Abstract
An effective tumoral delivery system should show minimal removal by the reticuloendothelial system (RES), promote tumor uptake and penetration, and minimize on-site clearance. This study reports the design and synthesis of advanced self-assembling peptide nanofiber precursor (NFP) analogues. The peptidic nature of NFP offers the design flexibility for on-demand customization with imaging agents and surface charges while maintaining a set size, allowing for real-time monitoring of kinetic and dynamic tumoral delivery by multimodal fluorescence/positron emission tomography/computed tomography (fluo/PET/CT) imaging, for formulation optimization. The optimized glutathione (GSH)-NFP displays a reduced capture by the RES as well as excellent tumor targeting and tissue invasion properties compared to naive NFP. Inside a tumor, GSH-NFP can structurally transform into ten times larger interfibril networks, serving as in situ depot that promotes weeks-long local retention. This nanofiber, which can further be designed to release the active pharmacophores within a tumor microenvironment, displays a superior therapeutic efficacy for inhibiting disease progression and improving the survival of animals bearing triple-negative breast cancer tumors compared to free drug and liposome formulation of the drug, in addition to a favorable toxicity profile.
Keywords: doxorubicin, interfibril network, peptide nanofibers, triple negative breast cancer, tumoral uptake, retention
1. Introduction
In the last decades, the development of many different nanocarriers for anticancer drug delivery has emerged. This technology takes advantage of the leakiness of tumoral tissues to deliver drugs to tumor sites.[1] Unfortunately, following systemic administration, nanocarriers are susceptible to removal by reticuloendothelial system (RES).[2] Even if a small amount of the carriers can avoid clearance and successfully reach a tumor, their deep-tissue targeting is inconsistent, resulting in a lack of tumor penetration to execute their full therapeutic potential.[3] On the other hand, a nanocarrier, whether it is composed of organic or inorganic material, may be intrinsically toxic and/or immunogenic,[2 a,4] display limited drug loading capacity,[5] release the drug prematurely, or incompletely dispense the drug at the tumor site,[6] leading to depreciation of safety and therapeutic efficacy. Inclusion of a layer of polyethylene glycol (PEG) on a nanocarrier surface can improve biocompatibility, minimize immunogenicity, and increase circulating half-life.[7] Many approaches have been used to enhance the tumoral uptake and penetration of nanocarriers, which include the modification of physicochemical properties (shape, size, and surface charge),[8] conjugation with targeting ligands to achieve active delivery,[9] application of magnetic fields to enhance tissue penetration,[10] and coad-ministration of adjuvants to increase the permeability of tumor vasculature or decrease the stromal barrier.[11] All of these approaches are normally aided by the poor lymphatic drainage that characterizes cancerous tissue, which has the effect of increasing the drug–tumor contact time.[12] Stimuli-responsive materials that react to exogenous (temperature, magnetic field, ultrasound, light, or electric pulses) or endogenous (pH, enzyme, or redox gradients) activators have also been designed to control drug release in a spatial-, temporal-, and dosage-controlled fashion.[13] Many nanocarriers specifically explore the slight difference in pH between healthy tissues and the extracellular environment of tumors to afford efficient drug release.[14]
Despite the tremendous efforts devoted to resolving the pitfalls of drug nanocarriers, a recent literature survey showed that, on average, 0.7% of a total injected dose is delivered to a solid tumor, suggesting there is a need to improve our current delivery strategy.[15] The rational design of a drug delivery system is complex; relying on a previously used approach to overcome a single tumor-associated barrier is ineffective. Various self-assembling peptide nanofibers have been employed as advanced materials in different biomedical applications.[16] For example, peptide and protein amyloids have been widely used for medical imaging, tissue engineering, and drug delivery.[17] Hybrids based peptide nanofibers have been utilized for targeting and imaging tumor cells.[18] Here, we employed a new biocompatible, nonimmunogenic formulation of self-assembling peptide nanofiber precursor (NFP) customized with multiple components to optimize the drug delivery efficiency (Figure 1a). The nanofiber 1) contains a high PEG content that can minimize the removal by the RES, 2) has a high aspect ratio to promote tumor uptake, 3) displays a singlelayered geometry to allow the escape from tumor blood vessel, 4) is surface-decorated with negative charges to infiltrate and saturate tumor, and 5) has the ability to structurally evolve into interfibril network for prolonging the retention on site; an enzyme-induced tumor retention (ETR) effect. Such a combinatorial approach to overcome multiple physiological barriers does not require a targeting ligand to achieve tumor-specific biodistribution, conferring to NFP a great potential as a carrier for treating cancers that lack cognate receptors. The therapeutic and safety benefits of an optimal NFP formulation that releases doxorubicin within the acidic tumoral environment are demonstrated in the present study.
Figure 1.
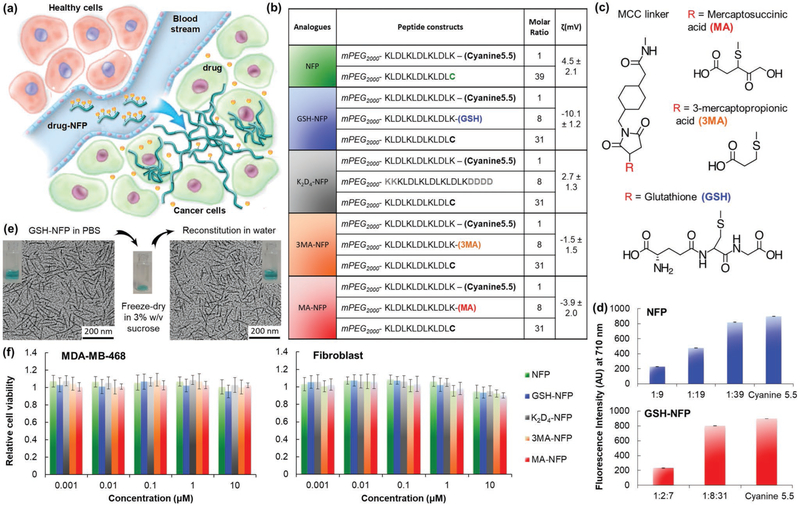
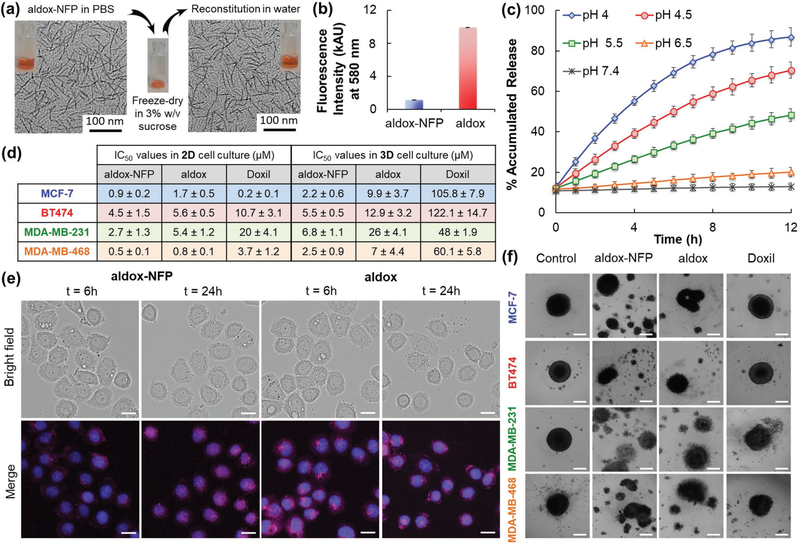
Design of advanced NFP analogues to enhance tumoral uptake, penetration, and local retention. a) NFP has a high aspect ratio that promotes its uptake by solid tumors. Multiple NFPs can penetrate tumor tissue and subsequently transform into larger interfibril networks via in situ activation by tumor-associated proteases, thus minimizing lymphatic clearance. When used for drug delivery, NFP prolongs the drug-tumor contact time to achieve more effective treatment. b) A table showing the peptide composition and surface charge (zeta potential) of the NFP analogues. The peptide derivatives were used to coassemble the nanofibers. c) Chemical structures of the electron donor glutathione or its derivatives (mercaptosuccinic or 3-mercaptopropionic acids) and the MCC linker used to conjugate them to the core peptide sequence. d) Charts showing the fluorescence intensities of naive NFP (top panel) and GSH-NFP (bottom) (0.1 × 10−6 m of fluorophore content) assembled from different ratios of the peptide constructs (b). At a 1:39 molar ratio of Cyanine5.5 and naked peptide constructs, the nanofibers displayed minimal self-quenching, as shown by the comparable fluorescence intensity to the free Cyanine5.5. e) GSH-NFP could be formulated in powder for long-term storage. Transmission electron microscopic images revealed that the nanofiber (10 × 10−6 m of peptide content) displayed the same size and morphology in sucrose (3% w/v) in a PBS buffer, before freeze-drying into powder or the reconstitution of lyophilized powder with water for injection. f) Graphs showing the cytotoxicity profile of the NFP analogues incubated for 3 days with the human triple-negative breast cancer MDA-MB-468 and fibroblast cell lines. Cell viability was determined using CellTiter Glo reagent (Promega), according to the manufacturer’s instructions.
