Abstract
While it is recognized that biocomputing can provide intelligent solutions to complex biosensing projects, it remains challenging to transform biomolecular logic gates into a convenient, portable, resettable and quantitative sensing systems for point-of-care (POC) diagnostics in a low-resource setting. To overcome these limitations, we herein report the first design of biocomputing on personal glucose meters (PGMs) by utilizing glucose and nicotinamide adenine dinucleotide as signal outputs, using DNAzymes and protein enzymes as building blocks, and demonstrating a general platform for installing logic-gate responses (YES, NOT, INHIBIT, NOR, NAND and OR) to a variety of biological species, such as cations (sodium), anions (citrate), organic metabolites (ADP, ATP) and enzymes (pyruvate kinase, alkaline phosphatase, alcohol dehydrogenases). A concatenated logical gate platform that is resettable is also demonstrated. Our system is highly modular and can be generally applied to POC diagnostics of many diseases, such as hyponatremia, hypernatremia and hemolytic anemia. In addition to broadening the clinical applications of the PGM, the method reported here opens a new avenue in biomolecular logic gates for the development of intelligent POC devices for on-site applications.
Keywords: Biocomputing, glucose meter, DNA, enzyme cascade, Point-of Care Testing
Entry for the Table of Contents
A new biocomputing platform that integrated PGMs with logic capability using metal-specific DNAzyme and native enzymes as building blocks was developed for the detection of multiple biological substances in clinical care.
Biocomputing has attracted significant interests across widespread scientific fields because it holds great potential for multi-parameter sensing, intelligent diagnostics, and advanced therapeutic applications.[1] Despite progress made so far, most biocomputing systems, performed by DNA, protein/enzymes, and other living organisms, represent only the proof of concept and demonstrate the possibility of performing logic operations, which are still far from practical realization.[2] In addition, these biomolecular logic gates often employ fluorescent,[3] colorimetric,[4] electrochemical,[5] or SERS[6] signals as their outputs, which often suffer from complicated handling procedures, expensive instrument-dependent readout, or a lack of portability.[7] To overcome these limitations, integrated logic gates performed on lateral flow strips, hydrogel, flow cell, pregnancy test strips and nanomaterials[8] have been described, but most of them either provide a semi-quantitative result or require sophisticated synthesis, and it is not easy to realize the logic system reset or scale them up for assembling large networking systems, which limits their application for field work or point-of-care (POC) testing. Therefore, it is still a challenge to transform biomolecular logic gates into a convenient, portable, resettable and quantitative sensing system for POC diagnostics in a low-resource setting.
Recently, we and others have taken advantage of the widely available pocket-sized personal glucose meter (PGM) and developed novel methods to repurpose the PGM for quantification of a wide range of targets beyond glucose.[9] Despite the progress, most studies can detect only a single target, which cannot meet the demand of modern biomedicine, as most applications require detection and quantification of multiple targets. One reason for the gap in this technology is that most of the commercialized PGMs have been designed to provide only a single signal output, glucose, which limits their application for advanced biocomputing that often needs multiple outputs. For example, signal processing through several concatenated logic gates represented by several biochemical reactions with multiple inputs but with only one output can result in significant loss of information at each step of the process.[10] To address these issues, we have recently explored a new PGM function, namely, dose-dependent response of nicotinamide coenzymes, such as the reduced form of nicotinamide adenine dinucleotide (NADH).[9c] This new discovery of PGM function offers good flexibility in terms of signal output for a variety of enzymatic reactions. To explore new dimensions in biomolecular logic gates, it would be of great interest to combine PGMs with logic gates to create intelligent POC devices for on-site applications.
