Abstract
We propose that phosphoproteomic-based studies will radically advance our knowledge about exercise-regulated signaling events. However, these studies use cutting-edge technologies that can be difficult for nonspecialists to understand. Hence, this review is intended to help nonspecialists 1) understand the fundamental technologies behind phosphoproteomic analysis and 2) use various bioinformatic tools that can be used to interrogate phosphoproteomic datasets.
Keywords: phosphopeptide, phosphorylation, mass spectrometry, liquid chromatography, bioinformatics
INTRODUCTION
Skeletal muscle exhibits remarkable plasticity and can rapidly adapt to changes in functional demand. For instance, repeated bouts of endurance exercise induce mitochondrial biogenesis and a concomitant increase in oxidative capacity. In contrast, repeated bouts of resistance exercise do not promote substantial changes in oxidative capacity but instead induce an increase in muscle mass and strength (1). Although the physiological and biochemical adaptations that occur in response to these distinct types of exercise have been well defined, the molecular mechanisms that drive the exercise-specific adaptations remain far from understood.
For skeletal muscle to undergo exercise-specific adaptations, it must both sense and transmit the information that is unique to each type of exercise stimulus. For the most part, the transmission of this information is mediated by signaling pathways that rely on the use of rapidly reversible posttranslational modifications to transmit information (e.g., phosphorylation, ubiquitination, acetylation, etc.). Phosphorylation is a considerably pervasive mechanism for signal transduction in the cell. In fact, current estimates indicate that more than 75% of all proteins are phosphorylated at some point during their life cycle, and phosphorylation-dependent signaling pathways have been implicated in the regulation of nearly all cellular processes (2,3). Thus, a comprehensive understanding of the phosphorylation events that are regulated by exercise (i.e., the exercise-regulated phosphoproteome) should provide critical insight into the mechanisms that drive exercise-specific adaptations.
Over the past 30 yr, the number of studies that have assessed the effects of exercise on phosphorylation-dependent signaling events has increased exponentially. For the most part, these studies have relied on the use of antibodies that allow for a targeted investigation of phosphorylation sites with known functions. With the rapid evolution of mass spectrometry (MS), we now have experimental evidence for nearly 300,000 unique phosphorylation sites in mammalian cells; however, only 134 of these are currently annotated with the term exercise (4). Thus, our knowledge about the potentially vast array of exercise-regulated phosphorylation events remains extremely limited. Indeed, using MS-based phosphoproteomics, Hoffman et al. (5) identified over 1000 different phosphorylation sites that were significantly altered by a bout of high-intensity aerobic exercise in human skeletal muscle. Likewise, a recent MS-based phosphoproteomics study from our lab led to the identification of nearly 700 different phosphorylation sites that were significantly altered by a bout of maximal intensity contractions in mouse skeletal muscle, and remarkably, only six of these sites had previously been annotated with the term exercise (6).
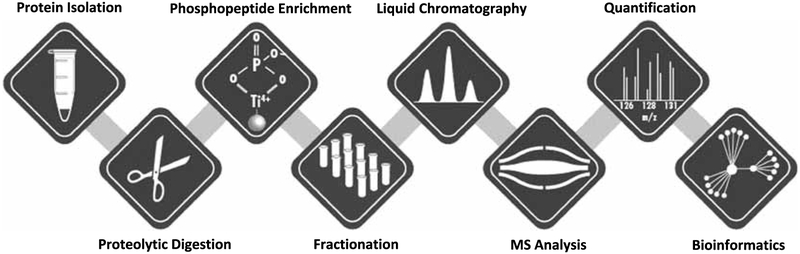
As illustrated by the aforementioned studies, advancements in the field of MS-based phosphoproteomics have made it possible to perform unbiased analyses on thousands of different phosphorylation events within a single experiment. This is an important point because MS-based phosphoproteomic technologies are rapidly becoming more accessible to exercise scientists. Furthermore, the National Institutes of Health (NIH) recently slated approximately $170 million for investment into the Molecular Transducers of Physical Activity Consortium (MoTrPAC), which will include extensive cataloging of the phosphoproteomic alterations that occur in skeletal muscle after different modes of exercise. Thus, it is our hypothesis that forthcoming MS-based phosphoproteomic studies will radically advance our understanding of exercise-induced signaling events. However, the technologies involved in MS-based phosphoproteomics are complex, and it is often very difficult for nonspecialists to readily interpret and use the resulting data. Therefore, the goal of this article is to assist exercise scientists in developing a fundamental understanding of MS-based phosphoproteomic technologies and the various bioinformatics approaches that can be used to extract biologically relevant information from phosphoproteomic datasets (Fig. 1).
Figure 1.
Overview of the core methodologies that are used in an mass spectrometry (MS)-based phosphoproteomic experiment.
THE FUNDAMENTAL TECHNIQUES OF MS-BASED PHOSPHOPROTEOMICS
Liquid Chromatography Tandem MS
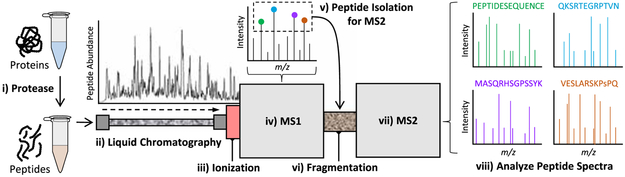
To appreciate the specific techniques that are used in MS-based phosphoproteomics, one must first have a basic understanding of the core analytical technology. In the vast majority of studies, the core technology is described as using a “shotgun/ bottom-up” approach, in which a complex mixture of peptides are analyzed in a large-scale, discovery-centric strategy. This objective is typically accomplished by coupling liquid chromatography (LC) with tandem MS (LC–tandem MS [LC-MS/MS]).