2. Results and Discussion
2.1. Design and Synthesis of NFP Analogues
We previously reported a peptide-based NFP composing with multiple β-sheet peptides (mPEG2000-KLDLKLDLKLDL-CONH2).[19] Each peptide comprises alternate hydrophobic (leucine) and hydrophilic (lysine and aspartic acid) residues conjugated to a 2 kDa methoxypolyethylene glycol (mPEG) chain. The mPEG (40% w/w) prevents interfibril aggregation.[19 b] NFP has a well-organized structure. The width and z-dimension are uniform because multiple peptides are stacked parallel to each other and are maintained by strong stabilizing hydrophobic and ionic interactions.[20] This structure offers the design flexibility for incorporating chemically disparate drug molecules and imaging agents while maintaining size and morphologic features. Formulations with on-demand payloads can be customized by simply adding a comixture of assemble-as-you-go peptide derivatives (e.g., drug and imaging agent-conjugates) in a phosphate-based buffer.[19a] NFP does not trigger innate immune responses, as no increases in inflammatory cytokines (interleukins IL-1β IL-2, IL-6, and IL-10, tumor necrosis factor alpha (TNF-α), and interferon gamma (INF-γ)) being detected in the sera of Balb/c mice overtime compared to the positive control, lipopolysaccharide (Figure S1a, Supporting Information). Changing the physicochemical properties of a nanocarrier including the size, shape, and surface charges can alter the in vivo behavior.[21] Smaller drug carriers (<8 nm) penetrate tumor better but can be rapidly removed by the kidneys.[22] Carriers with larger sizes (>200 nm) are more likely to be captured by the RES. Cationic carriers are taken up by tumors more efficiently than the anionic counterparts but are less likely to penetrate tumors.[23] The optimal length of NFP (100 nm) for tumor uptake was previously determined.[19b] In the present studies, we investigated the impact of surface charge on NFP delivery. To achieve this, we synthesized a panel of new advanced NFP analogues with different zeta potentials, by introducing various electron donors such as glutathione (GSH) or its derivatives, or additional amino acids into the nanofibers. The individual NFP analogues were coassembled from multiple derivatives of the core peptide sequence (Figure 1b). Each peptide was chemically modified to introduce: 1) a Cyanine5.5 fluorophore (Figure S1b, Supporting Information) for screening of the tumoral uptake by optical imaging; 2) a C-terminal cysteine to provide a drug conjugation site;[24] 3) extra lysine and aspartic residues; or 4) a GSH or derivatives, mercaptosuccinic acid (MA) or 3-mercaptopropionic acid (3MA) (Figure 1c), which are expected to enhance tumoral delivery and minimize the removal by the RES.[25] All NFPs, homogenized to 100 nm in length using extruder, carried the same amount of fluorophore, consisting of 1 equivalent of Cyanine5.5-labeled peptide to 39 equivalents of unlabeled-peptide. At this particular ratio, the nanofibers displayed minimal fluorescence quenching (Figure 1d),[26] ideal for comparing different nanofiber analogues for tumoral uptake by optical imaging. The assembled NFPs, e.g., GSH-NFP, showed a similar morphology as naïve NFP (Figure S1c, Supporting Information), and were stable in both human and mouse sera (Figure S1d, Supporting Information). When suspended in phosphate-buffered saline (PBS) containing 3% w/v sucrose, the nanofibers could be lyophilized for long-term storage in solid form and then reconstituted using sterile water for injection to recover the intact nanofibers when required. This was done to ease the dispensing process (Figure 1e). Prior to study the tumoral uptake, we confirmed that the NFP analogues were nontoxic in both cancer and normal cells with a cell viability assay (Figure 1f).
2.2. Screening of NFP Analogues for Tumoral Uptake by Optical Imaging
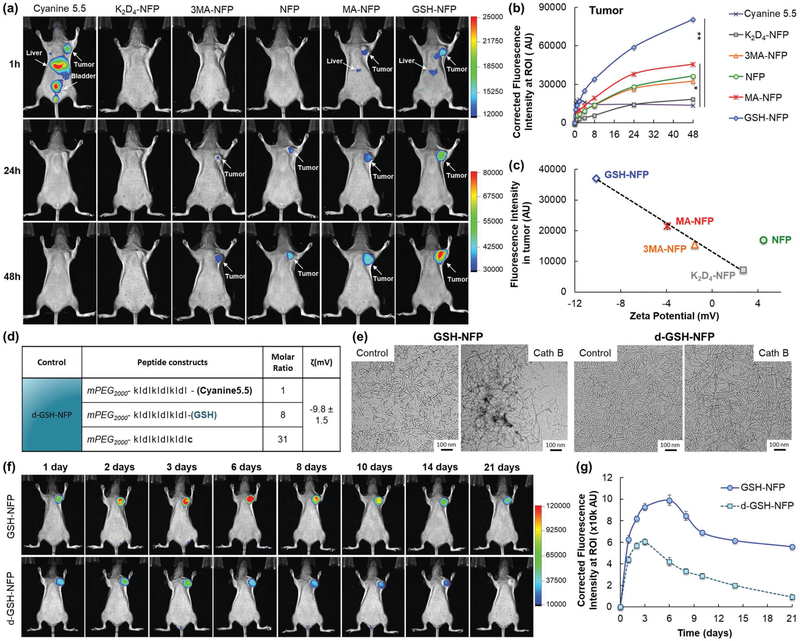
Optical imaging studies showed that GSH-NFP outperformed other NFP analogues in terms of tumoral uptake. The fluorescence intensity (nanofibers) taken up by orthotopically implanted human triple-negative MDA-MB-468 breast tumors in SCID mice was higher compared to other NFP analogues after intravenous (IV) administration (Figure 2a). A plot of the dynamic fluorescence changes at the tumor sites showed that all NFP analogues could enhance the tumoral uptake of Cyanine5.5. Among all the analogues, GSH-NFP was the most effective in terms of tumoral delivery and accumulation (Figure 2b). The tumor-specific delivery of GSH-NFP was further confirmed by ex vivo imaging of the organs (Figure S2a,b, Supporting Information), as shown by the higher fluorescence accumulation in the tumors compared to the livers and other organs. The GSH-NFP displayed a significant improvement of its tumor targeting properties compared to the previously described naive NFP.[19 b] The enhanced tumoral uptake of GSH-NFP was contributed to the negative surface charges supplemented by the irreversible conjugation of GSH, leading to a zeta potential value of −10 mV (Figure 1b). A plot of the observed fluorescence intensity in tumors versus the zeta potential of the NFP analogues revealed a linear relationship (Figure 2c) with the exception of naïve NFP, which does not consist of any GSH, its derivatives, or additional amino acid residues. The fluorescence intensity proportionally decreased with increasing the overall charge of the analogues (from −10 mV for GSH-NFP to +2.7 mV for K2D4-NFP), which strongly indicated that the tumor uptake was charge-correlated. The contribution of negatively charged nanocarriers to enhance tumoral uptake was supported by other studies.[27] We further confirmed the results in 3D-cell cultures. GSH-NFP showed higher accumulation inside MDA-MB-468 spheroids compared to the other analogues (Figure S2c, Supporting Information). Interestingly, GSH-NFP also exhibited the same in vivo distributions in animals bearing MDA-MB-231 tumors (Figure S3, Supporting Information), which suggests that the nanofiber could be applied for delivery to other solid tumors. A long-term optical imaging study showed that GSH-NFP has an excellent tumoral retention for more than 3 weeks (Figure 2f,g). The prolonged retention time was attributed to the ability of the nanofibers to transform into larger interfibril networks upon activation by tumor-associated proteases such as cathepsin B (Figure 2e). Immediately after the cleavage of hydrophilic mPEG by proteases (via the N-terminal lysine of the peptide construct),[19a] NFP became extremely hydrophobic. To improve solubility, multiple nanofibers assembled into 10× larger interfibril networks.[19a,20b] Here, the ETR effect in vivo was supported by comparing the tumoral delivery of GSH-NFP and its D-configuration counterpart (d-GSH-NFP) (Figure 2d). Both GSH-NFP and d-GSH-NFP shared the same physicochemical properties, except d-GSH-NFP could not be activated by proteases to form a large interfibril network (Figure 2e), and thus showing less tumoral uptake and a shorter local retention time (Figure 2f,g).
Figure 2.
GSH-NFP displays excellent tumor-targeting properties. a) Whole body fluorescence images were acquired at various time intervals (n = 3 per group) after injecting the nanofiber (0.5 nmol of fluorophore content) or Cyanine5.5 into SCID mice bearing human triple-negative MDA-MB-468 tumors via the tail vein. b) The fluorescence intensity at the tumor site was plotted to reveal the kinetic uptake of the nanofiber analogues or free Cyanine5.5. c) A plot of the fluorescence intensities of excised MDA-MB-468 tumors 48 h after the administration of NFP analogues versus the surface charges (zeta potential) of the nanofibers. d) Composition and surface charge of the nonactivatable control of GSH-NFP (d-GSH-NFP). e) TEM images of GSH-NFP and d-GSH-NFP before and 24 h after Cath B activation (0.3 U) in sodium acetate buffer (50 × 10−6 m, pH 4). f) Whole body fluorescence images of SCID mice bearing the tumors were acquired at various time intervals (n = 3 per group) after IV injection of the nanofibers. g) The fluorescence intensities at the tumor sites were plotted to compare the kinetic uptake and retention profiles between GSH-NFP and d-GSH-NFP.
2.3. GSH-NFP Displayed Excellent Tumor Penetration Accompanied with High Local Retention
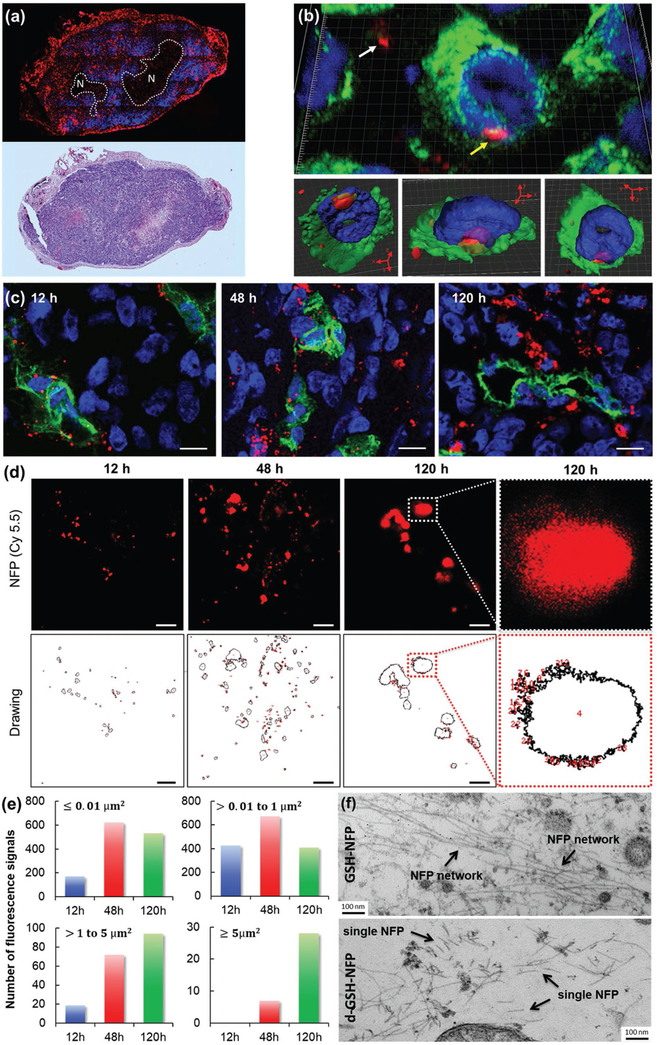
The favorable tumoral delivery of GSH-NFP was also contributed by its ability to penetrate tumor and then transform into interfibril networks. Epifluorescence microscopic imaging of the tumor sections showed that the nanofibers (red) delivered could initially escape from blood vessels (green), progressively spread out to infiltrate tumor tissues (Figure S4a, Supporting Information), and eventually invade the whole tumor, except for the necrotic areas (Figure 3a). Confocal microscopic studies showed that the nanofibers were located both between and within the tumor cells (Figure 3b; Movie S1, Supporting Information). The tissue penetration was confirmed by stimulated emission depletion (STED) microscopy (Figure 3c). With time, the nanofibers (in red) leaked from the blood vessels (in green) to spread out and penetrate the tumoral tissue. Here, high-resolution STED images were also acquired to study the kinetic and dynamic transformation of the nanofibers in tumor (Figure 3d). Both the areas and numbers of the Cyanine5.5 fluorescence (nanofiber networks) progressively increased overtime, suggesting that the GSH-NFP structurally evolved from single nanofibers to large interfibril networks (Figure 3e). We further corroborated the detailed structure of the networks with transmission electron microscopy (TEM) analysis, which appeared to be large micrometer-sized interfibril networks residing in the interstitial spaces and around the microvessels (Figure 3f; Figure S4b,c, Supporting Information). These networked fibers were much thinner and less organized than the natural collagen fibers (Figure S4d, Supporting Information), and represented the structural transformation of multiple NFPs induced by tumor-associated proteases. On the contrary, we could only find single nanofiber in the tumor sections of the animals injected with d-GSH-NFP (Figure 3f; Figure S4b, Supporting Information).