Herein, we present the first report of biocomputing on PGMs by utilizing glucose and NADH as signal outputs and demonstration of a general platform for installing logic-gate responses to a variety of biological substances. To test the feasibility of PGM-based logic gates, we first designed a YES gate for sodium ions using a Na+-specific DNAzyme (43E) and its substrate (43S).[11] As shown in Figure 1a, a DNA sandwich structure can be assembled on magnetic beads by connecting the DNA-invertase conjugate to a biotinylated capture DNA (biotin-DNA) through simultaneously hybridization with 43S. Upon addition of Na+, the DNAzyme can catalyze the cleavage of its substrate at the 3’-phosphoester bond of the ribonucleotide A (rA) and subsequent release of the DNA–invertase conjugates. The released DNA-invertase conjugates can then catalyze the hydrolysis of sucrose to glucose, producing a detectable PGM signal. In this design, we have achieved the proper execution of a single-input YES gate that produces a PGM signal in response to an input (Na+), i.e., an output 0 from an input 0 and an output 1 from an input 1. Using this YES gate, the target Na+ can be quantified in the range from 8.0 mM to 120 mM in human serums, with a detection limit of 1.9 mM (Fig.1b), which is much lower than the normal sodium level in human serum. A threshold value of 10 mg/dL was defined to separate the OFF and ON logic states of the output PGM signal, whereby only the presence of the input switches the output of the system to the ON state. In addition, the sensor maintained excellent selectivity for Na+ over other biologically relevant metal ions at physiologically relevant concentrations (Figure S1). To verify the accuracy and reliability of our system, we further compared it with the inductively coupled plasma mass spectrometry (ICP-MS) method for Na+ detection in human serum, and a strong positive correlation between these two methods was obtained (Fig. S2). Encouraged by the above successful Na+ assays in biological samples, we further explored potential applications in clinical evaluation of pathophysiological conditions hyponatremia (Na+ level < 135 mM) and hypernatremia (Na+ level > 145 mM). Two serum samples with the Na+ concentration at 135 mM and 145 mM were prepared by adjusting the standard serum samples via dilution with buffer or spiking with external sodium ions, respectively. To analyze the serum samples with a high concentration of Na+, some experimental parameters were also optimized (see details in SI). As shown in Figure 1c, a significant differentiation between hyponatremia and hypernatremia logic levels can be observed, indicating the potential use of a PGM-based YES gate for early evaluation of clinical parameters by binary reading.
Figure 1.
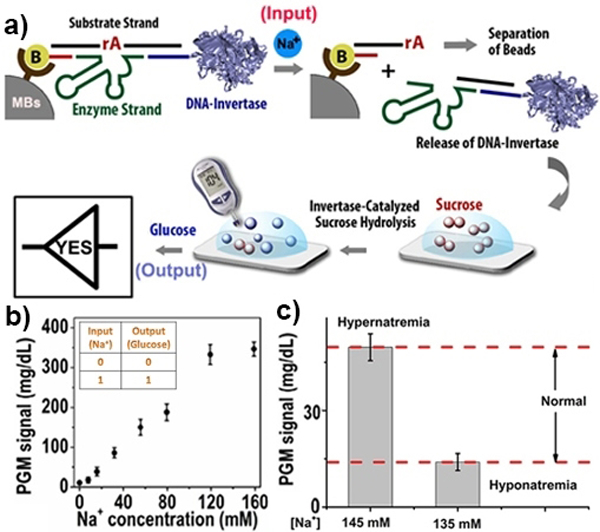
A “YES” logic gate using glucose as the signal output. a) Na+ detection based on Na+-DNAzyme-invertase conjugate and a PGM. Schematic showing the release of the invertase in the presence of Na+ and detection using a PGM. b) Calibration curve of Na+ detection in human serum using a PGM. Inset of b is the truth table for the YES gate. A threshold value of 10 mg/dL was defined to separate the OFF and ON logic states of the output PGM signal. c) A clinical application of a PGM-based YES gate for diagnosis of hyponatremia and hypernatremia.