Figure 2 summarizes this shotgun proteomics strategy in eight steps. (i) A protease, typically trypsin, cleaves the sample proteins into peptides. (ii) The complex peptide mixture is loaded onto a reversed-phase LC (RPLC) column, which, upon application of an elution gradient, separates the peptides according to their hydrophobicity. (iii) As the peptides elute from the LC column, an ionization source converts them into a gas phase and directs them into the MS. (iv) The MS measures the mass-to-charge ratio (m/z) of all the eluting “parent” peptide ions about once per second (MS1), thereby creating m/z spectral records of the parent peptides throughout the course of the elution period. (v) The most intense/abundant parent peptides identified in MS1 are isolated for further analysis. (vi) The isolated peptides undergo an additional fragmentation process. (vii) The fragmentation products are analyzed during a second MS scan (MS2). Steps (iii)–(vii) are continually repeated as peptides elute from the LC column. (viii) The m/z spectra for each peptide analyzed in MS2 serve as a sequence-specific fingerprint, allowing for determination of the precursor peptide’s primary amino acid sequence and knowledge of any associated posttranslational modifications.
Figure 2.
Typical steps in the shotgun analysis of peptides with liquid chromatography (LC) with tandem MS (LC–tandem MS [LC-MS/MS]). m/z, mass-to-charge ratio.
When considering a proteomics experiment, one should be mindful of three critical points: First, most samples comprise hundreds of thousands of unique peptide species. Second, the number of peptides that can be detected and quantified during an MS analysis is finite and proportional to the experiment duration. Third, peptides are selected for MS2 by order of decreasing intensity, and, therefore, low-intensity/abundance peptides often are not sampled. In the subsequent sections, we will describe how these points influence the individual steps that go into a large-scale phosphoproteomic analysis.
Phosphopeptide Enrichment
In general, phosphorylated proteins are present at low stoichiometric levels with the average ratio being around 6% (7). In other words, the abundance of phosphopeptides is quite low when compared with nonphosphorylated peptides. For this reason, one of the key steps in MS-based phosphoproteomics is the use of an enrichment procedure for isolating phosphopeptides. Over the past 15 yr, a number of different phosphopeptide enrichment procedures have been described and, in the remainder of this section, we will briefly describe the two most popular methods. For more comprehensive reviews of this topic the reader is referred to Leitner (8).
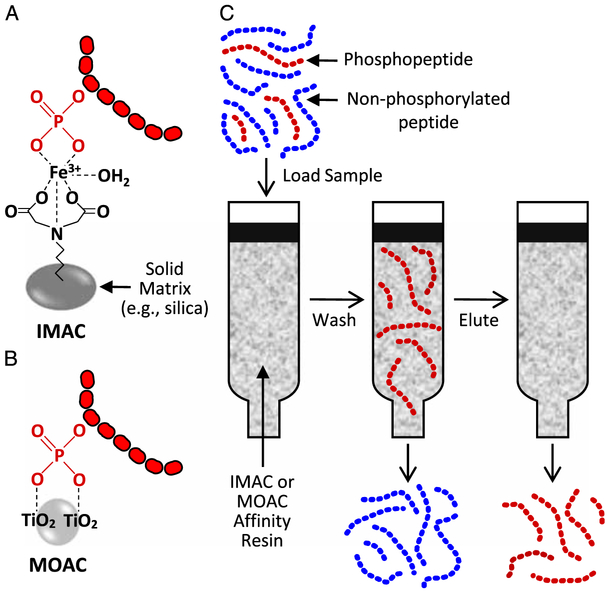
Currently, the most popular phosphopeptide enrichment strategies rely on metal-based affinity procedures that are performed within a magnetic bead or column-based format. One such strategy is called immobilized metal affinity chromatography (IMAC) and is based on the affinity of negatively charged phosphate groups for positively charged metal ions such as Fe3+ or Ga3+. In IMAC, the metal ions are chelated to a solid phase matrix (e.g., silica or magnetic beads) to create the affinity resin (Fig. 3A). Another popular strategy is called metal oxide affinity chromatography, and it relies on the affinity of negatively charged phosphate groups for metals in metal oxides (e.g., titanium dioxide [TiO2]). Unlike IMAC, the metal oxides do not require an additional solid support, but instead, the particles of the metal oxide itself serve as the affinity resin (Fig. 3B).
Figure 3.
Overview of phosphopeptide enrichment procedures using immobilized metal affinity chromatography (IMAC) or metal oxide affinity chromatography (MOAC) TiO2, titanium dioxide.
As illustrated in Figure 3C, the general phosphopeptide enrichment procedure begins with the loading of a peptide/phosphopeptide mixture onto a column that has been packed with the affinity resin. As the mixture moves through the column, the phosphopeptides preferentially bind to the affinity resin and then the nonphosphorylated peptides are removed with a series of washes. The phosphopeptides are then eluted to produce a semi-pure phosphopeptide mixture. Because the majority of the highly abundant nonphosphorylated peptides has been removed from the mixture, a shotgun LC-MS/MS analysis of this mixture will result in a large percentage of the sampled peptides being attributable to phosphopeptides. For instance, work from our lab has revealed that only a few of the peptides identified in an unenriched skeletal muscle sample are phosphopeptides, whereas, after IMAC, more than 50% of the identified peptides are phosphopeptides.
Fractionation
As described previously, enrichment procedures can markedly increase the number of phosphopeptides that are identified within a given sample. However, phosphopeptide-enriched mixtures typically remain highly complex and phosphopeptides with a low concentration can fall below an instrument’s limit of detection. As such, additional fractionation techniques are used often to separate a single complex sample into multiple less-complex mixtures, which, when analyzed separately, can allow for the analysis of phosphopeptides at higher concentrations. For example, if a sample is separated into 10 fractions, and each fraction is analyzed at the loading capacity of the LC-MS/MS, all phosphopeptides will be analyzed at 10-fold higher concentration compared with a single-shot analysis. Consequently, many of the phosphopeptides that may have fallen below the instrument’s limit of detection will now be identified.