Figure 3.
GSH-NFP displayed tumor penetration, infiltration, and invasion properties. a) Histologic studies showed that the GSH-NFP (red) had invaded the tumor 48 h after a single IV injection of GSH-NFP. The representative tumor section was stained with DAPI (blue) prior to fluorescence imaging. H&E staining was also performed on the adjacent tumor section to confirm the necrotic region (N). b) Confocal microscopic images (using Panoramic Confocal) of GSH-NFP (red) in tumor tissue 48 h post injection. The yellow arrow indicates nanofibers within cancer cells, whereas the white arrow indicates nanofibers in the interstices between the cells. DAPI (blue) and FITC-labeled anti-vimentin antibody (green) were used to stain the nucleus and the cytoskeleton, respectively. The same cell image was processed as a density map and observed in different projection views (bottom panel). c) Confocal microscopic images (using Leica TCS SP8 STED) of the tumor sections collected 12, 48, and 120 h after IV injection of GSH-NFP (5 nmol of fluorophore content). The nanofibers (Cy5.5 fluorescence channel) were imaged using STED 775 nm depletion laser. The blood vessels and cellular nucleus were stained with CD31 (green) and DAPI (blue), respectively. The scale bar represents 10 μm. The image resolution is 0.2 × 0.2 μm2 per pixel. d) Representative high-resolution (35 × 35 nm2 per pixel) STED images of the nanofiber structures (Cy5.5 fluorescence channel) in tumors, and their corresponding computer-generated drawings for measuring the number and size of the fluorescence using ImageJ Software. The scale bar represents 5 μm. e) A plot of the number and size of the fluorescence in the tumor sections at different time points. The images were first acquired using a high-resolution (35 × 35 nm2 per pixel) STED microscope, which were then analyzed for the size and number using ImageJ software. f) TEM analysis of the tumor sections 48 h after IV injection of GSH-NFP or d-GSH-NFP.
2.4. The Biodistribution of GSH-NFP
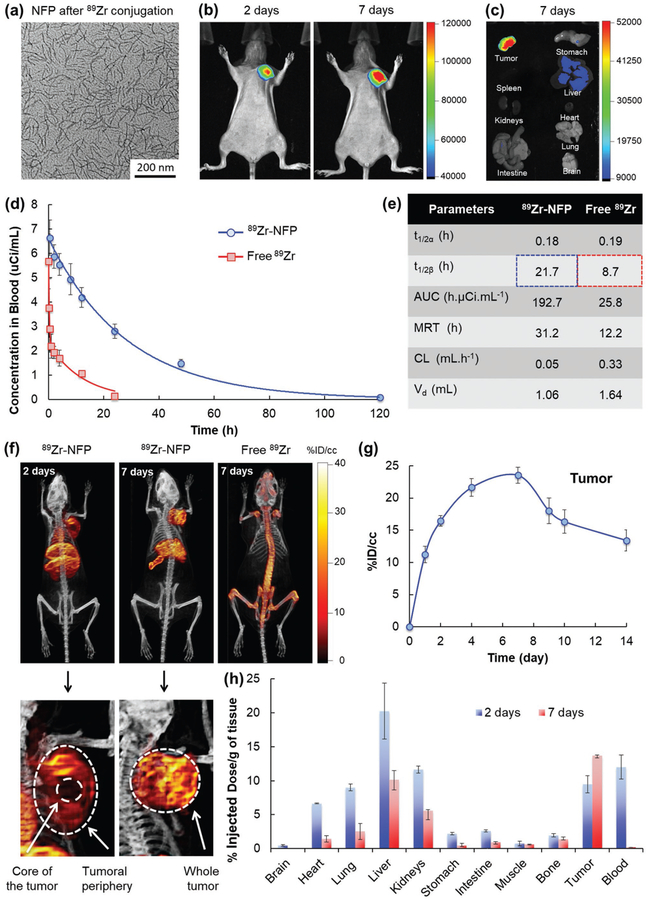
Optical imaging offered a rapid semi-quantitative method for screening the tumoral uptake of GSH-NFP. However, the amount of nanofiber accumulation in organs, e.g., liver and spleen, may be underestimated due to limited light penetration into the tissue, light scattering, nonspecific absorption, and fluorescence quenching by red blood cells. To obtain a quantitative readout, a dually Cyanine5.5 and 89Zr-labeled nanofiber (89Zr-NFP) was prepared for studying the biodistribution of GSH-NFP using multimodal fluorescence/positron emission tomography/computed tomography (fluo/PET/CT) imaging (Figure S5a,b, Supporting Information). Incorporating 89Zr into GHS-NFP neither alters the morphology of the nanofibers nor its tumoral uptake, as confirmed by TEM and optical imaging (Figure 4a-c). The pharmacokinetics of 89Zr-NFP fitted into a two-compartmental model, with a terminal half-life close to 22 h (Figure 4d,e). The long circulation time of the nanofibers also contributed to a progressive tumoral uptake and accumulation over several days. PET/CT imaging showed that 89Zr-NFP accumulated at the tumor periphery and in the main organs such as the heart, the liver, the spleen, and the kidneys 2 days postinjection (Figure 4f). After 7 days, the nanofibers had invaded and saturated the whole tumor (Movie S2, Supporting Information). A plot of the changes in the radioactivity showed that 89Zr-NFP progressively accumulated in tumor overtime and started being washed out slowly after 7 days (Figure 4g). However, the undelivered 89Zr-NFP was found accumulating in liver and spleen (Figure 4f). Further researches are undergoing to promote the hepatic clearance of the nanofibers. To confirm our results, we performed a serial end-point 89Zr-scintillated biodistribution study. Significant radioactivities were observed in different organs collected from animals euthanized 2 days after the injection of 89Zr-NFP, as the nanofibers were still circulating in the blood stream (Figure 4h). The amount of 89Zr-NFP was dramatically reduced in the heart, lungs, liver, and kidneys after 7 days, which suggests that the 89Zr-NFP was being cleared from the organs. No radioactivity could be detected in the blood at this time point. Between 2 and 7 days, the nanofibers were continuously being taken up by the tumors, achieving a peak of 13%ID g−1 that is higher than the previously reported 3–6%ID g−1 of pegylated liposome.[28] Here, we detected a trace amount of radioactivity in the bones over time (1.4% ID g−1) presumably because of the free Zr4+ osteophilic ions, which were liberated after the hepatic metabolism of 89Zr-NFP, strongly bound to the bones (Figure S6a, Supporting Information).[29] We also observed an abnormally high %ID g−1 in the spleen, as a result of a direct radiation-induced shrinkage of the organ (Figure S6b–d, Supporting Information)[30]
Figure 4.
Biodistribution of GSH-NFP. a) TEM analysis of GSH-NFP after DFO-89Zr conjugation. b) Representative fluorescence whole body images of SCID mice bearing MDA-MB-468 tumors (n = 4) 2 and 7 days after IV injection of 89Zr-NFP (100 μCi). c) Representative ex vivo fluorescence image of organs isolated from the animals 7 days after administration of 89Zr-NFP. d) In vivo PK profile and e) the calculated PK parameters of 89Zr-NFP and free 89Zr. (t1/2α = half-live of distribution phase, t1/2β = half-live of elimination phase, AUC = area under the curve, MRT = mean residence time, CL = clearance, Vd = volume of distribution). f) Representative PET/CT whole body images of SCID mice bearing MDA-MB-468 tumors were acquired 2 and 7 days after injection of 100 μCi of 89Zr-NFP or free 89Zr-oxylate as the control (n = 3 per group). g) Quantification of nanofibers (radioactivities) at the region of interest (ROI; tumor) was plotted to reveal the kinetic uptake and the retention properties of GSH-NFP at tumor sites. h) The biodistribution of 89Zr-NFP was determined in separate groups of animals (n = 3 per time point), with results being expressed as a percentage of the injected dose per gram of tissue (%ID/g).
2.5. GSH-NFP is an Effective Drug Carrier
The tumoral uptake, penetration, invasion, and ETR properties of GSH-NFP prompted us to evaluate its application for delivering chemotherapeutic treatments. For proof-of-principle studies, we selected aldoxorubicin (aldox), a derivative of doxorubicin (dox) modified by the introduction of maleimide functional group via a hydrazone linker that is sensitive to acidic conditions (Figure S7a, Supporting Information). Aldox relies on serum albumin as an intrinsic carrier through covalent binding to reach tumor site, which shows a superior clinically efficacy over dox.[31] Instead of albumin as the carrier, we prepared an aldox-loaded GSH-NFP (aldox-NFP) with 18% w/w of drug loading (Figure S7b,c, Supporting Information) to compare the cytotoxicity with free aldox and Doxil, an FDA-approved liposomal formulation of doxorubicin. The incorporation of the drug did not alter the morphology of the NFP (Figure S7d, Supporting Information) and the assembled nanofibers could be freeze-dried for long-term storage and then quickly reconstituted by simply adding water (Figure 5a). Successful drug loading was evidenced by a significant fluorescence quenching of the aldox (Figure 5b), which also allowed us to monitor the drug release according to an increase (recovery) of the fluorescence signal. Drug release from aldox-NFP was pH-dependent, given that the hydrazone linker connecting the nanofiber with the drug was sensitive to acidic conditions.[32] Aldox-NFP releases the active drug metabolite (dox) rapidly because it has a single layer structure with a large surface-to-volume ratio. The drug release kinetics were faster compared to other reported delivery platforms for dox,[33] with more than 80% of the accumulated payloads released within hours at acidic pH 4 (Figure 5c).