To expand the logic gate capability of PGMs to many other targets, we extended our building blocks from DNAzymes to protein enzymes for the construction of an INHIBIT gate, which is another notable type of logic operation due to its noncommutative behavior.[12] In an INHIBIT logic gate, one input serves as a veto which has the power to disable the whole system. This logic gate provides an important decision point for POC users. As illustrated in Figure S3a, based on the principle of metal-specific activation of enzymes, a logic gate is installed on PGMs using similar components as the YES gate but adding a downstream enzyme, Hexokinase, which requires Mg2+ for activity. As mentioned in the above YES gate, glucose signal could be produced in the presence of Na+; however, in the presence of Mg2+, hexokinase could be activated and catalyze the further hydrolysis of glucose to glucose-6-phosphate, which is PGM inert. As shown in Figure S3b, by employing Na+ and Mg2+ as inputs, a stronger glucose signal was obtained with Na+ input (1/0) than with other inputs (0/0, 0/1, 1/1). A threshold value of 10 mg/dL was defined to separate the OFF and ON logic states of the output PGM signal. These results indicated the successful construction of the Boolean INHIBIT logic gate, which is represented by the situation where the output is “1” and only if the input is “1” and the others are “0”. In addition, the coexistence of Na+ and Mg2+ in biological samples (e.g. normal human serum) did not significantly influence the logic output value (Figure S4); therefore, our system could potentially be applied for the clinical analysis of hypermagnesemia (Figure S4).
Having demonstrated the YES and INHIBIT logic gates, we investigated the design of a NOT logic gate and its applications in diagnosis of pyruvate kinase (PK) deficiency, a common cause for hemolytic anemia.[13] To achieve this goal, we used nicotinamide adenine dinucleotide (NAD), which is an enzyme cofactor involved in many enzymatic reactions that are essential for human metabolism.[14] Many disease-related enzymes and metabolites, including PK, are capable of facilitating the inter-conversion between NAD+ and NADH. As shown in Figure 2a, PK catalyzes the transphosphorylation of phosphoenolpyruvate to ADP, yielding pyruvate and ATP. The product pyruvate can be further converted into lactate in the presence of lactate dehydrogenase (LDH) by consuming NADH. Thus, in the presence of a PK input, an enzymatic cascade will be triggered, resulting in a decreased NADH signal (Figure 2c), and indicating the NOT logic operation on the PGM. Utilizing this assay, the input PK can be sensitively detected as low as 0.18 U/mL (Figure 2c), which is well below the clinic cut-off range of 2.2–4.4 U/mL for the diagnosis of PK deficiency.[15] To demonstrate that the NOT gate design can be generally applied to detection of other targets, another NOT gate was constructed for the detection of citrate, an important metabolite related to many diseases, such as cancer,[16] using citrate lyase (CL) and malate dehydrogenase (MDH) as the logic units. As shown in Figure 2b, a new enzyme cascade is triggered to convert citrate into oxaloacetate and then into NADH consumption. A quantitative relationship is established between the NADH signal and citrate concentration in the range of 0–12 mM (Figure 2d), with a detection limit of 0.2 mM, which is lower than the normal cut-off concentration of citrate in human urine (1.75 mM) for indicating the risk of kidney stone formation. For the above two NOT gate systems, a threshold value of 325 mg/dL was defined to separate the ON and OFF logic states of the output PGM signal, whereby only the presence of the input (PK or citrate) switches the output of the system to the OFF state.
Figure 2.
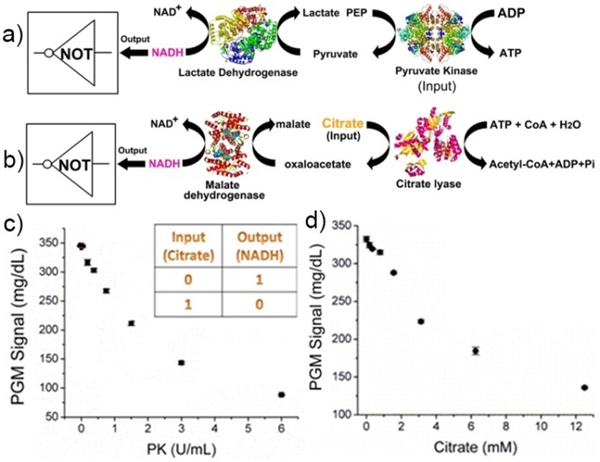
A “NOT” logic gate using NADH as the signal output. Schematic showing the biocatalytic cascade for the detection of pyruvate kinase (a) and citrate (b), and the calibration curves for PK (c) and citrate (d). Inset of c is the truth table for the NOT gate. A threshold value of 325 mg/dL was defined to separate the ON and OFF logic states of the output PGM signal.