To date, a variety of different fractionation techniques have been reported, and although some of these techniques involve the fractionation of samples at the level of proteins or subcellular organelles (9), the vast majority involve the fractionation of peptides. Commonly used peptide fractionation techniques include strong cation exchange chromatography, hydrophilic interaction chromatography (HILIC), electrostatic repulsion-hydrophilic interaction chromatography, and high pH RPLC. As reviewed by Yang et al. (10), each of these fractionation techniques carries its own set of advantages and disadvantages and, to date, no consensus has been reached over which technique is the best.
Regardless of the specific technique used, the overarching goal of fractionation is to produce multiple mixtures of lower complexity for the subsequent LC-MS/MS analysis, and this can produce an impressive boost to the number of identified phosphopeptides. For instance, a study by Zhou et al. demonstrated that 3726 unique phosphopeptides could be identified in a single 2-h LC-MS/MS run of an IMAC-enriched phospho-peptide mixture derived from HELA cells (11). However, when the IMAC-enriched phosphopeptide mixture was separated into 20 less complex fractions with HILIC, and then each fraction subjected to a 2-h LC-MS/MS run, the combined results led to the identification of 9066 unique phosphopeptides (a gain of more than 5000 phosphopeptides). Clearly, the fractionation procedure led to an increase in the number of identified phosphopeptides; however, it is important to consider that it also required a 20-fold increase in the amount of highly valuable time on the instrument. Thus, although fractionation steps can produce substantial gains in the number of identified phosphopeptides, the gains in phosphopeptide identifications have to be carefully weighed against the cost of the subsequent LC-MS/MS analysis.
A final point of consideration is that skeletal muscle is the most widely studied tissue by exercise scientists, and it is composed of highly abundant contractile proteins (e.g., myosin, titin, etc.) along with a plethora of lower abundance proteins (e.g., transcription factors). Indeed, a recent study by Deshmukh et al. (12) revealed that the dynamic range of the skeletal muscle proteome is spread over eight orders of magnitude, with myosin and titin alone accounting for approximately 34% of the total protein mass. As explained previously, the presence of peptides from the highly abundant contractile proteins can severely limit the detection of lower abundance peptides. As such, the use of peptide fractionation techniques before LC-MS/MS is particularly important for the phosphoproteomic analysis of skeletal muscle. In addition, simple centrifugation-based techniques can also be used to deplete homogenates of the contractile proteins before peptide digestion (6). However, even with these technical interventions, the ability to achieve deep phosphoproteomic coverage in skeletal muscle remains a challenge. For a more comprehensive review of this topic, the reader is referred to Deshmukh (13).
Quantification
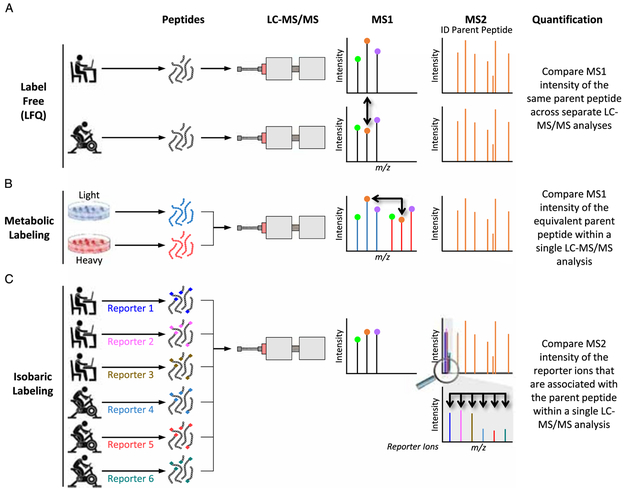
The quantification of phosphopeptides in MS-based phosphoproteomic experiments can be achieved in several ways, three of which we will explain. The simplest approach, coined label-free quantification (LFQ), uses the intensity of parent peptides in MS1 (14). As illustrated in Figure 4A (bold arrows), relative quantitation is achieved by comparing the intensities of the same parent peptides across separate LC-MS/MS analyses. This approach uses a relatively simple method of sample preparation that does not require additional costly reagents, and it has been successfully used in a number of phosphoproteomic experiments. However, with LFQ, each sample must be analyzed individually by the mass spectrometer, and thus, LFQ-based experiments can demand a large amount of LC-MS/MS time.
Figure 4.
The common strategies for relative quantification in liquid chromatography (LC) with tandem MS (LC–tandem MS [LC-MS/MS]). m/z, mass-to-charge ratio.
One approach to maximize sample throughput and to control for run-to-run variability is to simultaneously analyze multiple samples within a single LC-MS/MS experiment (i.e., to multiplex). However, for multiplexing to work, the peptides from each sample have to contain a tag that can be traced back to the originating sample. One type of tagging approach involves the metabolic labeling of sample proteins with heavy isotope-labeled amino acids and is referred to as stable isotope labeling with amino acids in cell culture (SILAC) or stable isotope labeling with amino acids in mammals (SILAM) (15). In SILAC, an isotope-labeled amino acid is substituted for the native amino acid in the culture media, whereas in SILAM, the animals’ food contains the isotope-labeled amino acid. When analyzed, the isotopically labeled peptides will have the exact same chromatographic properties as the native peptides but provide a unique m/z in the MS1 survey scan that is shifted by the number of heavy atoms in the labeled amino acid (Fig. 4B). Relative quantification can then be performed by directly comparing the intensity of each differently labeled peptide in the MS1 survey scan. Because the samples are pooled and analyzed during the same LC-MS/MS analysis, this method controls for run-to-run variability (16). However, SILAC and SILAM experiments increase the complexity of the MS1 spectrum, and redundant sampling of the same peptides (e.g., the heavy and light forms) can limit the total number of unique peptides that are analyzed at the level of MS2. Recently, neutron encoded (NeuCode) amino acids, which differ in mass by as little as 6 mDa, have offered a robust solution to the aforementioned issues (17,18). However, and perhaps of most relevance to exercise scientists, SILAM and NeuCode technologies are not easily extended to human samples because the label must be metabolically introduced before the sample collection takes place.