Figure 5.
GSH-NFP is an effective drug carrier. a) TEM analysis of aldox-NFP before freeze-drying and after reconstitution. b) Comparison of the fluorescence intensity of aldox-NFP and free aldox, based on the same drug content (10 × 10−6 m). c) Plot showing the percentage of accumulated drug release from the nanofiber (10 × 10−6 m) over time in PBS buffers with different pH values. d) Table showing the IC50 values of aldox-NFP, aldox, and Doxil determined using cell viability assay on 2D and 3D human breast cancer cell cultures. Experiments were performed in triplicate and the data were presented as a mean ± standard deviation. e) Fluorescence images of MDA-MB-468 cells incubated with either a suboptimal concentration of aldox-NFP or aldox (10 × 10−6 m of drug content) for 6 h and washed three times with culture media. Bright field and fluorescence images of the cells were acquired 6 and 24 h later. DAPI (blue) was used to stain the nuclei. The bar scale is 20 μm. f) Microscopic images of spheroids of human ER-positive (MCF-7), HER2-positive (BT474), and triple negative (MDA-MB-231 and MDA-MB-468) breast cancer lines incubated for 72 h with aldox-NFP, aldox, or Doxil (50 × 10−6 m of drug content). The scale bar is 200 μm.
In terms of cytotoxicity, aldox-NFP exhibited a similar potency to the free aldox on different human ER-positive (MCF-7), HER2-positive (BT474), and triple-negative (MDA-MB-231 and MDA-MB-468) breast cancer cells. Comparable half maximal inhibitory concentration (IC50) values were determined by a cell viability assay in 2D cell cultures (Figure 5d; Figure S7e, Supporting Information). The presence of glutathione did not affect the cellular uptake of the nanofibers via endocytosis, as shown by a similar degree of uptake by MDA-MB-468 cells between the naïve NFP and GSH-NFP (Figure S8a, Supporting Information).[19a] Both aldox-NFP and aldox were more effective than Doxil in killing cells, except when tested on a MCF-7 cell line, presumably using NFP as a carrier could enhance the intracellular uptake of aldox (Figure S8b, Supporting Information). Fluorescence microscopic studies confirmed that the drug release from aldox-NFP occurred intracellularly. MDA-MB-468 cells, preincubated with aldox-NFP, showed an increase (dequenching) in dox fluorescence over time, which indicates that the drugs were released from the nanofiber in the acidic lysosomal environment (Figure 5e). The released aldox was able to translocate to the targeted site (nucleus), as shown by colocalized nuclei staining and drug fluorescence. The therapeutic potential of aldox-NFP was highlighted in the 3D cell culture testing. The 3D cell culture provides the optimal environment for cancer cells to form a single organotypic spheroid that mimics the physiology of a tumor.[34] Depending on the tested cell lines, aldox-NFP displayed comparable cytotoxicity to aldox and was 8–50 fold more potent than Doxil (Figure 5d; Figure S7f, Supporting Information). The superior cytotoxicity of aldox-NFP was attributed to its ability to break different breast tumor spheroids into pieces more efficiently over time (Figure 5f; Figure S8c, Supporting Information).
2.6. The Diffusion of GSH-NFP into a Tumor
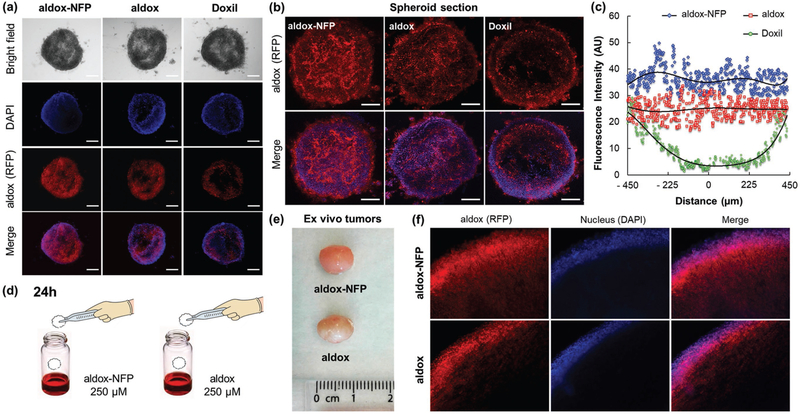
The superior cytotoxicity of aldox-NFP observed in 3D cell culture suggested that NFP could diffuse better into a spheroid compared to Doxil. To confirm this, we assembled a fluorescent aldox-NFP (Figure S7b, Supporting Information) and studied the intraspheroid uptake. Epifluorescence imaging revealed that aldox-NFP distributed evenly inside a spheroid after 3 h of incubation (Figure 6a). We further analyzed the spheroids using multiphoton microscopy (Figure 6b). A plot of the fluorescence intensity with the distance measured from the spheroid center confirmed the intraspheroid distribution of aldox-NFP (Figure 6c). In the case of Doxil, the diffusion was poor. The fluorescence signal only presented around the treated spheroid. To confirm the comparable diffusion of aldox and aldox-NFP, we first incubated tumors that were freshly harvested from the animals in a solution containing aldox or aldox-NFP (Figure 6d) and then prepared the tissue sections for comparison by imaging. As expected, the diffusion depth of aldox and aldox-NFP was similar (Figure 6f). Smaller particles with higher aspect can traverse narrow tumor vasculature via Brownian motion, whereas conventional spherical particles of the same size cannot pass through.[19b,35] The excellent diffusion property of aldox-NFP could be explained by the combined effect of the unique single layer structure and the presence of zwitterionic moiety from GSH, that are known to enhance tumor permeability.[27c,36] Further studies are needed to investigate the parameters that contribute to the tumoral diffusion of aldox-NFP.
Figure 6.
GSH-NFP is able to deliver the drugs inside a tumor. a) Representative fluorescence images of MDA-MB-468 spheroids were acquired 3 h after treatment (10 × 10−6 m of drug content). DAPI (blue) was used as a nuclear counterstain. The scale bar represents 100 μm. b) Multiphoton microscopic images represent the single, middle sections of the treated tumor spheroids. The scale bar represents 100 μm. c) Plots of the diffusion depth of aldox-NFP, aldox, and Doxil into MDA-MB-468 spheroids. d) To compare the tumoral diffusion profile of aldox-NFP and aldox, freshly harvested MDA-MB-468 tumors (isolated from the animals) were dipped into an aldox-NFP or aldox solution (250 × 10−6 m of drug content) for 24 h and then sectioned for histological analysis. e) Photos of the tumors 24 h after immersion into aldox-NFP and aldox solutions. f) Histology sections of the aldox- and aldox-NFP-treated tumors. DAPI (9 × 10−6 m) was added to the samples 1 h prior tumor fixation and sectioning.
2.7. The Therapeutic Efficacy and Toxicity Profile of Aldox-NFP
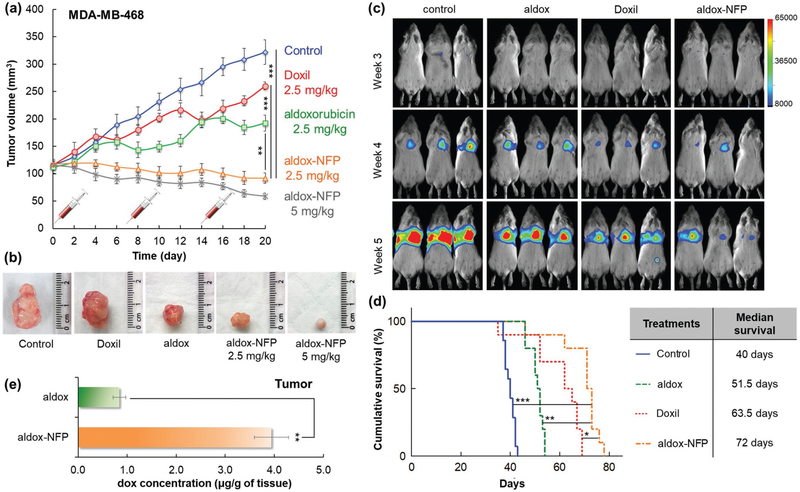
Aldox-NFP does not require a targeting ligand for tumoral delivery, suggesting that it could be an ideal platform for delivery of chemotherapeutics to tumors lacking clinically relevant receptors, such as triple negative breast cancer (TNBC). Thus, we evaluated the therapeutic efficacy of aldox-NFP. We first confirmed that incorporating aldox into GSH-NFP did not affect the tumor-preferential biodistribution (Figure S9a,b; Supporting Information), which suggests that our NFP platform has the desired flexibility for delivering a variety of customized payloads (drugs and imaging agents). We then compared the therapeutic efficacy of aldox-NFP, aldox, and Doxil. When administered at a low dose (2.5 mg kg−1 of drug content once a week for 3 weeks), aldox-NFP was more effective in inhibiting the growth of orthotopically implanted TNBC tumors compared to the free aldox and Doxil (Figure 7a,b). Using a higher dosage of aldox-NFP (5 mg kg−1) for the treatment could further reduce the tumor size. By contrast, Doxil treatment outcome was disappointing; the liposome formulation showed minimal tumor growth inhibition at this posology. The survival benefit of aldox-NFP over aldox and Doxil was shown in animals bearing MDA-MB-231 tumor lesions in the lungs. After confirming the animals developed lung metastasis (Figure S10a-c, Supporting Information), we treated the animals with aldox-NFP, aldox, or Doxil (5 mg kg−1 of drug content) weekly for 3 weeks. Bioluminescence imaging revealed that aldox-NFP treatment could delay the tumor progression in the lungs (Figure 7c), and showed a survival benefit over the free aldox and Doxil with a median survival of 72 days (Figure 7d). The remarkable therapeutic efficacy was attributed to its ability to carry and deliver more drugs to the tumors. A liquid chromatography-mass spectrometry (LC-MS)/mass spectrometry (MS) analysis confirmed that 2 days after treatment, the concentration of active drug metabolite (dox) detected in the tumor xenograft was significantly higher in animals injected with aldox-NFP (Figure 7e).
Figure 7.