To test the feasibility of installing logic gates with multiple inputs on our PGM system, we further designed a PGM sensor for ADP and ATP, which are important coenzymes in various biological processes. We employed ATP (0.5 mM) and ADP (0.5 mM) as inputs and defined the presence and absence of ATP and ADP as “1” and “0”, respectively. As shown in Figure S5a, an enzyme cascade reaction containing creatine kinase (CK), PK, and LDH, was triggered in the presence of either ATP or ADP, resulting the consumption of NADH in the enzyme solution. In a typical assay using 0.5 mM ATP or ADP as the inputs, a threshold value of 50 mg/dL was defined to separate the OFF and ON logic states of the output PGM signal. The PGM’s NADH signal was distinctly high (output = 1) only when the input was in a (0/0) state, while the output was 0 when inputs were (1/0), (0/1), and (1/1) (Fig. S5b), which indicated the successful construction of the Boolean NOR gate that is “On” only when both inputs are “Off”. In addition to the use of coenzymes as inputs, we also performed the logic gate on a PGM using native enzymes as the inputs. To this aim, PK (2 U/mL) and LDH (10 U/mL) were employed as inputs (Figure S6a), and the presence and absence of PK and LDH were defined as “1” and “0”, respectively. Under a typical assay conditions, a threshold value of 50 mg/dL was defined to separate the OFF and ON logic states of the output PGM signal. As shown in Figure S6b, the PGM’s NADH signal is distinctly low (output = 0) only when the input was in a (1/1) state, while the output was 1 when inputs were (0/0), (0/1), and (1/0) as the enzyme cascade reaction that consumes the NADH could be triggered only in the presence of both PK and LDH. These results indicated the successful construction of the Boolean NAND logic gate on our PGM system using native enzymes.
Taking advantage of the dose-dependent response of PGMs to both glucose and NADH, we further challenged our PGM system to perform logic-gate responses to native enzymes that are often detected in a panel but generally not involved in any cascade reactions. To this aim, alkaline phosphatase (ALP) and alcohol dehydrogenases (ADH) were employed as inputs, because ALP and ADH are related to human liver function and abnormal levels of ALP and ADH are observed in alcoholic liver disease.[17] As shown in Figure 3a, the presence of ALP could catalyse the hydrolysis of glucose-6-phosphate to glucose, while the presence of ADH could catalyse the interconversion between alcohols and acetaldehydes with the production of NADH, and, therefore, both generated a detectable signal on a PGM. A threshold value of 10 mg/dL was defined to separate the OFF and ON logic states of the output PGM signal. As shown in Figure 3b, the output signal of a PGM is higher than 10 mg/dL (defined as 1) when either one enzyme or both enzymes were present, which indicated a representative OR gate on the PGM.
Figure 3.
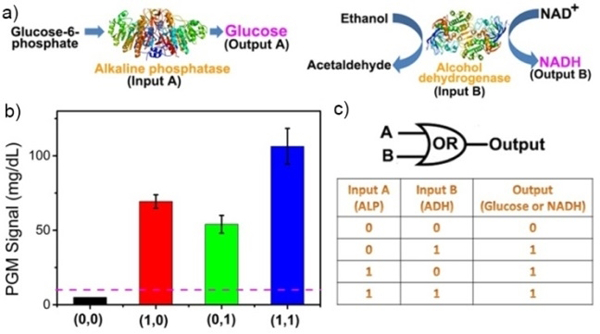
An “OR” logic gate using both NADH and glucose as the signal output. a) Schematic showing the biocatalytic reaction for the detection of alkaline phosphatase (ALP) and alcohol dehydrogenases (ADH). b) PGM signal for the Boolean OR logic functions operating with ALP and ADH. c) The truth table for the OR gate.