For human samples, multiplexing strategies that label peptides postdigestion are a viable option. Two common commercial options include “isobaric tags for relative and absolute quantification” (iTRAQ, Applied Biosystems) and “tandem mass tags” (TMT, Thermo Scientific) that allow for the multiplexing of up to 11 different samples (19). Peptides that are labeled with these chemical tags display identical chromatographic behavior, have the same m/z in the MS1 survey scan, and provide a unique reporter ion signal in the MS2 fragmentation spectra that is used for relative quantitation (Fig. 4C). Parent peptide ions from each experimental condition are co-isolated for MS2, providing a quantifiable signal for each experimental condition within the same scan.
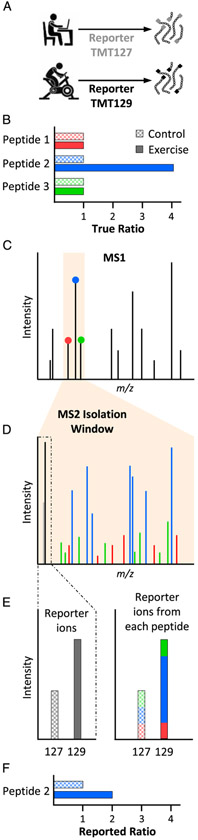
One notable downside to isobaric tags such as iTRAQ and TMT is that the quantitation often provides an underestimation of the magnitude of change that occurred in response to the experimental perturbation (20). This issue is commonly referred to as “ratio suppression,” and as illustrated in Figure 5, it originates from the co-isolation of “contaminating” peptides along with the target parent peptide that has been selected for MS2 analysis. Specifically, Figure 5 represents a hypothetical experiment in which the reporter ions from two different TMT tags were used to compare a control and exercised sample (Fig. 5A). The experiment assumes that, before performing the LC-MS/MS analysis, the absolute abundance (i.e., true ratio) of three different co-eluting peptides was already known (Fig. 5B). During the MS1 analysis of these peptides, the highly abundant (blue) peptide is selected for further interrogation at the level of MS2. However, the m/z isolation window that is used to select the blue peptide also captures the lower abundance green and red peptides (Fig. 5C). During MS2, only the highly abundant blue peptide is identified as being present, but, in reality, the spectra contain fragments from all three of the peptides along with their associated reporter ions (Fig. 5D). Because the specific source of the reporter ions is not discerned, all of the reporter ions are assigned to the blue peptide (Fig. 5E). Consequently, the reported ratio for the blue peptide is suppressed when compared with the true ratio (compare Fig. 5F with 5B). The problem of ratio suppression can be solved with more complex MS scan sequences (20,21), but these approaches are slower and only available on certain instrument platforms. Therefore, ratio suppression is often accepted as a necessary sacrifice in such experiments.
Figure 5.
Ratio suppression. MS, mass spectrometry; m/z, mass-to-charge ratio; tandem mass tags, TMT.
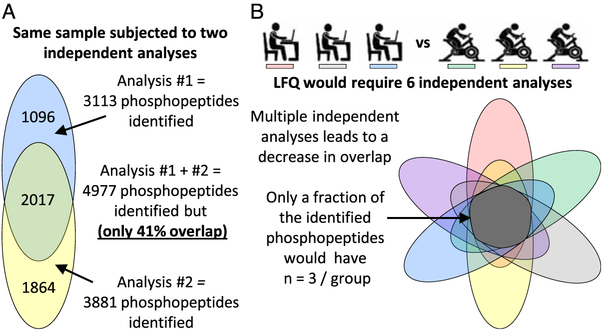
Lastly, all MS quantitation methods can suffer from an issue referred to as identification overlap. To appreciate the problem of identification overlap, it is first important to recognize that current LC-MS/MS technologies can only capture a fraction of the entire phosphoproteome. Moreover, the fraction of the phosphoproteome that is captured from one experiment to the next can vary (22). For example, consider the data shown in Figure 6A that summarize the number of phosphopeptides that were identified during two independent LC-MS/MS analyses of the same sample. In total, nearly 5000 different phosphopeptides were identified; however, only 41% of those phosphopeptides were identified in both analyses. Now imagine an LFQ-based experiment that involves three sedentary, and three exercised, subjects (Fig. 6B). This type of an LFQ-based experiment would require six independent LC-MS/MS analyses, and, although the entire experiment might lead to the identification of a very large number of phosphopeptides, only a subset of those phosphopeptides would have been identified, and, thus, quantified, in all six samples. As such, only a fraction of the identified phosphopeptides would be amenable to traditional downstream statistical analyses. The issue of identification overlap can largely be overcome when multiple samples are analyzed within the same multiplexed experiment (e.g., if the six samples described in Fig. 6B were analyzed within a single TMT-based experiment). However, identification overlap will reemerge as an issue if multiple multiplexed experiments are needed within the same project.
Figure 6.
Identification overlap. LFQ, label-free quantification.