Aldox-NFP shows superior in vivo therapeutic efficacy and survival benefit compared to free drug or Doxil. a) Comparison of the volume of MDA-MB-468 tumors in animals treated with PBS (control), Doxil, aldox, or aldox-NFP (n = 6 per group) weekly for three weeks overtime (**P < 0.01, ***p < 0.001). b) Representative photomicrographs of the tumors excised from the animals at the conclusion of treatment. c) Comparison of the progression of MDA-MB-231 tumor lesions in the lungs using bioluminescence imaging. The animals were treated with PBS (control), aldox, Doxil, or aldox-NFP (5 mg kg−1 of drug content) weekly for three weeks and monitored for survival. d) Kaplan-Meier cumulative survival plot of the treated animals (n = 10 per group). The difference in the survival between the mice treated with aldox-NFP and the other animal groups was evaluated using Mantel-Cox log-rank test (*P < 0.05, **P < 0.01, ***P < 0.001). A table listing the median survival time of each drug treatment group. e) The amount of drug delivered to the tumors were determined by LC-MS/MS analysis (**P < 0.01).
2.8. The Toxicity Profile of Aldox-NFP
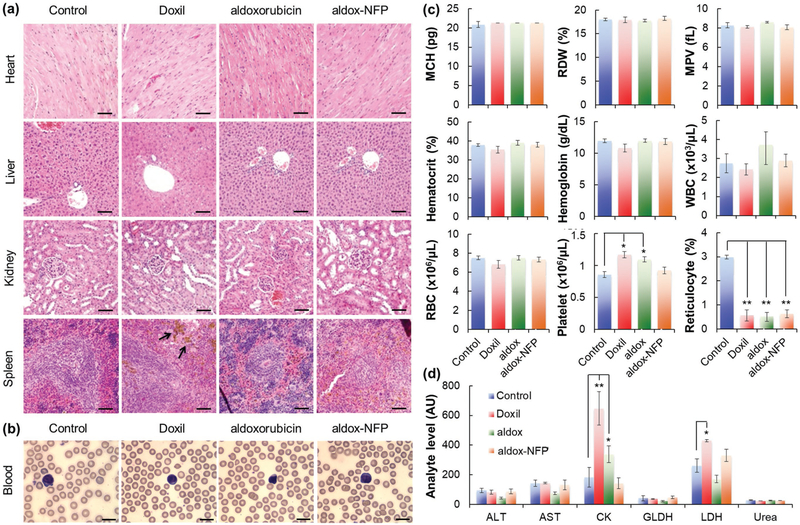
Despite PET scanner revealed the presence of some nanofibers in the major organs of animals treated with aldox-NFP (Figure S9b, Supporting Information), no toxicity was observed after the three-week treatment cycle. Histopathologic studies showed that there was no morphological evidence of injury in the heart, liver, kidneys, or spleen (Figure 8a), but brown hemosiderin granules were observed in the spleens of animals treated with Doxil. It is presumed that the liposomal formulation caused damage to erythrocytes, which accelerated their removal by macrophages in the spleen.[37] To assess acute toxicity, we performed hemograms and a blood biochemical analysis for the liver, renal, and muscle parameters in Balb/c mice after the administration of frequent dosages of aldox-NFP, aldox, and Doxil (2.5 mg kg−1 of drug content three times a week). None of the three treatments changed the number and morphology of the blood cells (Figure 8b). Hemograms corroborated that the drugs did not affect red blood cell number or hemoglobin concentration (Figure 8c). However, the reticulocyte counts decreased in all the animals injected with the three treatments but not the PBS control, which indicates the expected effects on erythropoietic activity (Figure 8c). Unlike aldox and Doxil, aldox-NFP did not increase the platelet or leukocyte counts. We also performed serum biochemistry to evaluate the organ functions of the animals. Neither renal, hepatic, nor muscle toxicity were observed in the animals treated with aldox-NFP. The activities of alanine aminotransferase (ALT), aspartate aminotransferase (AST), and glutamate dehydrogenase (GLDH) (for liver injury), creatine kinase (CK), and lactate dehydrogenase (LDH) (for muscle injury), as well as the concentration of urea nitrogen (for liver function; data not shown), were comparable to the results observed in control animals (Figure 8d). Interestingly, animals treated with Doxil and aldox, both of which are known to exhibit a superior safety profile over dox,[38] were susceptible to muscle injury, and possibly cardiotoxicity, as shown by the increased CK level in the serum. Overall, our results showed that aldox-NFP, compared to aldox and Doxil, displays strong antitumor activity with minimal host toxicity, thus conferring a great potential for triple negative breast cancer treatment.
Figure 8.
Aldox-NFP displays a low in vivo toxicity profile. a) Histopathologic evaluation of the major organs after any treatment. Black arrows indicate brown hemosiderin granules in the spleen. The scale bar represents 50 μm. b–d) Blood was withdrawn from the animals at the conclusion of each treatment for analysis. b) Blood smear test used to evaluate the number and shape of the blood cells. The scale bar represents 10 μm. c) Whole blood biochemistry indexes. Data were presented as mean ± standard deviation (SD). (MCV = mean corpuscular volume, MCH = mean corpuscular hemoglobin, RDW = red cell distribution width, MPV = mean platelet volume, WBC = white blood cell, RBC = red blood cell). d) Serum analyte activity for liver (ALT, AST, GLDH, LDH) or muscle injury (CK, AST, LDH). Data were presented as mean ± standard deviation (SD) (*P < 0.05, **P < 0.01). (ALT = alanine aminotransferase, AST = aspartate aminotransferase, CK = creatine kinase, GLDH = glutamate dehydrogenase, LDH = lactate dehydrogenase).
3. Conclusions
The design of a drug delivery system is complex, and identifying one that can overcome multiple physiological barriers is complicated. We have developed a self-assembling NFP that is customized with multiple components to optimize tumoral delivery efficiency. This well-controlled GSH-NFP system utilizes a combination of shape-controlled tumoral delivery, charge-assisted penetration, and enzyme-induced drug retention approach to improve the safety and efficacy of chemotherapeutic treatment. To the best of our knowledge, there is no other delivery system using this combinatorial approach. The excellent biocompatibility and stability of NFP offers design flexibility in assembling analogues of different lengths, charge distributions, drugs, imaging agents, chemical moieties, amino acid mutations, and functional domains, which allows us to rapidly investigate tumoral uptake through optical imaging and the subsequent in-depth study of biodistribution in real-time through 89Zr-PET/CT imaging. More specifically, this study reveals that the surface charge of NFP plays a substantial role in terms of tumor biodistribution. The optimal formulation, GSH-NFP, was able to escape from the blood vessel, infiltrate tumor, and then transform into larger interfibril networks on site that minimized the washout by the local lymphatic system to significantly improve retention time on site. Importantly, our PET imaging studies showed that GSH-NFP could eventually saturate the entire tumor over time. Unlike other stimuli-responsive organic and inorganic nanocarriers designed to aggregate at the tumor microenvironment to improve the retention on site and play the role of drug-delivery depots,[39] our NFPs can structurally evolve into more than ten times larger interfibril networks via tumor-associated protease activation to prolong local retention for weeks. Overall, the unique combination of tumor uptake, penetration, infiltration, residing, and retention properties confer great potential on advanced GSH-NFP as a carrier of chemotherapeutic agents. We expect that additional advancements to the NFP technology to carry combination therapies for treatment of other cancers and infectious diseases would be entirely feasible.
4. Experimental Section
Chemicals and Supplies:
Rink-amide resin, protected amino acids, and solvents used for peptide synthesis were obtained from Protein Technologies Inc. (Tucson, AZ). Fluorescein isothiocyanate (FITC), trifluoroacetic acid, thioanisole, anisole, 1,2-ethanedithiole, methyl-tert-butyl ether, N,N-diisopropylethylamine, hydrazine, GSH, MA, and 3MA were purchased from Sigma-Aldrich (Saint-Louis, MO). Sulfosuccinimidyl 4-[N-maleimidomethyl]cyclohexane-1-carboxylate (sulfo-SMCC) was obtained from Novachem (Calgary, Canada) and p-SNC-deferoxamine (DFO) was from Macrocyclics Inc. (Plato, TX). Aldoxorubicin was purchased from Medkoo Biosciences (Chapel Hill, NC). Doxorubicin was from LC Laboratories (Woburn, MA), and N2′- deacetyl-N2′-(3-mercapto-1-oxopropyl)-maytansine was supplied by Carbosynth Ltd (Compton, WB). Uranyl formate was purchased from Electron Microscopy Sciences (Hatfield, PA), and 4′6-diaminidino-2-pheylindole (DAPI) and LysoTracker Red were purchased from Life Technologies Inc. (Norwalk, CT). Cyanine5.5 NHS ester was obtained from Lumiprobe Corporation (Hallandale Beach, FL) and luciferin was supplied by Caliper LifeSciences (Hopkinton, MA).
Peptide Synthesis:
Peptides were synthesized using the traditional N-α-Fmoc methodology on rink-amide resin as previously described.[40] After peptide elongation, mPEG was attached to the peptide N-terminus in a solid phase and the (4,4-dimethyl-2,6-dioxo-cyclohexylidene)3-methylbutyl (ivDde) side-chain protection group of C-terminal lysine was selectively deprotected using hydrazine (2% v/v) in dimethylformamide. Subsequently, a fluorophore (FITC or Cyanine5.5 NHS ester), linker (sulfo-SMCC), or chelator (DFO) was conjugated to the peptides, as previously described.[19a] The peptides were obtained after precipitation in methyl-tert-butyl ether and then purified to >98% purity using reverse-phase high performance liquid chromatography (rp-HPLC). All the peptides were characterized by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) analysis (Tufts Medical School, Boston, MA).
Modification of the Peptide Constructs via Conjugation with Specific Groups:
GSH (5 mg, 16 μmol), MA (2.4 mg, 16 μmol), and 3MA (1.7 mg, 16 μmol) were added to the purified sulfo-SMCC-conjugated peptide (30 mg, 8 μmol) in a cosolvent of N-methylpyrrolidone (NMP) (3 mL) and PBS (10 × 10−3 m, pH 7, 3 mL) and allowed to react for 2 days at room temperature. The final GSH-MCC-peptide, MA-MCC-peptide, and 3MA-MCC-peptide constructs were purified by rp-HPLC and characterized with a MALDI-TOF analysis.