For most of the molecular logic gates reported to date, it is difficult to reset the logic system, which is a crucial property for practical applications. To address this issue, we have designed a concatenated logic gate system that can be reset and scaled up (Figure 4a). The system includes hydrolysis of sucrose (Input 1) by invertase to produce glucose (Output 1), hydrolysis of glucose by hexokinase with the addition of ATP (Input 2) to yield glucose-6-phosphate (Output 2), oxidation of glucose-6-phosphate by glucose-6-phosphate dehydrogenase (G6PD) with the addition of NAD+ (Input 3) to yield NADH (Output 3), and, finally, conversion of NADH to NAD+ (Output 4) by LDH with the addition of pyruvate (Input 4). This biocatalytic cascade combines one YES gate and three AND gates to form one subunit cycle. The output threshold value was defined as “0” when the PGM signal was lower than 10 mg/dL, while the threshold value of “1” was defined when the PGM signal was higher than 150 mg/dL. Figure 4b shows the corresponding PGM signal in response to the above biochemical reaction chain. Repeated switches between “off” (< 10 mg/dL) and “on” (defined as > 150 mg/dL) states on a PGM were obtained through the addition of appropriate inputs of enzyme substrates and coenzymes. More importantly, after one subunit cycle (Figure 4a), the PGM platform could, to an extent, be reset to the original state, and another biocatalytic cascade could be triggered through the supplement of original input 1 (sucrose). The above inputs of sucrose, ATP, NAD+, and pyruvate are important cofactors or metabolites in glycolytic pathway, and abnormal levels of these species are often related to numerous genetic diseases.[18] Therefore, these results indicated that our PGM-based logic gate platform can be readily scaled up to generate sophisticated networks of logic circuits and mimic natural biological pathways, which is extremely valuable in the practical application of logic devices.
Figure 4.
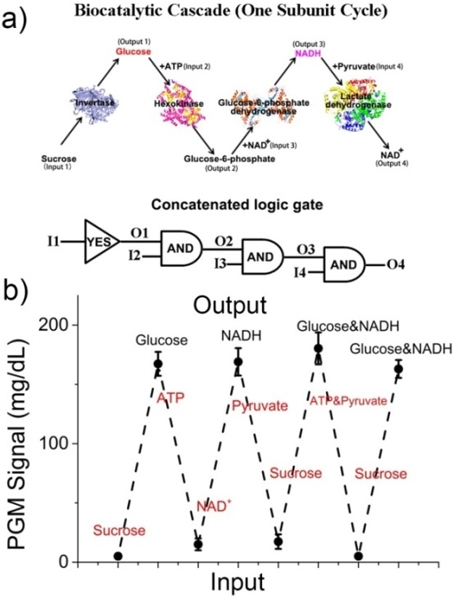
“Concatenated” logic gate using both NADH and glucose as the output. a) Scheme of the biocatalytic cascade with multiple inputs and outputs. b) The PGM response of repeated switches between “off” and “on” states by adding appropriate inputs of enzyme substrates and coenzymes.
In summary, we have demonstrated a new biocomputing platform that integrates PGMs with logic capability using metal-specific DNAzymes and protein enzymes as building blocks. A complete set of seven binary logic gates (YES, NOT, OR, NOR, NAND, INHIBIT, and Concatenated) has been constructed on PGMs for the detection of multiple biological substances. A series of metal ions, disease-related metabolites, coenzymes, and native enzymes have been employed as inputs, using glucose and/or NADH signal on a PGM as outputs. Potential applications of these methods for POC diagnostics of diseases (e.g. hyponatremia, hypernatremia) have also been demonstrated. In addition, our PGM-based platform offers unique advantages compared to existing methods for biocomputing applications: 1) it does not require calibration for every measurement once the calibration curves are obtained for the non-glucose targets, as the glucose readout by PGMs has been demonstrated to have minimal interference from the human sample matrix;[19] 2) this platform architecture is modular and could be repurposed for various applications. For instance, the DNAzyme sequences can be interchanged to enable responsiveness to other input monovalent (e.g. Ag+),[20] divalent (e.g. Pb2+),[21] and trivalent metal ions (e.g. Cr3+),[22] while the modulation of native enzyme-based logic systems can be achieved by switching to other biologically/clinically relevant targets involved in the cascade enzymatic reactions (e.g. lactate/pyruvate, G6PD).[9c] We anticipate that such PGM-based logic biosensors, which are capable of in vitro computation, can be tailored according to medical knowledge and used as expert biosensing devices for POC diagnostics.
