed] [Google Scholar]
- [11].Gubareva LV, Nedyalkova MS, Novikov DV, Murti KG, Hoffmann E, Hayden FG, a release-competent influenza A virus mutant lacking the coding capacity for the neuraminidase active site, J. Gen. Virol 83 (2002) 2683–2692. [DOI] [PubMed] [Google Scholar]
- [12].Cheng LS, Amaro RE, Xu D, Li WW, Arzberger PW, McCammon JA, Ensemble-based virtual screening reveals potential novel antiviral compounds for avian influenza neuraminidase, J. Med. Chem 51 (2008) 3878–3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Cieśla Ł, Moaddel R, Comparison of analytical techniques for the identification of bioactive compounds from natural products, Nat. Prod. Rep 33 (2016) 1131–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Li F, Zhang Y, Qiu D, Kang J, Screening of epidermal growth factor receptor inhibitors in natural products by capillary electrophoresis combined with high performance liquid chromatography–tandem mass spectrometry, J. Chromatogr. A 1400 (2015) 117–123. [DOI] [PubMed] [Google Scholar]
- [15].Marszałł MP, Sroka WD, Sikora A, Chełminiak D, Ziegler-Borowska M, Siódmiak T, Moaddel R, Ligand fishing using new chitosan based functionalized androgen receptor magnetic particles, J. Pharm. Biomed. Anal 127 (2016) 129–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Singh N, Ravichandran S, Spelman K, Fugmann SD, Moaddel R, The identification of a novel SIRT6 modulator from Trigonella foenum-graecum using ligand fishing with protein coated magnetic beads, J. Chromatogr. B 968 (2014) 105–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wang L, Zhao Y, Zhang Y, Zhang T, Kool J, Somsen GW, Wang Q, Jiang Z, Online screening of acetylcholinesterase inhibitors in natural products using monolith-based immobilized capillary enzyme reactors combined with liquid chromatography-mass spectrometry, J. Chromatogr. A (2018). [DOI] [PubMed] [Google Scholar]
- [18].Marszałł MP, Moaddel R, Kole S, Gandhari M, Bernier M, Wainer IW, Ligand and protein fishing with heat shock protein 90 coated magnetic beads, Anal. Chem 80 (2008) 7571–7575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Moaddel R, Marszałł MP, Bighi F, Yang Q, Duan X, Wainer IW, Automated ligand fishing using human serum albumin-coated magnetic beads, Anal. Chem 79 (2007) 5414–5417. [DOI] [PubMed] [Google Scholar]
- [20].Vanzolini KL, Jiang Z, Zhang X, Vieira LCC, Corrêa AG, Cardoso CL, Cass QB, Moaddel R, Acetylcholinesterase immobilized capillary reactors coupled to protein coated magnetic beads: a new tool for plant extract ligand screening, Talanta 116 (2013) 647–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yasuda M, Wilson DR, Fugmann SD, Moaddel R, Synthesis and characterization of SIRT6 protein coated magnetic beads: identification of a novel inhibitor of SIRT6 deacetylase from medicinal plant extracts, Anal. Chem 83 (2011) 7400–7407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wubshet SG, Brighente IMC, Moaddel R, Staerk D, Magnetic ligand fishing as a targeting tool for HPLC-HRMS-SPE-NMR: α-glucosidase inhibitory ligands and alkylresorcinol glycosides from Eugenia catharinae, J. Nat. Prod 78 (2015) 2657–2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zhu YT, Ren XY, Yuan L, Liu YM, Liang J, Liao X, Fast identification of lipase inhibitors in oolong tea by using lipase functionalised Fe3O4 magnetic nanoparticles coupled with UPLC-MS/MS, Food Chem 173 (2015) 521–526. [DOI] [PubMed] [Google Scholar]
- [24].Ren MT, Chen J, Song Y, Sheng LS, Li P, Qi LW, Identification and quantification of 32 bioactive compounds in Lonicera species by high performance liquid chromatography coupled with time-of-flight mass spectrometry, J. Pharm. Biomed. Anal 48 (2008) 1351–1360. [DOI] [PubMed] [Google Scholar]