NFP Assembly:
To assemble the NFP analogues, a mixture of the peptide constructs in the desired ratios (1 mg total) in dimethyl sulfoxide (DMSO) (20 μL) was added to a cosolvent of acetonitrile and water (1 mL, pH 7). The resulting NFPs were purified by size exclusion chromatography (Sephadex G-25), which were then homogenized into 100 nm in length using a mini-extruder (Avanti Polar Lipids, Alabaster, AL). To synthesize aldox-NFP, aldox (0.18 mg, 0.8 equivalent) was added to the peptide constructs (1 mg total, 1 equivalent) during nanofiber assembling. The concentration of NFP was measured by UV absorbance according to the predetermined extinction coefficient of Cyanine5.5 (209 000 cm−1 m−1) or aldox (13000 cm−1 m−1) in 5% (v/v) PBS in methanol.
Radiochemistry:
89Zr was obtained from the Memorial Sloan-Kettering Cancer Center. 89Zr-oxalate (500 μCi), neutralized with an equivalent volume of sodium carbonate solution (2 m), was mixed with NFP (20 × 10−6 m of DFO content, 250 μL) and incubated at room temperature for 1.5 h to form radiolabeled-nanofibers.[41] The resulting 89Zr-NFP was purified by size exclusion chromatography using Sephadex G-25 gel to remove the free zirconium. Aliquots of 89Zr-NFP (100 μCi, 100–150 μL) were prepared for in vivo injection.
Pharmacokinetic Study:
89Zr-NFP and free 89Zr (10 μCi, 100 μL) were administered to Balb/c mice via tail vein injection (n = 3 per condition). Using retro-orbital sinus puncture technique, blood samples (20 μL) were collected at various time intervals and the radioactivities were measured on Wizard 2 gamma counter (PerkinElmer). The serum halflife and the pharmacokinetic parameters of 89Zr-NFP and free 89Zr were estimated by fitting the data to a compartmental analysis using PKSolver 2.0 software.
Transmission Electron Microscopy (TEM):
NFP (10 × 10−6 m, 20 μL) was dropped onto formvar/carbon-coated 400 mesh copper grids (Electron Microscopy Sciences, Hatfield, PA) and an excess sample was removed with filter paper. After uranyl formate staining (0.5% v/v, 20 μL), the grids were examined under TEM (JEOL JEM-1400 LaB6 TEM operating at 120 kV). For the TEM analysis of the tumor sections, tissues were first fixed in modified Karmovsky’s fix solution (2.5% glutaraldehyde, 4% paraformaldehyde, and 0.02% picric acid in 0.1 m buffer) and then in reduced osmium tetroxide (1% OsO4− and 1.5% K-ferricyanide (aqueous). Following dehydration, the samples were embedded in an Epon analog resin (Embed812) and ultrathin sections (65 nm) were cut using a Diatome diamond-knife on Ultracut T ultramicrotone (Leica Microsystems, Wetzlar, Germany). The sections were contrasted with lead citrate and observed on the JEM 1400 electron microscope.
Atomic Force Microscopy (AFM):
NFP (3 × 10−6 m, 20 μL) were dropped onto a freshly cleaved mica surface. After 2 min, the samples were rinsed twice with deionized water (2 × 1 mL) and air-dried. AFM images were immediately acquired in air, at ambient temperature, on a Multimode 8 atomic force microscope (Brucker, Billerica, MA) operating in PeakForce tapping mode and using a super sharp AFM probe for high resolution imaging (SAA-HPI-SS, Brucker). Data were collected in height channel and topographic images were treated using NanoScope Analysis software.
Cell Lines:
Cell lines were obtained from ATCC (Manassas, VA) and were cultured according to the company’s instructions. All the supplies were purchased from Corning Cellgro Inc. (Tewksbury, MA).
Fluorescence Microscopy:
Cells (5 × 103 per well) were seeded on 8-well chamber slides (Ibidi, Madison, WI) until 75% of confluency was achieved. To study the real time cellular distribution of the nanofibers, NFPs (10 × 10−6 m of peptide content) were incubated with the cells for 6 h at 37 °C, washed twice with PBS, and then imaged using an EVOS FL Auto fluorescence microscope (Life Technology) with the appropriate excitation and emission filters. DAPI (9 × 10−6 m) was added to the culture media for nuclear counterstaining 30 min prior to imaging.
To study the intracellular release of the fluorophores from NFP, the culture media was replaced with fresh media after 6 h of NFP incubation. Subsequently, images were acquired at 0 and 24 h. DAPI (9 × 10−6 m) and LysoTracker Red (1 × 10−6 m) were added to the culture media to stain the nuclei and lysosomes, respectively, for 30 min prior to microscopic imaging.
Cell Viability and Cytotoxicity Assay:
To evaluate the cytotoxicity of aldox-NFP, cancer cells (2 × 103 per well) were seeded in 2D on a flat bottom 96-well plate overnight. Cells were then treated with different concentrations of aldox, Doxil, or aldox-NFP for 72 h. CellTiter Glo reagent was added in each well and the luminescence generated was recorded using a microplate reader (Tecan US Inc., Morrisville, NC). The dose response curves were plotted using Graph Pad Prism 6.0 software, and the half maximal inhibitory concentrations (IC50 values) were calculated.
To prepare the 3D cell cultures, cancer cells were suspended in RPMI 1640, supplemented with 2.5% of matrigel matrix basement membrane, which were then seeded on an ultralow attachment black with clear round bottom 96-well plates (104 cells per well). After centrifugation (10 min, 1000 rpm), the cells were subjected to further incubation for 72 h to form spheroids, as previously reported.[42] Cytotoxicity was then evaluated as described above using the CellTiter Glo 3D cell viability assay (Promega, Madison, WI). 3D spheroids were also imaged using EVOS FL Auto fluorescence microscope (Life Technology) and FluoView FV1000MPE Multiphoton microscope (Olympus). Images were acquired 3 h after incubation with aldox-NFP, aldox, and Doxil (10 × 10−6 m of drug content). DAPI (9 × 10−6 m) was added to the culture media for nuclear counterstaining 1 h prior to imaging.
In Vitro Drug Release Study:
aldox-NFP (10 × 10−6 m of drug content) in a PBS buffer of different pH was added to a 96-well plate (200 μL per well) to monitor the drug release from the nanofiber through the dequenching of dox fluorescence intensity. The fluorescence intensity of the samples was measured at excitation and emission wavelengths of 538 and 580 nm, respectively, using the microplate reader (Tecan US Inc., Morrisville, NC) the experiments were performed in triplicate.
Tumor Xenografts:
Five-to seven-weeks old female SHO mice (Charles River Laboratories, Wilmington, MA) were housed in a pathogen-free barrier room in the Animal Core Facility at WCM. All the procedures conducted on the mice were approved by the Weill Cornell Medical Center Institutional Animal Care and Use Committee (#2014–0030, #2014–0005, and #2015–0014) and were consistent with the recommendations of the American Veterinary Medical Association and the National Institutes of Health Guide for the Care and Use of Laboratory Animals. The mice were allowed to acclimate for at least 3 days and were fed with an AIN93 diet (Harlan Laboratories, Indianapolis, IN). MDA-MB-468 or MDA-MB-231 cells (7.5 × 106) suspended in PBS (150 μL) were inoculated into the mammary fat pads of the animals. Tumor growth was monitored weekly using a caliper. The tumor volume (V) was calculated according to the formula, V = (L ×I2)/2, where L is the longest diameter and I is the shortest perpendicular diameter.
Fluorescence Imaging:
When the tumor had progressed to 150 mm3, the animals were randomly assigned into six groups and were given Cyanine5.5 or NFP analogues (0.5 nmol of Cy5.5 content) in a PBS buffer (150 μL) via tail vein injections (n = 3 per group). Real-time optical imaging was performed using In vivo Xtreme imaging system (Bruker, Billerica, MA). Whole body fluorescence images were acquired at different time intervals using the appropriate excitation (670 nm) and emission (750 nm) filters. Bruker MI software was used to process the images and measure the fluorescence intensity in different region of interest (ROI). All the data were corrected using the organ autofluorescence measured prior to the treatment. The animals were euthanized and the organs were excised and washed with PBS prior to imaging in order to study end-point biodistribution.
Survival Study:
An experimental animal model was established in NOD scid gamma (NSG) mice (Jackson Laboratory, Bar Harbor, ME) by injecting MDA-MB-231 cancer cells that are transfected with plasmids carrying both luciferase and green fluorescent protein (GFP) genes (0.25 × 106 cells per animal) via the tail vein. All the animals developed tumor metastasis in the lungs over three weeks, as confirmed by bioluminescence imaging. The images were acquired using In vivo Xtreme imaging system (Bruker, Billerica, MA) 15 min after injection of the light-emitting luciferin compound (100 μL; 30 mg mL−1) via intraperitoneal (IP) injection. Mice were weekly treated with PBS (control), aldox, Doxil, or aldox-NFP (5 mg kg−1 of drug content) for three weeks and further maintained and monitored to record the survival.
PET Imaging:
To study the specific tumoral uptake and organ distribution of GSH-NFP, SCID mice bearing MDA-MB-468 tumors (n = 4) were injected with 89Zr-labeled NFP (100 μCi) via the tail vein. PET images were then acquired after 2 and 7 days, using an Inveon μPET/CT scanner (Siemens Medical Solutions, Malvern, PA). PET/CT maximum energy projections were processed with Amide v1.0.4 and Inveon Research Workplace software. Scintillation measurements were taken using a Wallac Wizard gamma counter. The mice were sacrificed by cervical dislocation before their tissues were harvested. PBS perfusion was not performed prior to tissue harvesting. The results were expressed as a percentage of the injected dose per gram of tissue (% ID g−1).
Confocal Microscopy:
The excised tumors were embedded in optimal cutting temperature (OCT) compound and freshly cut. Five micrometers sections were stained for immunofluorescence on a Leica Bond RX (Leica Biosystems, Buffalo Grove, IL) with anti-vimentin mouse monoclonal (0.1 μg mL−1) for 1 h and using 10 min of 1:200 Tyramide Alexa Fluor488 detection (Life Technologies, Carlsbad, CA). The sections were pretreated with a Leica Bond ER2 Buffer for 20 min at 100 °C before each staining. The sections were then mounted with Mowiol antifade mounting media for digital scanning with Pannoramic Confocal (3dHistech Ltd.) using 40× water objective.