Supplementary Material
Acknowledgements
We wish to thank the U.S. National Institutes of Health (MH110975, MH111337 and HD092155) for financial supports.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/anie.2017xxxxx
References
- [1].a) Zhan W, Crooks RM, J. Am. Chem. Soc. 2003, 125, 9934–9935; [DOI] [PubMed] [Google Scholar]; b) Zhou M, Du Y, Chen CG, Li BL, Wen D, Dong SJ, Wang EK, J. Am. Chem. Soc. 2010, 132, 2172–2174; [DOI] [PubMed] [Google Scholar]; c) Chang B-Y; Crooks JA; Chow K-F; Mavré F; Crooks RM, J. Am. Chem. Soc. 2010, 132, 15404–15409; [DOI] [PubMed] [Google Scholar]; d) Pei H, Liang L, Yao GB, Li J, Huang Q, Fan CH, Angew. Chem. Int. Ed. 2012, 51, 9020–9024; [DOI] [PubMed] [Google Scholar]; e) Katz E, Minko S, Chem. Commun. 2015, 51, 3493–3500; [DOI] [PubMed] [Google Scholar]; f) He KY, Li Y, Xiang BB, Zhao P, Hu YF, Huang Y, Li W, Nie Z, Yao SZ, Chem. Sci. 2015, 6, 3556–3564; [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Li DD, Cheng W, Li YJ, Xu YJ, Li XM, Yin YB, Ju HX, Ding SJ, Anal. Chem. 2016, 88, 7500–7506; [DOI] [PubMed] [Google Scholar]; h) Molinnus D, Poghossian A, Keusgen M, Katz E, Schoning MJ, Electroanalysis 2017, 29, 1840–1849; [DOI] [PMC free article] [PubMed] [Google Scholar]; i) Li Y, Sun SJ, Fan L, Hu SF, Huang Y, Zhang K, Nie Z, Yao SZ, Angew. Chem. Int. Ed. 2017, 56, 14888–14892; [DOI] [PubMed] [Google Scholar]; j) Liang JY, Yu X, Yang TG, Li ML, Shen L, Jin Y, Liu HY, Phys. Chem. Chem. Phys. 2017, 19, 22472–22481; [DOI] [PubMed] [Google Scholar]; k) Qu XM, Zhu D, Yao GB, Su S, Chao J, Liu HJ, Zuo XL, Wang LH, Shi JY, Wang LH, Huang W, Pei H, Fan CH, Angew. Chem. Int. Ed. 2017, 56, 1855–1858; [DOI] [PubMed] [Google Scholar]; l) Li J, Green AA, Yan H, Fan CH, Nat. Chem. 2017, 9, 1056–1067; [DOI] [PMC free article] [PubMed] [Google Scholar]; m) Peng HY, Newbigging AM, Wang ZX, Tao J, Deng WC, Le XC, Zhang HQ, Anal. Chem. 2018, 90, 190–207. [DOI] [PubMed] [Google Scholar]
- [2].a) Lake A, Shang S, Kolpashchikov DM, Angew. Chem. Int. Ed. 2010, 49, 4459–4462; [DOI] [PubMed] [Google Scholar]; b) Liu Y, Offenhausser A, Mayer D, Angew. Chem. Int. Ed. 2010, 49, 2595–2598; [DOI] [PubMed] [Google Scholar]; c) Benenson Y, Nat. Nanotech. 2011, 6, 465–467; [DOI] [PubMed] [Google Scholar]; d) de Silva AP, Chem. Asian J. 2011, 6, 750–766; [DOI] [PubMed] [Google Scholar]; e) Bel-Enguix G, Jiménez-López MD, Natural Computing 2012, 11, 131–139; [Google Scholar]; f) Prokup A, Hemphill J, Deiters A, J. Am. Chem. Soc. 2012, 134, 3810–3815; [DOI] [PubMed] [Google Scholar]; g) Gao JT, Liu YQ, Lin XD, Deng JK, Yin JJ, Wang S, Sci. Rep. 2017, 7 14014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].a) Bi S, Ji B, Zhang ZP, Zhu JJ, Chem. Sci. 2013, 4, 1858–1863; [Google Scholar]; b) Zhang M, Ye BC, Chem. Commun. 