BIOINFORMATICS
Data Processing
As stated before, the identification and relative abundance of peptides are extracted from the information present in both the MS1 and MS2 spectra. Modern instrumentation is capable of collecting individual spectra at speeds of over 20 Hz, and, as such, raw data can easily contain hundreds of thousands of different spectra. Various algorithms (e.g., Andromeda/MaxQuant, OMSSA, SEQUEST, etc.) have been developed to automate the process of matching spectra to possible peptide assignments. The peptide search database is produced by the in silico digestion of an organism’s reference protein database. The peptide spectra are also searched against a decoy database that typically consists of reversed protein sequences to provide a measure of false-positive assignments and to set a confidence threshold for peptide-spectrum matches (23). Finally, each putatively phosphorylated site is assigned a score, which represents the probability that the correct site of phosphorylation was appropriately identified (24,25).
Ultimately, the initial processing will produce a dataset with at least three levels of important information: 1) the amino acid sequence for each of the identified phosphopeptides along with the scores for the putative site(s) of phosphorylation, 2) the proteins on which each of the phosphorylation site(s) reside (i.e., the phosphoprotein), and 3) the quantitative information for each phosphopeptide. In large-scale projects, these datasets can easily contain information for more than 10,000 phosphopeptides and, as such, they can be quite intimidating and difficult for nonspecialists to further interrogate. Indeed, the downstream analysis of phosphoproteomic data currently stands as one of the largest bottlenecks in the field. Fortunately, programs that are aimed at facilitating downstream analyses have started to emerge. For instance, a program called Perseus can be freely downloaded, and it offers a wide array of utilities for navigating, annotating, filtering, analyzing, and visualizing phosphoproteomic data (http://www.perseus-framework.org) (26). Moreover, by importing information from external databases, Perseus can populate datasets with phosphorylation site specific information including, predicted kinases, experimentally validated kinases, and the known effects of phosphorylation on cellular processes, molecular functions, and protein-protein interactions.
In our opinion, Perseus is ideally suited for scientists that do not have previous experience with the analysis of phosphoproteomic data. Therefore, to help the reader with using Perseus, we have included a supplemental file that contains a reformatted version of the data from our recent study that mapped the phosphoproteomic alterations that occur after a bout of maximal intensity contractions (Supplemental Digital Content — Dataset 1, http://links.lww.com/ESSR/A45) (6). This file, along with the associated instructions, will enable the reader to install Perseus and then upload the dataset into its user interface. Once complete, the reader can then begin to interrogate the dataset by following our step-by-step instructions (See Supplemental Digital Content — Supplemental Methods, http://links.lww.com/ESSR/A44) or the tutorials that are available through the Perseus Web site. Importantly, many of the skills that can be learned through these tutorials are not specific to phosphoproteomic data but instead can be extended to the interrogation of unmodified or alternatively modified proteomic datasets (e.g., acetylation, ubiquitination, etc.).
Identifying the Regulated Phosphorylation Sites
Whether working in Perseus or another software platform, one of the first steps that is taken when attempting to gain insight from phosphoproteomic data is the creation of a list of the regulated phosphorylation sites and phosphoproteins. The development of such lists usually begins with statistical tests (e.g., Student’s t-test, unpaired t-test, moderated t-test, analysis of variance, etc.) that can be used to identify which sites experienced a significant alteration in their phosphorylation state. Importantly, all of the aforementioned statistical tests assume that two criteria are met: 1) the data are normally distributed and 2) there is equal variance among the different groups within the dataset. However, when changes in phosphorylation are calculated on a linear scale, the resulting data often will fail to meet these criteria. Thus, to overcome this issue, statistical tests are usually performed on data that has been log2 transformed (e.g., an eightfold change is equivalent to a log2 value of 3, i.e., 23 = 8).
Once P values have been calculated for each site, a secondary step is typically used to correct for the multitude of comparisons that were made within the dataset. This can be particularly important when analyzing large-scale datasets because a P value of 0.05 theoretically means that there is a 5% chance of making a type 1 error (i.e., a false-positive conclusion). Although a 5% chance of making a false-positive conclusion might be acceptable when analyzing a small number of conditions, it can be highly problematic in large-scale datasets. For example, analyzing a dataset with 10,000 phosphorylation sites at a P value of 0.05 would, in theory, lead to approximately 500 falsepositive conclusions. Therefore, to control for this false discovery rate (FDR), a variety of multiple hypothesis correction procedures can be used (e.g., the Benjamini-Hochberg method, permutation-based estimator of FDR, etc.) (27). All multiple hypothesis correction procedures provide an FDR-adjusted P value called the q value, with a q value of 0.05 implying that approximately 5% of significant results will be due to false positives. However, it bears mentioning that, despite their theoretical appeal, multiple hypothesis corrections procedures can produce overly conservative results when applied to phosphoproteomic data. The basis for this point goes beyond the scope of this article, and, thus, the reader is referred to Pascovici et al. (28) for a comprehensive review of this important topic.
A final point of consideration when developing a list of the regulated sites/phosphoproteins is whether a fold-change cutoff should be used. As mentioned previously, phosphoproteomic studies can suffer from ratio suppression, and the degree of ratio suppression can vary dramatically from one phosphopeptide to the next. As such, even very small fold changes could be of biological relevance. To date, there has been no consensus over the most appropriate fold-change cutoff, but when fold-change cutoffs are used, a value of 1.5-fold tends to reign in popularity (5,22). However, numerous studies have also been published in which additional fold-change cutoffs were not used (29). Again, the reader is referred to Pascovici et al. (28) for a further discussion of this topic.