Confocal and STED Microscopy:
Freshly harvested tumors were immediately embedded in OCT compound and frozen on dry ice prior sectioning. Tumor sections were stained by immunohistochemistry for CD31 (blood vessel) and DAPI (nucleus) on a Leica Bond RX automated staining platform (Leica Biosystems, Buffalo Grove, IL). The tumor sections were then observed using a Leica TCS SP8 STED 3× microscope (Leica Microsystems, Wetzlar, Germany). All images were acquired using 100× oil objective. STED 775 nm laser (Cy5.5 fluorescence channel) was employed for imaging the nanofibers (red). The nuclei (DAPI) and blood vessels (FITC-CD31) were imaged using only confocal setting. The images were analyzed using the Leica Application Suite X (LAS X) software. Computer-generated drawings from the STED images were used for analyzing the number and the area of the GSH-NFP structures (Cyanine5.5. fluorescence) using ImageJ software (n = 13 images per condition).
Antitumor Effect:
Mice bearing 120 mm3 MDA-MB-468 tumors[43] were randomly assigned into four groups and treated with PBS (control), aldox-NFP, free aldox, or doxil (n = 6 per group) via tail vein injections (2.5 or 5 mg of active pharmacophore kg−1) weekly for 3 weeks. The tumor size was measured with calipers every two days. At the conclusion of the experiment, the animals were euthanized and the tumors and organs were harvested, washed with PBS, and preserved in formaldehyde (10% in PBS v/v) for histopathological analysis.
Tissue Distribution of Doxorubicin Metabolites Released from Aldox-NFP:
The quantification of doxorubicin was performed with liquid chromatography-mass spectrometry analysis. Briefly, tumors were excised from nude mice bearing MDA-MB-468 tumors (n = 4 per condition) 48 h after the administration of aldox and aldox-NFP (2.5 mg kg−1) and then homogenized in 20% (v/v) acetic acid in acetonitrile (100 mg of tissue in 1 mL of extraction solution). Daunorubicin (Sigma, Saint-Louis, MO) was added to each sample mixture (1 μg mL−1) as an internal standard. After 16 h of extraction at 4°C, the samples were centrifuged at 10 000 rpm for 10 min and the organic phase containing the doxorubicin metabolites was collected for analysis by LC-MS/MS.
Histologic and Clinical Pathologic Analysis:
For histologic analysis, the excised tumors and main organs of interest (heart, liver, kidneys, and spleen) in formaldehyde (10%) were dehydrated with ethanol, embedded in paraffin, and then slides containing 5 μm sections were prepared. The slides were subsequently stained with hematoxylin and eosin Y solution (H&E) to assess histological alterations via light microscopy conducted by a single blinded pathologist. The blood samples were collected via retro-orbital sinus puncture into no anticoagulant or 3.2% citrate anticoagulant for the analysis of biochemical analytes (renal tests, liver, and muscle enzyme activity) and hemograms, respectively. The analysis was conducted in the Clinical Pathology Laboratory in the Animal Health Diagnostic Center at Cornell University using automated equipment (Hitachi P modular chemistry analyzer, Roche Diagnostics, Indianapolis, IN, USA and ADVIA 2120 hematology analyzer, Siemens Healthcare Diagnostics Inc., Tarrytown, NJ) using manufacturer reagents, with the exception of GLDH (Randox Laboratories Ltd, Antrim, UK).
Immunogenicity Assay:
The concentration of the innate immune and inflammatory cytokines IL1β, IL2, IL6, IL10, TNF-α, and INF-γ in serum was measured with commercial sandwich enzyme-linked immunosorbent assay kits (Invitrogen, Carlsbad, CA). ELISA assays were performed following the manufacturer’s instructions to measure the cytokine release in serum collected from female Balb/c mice (n = 3 per group) 24 and 48 h post IV injection of 5 mg kg−1 of NFP or lipopolysaccharide (5 mg kg−1) used as a positive control. Mice injected with PBS were used as a negative control.
Supplementary Material
Acknowledgements
Research reported in this publication was supported in part by the National Center for Advancing Translational Sciences (Grant No. UL1TR000457), National Cancer Institute (Grant Nos. R03 CA219718 and R01 CA222802 to B.L.), and Radiology Department, WCM. The authors would like to thank the Memorial Sloan Kettering Radiochemistry and Molecular Imaging Probe Core Facility for supplying 89Zr-oxylate. The production of the radioisotope at MSKCC was supported in part through the NIH/NIC Cancer Center Support Grant (P30 CA008748). The authors also thank Mesru Turkekul and Yevgeniy Romin from the Molecular Cytology Facility (MSKCC) for help with confocal imaging of the tumor and the Optical Microscopy and Imaging Core Facility (Weill Cornell Medicine) for help with TEM and multiphoton microscopy analyses. The authors finally acknowledge the Laboratory of Comparative Pathology Core Facility (MSKCC) partially funded by NCI grant P30 CA008748 for the preparation of the tumor sections for histological and fluorescence analyses as well as the Imaging and Surface Science Cores Facility at Advanced Science Research Center of the City University of New York (CUNY) for help with STED microscopic and AFM imaging.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Conflict of Interest
The authors declare no conflict of interest.
Contributor Information
Dr. Vanessa Bellat, Molecular Imaging Innovations Institute, Department of Radiology, Weill Cornell Medicine, 413 East 69th Street, New York, NY 10021, USA, sbl2004@med.cornell.edu
Dr. Richard Ting, Molecular Imaging Innovations Institute, Department of Radiology, Weill Cornell Medicine, 413 East 69th Street, New York, NY 10021, USA, sbl2004@med.cornell.edu
Dr. Teresa L. Southard, Department of Biomedical Sciences, College of Veterinary Medicine, Cornell University, Upper Tower Road, Ithaca, New York, NY 14853, USA
Dr. Linda Vahdat, Breast Medicine, Memorial Sloan-Kettering Cancer Center, 300 East 66th Street, New York, NY 10065, USA
Dr. Henrik Molina, Proteomic Resource Center, Rockefeller University, 1230 York Avenue, New York, NY 10065, USA
Dr. Joseph Fernandez, Proteomic Resource Center, Rockefeller University, 1230 York Avenue, New York, NY 10065, USA
Dr. Omer Aras, Department of Radiology, Memorial Sloan-Kettering Cancer Center, 1275 York Avenue, New York, NY 10065, USA
Dr. Tracy Stokol, Department of Population Medicine and Diagnostic Sciences, College of Veterinary Medicine, Cornell University, Upper Tower Road, Ithaca, New York, NY 14853, USA
Dr. Benedict Law, Molecular Imaging Innovations Institute, Department of Radiology, Weill Cornell Medicine, 413 East 69th Street, New York, NY 10021, USA, sbl2004@med.cornell.edu
References
- [1]a).Baeza A, Manzano M, Colilla M, Vallet-Regi M, Biomater. Sci 2016, 4, 803; [DOI] [PubMed] [Google Scholar]; b) Maeda H, Nakamura H, Fang J, Adv. Drug Delivery Rev 2013, 65, 71; [DOI] [PubMed] [Google Scholar]; c) Na JH, Koo H, Lee S, Han SJ, Lee KE, Kim S, Lee H, Lee S, Choi K, Kwon IC, Kim K, Mol. Pharm 2016, 13, 3700; [DOI] [PubMed] [Google Scholar]; d) Xiong H, Zhou D, Qi Y, Zhang Z, Xie Z, Chen X, Jing X, Meng F, Huang Y, Biomacromolecules 2015, 16, 3980. [DOI] [PubMed] [Google Scholar]
- [2] a).Dobrovolskaia MA, McNeil SE, Nat. Nanotechnol 2007, 2, 469; [DOI] [PubMed] [Google Scholar]; b) Nagesetti A, McGoron AJ, Colloids Surf., B 2016, 147, 492; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Shao D, Lu MM, Zhao YW, Zhang F, Tan YF, Zheng X, Pan Y, Xiao XA, Wang Z, Dong WF, Li J, Chen L, Acta Biomater 2017, 49, 531. [DOI] [PubMed] [Google Scholar]
- [3] a).Maeda H, Adv. Drug Delivery Rev 2015, 91, 3; [DOI] [PubMed] [Google Scholar]; b) Nichols JW, Bae YH, J. Controlled Release 2014, 190, 451; [DOI] [PubMed] [Google Scholar]; c) Prabhakar U, Maeda H, Jain RK, Sevick-Muraca EM, Zamboni W, Farokhzad OC, Barry ST, Gabizon A, Grodzinski P, Blakey DC, Cancer Res 2013, 73, 2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Xue HY, Liu S, Wong HL, Nanomedicine 2014, 9, 295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5] a).Jiang SH, Sun YA, Cui X, Huang X, He Y, Ji S, Shi W, Ge DT, Synth. Met 2013, 163, 19; [Google Scholar]; b) Mukherjee S, Ray S, Thakur RS, Indian J. Pharm. Sci 2009, 71, 349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6] a).Joo KI, Xiao L, Liu S, Liu Y, Lee CL, Conti PS, Wong MK, Li Z, Wang P, Biomaterials 2013, 34, 3098; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Liu KC, Yeo Y, Arch. Pharm. Res 2014, 37, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7] a).Baumhover NJ, Duskey JT, Khargharia S, White CW, Crowley ST, Allen RJ, Rice KG, Mol. Pharm 2015, 12, 4321; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Jokerst JV, Lobovkina T, Zare RN, Gambhir SS, Nanomedicine 2011, 6, 715; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Mishra P, Nayak B, Dey RK, Asian J. Pharm. Sci 2016, 11, 337. [Google Scholar]