- [25].Ishimoto T, Jigawa K, Henares TG, Endo T, Hisamoto H, Integration of neuraminidase inhibitor assay into a single-step operation using a combinable poly (dimethylsiloxane) capillary sensor, Analyst 138 (2013) 3158–3162. [DOI] [PubMed] [Google Scholar]
- [26].Megías C, Pedroche J, Yust MDM, Alaiz M, Girón-Calle J, Millán F, Vioque J, Immobilization of angiotensin-converting enzyme on glyoxyl-agarose, J. Agric. Food. Chem 54 (2006) 4641–4645. [DOI] [PubMed] [Google Scholar]
- [27].de Moraes MC, Santos JB, dos Anjos DM, Rangel LP, Vieira TCRG, Moaddel R, da Silva JL, Prion protein-coated magnetic beads: synthesis, characterization and development of a new ligands screening method, J. Chromatogr. A 1379 (2015) 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zhao H, Chen Z, Screening of neuraminidase inhibitors from traditional Chinese medicines by integrating capillary electrophoresis with immobilized enzyme microreactor, J. Chromatogr. A 1340 (2014) 139–145. [DOI] [PubMed] [Google Scholar]
- [29].Talbert JN, Goddard JM, Enzymes on material surfaces, Colloid Surf. B Biointerfaces 93 (2012) 8–19. [DOI] [PubMed] [Google Scholar]
- [30].Li A, Wang W, Xu W, Gong J, A microplate-based screening assay for neuraminidase inhibitors, Drug Discov. Ther 3 (2009) 260–265. [PubMed] [Google Scholar]
- [31].Zhang J, Wang Q, Fang H, Xu W, Liu A, Du G, Design, synthesis, inhibitory activity, and SAR studies of pyrrolidine derivatives as neuraminidase inhibitors, Bioorg. Med. Chem 15 (2007) 2749–2758. [DOI] [PubMed] [Google Scholar]
- [32].Liu AL, Wang HD, Lee SM, Wang YT, Du GH, Structure–activity relationship of flavonoids as influenza virus neuraminidase inhibitors and their in vitro anti-viral activities, Bioorg. Med. Chem 16 (2008) 7141–7147. [DOI] [PubMed] [Google Scholar]
- [33].Karar MGE, Matei MF, Jaiswal R, Illenberger S, Kuhnert N, Neuraminidase inhibition of dietary chlorogenic acids and derivatives-potential antivirals from dietary sources, Food Funct 7 (2016) 2052–2059. [DOI] [PubMed] [Google Scholar]
- [34].Ge H, Wang YF, Xu J, Gu Q, Liu HB, Xiao PG, Zhou J, Liu Y, Yang Z, Su H, Anti-influenza agents from traditional Chinese medicine, Nat. Prod. Rep 27 (2010) 1758–1780. [DOI] [PubMed] [Google Scholar]
- [35].Kim JH, Ryu YB, Lee WS, Kim YH, Neuraminidase inhibitory activities of quaternary isoquinoline alkaloids from corydalis turtschaninovii rhizome, Bioorg. Med. Chem 22 (2014) 6047–6052. [DOI] [PubMed] [Google Scholar]
- [36].Wang X, Jia W, Zhao A, Wang X, Anti-influenza agents from plants and traditional Chinese medicine, Phytother. Res 20 (2006) 335–341. [DOI] [PubMed] [Google Scholar]
- [37].Qi LW, Chen CY, Li P, Structural characterization and identification of iridoid glycosides, saponins, phenolic acids and flavonoids in flos lonicerae japonicae by a fast liquid chromatography method with diod-array detection and time-of-flight mass spectrometry, Rapid Commun. Mass Spectrom 23 (2009) 3227–3242. [DOI] [PubMed] [Google Scholar]
- [38].Liu S, Yan J, Xing J, Song F, Liu Z, Liu S, Characterization of compounds and potential neuraminidase inhibitors from the n-butanol extract of compound indigowoad root granule using ultrafiltration and liquid chromatography-tandem mass spectrometry, J. Pharm. Biomed. Anal 59 (2012) 96–101. [DOI] [PubMed] [Google Scholar]
- [39].Ding Y, Cao Z, Cao L, Ding G, Wang Z, Xiao W, Antiviral activity of chlorogenic acid against influenza A (H1N1/H3N2) virus and its inhibition of neuraminidase, Sci. Rep 7 (2017) 45723–45733. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.