Extracting Biologically Relevant Information
Many of the bioinformatics tools that can be used to gain insight from phosphoproteomics datasets are based on enrichment analyses. In brief, these methods test whether specific features of interests (biological processes, signaling pathways, cellular compartments, etc.) are over- or underrepresented within the list of regulated phosphorylation sites/phosphoproteins when compared with a background list. For phosphoproteomics studies, the background list is typically composed of all of the phosphorylation sites or phosphoproteins that were identified within the given dataset. The reason for this is that various steps in the phosphoproteomics workflow can introduce biases into the dataset. For instance, phosphoproteomic experiments are biased toward the identification of the most abundant phosphopeptides. Furthermore, phosphopeptide enrichment protocols can display a bias toward enriching for single versus doubly phosphorylated peptides or toward enriching for hydrophobic versus hydrophilic phosphopeptides. Thus, using only the phosphorylation sites/phosphoproteins identified within the given dataset can be critical for accurate enrichment analyses. As such, care should be taken when making conclusions from enrichment analysis programs that do not allow for the use of this type of custom background. For an excellent example of how an inappropriate background can influence the outcomes of an enrichment analysis, see Munk et al. (30).
Gene Ontology and Pathway Enrichment Analyses
Several programs (e.g., Perseus, Database for Annotation, Visualization, and Integrated Discovery [DAVID], PANTHER, Gorilla, etc.) can be used to annotate individual phosphoproteins with their associated descriptors such as gene ontology (GO) terms or membership in pathways such as those described by the Kyoto Encyclopedia of Genes and Genomes (KEGG) or Reactome. Briefly, GO terms describe the biology of individual gene products in terms of their 1) molecular functions, 2) biological processes, and 3) cellular components, whereas KEGG and Reactome pathways represent manually curated maps of various signaling networks (e.g., the insulin signaling pathway). Once annotated, the aforementioned programs can perform statistical tests to determine whether specific descriptors are over- or underrepresented within the regulated versus background list.
It bears mentioning that DAVID is one of the most popular GO and pathway enrichment programs (31). Part of DAVID’s popularity stems from the fact that it runs through a freely available Web-based application (http://david.abcc.ncifcrf.gov/home.jsp), it has a user-friendly interface, and it can query a very large repertoire of recently updated databases for the annotation of both regulated and background lists. Once annotated, DAVID can then calculate the fold enrichment and the associated P and q values for each annotation term. For a step-by-step description of how to use DAVID, see Huang et al. (32).
Prediction of Regulated Kinases
One of the goals in most phosphoproteomic experiments is to identify which kinase(s) drive the changes in phosphorylation. Over the past decade, a number of different approaches have been developed to accomplish this goal; however, a comprehensive description of these approaches is beyond the scope of this review. As such, we will only briefly describe some of the different approaches that are available. Also note that none of these approaches provide definitive proof that a kinases activity has been altered. Instead, these approaches are intended to help investigators develop informed hypotheses that can be further tested with more direct approaches. For a comprehensive review of this topic, the reader is referred to Munk et al. (33).
One of the most direct ways to predict which kinases are regulated is to assess whether changes have occurred on phosphorylation site(s) that are known to modulate kinase activity. Moreover, datasets can be directly searched for phosphorylation sites that have known kinases. When combined, the results from these approaches can lead to high confidence predictions; however, the number of sites with known functions or kinases is usually quite limited. For instance, we combined the use of Perseus and PhosphoSitePlus databases (Version 7-10-2017) (4) to identify the phosphorylation sites in Supplemental Dataset 1 that have known functions or kinases, and the outcomes revealed that only 93 of the nearly 6000 identified phosphorylation sites had known kinases, and just 61 had a known function. Furthermore, only 19 of the aforementioned sites experienced a significant alteration in their phosphorylation state after a bout of maximal-intensity contractions (moderated t-test with Benjamini-Hochberg adjusted P value of ≤0.05). Nonetheless, an examination of these 19 sites strongly suggests that maximal-intensity contractions activate ERK1/2, which then phosphorylates the T320 on Mapkapk2 and, inturn, promotes the activation of Mapkapk2.
Another approach that can be used to predict regulated kinases relies on the notion that most kinases contain a well-conserved activation loop phosphorylation site that, when phosphorylated, promotes activation of the kinase (34). Recently, the Olsen group from Novo Nordisk Foundation released a Web-based application that can be used to identify these sites (http://phomics.jensenlab.org/). Detailed instruction for using this Web-based application can be found in Munk et al. (33), and, with their approach, one can rapidly gain additional insight into which kinases might be regulated by the experimental perturbation.
Phosphoproteomic data are also amenable to determining whether specific phosphorylation motifs (i.e., the amino acid sequence that surrounds the site of phosphorylation) are over- or underrepresented within the list of regulated sites. This type of analysis can be informative because the phosphorylation motif plays a major role in determining which kinase(s) will phosphorylate it. For instance, the mitogen-activated protein kinases (MAPK) strongly prefer to phosphorylate serine and threonine residues that are immediately followed by a proline (i.e., xxxSPxxx or xxxTPxxx motifs, where x represents any amino acid) (35). Thus, if an enrichment analysis reveals overrepresentation of these motifs, then one should begin to suspect that the experimental perturbation may have promoted an increase in the activity of the MAPK. A number of different motif enrichment analysis programs have been described (e.g., MoDL, FMotif, Motif-All, and Motif-X) with Motif-X reigning as the most widely used (36). Motif-X can be run through a free Web-based application (http://motif-x.med.harvard.edu/), and detailed protocols for running Motif-X can be found in Chou and Schwartz (37).