- [8] a).Huang KY, Ma HL, Liu J, Huo SD, Kumar A, Wei T, Zhang X, Jin SB, Gan YL, Wang PC, He ST, Zhang XN, Liang XJ, ACS Nano 2012, 6, 4483; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Huo S, Ma H, Huang K, Liu J, Wei T, Jin S, Zhang J, He S, Liang XJ, Cancer Res 2013, 73, 319; [DOI] [PubMed] [Google Scholar]; c) Stylianopoulos T, Poh MZ, Insin N, Bawendi MG, Fukumura D, Munn LL, Jain RK, Biophys. J 2010, 99, 1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9] a).Sugahara KN, Teesalu T, Karmali PP, Kotamraju VR, Agemy L, Greenwald DR, Ruoslahti E, Science 2010, 328, 1031; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Shin MC, Zhang J, Min KA, Lee K, Byun Y, David AE, He HN, Yang VC, J. Biomed. Mater. Res., Part A 2014, 102, 575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kong SD, Zhang W, Lee JH, Brammer K, Lal R, Karin M, Jin S, Nano Lett 2010, 10, 5088. [DOI] [PubMed] [Google Scholar]
- [11] a).Ji T, Li S, Zhang Y, Lang J, Ding Y, Zhao X, Zhao R, Li Y, Shi J, Hao J, Zhao Y, Nie G, ACS Appl. Mater. Interfaces 2016, 8, 3438; [DOI] [PubMed] [Google Scholar]; b) Pandey A, Sarangi S, Chien K, Sengupta P, Papa AL, Basu S, Sengupta S, Nanotechnology 2014, 25, 445101. [DOI] [PubMed] [Google Scholar]
- [12] a).Greish K, J. Drug Targeting 2007, 15, 457; [DOI] [PubMed] [Google Scholar]; b) Greish K, Methods Mol. Biol. (N. Y.) 2010, 624, 25; [DOI] [PubMed] [Google Scholar]; c) Leu AJ, Berk DA, Lymboussaki A, Alitalo K, Jain RK, Cancer Res 2000, 60, 4324. [PubMed] [Google Scholar]
- [13] a).Ganta S, Devalapally H, Shahiwala A, Amiji M, J. Controlled Release 2008, 126, 187; [DOI] [PubMed] [Google Scholar]; b) Mura S, Nicolas J, Couvreur P, Nat. Mater 2013, 12, 991. [DOI] [PubMed] [Google Scholar]
- [14] a).Cai XL, Luo YN, Zhang WY, Du D, Lin YH, ACS Appl. Mater. Interfaces 2016, 8, 22442; [DOI] [PubMed] [Google Scholar]; b) Dong Z, Feng L, Zhu W, Sun X, Gao M, Zhao H, Chao Y, Liu Z, Biomaterials 2016, 110, 60; [DOI] [PubMed] [Google Scholar]; c) Han L, Tang C, Yin C, ACS Appl. Mater. Interfaces 2016, 8, 23498; [DOI] [PubMed] [Google Scholar]; d) Liu N, Han JM, Zhang XC, Yang Y, Liu Y, Wang YM, Wu GL, Colloids Surf., B 2016, 145, 401. [DOI] [PubMed] [Google Scholar]
- [15].Wilhelm S, Tavares AJ, Dai Q, Ohta S, Audet J, Dvorak HF, Chan WCW, Nat. Rev. Mater 2016, 1, 16014. [Google Scholar]
- [16].Zhang WS, Yu XQ, Li Y, Su ZQ, Jandt KD, Wei G, Prog. Polym. Sci 2018, 80, 94. [Google Scholar]
- [17].Wei G, Su Z, Reynolds NP, Arosio P, Hamley IW, Gazit E, Mezzenga R, Chem. Soc. Rev 2017, 46, 4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Su ZQ, Shen HY, Wang HX, Wang JH, Li JF, Nienhaus GU, Shang L, Wei G, Adv. Funct. Mater 2015, 25, 5472. [Google Scholar]
- [19] a).Bellat V, Lee HH, Vahdat L, Law B, Biomacromolecules 2016, 17, 2040; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wagh A, Singh J, Qian S, Law B, Nanomedicine 2013, 9, 449. [DOI] [PubMed] [Google Scholar]
- [20] a).Law B, Tung CH, Biomacromolecules 2008, 9, 421; [DOI] [PubMed] [Google Scholar]; b) Law B, Quinti L, Choi Y, Weissleder R, Tung CH, Mol. Cancer Ther 2006, 5, 1944. [DOI] [PubMed] [Google Scholar]
- [21] a).Chakraborty S, Dhakshinamurthy GS, Misra SK, J. Biomed. Mater. Res., Part A 2017, 105, 2906; [DOI] [PubMed] [Google Scholar]; b) Duan XP, Li YP, Small 2013, 9, 1521;23019091 [Google Scholar]; c) Gong NQ, Chen SZ, Jin SB, Zhang JC, Wang PC, Liang XJ, Regener. Biomater 2015, 2, 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Nel AE, Madler L, Velegol D, Xia T, Hoek EM, Somasundaran P, Klaessig F, Castranova V, Thompson M, Nat. Mater 2009, 8, 543. [DOI] [PubMed] [Google Scholar]
- [23] a).Campbell RB, Fukumura D, Brown EB, Mazzola LM, Izumi Y, Jain RK, Torchilin VP, Munn LL, Cancer Res 2002, 62, 6831; [PubMed] [Google Scholar]; b) Krasnici S, Werner A, Eichhorn ME, Schmitt-Sody M, Pahernik SA, Sauer B, Schulze B, Teifel M, Michaelis U, Naujoks K, Dellian M, Int. J. Cancer 2003, 105, 561. [DOI] [PubMed] [Google Scholar]
- [24] a).Kang YJ, Park DC, Shin HH, Park J, Kang S, Biomacromolecules 2012, 13, 4057; [DOI] [PubMed] [Google Scholar]; b) Alavizadeh SH, Akhtari J, Badiee A, Golmohammadzadeh S, Jaafari MR, Expert Opin. Drug Delivery 2016, 13, 325. [DOI] [PubMed] [Google Scholar]
- [25] a).Vinluan RD 3rd, Liu J, Zhou C, Yu M, Yang S, Kumar A, Sun S, Dean A, Sun X, Zheng J, ACS Appl. Mater. Interfaces 2014, 6, 11829; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zhang XD, Luo ZT, Chen J, Song SS, Yuan X, Shen X, Wang H, Sun YM, Gao K, Zhang LF, Fan SJ, Leong DT, Guo ML, Xie JP, Sci. Rep 2015, 5, 8669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Malik R, Qian S, Law B, Anal. Biochem 2011, 412, 26. [DOI] [PubMed] [Google Scholar]
- [27] a).Kim B, Han G, Toley BJ, Kim CK, Rotello VM, Forbes NS, Nat. Nanotechnol 2010, 5, 465; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Tian L, Lu L, Qiao Y, Ravi S, Salatan F, Melancon MP, J. Funct. Biomater 2016, 7, E19; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Zhang YJ, Chen WZ, Yang CC, Fan QL, Wu W, Jiang XQ, J. Controlled Release 2016, 237, 115. [DOI] [PubMed] [Google Scholar]
- [28] a).Petersen AL, Henriksen JR, Binderup T, Elema DR, Rasmussen PH, Hag AM, Kjaer A, Andresen TL, Eur. J. Nucl. Med. Mol. Imaging 2016, 43, 941; [DOI] [PubMed] [Google Scholar]; b) Abou DS, Thorek DL, Ramos NN, Pinkse MW, Wolterbeek HT, Carlin SD, Beattie BJ, Lewis JS, Pharm. Res 2013, 30, 878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29] a).Chang AJ, Desilva R, Jain S, Lears K, Rogers B, S. Lapi, Pharmaceuticals 2012, 5, 79; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Chen F, Goel S, Valdovinos HF, Luo H, Hernandez R, Barnhart TE, Cai W, ACS Nano 2015, 9, 7950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Trip AK, Sikorska K, van Sandick JW, Heeg M, Cats A, Boot H, Jansen EP, Verheij M, Radiother. Oncol 2015, 116, 239. [DOI] [PubMed] [Google Scholar]
- [31] a).Chawla SP, Chua VS, Hendifar AF, Quon DV, Soman N, Sankhala KK, Wieland DS, Levitt DJ, Cancer - Am. Cancer Soc 2015, 121, 570; [DOI] [PubMed] [Google Scholar]; b) Chawla SP, Papai Z, Mukhametshina G, Sankhala K, Vasylyev L, Fedenko A, Khamly K, Ganjoo K, Nagarkar R, Wieland S, Levitt DJ, JAMA Oncol 2015, 1, 1272. [DOI] [PubMed] [Google Scholar]
- [32] a).Prabaharan M, Grailer JJ, Pilla S, Steeber DA, Gong S, Biomaterials 2009, 30, 5757; [DOI] [PubMed] [Google Scholar]; b) Quader S, Cabral H, Mochida Y, Ishii T, Liu X, Toh K, Kinoh H, Miura Y, Nishiyama N, Kataoka K, J. Controlled Release 2014, 188, 67. [DOI] [PubMed] [Google Scholar]
- [33].Xu R, Zhang G, Mai J, Deng X, Segura-Ibarra V, Wu S, Shen J, Liu H, Hu Z, Chen L, Huang Y, Koay E, Huang Y, Liu J, Ensor JE, Blanco E, Liu X, Ferrari M, Shen H, Nat. Biotechnol 2016, 34, 414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Abbott A, Nature 2003, 424, 870. [DOI] [PubMed] [Google Scholar]
- [35] a).Smith BR, Kempen P, Bouley D, Xu A, Liu Z, Melosh N, Dai H, Sinclair R, Gambhir SS, Nano Lett 2012, 12, 3369; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Mier W, Babich J, Haberkorn U, Eur. J. Nucl. Med. Mol. Imaging 2014, 41, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Choi HS, Gibbs SL, Lee JH, Kim SH, Ashitate Y, Liu F, Hyun H, Park G, Xie Y, Bae S, Henary M, Frangioni JV, Nat. Biotechnol 2013, 31, 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wang Y, Juan LV, Ma X, Wang D, Ma H, Chang Y, Nie G, Jia L, Duan X, Liang XJ, Curr. Drug Metab 2010, 11, 507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38] a).O’Brien MER, Wigler N, Inbar M, Rosso R, Grischke E, Santoro A, Catane R, Kieback DG, Tomczak P, Ackland SP, Orlandi F, Mellars L, Alland L, Tendler C, C. B. C. S. Grp, Ann. Oncol 2004, 15, 440; [DOI] [PubMed] [Google Scholar]; b) Lebrecht D, Geist A, Ketelsen UP, Haberstroh J, Setzer B, Kratz F, Walker UA, Int. J. Cancer 2007, 120, 927. [DOI] [PubMed] [Google Scholar]
- [39] a).Hu Q, Sun W, Lu Y, Bomba HN, Ye Y, Jiang T, Isaacson AJ, Gu Z, Nano Lett 2016, 16, 1118; [DOI] [PubMed] [Google Scholar]; b) Ruan S, Hu C, Tang X, Cun X, Xiao W, Shi K, He Q, Gao H, ACS Nano 2016, 10, 10086; [DOI] [PubMed] [Google Scholar]; c) Ai X, Ho CJ, Aw J, Attia AB, Mu J, Wang Y, Wang X, Wang Y, Liu X, Chen H, Gao M, Chen X, Yeow EK, Liu G, Olivo M, Xing B, Nat. Commun 2016, 7, 10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Law B, Weissleder R, Tung CH, Bioconjugate Chem 2007, 18, 1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zeglis BM, Lewis JS, J. Visualized Exp 2015, 96, e52521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ivascu A, Kubbies M, J Biomol Screen 2006, 11, 922. [DOI] [PubMed] [Google Scholar]
- [43].Morton CL, Houghton PJ, Nat. Protoc 2007, 2, 247. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.