2012, 48, 3647–3649; [DOI] [PubMed] [Google Scholar]; c) Guo YH, Wu J, Ju HX, Chem. Sci. 2015, 6, 4318–4323; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Wang WJ, Huang S, Li JJ, Rui K, Bi S, Zhang JR, Zhu JJ, Chem. Sci. 2017, 8, 174–180; [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Tang YD, Lu BY, Zhu ZT, Li BL, Chem. Sci. 2018, 9, 760–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].a) Xianyu YL, Wang Z, Sun JS, Wang XF, Jiang XY, Small 2014, 10, 4833–4838; [DOI] [PubMed] [Google Scholar]; b) Wu CT, Fan DQ, Zhou CY, Liu YQ, Wang EK, Anal. Chem. 2016, 88, 2899–2903; [DOI] [PubMed] [Google Scholar]; c) Chen JH, Zhou SG, Wen JL, Angew. Chem. Int. Ed. 2015, 54, 446–450; [DOI] [PubMed] [Google Scholar]; d) Huang YY, Pu F, Ren JS, Qu XG, Chem. Eur. J. 2017, 23, 9156–9161. [DOI] [PubMed] [Google Scholar]
- [5].a) Tang LH, Wang Y, Li JH, Chem. Soc. Rev. 2015, 44, 6954–6980; [DOI] [PubMed] [Google Scholar]; b) Ge L, Wang WX, Sun XM, Hou T, Li F, Anal. Chem. 2016, 88, 9691–9698; [DOI] [PubMed] [Google Scholar]; c) Zhai QF, Fan DQ, Zhang XW, Li J, Wang EK, Npg Asia Mater. 2017, 9; [Google Scholar]; d) Du Y, Han X, Wang CX, Li YH, Li BL, Duan HW, ACS Sens. 2018, 3, 54–58. [DOI] [PubMed] [Google Scholar]
- [6].a) Witlicki EH, Johnsen C, Hansen SW, Silverstein DW, Bottomley VJ, Jeppesen JO, Wong EW, Jensen L, Flood AH, J. Am. Chem. Soc. 2011, 133, 7288–7291; [DOI] [PubMed] [Google Scholar]; b) Wu ZT, Dong BR, Zhou XD, Shen AG, Hu JM, Chem. Eur. J. 2015, 21, 14301–14304. [DOI] [PubMed] [Google Scholar]
- [7].a) Elbaz J, Moshe M, Willner I, Angew. Chem. Int. Ed. 2009, 48, 3834–3837; [DOI] [PubMed] [Google Scholar]; b) Mu L, Shi W, She G, Chang JC, Lee ST, Angew. Chem. Int. Ed. 2009, 48, 3469–3472; [DOI] [PubMed] [Google Scholar]; c) Frasconi M, Tel-Vered R, Elbaz J, Willner I, J. Am. Chem. Soc. 2010, 132, 2029–2036. [DOI] [PubMed] [Google Scholar]
- [8].a) Liu D, Chen W, Sun K, Deng K, Zhang W, Wang Z, Jiang X, Angew. Chem. Int. Ed. 2011, 50, 4103–4107; [DOI] [PubMed] [Google Scholar]; b) Liu DB, Chen WW, Sun K, Deng K, Zhang W, Wang Z, Jiang XY, Angew. Chem. Int. Ed. 2011, 50, 4103–4107; [DOI] [PubMed] [Google Scholar]; c) Moon TS, Lou C, Tamsir A, Stanton BC, Voigt CA, Nature 2012, 491, 249–253; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Wang L, Zhu JB, Han L, Jin LH, Zhu CZ, Wang EK, Dong SJ, ACS nano 2012, 6, 6659–6666; [DOI] [PubMed] [Google Scholar]; e) Bi S, Ji B, Zhang Z, Zhu J-J, Chem. Sci. 2013, 4, 1858–1863; [Google Scholar]; f) Miyamoto T, Razavi S, DeRose R, Inoue T, ACS Synth. Biol. 2013, 2, 72–82; [DOI] [PMC free article] [PubMed] [Google Scholar]; g) He XW, Li Z, Chen MZ, Ma N, Angew. Chem. Int. Ed. 2014, 53, 14447–14450; [DOI] [PubMed] [Google Scholar]; h) Qin CY, Gao Y, Wen W, Zhang XH, Wang SF, Biosens. Bioelectron. 