As alluded previously, predictions of whether a kinase will phosphorylate a given substrate can be derived from how well the substrate conforms to the preferred phosphorylation motif on the kinase. Importantly, the strength of these predictions can be further improved by considering whether the kinase and substrate interact (38). Such interactions can be direct (e.g., physical association between the kinase and substrate) or indirect (e.g., the name of the kinase and substrate is frequently found together in the same articles). Consideration of these interactions stands at the foundation of a kinase-substrate prediction program called NetworKIN (39). With NetworKIN, a list of the most likely kinases for each individual phosphorylation site can be generated. The output for each site can then be clustered according to whether the sites showed an increase, decrease, or no change in phosphorylation. Additional statistical analyses can then be performed to determine whether any of the predicted kinases are over- or underrepresented within each group. For details on how to perform this type of analysis, see Munk et al. (33).
Using Protein-Protein Interaction Networks to Identify Signaling Pathways
Recently, Rudolph et al. (40) described an innovative method for identifying functionally relevant signaling proteins and pathways in phosphoproteomic datasets. Specifically, their approach was based on the premise that the function of a signaling protein can be determined by assessing changes in the phosphorylation of the proteins that it interacts with. As such, their approach uses a program called PHOTON to analyze phosphoproteomic data within the context of a protein interaction network. This is quite different from traditional approaches that attempt to identify functionally relevant signaling proteins on the basis of whether they experienced a significant alteration, or set fold change, in their phosphorylation state. Importantly, within PHOTON, a functionally relevant signaling protein does not necessarily have to experience a change in its phosphorylation state but instead could indirectly partake in the process of signaling transduction (e.g., act an adaptor protein such as Grb2). With this point in mind, it is not surprising that PHOTON outperformed traditional approaches when using phosphoproteomic datasets to identify known components of the insulin and EGF signaling pathways (40).
In addition to identifying functionally relevant signaling proteins, PHOTON can also be used to generate a visual and interactive display of the predicted signaling network. Moreover, PHOTON can be used to predict the most functionally relevant phosphorylation events within the signaling network. Thus, in our opinion, PHOTON represents a highly valuable addition to the repertoire of bioinformatics tools that can be used to interrogate phosphoproteomic datasets, and the reader is strongly encouraged to consider the use of this innovative approach. To facilitate this effort, an additional dataset (Supplemental Digital Content — Supplemental Dataset 2, http://links.lww.com/ESSR/A41) along with instructions for moving data from Perseus into the user interface of PHOTON has been included in the supplemental methods, http://links.lww.com/ESSR/A44.
SUMMARY
Phosphorylation-mediated signaling pathways play a central role in the ability of cells to respond to environmental stimuli, yet our knowledge about the signaling pathways that are regulated by exercise remains incomplete. Fortunately, advancements in phosphoproteomic technologies have made it possible to profile thousands of different phosphorylation events within a single experiment. With NIH’s investment in MoTrPAC, these technologies will enable an extensive cataloging of the phosphoproteomic alterations that occur in response to different modes of exercise. The results from these studies could radically advance our understanding of exercise-induced signaling events; however, for this to occur, the field will have to be armed with researchers that can both identify, and extract, the most biologically relevant results. Undoubtedly, this will require a collaborative effort between systems biologists, bioinformaticians, and exercise scientists. Only with such a team science approach will we successfully fulfill the potential that exists within MoTrPAC and related projects. We hope that this review will help prepare exercise scientists, and the aligned communities, for the type of data that is soon to come.
Supplementary Material
Key Points.
Phosphorylation-mediated signaling pathways play a central role in the ability of cells to respond to environmental stimuli, yet our knowledge about the signaling pathways that are regulated by exercise remains incomplete.
Advancements in the field of phosphoproteomics have made it possible to profile thousands of different phosphorylation events within a single experiment.
This review is intended to help nonspecialists understand the cutting-edge technologies that are used during a typical phosphoproteomic experiment.
Various bioinformatics tools can be used to interrogate large-scale phosphoproteomic datasets. To assist the reader in becoming proficient with the use of these tools, an example dataset, along with step-by-step instructions, is provided as a supplement material.
Acknowledgment
The work in this publication was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health (NIH) under award no. AR057347 to T.A.H. as well as NIH grant R35 GMH8H0 to J.J.C. and NIH traineeship (5 T32 GM008349) to G.M.W.
Footnotes
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s Web site (www.acsm-essr.org).
References
- 1.Hoppeler H, Baum O, Lurman G, Mueller M. Molecular mechanisms of muscle plasticity with exercise. Compr. Physiol 2011; 1 (3):1383–412. [DOI] [PubMed] [Google Scholar]
- 2.Cohen P The origins of protein phosphorylation. Nat. Cell Biol 2002; 4(5):E127–30. [DOI] [PubMed] [Google Scholar]
- 3.Sharma K, D'Souza RC, Tyanova S, et al. Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Rep. 2014; 8(5):1583–94. [DOI] [PubMed] [Google Scholar]
- 4.Hornbeck PV, Kornhauser JM, Tkachev S, et al. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012; 40(Database issue):D261–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffman NJ, Parker BL, Chaudhuri R, et al. Global phosphoproteomic analysis of human skeletal muscle reveals a network of exercise-regulated kinases and AMPK substrates. Cell Metab. 2015; 22(5):922–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Potts GK, McNally RM, Blanco R, et al. A map of the phosphoproteomic alterations that occur after a bout of maximal-intensity contractions. J. Physiol 2017; 595(15):5209–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu R, Haas W, Dephoure N, et al. A large-scale method to measure absolute protein phosphorylation stoichiometries. Nat. Methods 2011; 8(8): 677–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leitner A Enrichment strategies in phosphoproteomics. Methods Mol. Biol 2016; 1355:105–21. [DOI] [PubMed] [Google Scholar]
- 9.Trost M, Bridon G, Desjardins M, Thibault P. Subcellular phosphoproteomics. Mass Spectrom. Rev 2010; 29(6):962–90. [DOI] [PubMed] [Google Scholar]
- 10.Yang C, Zhong X, Li L. Recent advances in enrichment and separation strategies for mass spectrometry-based phosphoproteomics. Electrophoresis. 2014; 35(24):3418–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou H, Di Palma S, Preisinger C, et al. Toward a comprehensive characterization of a human cancer cell phosphoproteome. J. Proteome Res. 2013; 12(1):260–71. [DOI] [PubMed] [Google Scholar]
- 12.Deshmukh AS, Murgia M, Nagaraj N, Treebak JT, Cox J, Mann M. Deep proteomics of mouse skeletal muscle enables quantitation of protein isoforms, metabolic pathways, and transcription factors. Mol. Cell. Proteomics: MCP 2015; 14(4):841–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deshmukh AS. Proteomics of skeletal muscle: focus on insulin resistance and exercise biology. Proteomes. 2016; 4(1):E6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics: MCP 2014; 13(9):2513–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ong SE, Blagoev B, Kratchmarova I, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics: MCP 2002; 1(5):376–86. [DOI] [PubMed] [Google Scholar]
- 16.Piehowski PD, Petyuk VA, Orton DJ, et al. Sources of technical variability in quantitative LC-MS proteomics: human brain tissue sample analysis. J. Proteome Res 2013; 12(5):2128–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hebert AS, Merrill AE, Bailey DJ, et al. Neutron-encoded mass signatures for multiplexed proteome quantification. Nat. Methods 2013; 10(4):332–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baughman JM, Rose CM, Kolumam G, et al. NeuCode proteomics reveals Bap1 regulation of metabolism. Cell Rep. 2016; 16(2):583–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wiese S, Reidegeld KA, Meyer HE, Warscheid B. Protein labeling by iTRAQ: a new tool for quantitative mass spectrometry in proteome research. Proteomics. 2007; 7(3):340–50. [DOI] [PubMed] [Google Scholar]
- 20.Wenger CD, Lee MV, Hebert AS, et al. Gas-phase purification enables accurate, multiplexed proteome quantification with isobaric tagging. Nat. Methods 2011; 8(11):933–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McAlister GC, Nusinow DP, Jedrychowski MP, et al. MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal. Chem 2014; 86(14):7150–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Humphrey SJ, Azimifar SB, Mann M. High-throughput phosphoproteomics reveals in vivo insulin signaling dynamics. Nat. Biotechnol 2015; 33(9):990–5. [DOI] [PubMed] [Google Scholar]
- 23.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 2007; 4(3):207–14. [DOI] [PubMed] [Google Scholar]
- 24.Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 2006; 24(10):1285–92. [DOI] [PubMed] [Google Scholar]
- 25.Taus T, Kocher T, Pichler P, et al. Universal and confident phosphorylation site localization using phosphoRS. J. Proteome Res 2011; 10(12):5354–62. [DOI] [PubMed] [Google Scholar]
- 26.Tyanova S, Temu T, Sinitcyn P, et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016; 13(9):731–40. [DOI] [PubMed] [Google Scholar]
- 27.Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003; 19(3):368–75. [DOI] [PubMed] [Google Scholar]
- 28.Pascovici D, Handler DC, Wu JX, Haynes PA. Multiple testing corrections in quantitative proteomics: a useful but blunt tool. Proteomics. 2016; 16(18): 2448–53. [DOI] [PubMed] [Google Scholar]
- 29.Lundby A, Andersen MN, Steffensen AB, et al. In vivo phosphoproteomics analysis reveals the cardiac targets of β-adrenergic receptor signaling. Sci. Signal 2013; 6(278):rs11. [DOI] [PubMed] [Google Scholar]
- 30.Munk S, Refsgaard JC, Olsen JV. Systems analysis for interpretation of phosphoproteomics data. Methods Mol. Biol 2016; 1355:341–60. [DOI] [PubMed] [Google Scholar]
- 31.Dennis G Jr, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003; 4(5):P3. [PubMed] [Google Scholar]
- 32.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc 2009; 4(1):44–57. [DOI] [PubMed] [Google Scholar]
- 33.Munk S, Refsgaard JC, Olsen JV, Jensen LJ. From phosphosites to kinases. Methods Mol. Biol 2016; 1355:307–21. [DOI] [PubMed] [Google Scholar]
- 34.Nolen B, Taylor S, Ghosh G. Regulation of protein kinases; controlling activity through activation segment conformation. Mol. Cell 2004; 15(5):661–75. [DOI] [PubMed] [Google Scholar]
- 35.Pearson G, Robinson F, Beers Gibson T, et al. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr. Rev 2001; 22(2):153–83. [DOI] [PubMed] [Google Scholar]
- 36.Schwartz D, Gygi SP. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol 2005; 23(11):1391–8. [DOI] [PubMed] [Google Scholar]
- 37.Chou MF, Schwartz D. Biological sequence motif discovery using motif-x. Curr. Protoc. Bioinformatics 2011. Chapter 13:Unit 13:15–24. [DOI] [PubMed] [Google Scholar]
- 38.Linding R, Jensen LJ, Ostheimer GJ, et al. Systematic discovery of in vivo phosphorylation networks. Cell. 2007; 129(7):1415–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horn H, Schoof EM, Kim J, et al. KinomeXplorer: an integrated platform for kinome biology studies. Nat. Methods 2014; 11(6):603–4. [DOI] [PubMed] [Google Scholar]
- 40.Rudolph JD, de Graauw M, van de Water B, Geiger T, Sharan R. Elucidation of signaling pathways from large-scale phosphoproteomic data using protein interaction networks. Cell Syst. 2016; 3(6):585–93.e3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.