2016, 79, 522–530; [DOI] [PubMed] [Google Scholar]; i) Du Y, Pothukuchy A, Gollihar JD, Nourani A, Li BL, Ellington AD, Angew. Chem. Int. Ed. 2017, 56, 992–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].a) Xiang Y, Lu Y, Nat. Chem. 2011, 3, 697–703; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Du Y, Hughes RA, Bhadra S, Jiang YS, Ellington AD, Li BL, Sci. Rep. 2015, 5; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Zhang JJ, Xiang Y, Wang M, Basu A, Lu Y, Angew. Chem. Int. Ed. 2016, 55, 732–736; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Xiang Y, Lu Y, Chem. Commun. 2013, 49, 585–587; [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Xiang Y, Lu Y, Anal. Chem. 2012, 84, 4174–4178; [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Xiang Y, Lu Y, Anal. Chem. 2012, 84, 1975–1980; [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Gu C, Lan T, Shi H, Lu Y, Anal. Chem. 2015, 87, 7676–7682; [DOI] [PubMed] [Google Scholar]; h) Lan T, Zhang J, Lu Y, Biotechnol Adv. 2016, 34, 331–341; [DOI] [PMC free article] [PubMed] [Google Scholar]; i) Zhang J, Shen Z, Xiang Y, Lu Y, ACS Sens. 2016, 1, 1091–1096 [Google Scholar]
- [10].a) Lai YH, Sun SC, Chuang MC, Biosensors 2014, 4, 273–300; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Grattieri M, Minteer SD, ACS Sens. 2018, 3, 44–53. [DOI] [PubMed] [Google Scholar]
- [11].a) Torabi SF, Wu PW, McGhee CE, Chen L, Hwang K, Zheng N, Cheng JJ, Lu Y, Proc. Natl. Acad. Sci. U.S.A. 2015, 112, 5903–5908; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wu ZK, Fan HH, Satyavolu NSR, Wang WJ, Lake R, Jiang JH, Lu Y, Angew. Chem. Int. Ed. 2017, 56, 8721–8725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wang S, Sun J, Zhao JH, Lu SS, Yang XR, Anal. Chem. 2018, 90, 3437–3442. [DOI] [PubMed] [Google Scholar]
- [13].Min-Oo G, Tam MF, Stevenson MM, Gros P, Blood Cell. Mol. Dis. 2007, 39, 63–69. [DOI] [PubMed] [Google Scholar]
- [14].Rajman L, Chwalek K, Sinclair DA, Cell Metab. 2018, 27, 529–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].a) Zanella A, Fermo E, Bianchi P, Valentini G, Br. J. Haematol. 2005, 130, 11–25; [DOI] [PubMed] [Google Scholar]; b) Titapiwatanakun R, Hoyer JD, Crain K, Arndt CAS, Pediatr. Blood Cancer 2008, 51, 819–821. [DOI] [PubMed] [Google Scholar]
- [16].Iacobazzi V, Infantino V, Biol. Chem. 2014, 395, 387–399. [DOI] [PubMed] [Google Scholar]
- [17].Khayrollah AA, Al-Tamer YY, Taka M, Skursky L, Ann. Clin. Biochem. 1982, 19, 35–42. [DOI] [PubMed] [Google Scholar]
- [18].Biochem Res Trends 2009, 1–196.
- [19].Tonyushkina K, Nichols JH, J Diabetes Sci. Technol. 2009, 3, 971–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Saran R, Kleinke K, Zhou WH, Yu TM, Lee JW, Biochemistry (N.Y.) 2017, 56, 1955–1962. [DOI] [PubMed] [Google Scholar]
- [21].a) Lan T, Furuya K, Lu Y, Chem. Commun. 2010, 46, 3896–3898; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) McGhee CE, Loh KY, Lu Y, Curr. Opin. Biotechnol. 2017, 45, 191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zhou WH, Vazin M, Yu TM, Ding JS, Liu JW, Chem. Eur. J. 2016, 22, 9835–9840. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


